In nature, the extracellular matrix (ECM) is finely tuned in mechanics and biochemical composition to support the rich function of the corresponding cells, tissues and organs it resides. Thus, materials that attempt to mimic the ECM must carefully select the composition and optimize it to favor a particular cell type, tissue or organ. Depending on the number of components to be introduced and the interactions present between the different components the composition that would be ideal to use to guide a specific cell fate is not obvious. However, far to often we resign ourselves to mixing equimolar amounts of each component or select some conditions to test using incomplete information. Thus, when designing hydrogel scaffolds with more than one component (e.g. multiple growth factors, peptides, etc), using traditional “one factor at a time” studies is not only inefficient due to the number of conditions needed to sample sufficient concentration space but also impractical due to the inability to understand complex interactions between multiple factors.
Herein, we show that for the purpose of differentiating neural progenitor cells into mature neurons the optimization of concentration of peptide components (e.i. RGD, YIGSR, IKVAV) is essential to arrive at a composition that leads to differentiation. We find that the “typical” equimolar concentration of each factor does not result in a matrix that can direct differentiation and that only the hydrogel with optimized concentration can. We find that the optimized hydrogel not only results in increased number of mature neurons but it also accelerates differentiation compared to 2D differentiation strategies observing differentiated cells at 7 days of culture.
We employed multifactorial experiments to efficiently and methodically understand the effects of three ECM derived peptide ligands, RGD, YIGSR and IKVAV, on neural progenitor survival and differentiation. These peptide ligands are known to be important in engineered materials for neuronal differentiation [1–2]. Response surface methods (RSM) are a type of multifactorial experiment designed to estimate interactions between different factors to predict the shape of the response surface investigated. [3] This method systematically modulates the factors of interest to better understand the effects of each individual factor as well as the interacting factor effects on the system response. Box-Behnken and central composite designs are the two most common RSM designs and are chosen and modified based on the amount of time and resources available as well as the surface response accuracy required. These strategies have recently gained more traction in the academic world where several groups have used this approach to optimize responses across various systems. [4–6] For example, central composite designs were used to optimize ligand concentrations within a peptide matrix for the two-dimensional proliferation of endothelial cells. [6] Statistical approaches like this are more common in the bioprocessing industry [7–8], and pharmaceuticals [9] where complex processes involve many input factors.
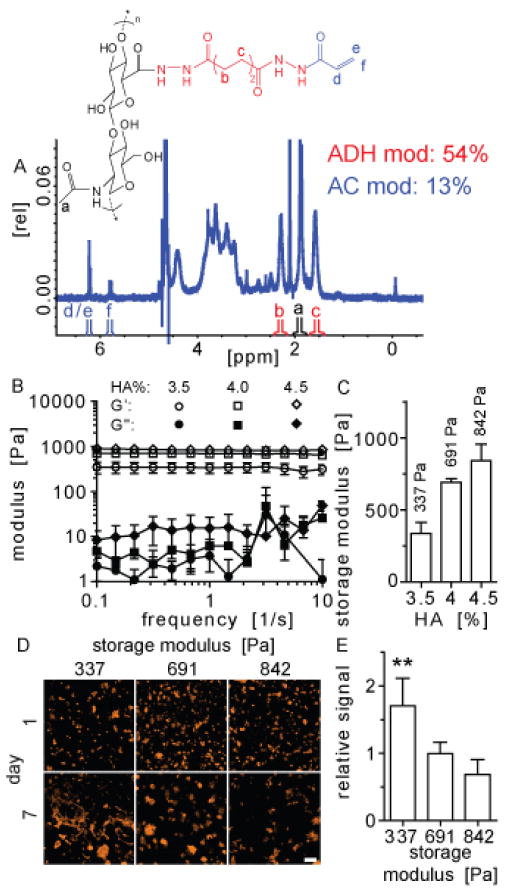
As the hydrogel backbone we utilized our established hyaluronic acid hydrogel, which is composed of 60kDa acrylated hyaluronic acid crosslinked using di-thiol containing peptides through Michael type addition. [10] This chemistry has been used extensively to covalently form hydrogels for tissue engineering applications. [11–13] Acrylate groups are added to the HA backbone through a two-step chemical reaction with a final carboxylic acid to acrylate modification of 13% confirmed via H-NMR (HA-AC, Figure 1A). Hyaluronic acid is a non-sulfated glycosaminoglycan composed of two repeat units (D-glucoronic acid and N-acetyl-D-glucosamine) that is naturally occurring and produced throughout the body. [14–15] The mechanical properties of the hydrogel can be modulated by adjusting the hyaluronic acid content. This is important as matrix mechanics have been shown to play a key role in stem cell fate. [16–17] We varied HA weight percent in the hydrogels from 3.5–4.5% and determined the storage modulus (G′) ranged from ~337 ± 76.22 Pa to 841.9 ± 112.5 Pa via a rheometry (Figure 1B–C).
Figure 1.
Hyaluronic acid was modified to contain an acrylate functional group and (A) degree of modification (16%) was confirmed via NMR spectroscopy. Hydrogel formulations with different hyaluronic acid concentrations (3.5–4.5%) were made and (B–C) the mechanical properties characterized. (D) F-actin of iPS-NPCs encapsulated in these three gel conditions were stained at days 1 and 7 and (E) cell spreading was greatest in the softest gel (3.5%). The symbol ** represents statistical significance to the level of p < 0.01. Statistical significance was determined using multiple comparisons with a Tukey post hoc test.
Induced pluripotent stem cells have great potential for clinical applications due to their ability to form all three germ layers. [18–19] We utilized neural progenitor cells derived from induced pluripotent stem cells (iPS-NPCs) to test our hypothesis that peptide component optimization through DOE would result in a hydrogel that is superior in its ability to support differentiation of these cells than adding one single peptide or equimolar concentrations of the three peptides. Before beginning the differentiation we determined what hydrogel storage modulus resulted in the most spreading. iPS-NPC cells were encapsulated in hydrogels with three different stiffnesses and the cells were fixed and stained with phalloidin at days 1 and 7 after gelation to investigate cell spreading (Figure 1D). The amount of phalloidin signal at day 7 was normalized to day 1 and the 337 Pa HA condition produced significantly more cell spreading than the 690.8 Pa and 841.9 Pa HA hydrogels (Figure 1E). The increased spreading could be due to the fact that this hydrogel has mechanical properties similar to the brain which could also play a role in guiding the differentiation of the NPCs. [20–21] Thus, a hydrogel with 337 Pa storage modulus was used for the remainder of the study.
To investigate the effect that the three adhesion ligands (RGD, YIGSR, IKVAV) have on encapsulated stem cell behavior, statistical design of experiments (DOE) was used. This approach enabled us to systematically modulate the multiple adhesion ligands to gain an understanding of not only their individual effects, but their interacting effects as well. For each iteration, a response surface methodology experiment was developed in which each variable factor (adhesion ligand) was given a defined range (μM) to be modulated within. Central composite on face designs were utilized because we were equally interested in the entire response surface and our system enabled us to test the end points of each factor. While our ultimate goal is to develop a matrix that promotes differentiation, a higher throughput-screening assay was needed. We decided to investigate cell survival one-week post-encapsulation by measuring DNA content within the gels. Data from hydrogels at day 7 were normalized to gels from the same condition at day 1 to account for any differences in the numbers of cells encapsulated in the gel precursor solution.
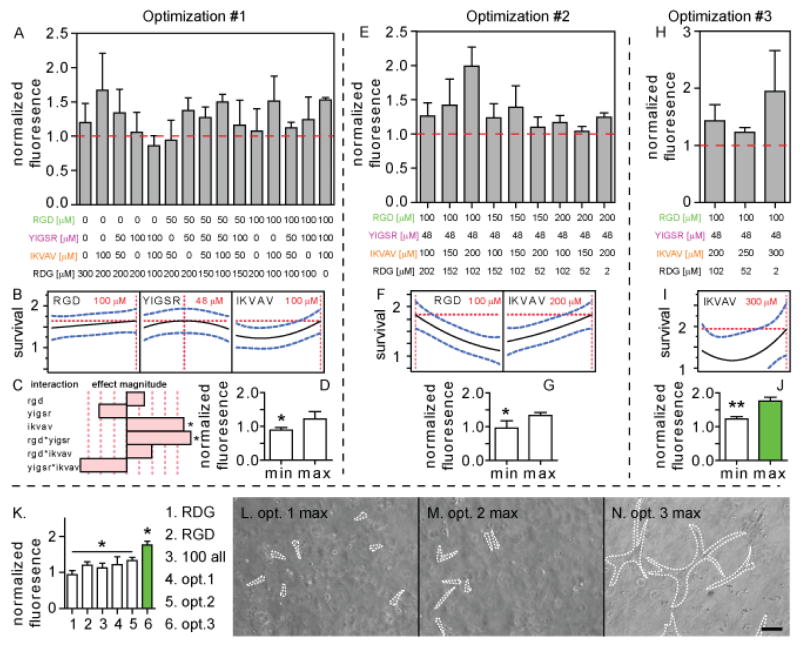
The initial central composite on face design contained 16 different hydrogel conditions in which the three-peptide concentrations were varied from 0–100 μM. To ensure that the total peptide concentration in each condition remained constant, a scrambled RGD sequence (RDG, Ac-GCGYRDGSP-NH2) was used as a “filler” peptide (Figure 2A). Because each condition was done in triplicate and the CCI design mandates the center point condition be duplicated we show the 15 true conditions. The results seen illustrate how difficult it would be to determine the effects of the peptides on survival without the use of this statistical design of experiments approach. After inputting these results into the DOE software, a least squares polynomial regression was done to estimate the effect of each peptide on survival. This model then predicted peptide concentrations that would give the greatest (RGD: 100 μM, YIGSR: 48 μM, IKVAV: 100 μM) and least cell survival (RGD: 0 μM, YIGSR: 100 μM, IKVAV: 55 μM, Figure 2B, Supplementary information, Figure S1A). The results from the first optimization indicate that the IKVAV peptide has the greatest effect on the survival (Figure 2C, Supplementary information, Figure S1B–D). The laminin-derived IKVAV motif has previously been found to have an effect on neuronal precursor cell adhesion to hydrogels. [22] For two-factor interactions, the RGD/YIGSR combination had a significant effect as well. The optimal maximum and minimum peptide conditions were used in a confirmation experiment to test their effect on cell survival with a significant difference observed (Figure 2D).
Figure 2.
Statistical designs of experiments approach was used to (A) investigate the interactions between RGD, YIGSR and IKVAV adhesion peptides (0–100 μM each) and their ability to promote cell survival. Experiments give a (B) predictive max peptide mixture and (C) the effect magnitude of all factors and two-factor interactions. Confirmation of (D) max and min predictive peptide mixture. (E–G) A second series of optimization experiments to test higher peptide concentrations for RGD and IKVAV (100–200 μM each). (H–J) A third series of optimization experiments to test higher peptide concentration for IKVAV (200–300 μM). (K) Validation of the three optimized conditions compared to RDG, RGD and 100 μM of each peptide. Phase images of the (L–N) three optimized conditions with spread cells outlined. Scale bar = 100 μm.
The maximum predicted survival condition for the RGD and IKVAV motif’s occurred at the upper limit of the initial range tested. Thus, we performed a second central composite on face design in which the concentration of those two peptides was raised from 100–200 μM while keeping the YIGSR motif constant at the concentration given during the first optimization (48 μM, Figure 2E). The same survival assay was performed and the results were fit into another least-squares polynomial regression. The results indicate that the maximum is at 100 μM RGD and 200 μM IKVAV and the minimum at 199 μM RGD and 166 μM IKVAV (Figure 2F, Supplementary information, Figure S2A). These optimal maximum and minimum peptide concentrations were used to confirm the predictive model (Figure 2G). While the predicted RGD maximum concentration was at the lower limit of 100 μM for this second optimization iteration, it was at the upper limit of 100 μM during the first optimization iteration. Thus, the RGD concentration was kept constant at 100 μM for the next optimization.
A third optimization was done for the IKVAV motif from 200–300 μM while keeping RGD and YIGSR constant at 100 and 48 μM, respectively (Figure 2H). The same protocol as the first two optimizations was performed with a least-squares polynomial regression fit to data obtained from the survival assay. The maximum concentration given was 300 μM IKVAV with a minimum of 235 μM (Figure 2I, Supplementary information, Figure S2C). These concentrations were used with the pre-set RGD (100 μM) and YIGSR (48 μM) concentrations, to perform a confirmation survival assay with the maximum condition producing significantly higher survival than the minimum (Figure 2J). It is also interesting to note that the predicted minimum formulations in each of the optimization iteration have been improving in addition to the predicted maximum formulations. This should be expected as each iteration has brought the “global” survival results higher. While the regression fit predicted an optimum survival formulation with over 300 μM of IKVAV, we did not want to overload the system with more peptides as that can affect the mechanical properties of the system and introduce another variable. [23]
A final round of confirmation experiments were performed to test whether the optimized formulations did increase encapsulated iPS-NPC survival. The maximum survival conditions from each optimization iteration were compared to a 450 μM RDG control, 300 μM RGD condition (with 150 μM RDG), and a 100 all condition (100 μM RGD, 100 μM YIGSR, 100 μM IKVAV, 50 μM RDG, Figure 2K). The last two conditions were added to test whether this design of experiments approach improved survival over more traditional hydrogel formulations that would include just RGD or would evenly distribute the concentrations of the active RGD, YIGSR and IKVAV motifs. While the average survival increased with each optimization maximum, there was no statistical difference in survival between the first two optimization maximums and the RGD or 100 all conditions. However, the third and final maximum condition was found to statistically improve encapsulated iPS-NPC survival over the traditional RGD and 100 all conditions as well as the first and second optimization maximums. This result confirms the benefit of this statistical approach of modulating motif concentrations to improve survival versus using a standard approach of evenly distributing the motifs. The optimized formulations also improved general cell spreading within the hydrogels (Figure 2L–N).
With an optimal hydrogel survival formulation, the next step was to investigate differentiation via immunohistochemistry. Cells were plated in 2D and cells were encapsulated in three hydrogel conditions: 100 all, RGD only, and the final optimized peptide formulation. Cell’s were cultured for one week, fixed and stained for an early marker of differentiation of NPCs towards neurons (DOUBLECORTIN, DCX, Figure 3A–D). DCX is a microtubule associated protein expressed in immature neurons and located along growing extensions. [24] DOUBLECORTIN positive cells were observed throughout all 4 conditions. However, the location of the expression is different, with the un-optimized conditions containing DCX only in the cell body, while the optimized condition containing DCX in their extensions consistent with immature neuron differentiation (Figure 3B–D). The DCX extension length was measured and found to be statistically higher for the optimized condition (Figure 3F). We proceeded to stain for a marker of mature neurons (tuj1) since all 4 conditions were positive for DOUBLECORTIN (Figure 3G–J). Tuj1 is a neuronal protein that helps stabilize microtubules in the cell body and axons of mature neurons. Virtually no tuj1 positive cells were observed in the 2D condition (<1%), while over 20–30% were observed across the 3D hydrogel conditions (Figure 3K). This data indicates that culturing NPCs in our 3D hydrogel environment promotes NPC differentiation to mature neurons faster than the traditional 2D culture. Mature neuronal markers were not observed for 2D cultured cells until 2 weeks of culture (Supplementary information, Figure S3). This strategy could be used to promote the differentiation of encapsulated pluripotent stem cells in different hydrogel transplantation based therapies. Similar to the DCX staining, Tuj1 was observed in the cell body only for the un-optimized condition, while it was present in the cell extensions for the optimized condition. The tuj1 extension length was measured and found to be statistically higher for the optimized condition (Figure 3L).
Figure 3.
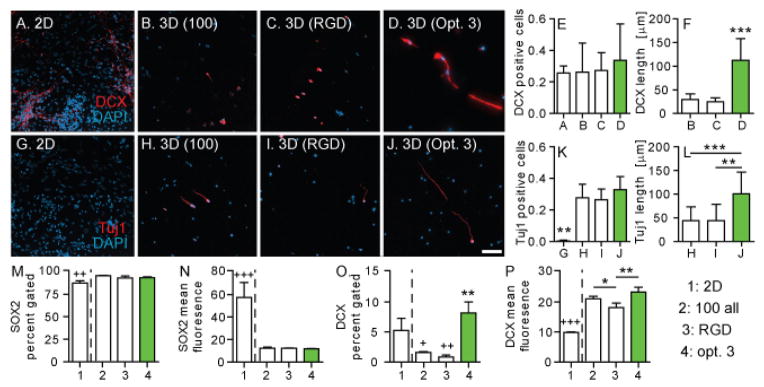
iPS-NPCs cultured for one week in (A) 2-D, and (B–D) hydrogels were stained for DOUBLECORTIN (DCX) and DAPI. Number of (E) DCX positive cells and (F) length were quantified. Cells were also stained for (G–J) tuj1 and DAPI with (K) tuj1 positive cells and (L) length quantified. Flow cytometry data for (M–N) SOX2 and (O–P) DCX indicate that the 3D hydrogels promote differentiation of the iPS-NPCs.
Flow cytometry was performed as an additional more quantitative measure of differentiation. SOX2, a neural progenitor marker, and DCX, an immature neuronal marker, were used. There was no significant difference between the hydrogel conditions for either the SOX2 percent gated or mean fluorescence values (Figure 3M–N). Compared to the 2D condition, all three hydrogel conditions produced more SOX2 positive cells (p<0.01) but significantly lower fluorescence levels (p<0.001). For the immature neuron marker (DCX), the 2D condition produced significantly more positive cells than the RGD and 100 all hydrogel conditions (p<0.05, Figure 3O). Within the hydrogel conditions the optimized hydrogel contained more DCX positive cells than the other two conditions tested (p<0.01). For mean fluorescence the 2D condition contained significantly less signal than all three hydrogel conditions (p<0.001). For the RGD hydrogel condition contained less intense signals than both the 100 all condition (p<0.05) and the optimized condition (p<0.01, Figure 3P). Optimizing the peptides for survival promoted more differentiation of the iPS-NPCs compared to the control hydrogels containing RGD only or 100 μM of each peptide. Compared to standard 2D culture, the hydrogels promoted more immature neuron differentiation, which indicates that the 3D environment is more conducive to promoting differentiation. [25]
In conclusion, we have demonstrated the benefit of using a statistical design of experiments methodology to approach systems with multiple factors. This approach enabled us to systematically modulate three bioactive signal motifs (RGD, YIGSR, IKVAV) to determine their individual and collective effects on encapsulated iPS-NPC survival. After three optimization rounds, the final iteration formulation was further investigated for its ability to promote differentiation of the NPCs. Compared to cells plated in 2D, more mature neurons were observed in our hydrogels. Furthermore, the optimized survival condition promoted more differentiation than the RGD and 100 all conditions. Overall, the data presented in this paper illustrates the benefits of using both the DOE approach in multi-factor systems and culturing NPCs in a three-dimensional hydrogel. Although we used survival to determine the optimal composition of the hydrogel, and for this system and cell type this led to a composition that promoted differentiation, ideally the optimization would be performed with the desired characteristic. However, having a system with sufficient throughput is critical. For example, the use of genetic markers that activate the expression of a reporter gene when the desired phenotype is achieved would be useful.
Experimental Section
Cell culture
iPSCs were generated using discarded anonymous human tissue (ESCRO approval: Lowry 2006-019-08) as previously described [ref 26 and others in next sentence]. hiPSC-NPCs were generated and cultured as in as in [Patterson et al 2011 Cell Research and Patterson et al 2014 Stem Cell Reports]. They were maintained in Dulbecco’s modified eagle’s medium:F12 (DMEM:F12, Sigma-Aldrich, St. Louis, MO) containing 1x B27 (Sigma-Aldrich), 1x N2, epidermal growth factor (50 ng/mL, EGF, Sigma-Aldrich), basic fibroblast growth factor (20 ng/mL, bFGF, Sigma-Aldrich), 1% penicillin/streptomycin (Invitrogen, Grand Island, NY), and 0.1% primocin (InVivoGen, San Diego, CA). They were cultured at 37 °C with 5% CO2 using standard protocols. Once encapsulated in hydrogels, the cells were cultured in the full maintenance media less the EGF and bFGF (hydrogel culture media).
Hyaluronic acid modification
Hyaluronic acid was functionalized with an acrylate group using a two-step synthesis as previously described. [10, 23] The product was analyzed with 1H-NMR (D20) and the degree of acrylation (16%) determined by dividing the multiplet peak at δ = 6.2 (cis and trans acrylate hydrogens) by the singlet peak at δ = 1.6 (singlet peak of acetyl methyl protons in HA, Figure 1A).
Hydrogel gelation
Lyophilized acrylated hyaluronic acid was dissolved in 4-(2-hydroxyethyl)-1-piperazine ethane-sulfonic acid (HEPES, 0.3 M) buffer for 15 minutes at 37 °C. The appropriate concentration of Ac-GCGYGRGDSPG-NH2 adhesion peptide (RGD, Genscript, Piscataway, NJ), Ac-GCGYGYIGSR-NH2 (YIGSR, Genscript), Ac-IKVAVGYGCG-NH2 (IKVAV, Genscript) was dissolved in 0.3 M HEPES and added to the dissolved HA-AC and allowed to react for 20 minutes at 37 °C. Hydrogel culture media, iPS-NPCs (3,000 cells/μL final concentration), were then added. An aliquot of a MMP-degradable peptide crosslinker (Ac-GCRDGPQGIWGQDRCG-NH2, Genscript) was dissolved in 0.3 M HEPES and added to the gel precursor solution. 10 μLs of this solution was pipetted onto, and sandwiched between two Sigmacote (Sigma-Aldrich) functionalized glass coverslips and placed in an incubator for 30 minutes at 37 °C to gel.
2D cell culture
Tissue culture plates were coated with poly-L-ornithine hydrobromide (100 μg/mL, Sigma-Aldrich) and incubated overnight at 37°C. The plates were washed with PBS and then coated with laminin (3 μg/mL) overnight at 37°C. After washing the plates with PBS, iPS-NPCs were seeded onto the plates.
Rheometry
Gels without cells were made as described above and the modulus was measured with a plate-to-plate rheometer (Physica MCR 301, Anton Paar, Ashland, VA) using a 8 mm plate with a frequency range of 0.1–10 rad/s under a constant strain of 1% at 37 °C.
Fixing/imaging
2D cells and 3D gels were rinsed in 1x PBS and fixed with 4% paraformaldehyde for 15 minutes at room temperature. The gels were blocked in 1x PBS + 0.1% Triton X-100 (Fisher-Scientific) + 2% normal donkey serum for 1 hour at room temperature. Next, they were incubated in primary antibody at the appropriate concentration overnight at 4°C. After the samples were incubated in the appropriate secondary antibody for 2 hours at room temperature the samples were counterstained with the nuclear marker 4′, 6-diamidino-2-phenylindole (DAPI, 1:500, Invitrogen) for 15 minutes at room temperature. For samples with phalloidin, a 90 minute incubation in rhodamine phalloidin (Invitrogen) was used. Primary antibodies were used as follows: guinea pig anti-DOUBLECORTIN (DCX, 1:2000, Milipore, Billerica, MA), chicken anti-tuj1 (1:1000, Milipore). Secondary antibodies, matching the desired primary antibody host, conjugated to cyanine 3 (1:200, Jackson Immuno Research, West Grove, PA) were used. Samples were imaged using a Nikon Confocal. Phalloiding cell spreading was quantified by dividing positive signal area at day 7 by positive signal area at day 1. DCX and Tuj1 positive cell fraction was quantified by counting the number of positive cells and normalizing to the total number of DAPI nuclei observed in same field of view. DCX and Tuj1 length was quantified by measuring the length of the longest cell dimension.
Cell survival
Cell survival was measured using the CyQUANT cell proliferation assay kit (Invitrogen). Gels were washed twice in PBS and frozen in a −80 freezer at days 1 and 7 after hydrogel gelation. After thawing the gels, they were degraded by incubation in TrypLE express (Invitrogen) at 37 °C for 15 minutes. The cells were isolated by centrifuging the solution at 250 rcf x 5 min. and re-suspended in 200 μL of the CyQUANT GR dye/cell-lysis buffer. After 5 minute incubation, the fluorescence was read using a plate reader at 480 nm.
Flow cytometry
Gels at day 1 or 7 after gelation were rinsed in PBS and degraded by incubating the cell in TrypLE and pipetting the gel up and down via a 1000 μL pipette. The solution was spun at 250 rcf for 10 min. and re-suspended in BD Cytofix (Becton Dickinson, New Jersey) for 30 minutes at room temperature. After spinning down at 250 rcf for 5 min, samples were re-suspended in 1x PBS + 1% bovine serum albumin (BSA, Fisher Scientific) + 0.2% saponin (Sigma-Aldrich) for 15 minutes at room temperature. Antibodies were then added to the solution for 30 minutes at room temperature. Following a spin at 250 rcf for 5 minuntes, the samples were re-suspended in 1% paraformaldehyde for FACS. Analysis was performed using a FACScan X and the data was analyzed using FLOWJO. Triplicates were done for each condition with 3000 events/sample. The day 1 SOX2 samples were gated such that it was 95% positive. The DCX samples were gated to contain the positive signal peak.
Design of experiments
JMP software (SAS, Cary, NC) was used to generate the hydrogel conditions and to analyze the subsequent data. A surface response methdodolgy (RSM) setup was used to vary the ligands of interest within a specific concentration range using a central composite inscribed design (CCI). Data from the cell survival assay for each condition recommended was inputted back into the software. The data was analyzed via a least squares regression model to determine significance of the factors and plot the predicted response surface.
Statistics
Statistical analysis was performed using Prism (GraphPad, San Diego, CA). Data was analyzed using either a 2-sample t test or a one way analysis of variance (ANOVA) test with a Tukey-Kramer post-test and a 95% confidence interval.
Supplementary Material
Acknowledgments
The authors would like to thank the California Institute for Regenerative Medicine (CIRM RT2-01881) for funding. JL would like to thank the NIH funded Biotechnology Training Grant (T32 GM067555) for a predoctoral fellowship. JL would also like to thank Jessica Cinkornpumin and Soheila Azghad from the Lowry Lab for preparing iPS-NPCs.
Footnotes
Supporting Information is available from the Wiley Online Library or from the author.
References
- 1.Silva GA, Czeisler C, Niece KL, Beniash E, Harrington DA, Kessler JA, Stupp SI. Science. 2004;303:1352. doi: 10.1126/science.1093783. [DOI] [PubMed] [Google Scholar]
- 2.Kam L, Shain W, Turner JN, Bizios R. Biomaterials. 2001;22:1049. doi: 10.1016/s0142-9612(00)00352-5. [DOI] [PubMed] [Google Scholar]
- 3.Collins LM, Dziak JJ, Li RZ. Psychol Methods. 2009;14:202. doi: 10.1037/a0015826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flaim CJ, Chien S, Bhatia SN. Nat Methods. 2005;2:119. doi: 10.1038/nmeth736. [DOI] [PubMed] [Google Scholar]
- 5.Decaris ML, Leach JK. Annals of Biomedical Engineering. 2011;39:1174. doi: 10.1007/s10439-010-0217-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jung JP, Moyano JV, Collier JH. Integr Biol-Uk. 2011;3:185. doi: 10.1039/c0ib00112k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumar V, Bhalla A, Rathore AS. Biotechnol Progr. 2014;30:86. doi: 10.1002/btpr.1821. [DOI] [PubMed] [Google Scholar]
- 8.Mandenius CF, Brundin A. Biotechnol Progr. 2008;24:1191. doi: 10.1002/btpr.67. [DOI] [PubMed] [Google Scholar]
- 9.Kettanehwold N. J Pharmaceut Biomed. 1991;9:605. [Google Scholar]
- 10.Lei YG, Gojgini S, Lam J, Segura T. Biomaterials. 2011;32:39. doi: 10.1016/j.biomaterials.2010.08.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lutolf MP, Tirelli N, Cerritelli S, Cavalli L, Hubbell JA. Bioconjugate Chem. 2001;12:1051. doi: 10.1021/bc015519e. [DOI] [PubMed] [Google Scholar]
- 12.Hubbell JA, Lutolf MP, Lauer-Fields JL, Schmoekel HG, Metters AT, Weber FE, Fields GB. P Natl Acad Sci USA. 2003;100:5413. doi: 10.1073/pnas.0737381100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adeloew C, Segura T, Hubbell JA, Frey P. Biomaterials. 2008;29:314. doi: 10.1016/j.biomaterials.2007.09.036. [DOI] [PubMed] [Google Scholar]
- 14.Allison DD, Grande-Allen KJ. Tissue Eng. 2006;12:2131. doi: 10.1089/ten.2006.12.2131. [DOI] [PubMed] [Google Scholar]
- 15.Fraser JRE, Laurent TC, Laurent UBG. J Intern Med. 1997;242:27. doi: 10.1046/j.1365-2796.1997.00170.x. [DOI] [PubMed] [Google Scholar]
- 16.Discher DE, Mooney DJ, Zandstra PW. Science. 2009;324:1673. doi: 10.1126/science.1171643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Discher DE, Sweeney L, Sen S, Engler A. Biophysical Journal. 2007:32a. [Google Scholar]
- 18.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Cell. 2007;131:861. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 19.Patterson M, Chan DN, Ha I, Case D, Cui YY, Van Handel B, Mikkola HKA, Lowry WE. Cell Research. 2012;22:178. doi: 10.1038/cr.2011.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Georges PC, Miller WJ, Meaney DF, Sawyer ES, Janmey PA. Biophysical Journal. 2006;90:3012. doi: 10.1529/biophysj.105.073114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aurand ER, Wagner JL, Shandas R, Bjugstad KB. Stem Cell Res. 2014;12:11. doi: 10.1016/j.scr.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng TY, Chen MH, Chang WH, Huang MY, Wang TW. Biomaterials. 2013;34:2005. doi: 10.1016/j.biomaterials.2012.11.043. [DOI] [PubMed] [Google Scholar]
- 23.Lam J, Segura T. Biomaterials. 2013;34:3938. doi: 10.1016/j.biomaterials.2013.01.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Friocourt G, Koulakoff A, Chafey P, Boucher D, Fauchereau F, Chelly J, Francis F. Cereb Cortex. 2003;13:620. doi: 10.1093/cercor/13.6.620. [DOI] [PubMed] [Google Scholar]
- 25.Li Y, Liu M, Yan Y, Yang ST. World J Stem Cells. 2014;6:11. doi: 10.4252/wjsc.v6.i1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lowry WE, Richter L, Yachechko R, Pyle AD, Tchieu J, Sridharan R, Clark AT, Plath K. Proc Natl Acad Sci U S A. 2008;105:2883. doi: 10.1073/pnas.0711983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.