Abstract
The title Schiff base compound, C22H19NO2S, crystallized with two independent molecules (A and B) in the asymmetric unit. Both molecules have an E conformation about the C=N bond. The two molecules differ in the orientation of the aromatic rings with respect to each other. The outer 4-methoxybenzene ring is inclined to the central benzene ring and the outer 4-acetylbenzene ring by 1.80 (19) and 63.73 (19)°, respectively, in molecule A, and by 6.72 (18) and 68.53 (19)°, respectively, in molecule B. The two outer benzene rings are inclined to one another by 63.77 (18) and 63.19 (18)° in molecules A and B, respectively. In the crystal, the individual molecules stack in columns along [010], and are linked by a number of C—H⋯π interactions, forming slabs lying parallel to (001).
Keywords: crystal structure, Schiff base, 4-amino-4-acetyldiphenyl sulfide, C—H⋯π interactions
Related literature
For the synthesis and structures of Schiff bases, see, for example: Kahwa et al. (1986 ▸). For their use as protein and enzyme mimics, see: Santos et al. (2001 ▸). For their use as corrosion inhibitors, see: Ahamad et al. (2010 ▸); Negm et al. (2010 ▸). For their coordination properties, see: Özkar et al. (2004 ▸); Hebbachi & Benali-Cherif (2005 ▸). For complexation of Schiff bases with transition metals, see: Izatt et al. (1995 ▸); Kalcher et al. (1995 ▸). For the crystal structure of a very similar Schiff base compound derived from 4-amino-4-acetyldiphenyl sulfide, see: Hebbachi et al. (2013 ▸).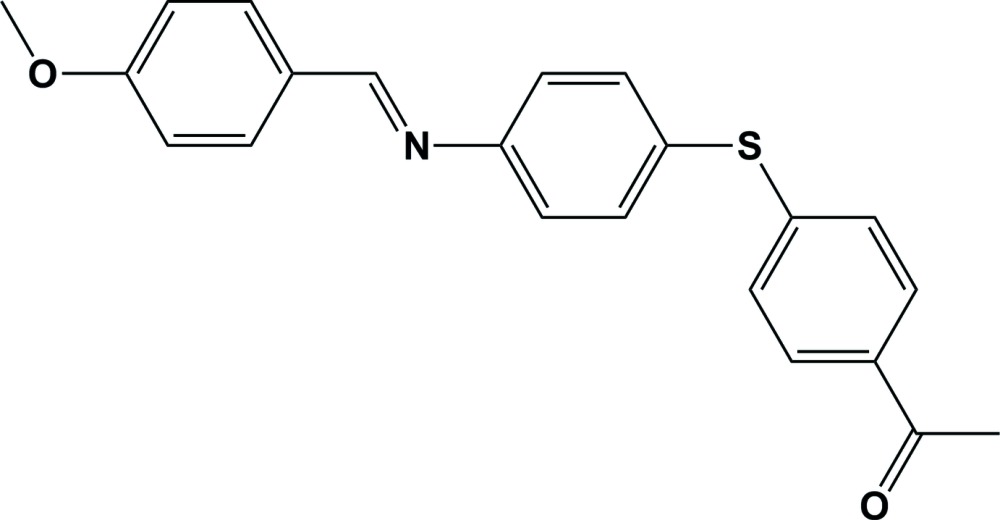
Experimental
Crystal data
C22H19NO2S
M r = 361.44
Triclinic,

a = 5.7708 (2) Å
b = 8.0867 (3) Å
c = 19.6929 (8) Å
α = 81.844 (2)°
β = 86.664 (3)°
γ = 85.662 (3)°
V = 906.05 (6) Å3
Z = 2
Mo Kα radiation
μ = 0.20 mm−1
T = 293 K
0.1 × 0.1 × 0.1 mm
Data collection
Bruker SMART 1K CCD area-detector diffractometer
19586 measured reflections
6013 independent reflections
4850 reflections with I > 2σ(I)
R int = 0.032
Refinement
R[F 2 > 2σ(F 2)] = 0.039
wR(F 2) = 0.076
S = 1.03
6013 reflections
473 parameters
3 restraints
H-atom parameters constrained
Δρmax = 0.14 e Å−3
Δρmin = −0.19 e Å−3
Absolute structure: Flack x determined using 1952 quotients [(I +)−(I −)]/[(I +)+(I −)] (Parsons et al., 2013 ▸)
Absolute structure parameter: 0.06 (3)
Data collection: SMART (Bruker, 2006 ▸); cell refinement: SAINT (Bruker, 2006 ▸); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▸); program(s) used to refine structure: SHELXL2014 (Sheldrick, 2015 ▸); molecular graphics: ORTEP-3 for Windows (Farrugia, 2012 ▸) and Mercury (Macrae et al., 2008 ▸); software used to prepare material for publication: WinGX (Farrugia, 2012 ▸) and SHELXL2014.
Supplementary Material
Crystal structure: contains datablock(s) I, Global. DOI: 10.1107/S205698901500033X/su5056sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S205698901500033X/su5056Isup2.hkl
Supporting information file. DOI: 10.1107/S205698901500033X/su5056Isup3.cml
. DOI: 10.1107/S205698901500033X/su5056fig1.tif
The molecular structure of the two independent molecules (A and B) of the title compound, with atom labelling. Displacement ellipsoids are drawn at the 50% probability level.
b . DOI: 10.1107/S205698901500033X/su5056fig2.tif
A view along the b axis of the crystal packing of the title compound. C-H⋯π interactions are shown as dashed lines (see Table 1 for details; molecule A is red; molecule B is blue; H atoms not involved in these interactions have been omitted for clarity).
CCDC reference: 1042562
Additional supporting information: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (, ).
Cg1, Cg2, Cg3 and Cg6 are the centroids of the C2C7, C9C14, C15C20 and C37C42 rings, respectively.
| DHA | DH | HA | D A | DHA |
|---|---|---|---|---|
| C17H17Cg6i | 0.93 | 3.00 | 3.734(4) | 137 |
| C26H26Cg1 | 0.93 | 2.96 | 3.763(4) | 146 |
| C32H32Cg2 | 0.93 | 2.98 | 3.706(4) | 136 |
| C41H41Cg3ii | 0.93 | 2.99 | 3.670(4) | 131 |
Symmetry codes: (i)  ; (ii)
; (ii)  .
.
Acknowledgments
The authors thank the Algerian Ministère de l’Enseignement Supérieur et de la Recherche Scientifique for financial support.
supplementary crystallographic information
S1. Comment
The synthesis and structures of Schiff bases have attracted much attention in biology and chemistry (Kahwa et al., 1986). One of the aims of investigating their structural chemistry is to develop protein and enzyme mimics (Santos et al., 2001). Structural information is useful in investigating the coordination properties of Schiff bases functioning as ligands (Özkar et al., 2004; Hebbachi & Benali-Cherif, 2005). They have a great capacity for complexation of transition metals (Izatt et al., 1995; Kalcher et al., 1995). They are also used as corrosion inhibitors (Ahamad et al., 2010; Negm et al., 2010). There are only a few reported crystal structures of Schiff bases derived from 4-amino-4-acetyldiphenyl sulfide (Hebbachi et al., 2013). As a part of our ongoing research, we have synthesized the title compound and report herein on its crystal structure.
The title compound, Fig. 1, crystallized with two independent molecules (A and B) in the asymmetric unit. Both molecules have an E conformation about the C═N bond, with torsion angles C2—C1═N1—C9 and C24—C23═ N2—C31 being -179.9 (3) and 177.2 (3), respectively.
The two molecules differ in the orientation of the aromatic rings with respect to one another. The outer 4-methoxybenzene ring is inclined to the central benzene ring and the outer 3-acetylbenzene ring by 1.80 (19) and 63.73 (19) °, respectively, in molecule A, and by 6.72 (18) and 68.53 (19) °, respectively in molecule B. The two outer benzene rings are inclined to one another by 63.77 (18) and 63.19 (18) ° in molecules A and B, respectively.
The bond lengths and angles are close to those observed for a very similar structure, viz. (E)-1-(4-((4-(((4-hydroxynaphthalen-1-yl)methylene)amino)phenyl)thio) phenyl)ethan-1-one (Hebbachi et al., 2013). For example, the sulfur atom has sp3 hybridization as indicated by the C—S—C angle of 106.01 (15) and 105.99 (15) ° in molecules A and B, respectively, compared to 104.88 (15) ° observed in the above mentioned compound.
In the crystal, molecules stack along [010] in columns composed of either A or B molecules, and are linked by a number of C-H···π interactions (Table 1 and Fig. 2) forming slabs lying parallel to (001).
S2. Experimental
The title Schiff base was prepared by condensation of 4-amino-4-acetyl diphenylsulfure and anisaldehyde in a 1:1 molar ratio, in an ethanol solution containing a few drops of dry piperidine. The mixture was stirred under reflux for 3 h. The mixture was then concentrated and cooled. Colourless prismatic crystals of title compound were obtained by recrystallization from a mixture of chloroform/hexane (1/1). They were collected by filtration and dried in air (yield: 64%; m.p.: 421 K).
S3. Refinement
H atoms were positioned geometrically and refined using a riding model: C—H = 0.93 - 0.98 Å with Uiso(H) = 1.5Ueq(C) for methyl H atoms and = 1.2Ueq(C) for other H atoms.
Figures
Fig. 1.

The molecular structure of the two independent molecules (A and B) of the title compound, with atom labelling. Displacement ellipsoids are drawn at the 50% probability level.
Fig. 2.

A view along the b axis of the crystal packing of the title compound. C-H···π interactions are shown as dashed lines (see Table 1 for details; molecule A is red; molecule B is blue; H atoms not involved in these interactions have been omitted for clarity).
Crystal data
| C22H19NO2S | Z = 2 |
| Mr = 361.44 | F(000) = 380 |
| Triclinic, P1 | Dx = 1.325 Mg m−3 |
| a = 5.7708 (2) Å | Mo Kα radiation, λ = 0.71073 Å |
| b = 8.0867 (3) Å | θ = 1.0–27.1° |
| c = 19.6929 (8) Å | µ = 0.20 mm−1 |
| α = 81.844 (2)° | T = 293 K |
| β = 86.664 (3)° | Prism, colourless |
| γ = 85.662 (3)° | 0.1 × 0.1 × 0.1 mm |
| V = 906.05 (6) Å3 |
Data collection
| Bruker SMART 1K CCD area-detector diffractometer | 4850 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.032 |
| Graphite monochromator | θmax = 25.0°, θmin = 2.6° |
| ω scan | h = −6→6 |
| 19586 measured reflections | k = −9→9 |
| 6013 independent reflections | l = −22→23 |
Refinement
| Refinement on F2 | Hydrogen site location: inferred from neighbouring sites |
| Least-squares matrix: full | H-atom parameters constrained |
| R[F2 > 2σ(F2)] = 0.039 | w = 1/[σ2(Fo2) + (0.0319P)2 + 0.0416P] where P = (Fo2 + 2Fc2)/3 |
| wR(F2) = 0.076 | (Δ/σ)max < 0.001 |
| S = 1.03 | Δρmax = 0.14 e Å−3 |
| 6013 reflections | Δρmin = −0.19 e Å−3 |
| 473 parameters | Absolute structure: Flack x determined using 1952 quotients [(I+)-(I-)]/[(I+)+(I-)] (Parsons et al., 2013) |
| 3 restraints | Absolute structure parameter: 0.06 (3) |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| S1 | 0.40421 (16) | 0.84717 (13) | 0.09943 (5) | 0.0521 (3) | |
| O1 | 1.2589 (5) | 0.9295 (3) | 0.65028 (13) | 0.0582 (8) | |
| O2 | 1.1549 (6) | 1.1965 (5) | −0.14550 (16) | 0.0981 (13) | |
| N1 | 0.7980 (6) | 0.9227 (4) | 0.36608 (16) | 0.0474 (8) | |
| C1 | 0.9871 (7) | 0.8570 (5) | 0.38833 (19) | 0.0467 (10) | |
| H1 | 1.0810 | 0.7913 | 0.3612 | 0.056* | |
| C2 | 1.0659 (6) | 0.8797 (5) | 0.45527 (18) | 0.0410 (9) | |
| C3 | 0.9313 (7) | 0.9756 (5) | 0.49822 (19) | 0.0473 (10) | |
| H3 | 0.7912 | 1.0293 | 0.4835 | 0.057* | |
| C4 | 1.0031 (7) | 0.9914 (5) | 0.5619 (2) | 0.0474 (10) | |
| H4 | 0.9124 | 1.0569 | 0.5898 | 0.057* | |
| C5 | 1.2101 (7) | 0.9106 (5) | 0.58501 (18) | 0.0418 (10) | |
| C6 | 1.3488 (7) | 0.8174 (5) | 0.54288 (19) | 0.0513 (11) | |
| H6 | 1.4893 | 0.7645 | 0.5577 | 0.062* | |
| C7 | 1.2765 (7) | 0.8037 (5) | 0.4784 (2) | 0.0519 (10) | |
| H7 | 1.3709 | 0.7422 | 0.4498 | 0.062* | |
| C8 | 1.4655 (8) | 0.8448 (6) | 0.6785 (2) | 0.0705 (13) | |
| H8A | 1.4721 | 0.8630 | 0.7255 | 0.106* | |
| H8B | 1.5993 | 0.8877 | 0.6526 | 0.106* | |
| H8C | 1.4638 | 0.7271 | 0.6764 | 0.106* | |
| C9 | 0.7237 (6) | 0.9003 (4) | 0.30085 (18) | 0.0395 (9) | |
| C10 | 0.5119 (6) | 0.9821 (5) | 0.28283 (19) | 0.0454 (10) | |
| H10 | 0.4293 | 1.0456 | 0.3132 | 0.054* | |
| C11 | 0.4219 (6) | 0.9703 (5) | 0.22036 (19) | 0.0446 (10) | |
| H11 | 0.2796 | 1.0260 | 0.2089 | 0.054* | |
| C12 | 0.5420 (6) | 0.8762 (5) | 0.17482 (18) | 0.0404 (9) | |
| C13 | 0.7546 (6) | 0.7946 (5) | 0.19237 (19) | 0.0462 (10) | |
| H13 | 0.8370 | 0.7312 | 0.1619 | 0.055* | |
| C14 | 0.8444 (6) | 0.8068 (5) | 0.25442 (19) | 0.0446 (10) | |
| H14 | 0.9876 | 0.7520 | 0.2655 | 0.054* | |
| C15 | 0.5884 (6) | 0.9284 (4) | 0.02990 (18) | 0.0392 (9) | |
| C16 | 0.7859 (6) | 1.0131 (4) | 0.03509 (18) | 0.0417 (9) | |
| H16 | 0.8307 | 1.0313 | 0.0779 | 0.050* | |
| C17 | 0.9151 (6) | 1.0700 (5) | −0.02354 (19) | 0.0474 (10) | |
| H17 | 1.0464 | 1.1273 | −0.0196 | 0.057* | |
| C18 | 0.8547 (6) | 1.0441 (5) | −0.08816 (19) | 0.0456 (10) | |
| C19 | 0.6550 (7) | 0.9614 (5) | −0.09213 (19) | 0.0489 (10) | |
| H19 | 0.6093 | 0.9443 | −0.1350 | 0.059* | |
| C20 | 0.5227 (7) | 0.9041 (5) | −0.03450 (19) | 0.0475 (10) | |
| H20 | 0.3894 | 0.8491 | −0.0386 | 0.057* | |
| C21 | 1.0007 (8) | 1.1056 (6) | −0.1499 (2) | 0.0569 (11) | |
| C22 | 0.9573 (9) | 1.0531 (6) | −0.2177 (2) | 0.0800 (15) | |
| H22A | 1.0659 | 1.1021 | −0.2522 | 0.120* | |
| H22B | 0.8016 | 1.0901 | −0.2302 | 0.120* | |
| H22C | 0.9767 | 0.9333 | −0.2144 | 0.120* | |
| S2 | −0.10116 (16) | 0.32763 (13) | 0.16371 (5) | 0.0510 (3) | |
| O3 | 0.8434 (5) | 0.4410 (4) | 0.69853 (14) | 0.0654 (8) | |
| O4 | 0.2586 (6) | 0.6780 (4) | −0.14986 (14) | 0.0845 (10) | |
| N2 | 0.4418 (5) | 0.4400 (4) | 0.40143 (16) | 0.0475 (8) | |
| C23 | 0.4230 (7) | 0.3483 (5) | 0.4586 (2) | 0.0528 (11) | |
| H23 | 0.3251 | 0.2609 | 0.4627 | 0.063* | |
| C24 | 0.5470 (7) | 0.3721 (5) | 0.51887 (19) | 0.0446 (10) | |
| C25 | 0.7361 (7) | 0.4704 (5) | 0.5146 (2) | 0.0490 (10) | |
| H25 | 0.7923 | 0.5194 | 0.4718 | 0.059* | |
| C26 | 0.8425 (7) | 0.4964 (5) | 0.57344 (19) | 0.0487 (10) | |
| H26 | 0.9686 | 0.5627 | 0.5701 | 0.058* | |
| C27 | 0.7591 (7) | 0.4226 (5) | 0.6370 (2) | 0.0466 (10) | |
| C28 | 0.5749 (7) | 0.3232 (5) | 0.6414 (2) | 0.0539 (11) | |
| H28 | 0.5203 | 0.2723 | 0.6840 | 0.065* | |
| C29 | 0.4705 (7) | 0.2986 (5) | 0.5829 (2) | 0.0557 (11) | |
| H29 | 0.3458 | 0.2310 | 0.5866 | 0.067* | |
| C30 | 1.0244 (8) | 0.5508 (6) | 0.6994 (2) | 0.0753 (14) | |
| H30A | 1.0722 | 0.5461 | 0.7456 | 0.113* | |
| H30B | 0.9687 | 0.6632 | 0.6826 | 0.113* | |
| H30C | 1.1544 | 0.5171 | 0.6706 | 0.113* | |
| C31 | 0.3062 (6) | 0.4115 (4) | 0.34626 (18) | 0.0399 (9) | |
| C32 | 0.3869 (6) | 0.4741 (4) | 0.28040 (18) | 0.0429 (9) | |
| H32 | 0.5214 | 0.5320 | 0.2746 | 0.052* | |
| C33 | 0.2704 (6) | 0.4515 (5) | 0.22338 (19) | 0.0422 (9) | |
| H33 | 0.3268 | 0.4939 | 0.1797 | 0.051* | |
| C34 | 0.0701 (6) | 0.3659 (4) | 0.23136 (17) | 0.0362 (9) | |
| C35 | −0.0163 (6) | 0.3070 (5) | 0.29683 (18) | 0.0428 (10) | |
| H35 | −0.1530 | 0.2516 | 0.3025 | 0.051* | |
| C36 | 0.1005 (6) | 0.3307 (5) | 0.35358 (18) | 0.0455 (10) | |
| H36 | 0.0405 | 0.2920 | 0.3973 | 0.055* | |
| C37 | 0.0526 (6) | 0.4036 (4) | 0.08628 (17) | 0.0385 (9) | |
| C38 | 0.2689 (6) | 0.3346 (5) | 0.06659 (18) | 0.0438 (9) | |
| H38 | 0.3481 | 0.2541 | 0.0969 | 0.053* | |
| C39 | 0.3663 (7) | 0.3849 (5) | 0.00259 (19) | 0.0434 (10) | |
| H39 | 0.5111 | 0.3369 | −0.0101 | 0.052* | |
| C40 | 0.2549 (6) | 0.5052 (5) | −0.04367 (17) | 0.0396 (9) | |
| C41 | 0.0402 (7) | 0.5757 (5) | −0.02320 (19) | 0.0476 (10) | |
| H41 | −0.0369 | 0.6583 | −0.0531 | 0.057* | |
| C42 | −0.0611 (7) | 0.5250 (5) | 0.04106 (19) | 0.0465 (10) | |
| H42 | −0.2059 | 0.5726 | 0.0539 | 0.056* | |
| C43 | 0.3520 (7) | 0.5607 (5) | −0.1144 (2) | 0.0515 (10) | |
| C44 | 0.5658 (8) | 0.4716 (6) | −0.1407 (2) | 0.0737 (14) | |
| H44A | 0.5995 | 0.5168 | −0.1877 | 0.111* | |
| H44B | 0.5421 | 0.3545 | −0.1378 | 0.111* | |
| H44C | 0.6939 | 0.4859 | −0.1136 | 0.111* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S1 | 0.0437 (6) | 0.0641 (8) | 0.0497 (6) | −0.0147 (5) | −0.0073 (5) | −0.0046 (5) |
| O1 | 0.064 (2) | 0.069 (2) | 0.0441 (17) | −0.0068 (16) | −0.0058 (15) | −0.0138 (14) |
| O2 | 0.085 (3) | 0.149 (4) | 0.062 (2) | −0.049 (3) | 0.0066 (19) | −0.001 (2) |
| N1 | 0.046 (2) | 0.052 (2) | 0.0449 (19) | −0.0003 (17) | −0.0026 (17) | −0.0098 (16) |
| C1 | 0.053 (3) | 0.044 (3) | 0.043 (2) | 0.002 (2) | 0.002 (2) | −0.0083 (18) |
| C2 | 0.042 (2) | 0.040 (2) | 0.040 (2) | −0.0033 (18) | 0.0031 (19) | −0.0058 (18) |
| C3 | 0.044 (2) | 0.046 (3) | 0.051 (3) | 0.0025 (19) | 0.001 (2) | −0.007 (2) |
| C4 | 0.048 (3) | 0.047 (3) | 0.048 (2) | −0.002 (2) | 0.008 (2) | −0.0152 (19) |
| C5 | 0.047 (3) | 0.041 (2) | 0.039 (2) | −0.011 (2) | −0.002 (2) | −0.0073 (18) |
| C6 | 0.046 (2) | 0.060 (3) | 0.047 (2) | 0.005 (2) | −0.007 (2) | −0.011 (2) |
| C7 | 0.053 (3) | 0.053 (3) | 0.050 (2) | 0.006 (2) | 0.000 (2) | −0.014 (2) |
| C8 | 0.073 (3) | 0.085 (4) | 0.056 (3) | −0.014 (3) | −0.017 (3) | −0.006 (2) |
| C9 | 0.039 (2) | 0.039 (2) | 0.040 (2) | −0.0044 (18) | 0.0025 (19) | −0.0066 (17) |
| C10 | 0.041 (2) | 0.044 (3) | 0.050 (2) | 0.0023 (19) | 0.003 (2) | −0.0083 (19) |
| C11 | 0.033 (2) | 0.047 (3) | 0.050 (3) | 0.0010 (18) | −0.001 (2) | 0.0019 (19) |
| C12 | 0.040 (2) | 0.040 (2) | 0.040 (2) | −0.0035 (19) | −0.0002 (19) | −0.0033 (18) |
| C13 | 0.045 (3) | 0.049 (3) | 0.046 (2) | 0.002 (2) | −0.001 (2) | −0.0119 (19) |
| C14 | 0.039 (2) | 0.045 (3) | 0.048 (2) | 0.0059 (19) | −0.006 (2) | −0.006 (2) |
| C15 | 0.041 (2) | 0.037 (2) | 0.041 (2) | 0.0025 (18) | −0.0080 (18) | −0.0080 (17) |
| C16 | 0.041 (2) | 0.047 (2) | 0.040 (2) | −0.0034 (19) | −0.0093 (19) | −0.0110 (18) |
| C17 | 0.040 (2) | 0.059 (3) | 0.046 (2) | −0.009 (2) | −0.006 (2) | −0.011 (2) |
| C18 | 0.045 (2) | 0.049 (3) | 0.042 (2) | 0.006 (2) | −0.0108 (19) | −0.0064 (19) |
| C19 | 0.054 (3) | 0.055 (3) | 0.039 (2) | −0.001 (2) | −0.015 (2) | −0.0109 (19) |
| C20 | 0.047 (3) | 0.050 (3) | 0.048 (3) | −0.004 (2) | −0.015 (2) | −0.012 (2) |
| C21 | 0.054 (3) | 0.065 (3) | 0.050 (3) | 0.005 (2) | −0.005 (2) | −0.003 (2) |
| C22 | 0.114 (4) | 0.078 (4) | 0.047 (3) | 0.001 (3) | −0.002 (3) | −0.010 (2) |
| S2 | 0.0423 (6) | 0.0712 (8) | 0.0421 (6) | −0.0166 (5) | −0.0022 (5) | −0.0097 (5) |
| O3 | 0.078 (2) | 0.073 (2) | 0.0477 (18) | −0.0194 (17) | −0.0165 (16) | −0.0049 (15) |
| O4 | 0.118 (3) | 0.082 (2) | 0.0437 (18) | 0.014 (2) | −0.0032 (18) | 0.0140 (17) |
| N2 | 0.051 (2) | 0.053 (2) | 0.041 (2) | −0.0030 (16) | −0.0040 (17) | −0.0137 (17) |
| C23 | 0.059 (3) | 0.045 (3) | 0.056 (3) | −0.007 (2) | −0.011 (2) | −0.008 (2) |
| C24 | 0.051 (3) | 0.037 (2) | 0.049 (2) | −0.001 (2) | −0.011 (2) | −0.0106 (19) |
| C25 | 0.052 (3) | 0.051 (3) | 0.043 (2) | −0.001 (2) | 0.002 (2) | −0.0062 (19) |
| C26 | 0.047 (2) | 0.053 (3) | 0.047 (3) | −0.009 (2) | −0.004 (2) | −0.008 (2) |
| C27 | 0.050 (3) | 0.048 (3) | 0.044 (2) | −0.001 (2) | −0.008 (2) | −0.0105 (19) |
| C28 | 0.061 (3) | 0.054 (3) | 0.046 (2) | −0.008 (2) | −0.009 (2) | 0.000 (2) |
| C29 | 0.058 (3) | 0.050 (3) | 0.059 (3) | −0.013 (2) | −0.012 (2) | 0.001 (2) |
| C30 | 0.066 (3) | 0.097 (4) | 0.070 (3) | −0.024 (3) | −0.018 (3) | −0.021 (3) |
| C31 | 0.041 (2) | 0.039 (2) | 0.040 (2) | 0.0026 (18) | −0.0019 (18) | −0.0089 (18) |
| C32 | 0.038 (2) | 0.046 (3) | 0.045 (2) | −0.0111 (18) | −0.0022 (19) | −0.0047 (18) |
| C33 | 0.040 (2) | 0.048 (2) | 0.037 (2) | −0.0061 (18) | 0.0042 (18) | −0.0023 (17) |
| C34 | 0.034 (2) | 0.038 (2) | 0.036 (2) | 0.0002 (18) | −0.0014 (17) | −0.0065 (17) |
| C35 | 0.036 (2) | 0.048 (3) | 0.045 (2) | −0.0100 (18) | 0.0006 (19) | −0.0041 (19) |
| C36 | 0.044 (2) | 0.057 (3) | 0.035 (2) | −0.003 (2) | 0.0018 (19) | −0.0033 (19) |
| C37 | 0.040 (2) | 0.042 (2) | 0.036 (2) | −0.0046 (18) | −0.0063 (18) | −0.0107 (18) |
| C38 | 0.043 (2) | 0.045 (2) | 0.040 (2) | 0.0057 (19) | −0.0080 (19) | 0.0012 (17) |
| C39 | 0.041 (2) | 0.047 (3) | 0.042 (2) | 0.0021 (19) | −0.0037 (19) | −0.0063 (19) |
| C40 | 0.047 (2) | 0.039 (2) | 0.035 (2) | −0.0056 (18) | −0.0084 (18) | −0.0067 (17) |
| C41 | 0.054 (3) | 0.045 (3) | 0.044 (2) | 0.005 (2) | −0.016 (2) | −0.0025 (18) |
| C42 | 0.041 (2) | 0.049 (3) | 0.049 (2) | 0.0074 (19) | −0.007 (2) | −0.011 (2) |
| C43 | 0.069 (3) | 0.050 (3) | 0.037 (2) | −0.009 (2) | −0.010 (2) | −0.006 (2) |
| C44 | 0.073 (3) | 0.092 (4) | 0.053 (3) | −0.004 (3) | 0.013 (3) | −0.005 (3) |
Geometric parameters (Å, º)
| S1—C15 | 1.765 (4) | S2—C37 | 1.775 (3) |
| S1—C12 | 1.776 (4) | S2—C34 | 1.779 (3) |
| O1—C5 | 1.363 (4) | O3—C27 | 1.362 (4) |
| O1—C8 | 1.430 (5) | O3—C30 | 1.424 (5) |
| O2—C21 | 1.210 (5) | O4—C43 | 1.207 (5) |
| N1—C1 | 1.252 (4) | N2—C23 | 1.260 (4) |
| N1—C9 | 1.417 (4) | N2—C31 | 1.429 (4) |
| C1—C2 | 1.459 (5) | C23—C24 | 1.464 (5) |
| C1—H1 | 0.9300 | C23—H23 | 0.9300 |
| C2—C7 | 1.391 (5) | C24—C29 | 1.378 (5) |
| C2—C3 | 1.392 (5) | C24—C25 | 1.389 (5) |
| C3—C4 | 1.369 (5) | C25—C26 | 1.391 (5) |
| C3—H3 | 0.9300 | C25—H25 | 0.9300 |
| C4—C5 | 1.386 (5) | C26—C27 | 1.385 (5) |
| C4—H4 | 0.9300 | C26—H26 | 0.9300 |
| C5—C6 | 1.381 (5) | C27—C28 | 1.372 (5) |
| C6—C7 | 1.380 (5) | C28—C29 | 1.376 (5) |
| C6—H6 | 0.9300 | C28—H28 | 0.9300 |
| C7—H7 | 0.9300 | C29—H29 | 0.9300 |
| C8—H8A | 0.9600 | C30—H30A | 0.9600 |
| C8—H8B | 0.9600 | C30—H30B | 0.9600 |
| C8—H8C | 0.9600 | C30—H30C | 0.9600 |
| C9—C10 | 1.385 (5) | C31—C36 | 1.388 (5) |
| C9—C14 | 1.391 (5) | C31—C32 | 1.391 (5) |
| C10—C11 | 1.381 (5) | C32—C33 | 1.382 (5) |
| C10—H10 | 0.9300 | C32—H32 | 0.9300 |
| C11—C12 | 1.381 (5) | C33—C34 | 1.382 (5) |
| C11—H11 | 0.9300 | C33—H33 | 0.9300 |
| C12—C13 | 1.386 (5) | C34—C35 | 1.387 (5) |
| C13—C14 | 1.374 (5) | C35—C36 | 1.382 (5) |
| C13—H13 | 0.9300 | C35—H35 | 0.9300 |
| C14—H14 | 0.9300 | C36—H36 | 0.9300 |
| C15—C20 | 1.389 (5) | C37—C42 | 1.382 (5) |
| C15—C16 | 1.389 (5) | C37—C38 | 1.385 (5) |
| C16—C17 | 1.379 (5) | C38—C39 | 1.370 (5) |
| C16—H16 | 0.9300 | C38—H38 | 0.9300 |
| C17—C18 | 1.387 (5) | C39—C40 | 1.383 (5) |
| C17—H17 | 0.9300 | C39—H39 | 0.9300 |
| C18—C19 | 1.387 (5) | C40—C41 | 1.386 (5) |
| C18—C21 | 1.486 (5) | C40—C43 | 1.491 (5) |
| C19—C20 | 1.374 (5) | C41—C42 | 1.384 (5) |
| C19—H19 | 0.9300 | C41—H41 | 0.9300 |
| C20—H20 | 0.9300 | C42—H42 | 0.9300 |
| C21—C22 | 1.498 (6) | C43—C44 | 1.485 (6) |
| C22—H22A | 0.9600 | C44—H44A | 0.9600 |
| C22—H22B | 0.9600 | C44—H44B | 0.9600 |
| C22—H22C | 0.9600 | C44—H44C | 0.9600 |
| C15—S1—C12 | 105.80 (17) | C37—S2—C34 | 105.96 (16) |
| C5—O1—C8 | 118.5 (3) | C27—O3—C30 | 118.7 (3) |
| C1—N1—C9 | 121.9 (3) | C23—N2—C31 | 119.6 (3) |
| N1—C1—C2 | 122.5 (3) | N2—C23—C24 | 123.6 (4) |
| N1—C1—H1 | 118.8 | N2—C23—H23 | 118.2 |
| C2—C1—H1 | 118.8 | C24—C23—H23 | 118.2 |
| C7—C2—C3 | 118.1 (4) | C29—C24—C25 | 118.2 (3) |
| C7—C2—C1 | 120.8 (3) | C29—C24—C23 | 119.2 (4) |
| C3—C2—C1 | 121.1 (3) | C25—C24—C23 | 122.6 (4) |
| C4—C3—C2 | 120.6 (4) | C24—C25—C26 | 120.9 (4) |
| C4—C3—H3 | 119.7 | C24—C25—H25 | 119.5 |
| C2—C3—H3 | 119.7 | C26—C25—H25 | 119.5 |
| C3—C4—C5 | 120.5 (4) | C27—C26—C25 | 119.4 (4) |
| C3—C4—H4 | 119.7 | C27—C26—H26 | 120.3 |
| C5—C4—H4 | 119.7 | C25—C26—H26 | 120.3 |
| O1—C5—C6 | 124.7 (4) | O3—C27—C28 | 114.8 (4) |
| O1—C5—C4 | 115.4 (3) | O3—C27—C26 | 125.4 (4) |
| C6—C5—C4 | 119.9 (3) | C28—C27—C26 | 119.8 (4) |
| C7—C6—C5 | 119.2 (4) | C27—C28—C29 | 120.3 (4) |
| C7—C6—H6 | 120.4 | C27—C28—H28 | 119.9 |
| C5—C6—H6 | 120.4 | C29—C28—H28 | 119.9 |
| C6—C7—C2 | 121.6 (4) | C28—C29—C24 | 121.4 (4) |
| C6—C7—H7 | 119.2 | C28—C29—H29 | 119.3 |
| C2—C7—H7 | 119.2 | C24—C29—H29 | 119.3 |
| O1—C8—H8A | 109.5 | O3—C30—H30A | 109.5 |
| O1—C8—H8B | 109.5 | O3—C30—H30B | 109.5 |
| H8A—C8—H8B | 109.5 | H30A—C30—H30B | 109.5 |
| O1—C8—H8C | 109.5 | O3—C30—H30C | 109.5 |
| H8A—C8—H8C | 109.5 | H30A—C30—H30C | 109.5 |
| H8B—C8—H8C | 109.5 | H30B—C30—H30C | 109.5 |
| C10—C9—C14 | 118.5 (3) | C36—C31—C32 | 118.3 (3) |
| C10—C9—N1 | 115.5 (3) | C36—C31—N2 | 125.3 (3) |
| C14—C9—N1 | 126.0 (3) | C32—C31—N2 | 116.4 (3) |
| C11—C10—C9 | 120.8 (3) | C33—C32—C31 | 121.1 (3) |
| C11—C10—H10 | 119.6 | C33—C32—H32 | 119.5 |
| C9—C10—H10 | 119.6 | C31—C32—H32 | 119.5 |
| C12—C11—C10 | 120.3 (3) | C34—C33—C32 | 120.0 (3) |
| C12—C11—H11 | 119.8 | C34—C33—H33 | 120.0 |
| C10—C11—H11 | 119.8 | C32—C33—H33 | 120.0 |
| C11—C12—C13 | 119.2 (3) | C33—C34—C35 | 119.6 (3) |
| C11—C12—S1 | 118.3 (3) | C33—C34—S2 | 125.6 (3) |
| C13—C12—S1 | 122.2 (3) | C35—C34—S2 | 114.7 (3) |
| C14—C13—C12 | 120.4 (3) | C36—C35—C34 | 120.0 (4) |
| C14—C13—H13 | 119.8 | C36—C35—H35 | 120.0 |
| C12—C13—H13 | 119.8 | C34—C35—H35 | 120.0 |
| C13—C14—C9 | 120.7 (3) | C35—C36—C31 | 120.9 (3) |
| C13—C14—H14 | 119.6 | C35—C36—H36 | 119.5 |
| C9—C14—H14 | 119.6 | C31—C36—H36 | 119.5 |
| C20—C15—C16 | 119.3 (3) | C42—C37—C38 | 119.2 (3) |
| C20—C15—S1 | 115.1 (3) | C42—C37—S2 | 117.6 (3) |
| C16—C15—S1 | 125.5 (3) | C38—C37—S2 | 122.8 (3) |
| C17—C16—C15 | 119.7 (3) | C39—C38—C37 | 120.1 (3) |
| C17—C16—H16 | 120.2 | C39—C38—H38 | 120.0 |
| C15—C16—H16 | 120.2 | C37—C38—H38 | 120.0 |
| C16—C17—C18 | 121.8 (3) | C38—C39—C40 | 121.7 (3) |
| C16—C17—H17 | 119.1 | C38—C39—H39 | 119.2 |
| C18—C17—H17 | 119.1 | C40—C39—H39 | 119.2 |
| C17—C18—C19 | 117.5 (3) | C39—C40—C41 | 117.9 (3) |
| C17—C18—C21 | 120.0 (4) | C39—C40—C43 | 123.4 (3) |
| C19—C18—C21 | 122.5 (3) | C41—C40—C43 | 118.7 (3) |
| C20—C19—C18 | 121.8 (3) | C42—C41—C40 | 121.0 (3) |
| C20—C19—H19 | 119.1 | C42—C41—H41 | 119.5 |
| C18—C19—H19 | 119.1 | C40—C41—H41 | 119.5 |
| C19—C20—C15 | 119.9 (4) | C37—C42—C41 | 120.1 (3) |
| C19—C20—H20 | 120.1 | C37—C42—H42 | 119.9 |
| C15—C20—H20 | 120.1 | C41—C42—H42 | 119.9 |
| O2—C21—C18 | 120.4 (4) | O4—C43—C44 | 120.1 (4) |
| O2—C21—C22 | 120.1 (4) | O4—C43—C40 | 120.3 (4) |
| C18—C21—C22 | 119.5 (4) | C44—C43—C40 | 119.6 (4) |
| C21—C22—H22A | 109.5 | C43—C44—H44A | 109.5 |
| C21—C22—H22B | 109.5 | C43—C44—H44B | 109.5 |
| H22A—C22—H22B | 109.5 | H44A—C44—H44B | 109.5 |
| C21—C22—H22C | 109.5 | C43—C44—H44C | 109.5 |
| H22A—C22—H22C | 109.5 | H44A—C44—H44C | 109.5 |
| H22B—C22—H22C | 109.5 | H44B—C44—H44C | 109.5 |
| C9—N1—C1—C2 | 179.7 (3) | C31—N2—C23—C24 | −177.4 (3) |
| N1—C1—C2—C7 | 179.5 (4) | N2—C23—C24—C29 | 162.3 (4) |
| N1—C1—C2—C3 | 0.6 (6) | N2—C23—C24—C25 | −15.8 (6) |
| C7—C2—C3—C4 | −1.2 (6) | C29—C24—C25—C26 | −1.2 (6) |
| C1—C2—C3—C4 | 177.8 (3) | C23—C24—C25—C26 | 176.9 (4) |
| C2—C3—C4—C5 | −0.8 (5) | C24—C25—C26—C27 | 0.3 (6) |
| C8—O1—C5—C6 | −1.3 (5) | C30—O3—C27—C28 | −175.9 (4) |
| C8—O1—C5—C4 | 177.8 (4) | C30—O3—C27—C26 | 3.3 (6) |
| C3—C4—C5—O1 | −177.2 (3) | C25—C26—C27—O3 | −178.3 (3) |
| C3—C4—C5—C6 | 2.0 (6) | C25—C26—C27—C28 | 0.8 (6) |
| O1—C5—C6—C7 | 178.0 (4) | O3—C27—C28—C29 | 178.3 (4) |
| C4—C5—C6—C7 | −1.2 (6) | C26—C27—C28—C29 | −0.9 (6) |
| C5—C6—C7—C2 | −0.9 (6) | C27—C28—C29—C24 | 0.0 (6) |
| C3—C2—C7—C6 | 2.0 (6) | C25—C24—C29—C28 | 1.1 (6) |
| C1—C2—C7—C6 | −177.0 (4) | C23—C24—C29—C28 | −177.1 (4) |
| C1—N1—C9—C10 | −179.3 (4) | C23—N2—C31—C36 | 22.6 (5) |
| C1—N1—C9—C14 | 0.0 (5) | C23—N2—C31—C32 | −159.0 (4) |
| C14—C9—C10—C11 | 0.4 (5) | C36—C31—C32—C33 | −2.3 (5) |
| N1—C9—C10—C11 | 179.7 (3) | N2—C31—C32—C33 | 179.2 (3) |
| C9—C10—C11—C12 | 0.1 (5) | C31—C32—C33—C34 | 0.1 (5) |
| C10—C11—C12—C13 | −0.4 (5) | C32—C33—C34—C35 | 1.8 (5) |
| C10—C11—C12—S1 | 173.4 (3) | C32—C33—C34—S2 | 179.4 (3) |
| C15—S1—C12—C11 | 121.4 (3) | C37—S2—C34—C33 | 5.3 (4) |
| C15—S1—C12—C13 | −65.0 (3) | C37—S2—C34—C35 | −176.9 (3) |
| C11—C12—C13—C14 | 0.2 (5) | C33—C34—C35—C36 | −1.5 (5) |
| S1—C12—C13—C14 | −173.3 (3) | S2—C34—C35—C36 | −179.4 (3) |
| C12—C13—C14—C9 | 0.3 (6) | C34—C35—C36—C31 | −0.7 (5) |
| C10—C9—C14—C13 | −0.6 (5) | C32—C31—C36—C35 | 2.6 (5) |
| N1—C9—C14—C13 | −179.8 (3) | N2—C31—C36—C35 | −179.0 (3) |
| C12—S1—C15—C20 | 174.3 (3) | C34—S2—C37—C42 | −122.2 (3) |
| C12—S1—C15—C16 | −6.2 (4) | C34—S2—C37—C38 | 64.7 (3) |
| C20—C15—C16—C17 | −0.7 (5) | C42—C37—C38—C39 | −1.0 (5) |
| S1—C15—C16—C17 | 179.9 (3) | S2—C37—C38—C39 | 172.0 (3) |
| C15—C16—C17—C18 | −0.5 (6) | C37—C38—C39—C40 | 0.5 (6) |
| C16—C17—C18—C19 | 1.4 (5) | C38—C39—C40—C41 | 0.6 (6) |
| C16—C17—C18—C21 | −179.2 (4) | C38—C39—C40—C43 | −178.5 (4) |
| C17—C18—C19—C20 | −1.1 (6) | C39—C40—C41—C42 | −1.1 (5) |
| C21—C18—C19—C20 | 179.5 (4) | C43—C40—C41—C42 | 178.0 (3) |
| C18—C19—C20—C15 | −0.1 (6) | C38—C37—C42—C41 | 0.4 (5) |
| C16—C15—C20—C19 | 1.0 (5) | S2—C37—C42—C41 | −173.0 (3) |
| S1—C15—C20—C19 | −179.5 (3) | C40—C41—C42—C37 | 0.7 (5) |
| C17—C18—C21—O2 | −9.5 (6) | C39—C40—C43—O4 | −172.4 (4) |
| C19—C18—C21—O2 | 169.9 (4) | C41—C40—C43—O4 | 8.5 (6) |
| C17—C18—C21—C22 | 170.0 (4) | C39—C40—C43—C44 | 7.1 (6) |
| C19—C18—C21—C22 | −10.6 (6) | C41—C40—C43—C44 | −172.0 (4) |
Hydrogen-bond geometry (Å, º)
Cg1, Cg2, Cg3 and Cg6 are the centroids of the C2–C7, C9–C14, C15–C20 and C37–C42 rings, respectively.
| D—H···A | D—H | H···A | D···A | D—H···A |
| C17—H17···Cg6i | 0.93 | 3.00 | 3.734 (4) | 137 |
| C26—H26···Cg1 | 0.93 | 2.96 | 3.763 (4) | 146 |
| C32—H32···Cg2 | 0.93 | 2.98 | 3.706 (4) | 136 |
| C41—H41···Cg3ii | 0.93 | 2.99 | 3.670 (4) | 131 |
Symmetry codes: (i) x+1, y+1, z; (ii) x−1, y, z.
Footnotes
Supporting information for this paper is available from the IUCr electronic archives (Reference: SU5056).
References
- Ahamad, I., Prasad, R. & Quraishi, M. A. (2010). Corros. Sci. 52, 933–942.
- Bruker (2006). APEX2 and SAINT. Bruker AXS Inc., Madison, Wisconsin, USA.
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Hebbachi, R. & Benali-Cherif, N. (2005). Acta Cryst. E61, m1188–m1190.
- Hebbachi, R., Mousser, H. & Mousser, A. (2013). Acta Cryst. E69, o67–o68. [DOI] [PMC free article] [PubMed]
- Izatt, R. M., Pawlak, K., Bradshaw, J. S. & Bruening, R. L. (1995). Chem. Rev. 95, 2529–2586.
- Kahwa, I. A., Selbin, J., Hsieh, T. C.-Y. & Laine, R. A. (1986). Inorg. Chim. Acta, 118, 179–185.
- Kalcher, K., Kauffmann, J. M., Wang, J., Švancara, I., Vytřas, K., Neuhold, C. & Yang, Z. (1995). Electroanalysis, 7, 5–22.
- Macrae, C. F., Bruno, I. J., Chisholm, J. A., Edgington, P. R., McCabe, P., Pidcock, E., Rodriguez-Monge, L., Taylor, R., van de Streek, J. & Wood, P. A. (2008). J. Appl. Cryst. 41, 466–470.
- Negm, N. A., Elkholy, Y. M., Zahran, M. K. & Tawfik, S. M. (2010). Corros. Sci. 52, 3523–3536.
- Özkar, S., Ülkü, D., Yıldırım, L. T., Biricik, N. & Gümgüm, B. (2004). J. Mol. Struct. 688, 207–211.
- Parsons, S., Flack, H. D. & Wagner, T. (2013). Acta Cryst. B69, 249–259. [DOI] [PMC free article] [PubMed]
- Santos, M. L. P., Bagatin, I. A., Pereira, E. M. & Da Costa Ferreira, A. M. (2001). J. Chem. Soc. Dalton Trans. pp. 838–844.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2015). Acta Cryst. C71, 3–8.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, Global. DOI: 10.1107/S205698901500033X/su5056sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S205698901500033X/su5056Isup2.hkl
Supporting information file. DOI: 10.1107/S205698901500033X/su5056Isup3.cml
. DOI: 10.1107/S205698901500033X/su5056fig1.tif
The molecular structure of the two independent molecules (A and B) of the title compound, with atom labelling. Displacement ellipsoids are drawn at the 50% probability level.
b . DOI: 10.1107/S205698901500033X/su5056fig2.tif
A view along the b axis of the crystal packing of the title compound. C-H⋯π interactions are shown as dashed lines (see Table 1 for details; molecule A is red; molecule B is blue; H atoms not involved in these interactions have been omitted for clarity).
CCDC reference: 1042562
Additional supporting information: crystallographic information; 3D view; checkCIF report


