SUMMARY
BCL-2 is a negative regulator of apoptosis implicated in homeostatic and pathologic cell survival. The canonical anti-apoptotic mechanism involves entrapment of activated BAX by a groove on BCL-2, preventing BAX homo-oligomerization and mitochondrial membrane poration. The BCL-2 BH4 domain also confers anti-apoptotic functionality, but the mechanism is unknown. We find that a synthetic α-helical BH4 domain binds to BAX with nanomolar affinity and independently inhibits the conformational activation of BAX. Hydrogen-deuterium exchange mass spectrometry demonstrated that the N-terminal conformational changes in BAX induced by a triggering BIM BH3 helix were suppressed by the BCL-2 BH4 helix. Structural analyses localized the BH4 interaction site to a groove formed by residues of α1, α1–α2 loop, and α2–α3 and α5–α6 hairpins on the BAX surface. These data reveal a previously unappreciated binding site for targeted inhibition of BAX and suggest that the BCL-2 BH4 domain may participate in apoptosis blockade by a noncanonical interaction mechanism.
INTRODUCTION
BCL-2 is the founding member of a family of homologous proteins that regulate apoptosis through protein-protein interaction. Thirty years of BCL-2 family research has revealed a complex signaling network governed by a large series of anti- and pro-apoptotic members, reflecting a physiologic balancing act essential to organism homeostasis. BCL-2 family deregulation provokes diseases of too many or too few cells, making these proteins ripe targets for therapeutic modulation. The structure of BCL-XL in complex with a single, α-helical killer domain from BAK demonstrated what is now the canonical paradigm for how anti-apoptotic BCL-2 family proteins bind and neutralize the death proteins of the BCL-2 family (Sattler et al., 1997). This discovery informed the development of high-fidelity BCL-2 inhibitors that are proving effective in overcoming apoptotic suppression in human cancer (Souers et al., 2013).
Despite the extensive body of BCL-2 family literature, many fundamental questions about these critical proteins remain unanswered, including just how the full-length forms of BCL-2 proteins explicitly engage as heterodimers or homo-oligomers in solution or the membrane environment. Among the unfinished business of BCL-2 family protein interactions is the longstanding curiosity that anti-apoptotic proteins lacking their N-terminal BH4 domain are at least partially defective in suppressing apoptosis (Hirotani et al., 1999; Huang et al., 1998). Yet, one study found that BH4-deleted BCL-2 remains fully competent at engaging a broad spectrum of pro-apoptotic members, including BAX (Huang et al., 1998), rendering the mechanistic role of the BH4 domain uncertain. Interestingly, select BH4 domain peptides can independently inhibit apoptosis in cells and in vivo when delivered as TAT fusions (Donnini et al., 2009; Hotchkiss et al., 2006; Sugioka et al., 2003), but how they exert this anti-apoptotic activity is also unknown.
To investigate the anti-apoptotic functionality of the BCL-2 BH4 domain, we applied all-hydrocarbon stapling to generate chemical probes based on the peptide sequence of the BCL-2 BH4 domain. Depending on the peptide sequence and the location and composition of the inserted staples, all-hydrocarbon stapling can refold peptides into their native and bioactive α-helical state (Walensky and Bird, 2014). Why staple BH4 domain sequence for this study? First, a majority of the defined BH4 region is α-helical in BCL-2 structures (Lee et al., 1996; Petros et al., 2001). Indeed, disruption of BCL-2 BH4 α-helical structure has recently been shown to abrogate its protein interaction capacity (Monaco et al., 2013). Second, out of context from the full-length protein, short α-helical peptides can unfold, resulting in loss of native structure and biological activity, and rendering them suboptimal probes. Finally, the ideal approach, structurally characterizing the full-length BAX/BCL-2 heterodimeric complexes in solution or in the membrane context, has defied 30 years of experimentation, so chemical biology strategies provide an opportunity to advance the inquiry until the structures of these “holy grail” full-length complexes can be solved. Thus, we developed and applied stabilized alpha-helices of BCL-2 domains (SAHBs) modeled after BCL-2 BH4 sequence in a series of multidisciplinary studies to interrogate (1) whether an α-helical BCL-2 BH4 domain directly engages pro-apoptotic BAX, (2) the functional implications and mechanism of action of this putative interaction, and (3) the location of the BH4 interaction site on BAX. Based on the results of these studies, which span biochemical, cellular, mass spectrometry, and NMR analyses, we report a mechanism for inhibition of pro-apoptotic BAX by a synthetic BH4 domain.
RESULTS
α-Helical BCL-2 BH4 Domain Peptides Directly Bind to BAX
The BH4 domain is shared among anti-apoptotic BCL-2 family proteins and, based on the structure of a BCL-2/BCL-XL chimera (Petros et al., 2001), comprises the first α-helix and a portion of the unstructured loop between α1 and α2 (Figure 1A). Of note, the true structure of the BCL-2 BH4 domain—either in solution or in the physiologically relevant membrane environment—is unknown, as the structure of full-length native BCL-2 remains unsolved. In considering the sequence template for BCL-2 BH4 SAHB synthesis, we included the reportedly unstructured residues for several reasons. First, prior studies demonstrated that discrete amino acids within this conserved N-terminal portion of the α1–α2 loop are critical to functional BH4-binding activity (Hirotani et al., 1999; Ma et al., 2013). Second, structure prediction software (e.g., Laser-gene) indicates that this sequence composition favors folding as a continuous α-helix with α1. Finally, hydrophobic-to-hydrophilic mutations localized to a putative hydrophobic surface of an amphipathic BH4 helix impaired BCL-2 function, whereas select alanine substitutions at the hydrophilic face had no effect (Hirotani et al., 1999; Huang et al., 1998; Lee et al., 1996). Taken together, these data suggest that the bioactivity of the BCL-2 BH4 domain relies both on sequences beyond the α1 helix and, potentially, an extended amphipathic α-helical fold. Thus, we installed all-hydrocarbon staples at a series of sequential (i, i+4) positions along the length of the classically defined BCL-2 BH4 domain sequence (Figure 1B; Table S1).
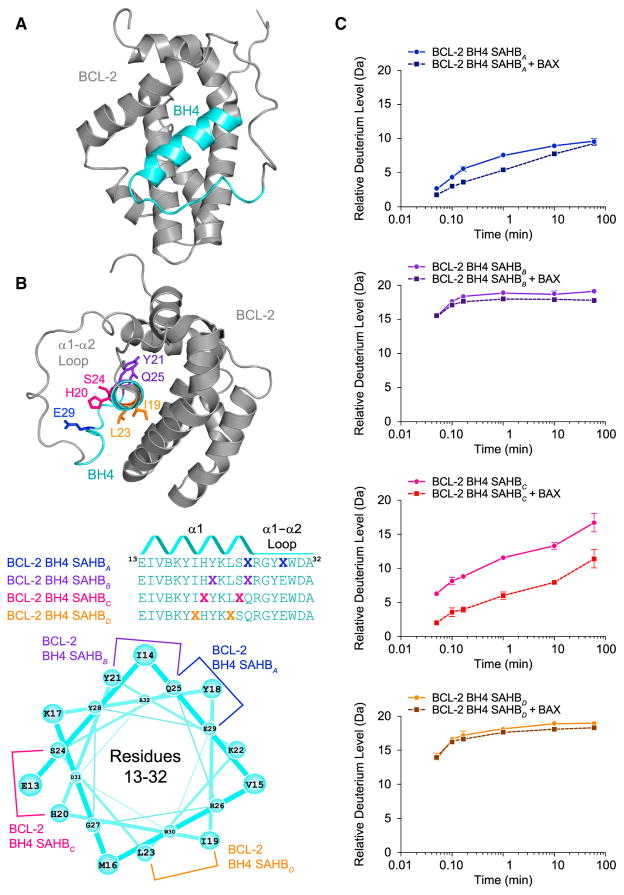
Figure 1. Sequence, Secondary Structure, and BAX-Binding Activity of BCL-2 BH4 SAHBs.
(A) The BH4 domain of BCL-2 comprises α1 and proximal residues of the α1–α2 loop, based on the structure of a BCL-2/BCL-XL chimera (PDB: 1G5M).
(B) A panel of stabilized α-helices of BCL-2 domains (SAHBs) was designed based on the BH4 sequence and differential placement of all-hydrocarbon staples (X) along the BH4 domain surfaces. B represents norleucine, which replaced the native methionine to avoid sulfur-based interference with the efficiency of the Grubb’s catalyst. An idealized helical wheel demonstrates the differential placement of all-hydrocarbon staples along the BH4 sequence.
(C) The secondary structural stability of BCL-2 BH4 SAHBs in solution as measured by the relative level of deuterium incorporation over time (solid lines) using mass spectrometry. Changes in the rate or overall level of deuterium incorporation by SAHBs in the presence of added recombinant BAX (dashed lines) reflect protein interaction with or without induced peptide folding. Error bars represent the spread of data for all charge states in duplicate experiments. See also Figure S1.
We evaluated the secondary structures of BCL-2 BH4 SAHBs by hydrogen-deuterium exchange mass spectrometry (HXMS), which probes protein structure by measuring both the solvent accessibility and hydrogen-bonding states of backbone amide hydrogens (Engen, 2009). When diluted into deuterated buffers, backbone amide hydrogens rapidly exchange with deuterium. In contrast, the hydrogen bonding of backbone amide hydrogens, as in α-helical peptides, can retard or prevent deuterium exchange (Shi et al., 2013). We found that different staple locations impacted the deuterium exchange profile of the BCL-2 BH4 peptide, and thus its secondary structure, to varying degrees.
Whereas BCL-2 BH4 SAHBB and SAHBD behaved as unstructured, linear peptides, as evidenced by full deuterium incorporation within 2–5 s of exposure to the deuterium-containing buffer, the SAHBA staple markedly slowed the kinetics of deuterium exchange and blocked complete incorporation (Figure 1C). The SAHBC staple exhibited an intermediate effect, in which the kinetics of deuterium incorporation were slowed, but near-complete exchange was ultimately achieved (Figure 1C). These data indicate that BCL-2 BH4 SAHBA and SAHBC are structurally stabilized by staple incorporation, with BCL-2 SAHBA notably more rigid than BCL-2 BH4 SAHBC.
We next examined the impact of BAX protein on the deuterium exchange profiles of BCL-2 BH4 SAHBs. Peptides that bind to a protein target can manifest decreased exchange relative to the unbound state due to sequestration of the exchangeable hydrogens at the protein binding interface and potential backbone hydrogen protection from induced peptide folding. The unstructured BCL-2 BH4 SAHBB and SAHBD peptides showed no change in their deuterium exchange profiles upon incubation with recombinant full-length BAX, reflective of their failure to engage BAX (Figure 1C). In contrast, the structured BCL-2 BH4 SAHBA peptide demonstrated even slower deuterium incorporation upon exposure to BAX, consistent with a direct binding interaction (Figure 1C). The partially structured BCL-2 BH4 SAHBC construct manifested a dramatic decrease in total deuterium incorporation, reflective of the peptide becoming more structured upon protein interaction (Figure 1C). These results indicate that there is a discrete structural requirement for BH4 interaction with BAX, as we observed previously for the BAX-activating BH3 domains of BID, BIM, and PUMA (Edwards et al., 2013; Walensky et al., 2006). To corroborate the HXMS results, we measured the BAX-binding activity of BCL-2 BH4 SAHBs A and C by fluorescence polarization assay. We observed a dose-responsive binding interaction between FITC-BCL-2 BH4 SAHBs A and C, and recombinant full-length BAX, in the nanomolar range (Figure S1). Notably, this binding activity was stable over time, which stands in stark contrast to the “hit-and-run” BAX-binding activity of activator BH3 SAHBs that promote the conformational transformation and oligomerization of BAX (Gavathiotis et al., 2010).
Sequence-Specific Inhibition of BAX Activation by BCL-2 BH4 SAHBs
We next sought to determine if the observed solution-phase binding interaction between BAX and BCL-2 BH4 SAHBA impacted the functional activity of BAX in a membrane context. Addition of an activator BH3 helix, such as BIM SAHBA (aa 145–164), to BAX in the presence of ANTS/DPX-encapsulated liposomes results in BAX-mediated membrane poration and release of dequenched fluorophore into the reaction buffer. The addition of BCL-2 BH4 SAHBA dose-responsively inhibited BIM SAHBA-triggered BAX activation (Figure 2A). We confirmed that the inhibitory effect of BCL-2 BH4 SAHBA did not derive from a disruptive interaction with BIM SAHBA itself (Figure S2A). In addition, we evaluated the capacity of BCL-2 BH4 SAHBA to inhibit BAX activation by an alternate stimulus, namely heat (Pagliari et al., 2005). BCL-2 BH4 SAHBA also suppressed heat-induced BAX activation in a dose-responsive manner (Figure 2B). To confirm that these effects were not exclusively dependent on the A staple position, we documented that BCL-2 BH4 SAHBC also inhibited BIM SAHBA- and heat-triggered BAX activation (Figures S2B and S2C).
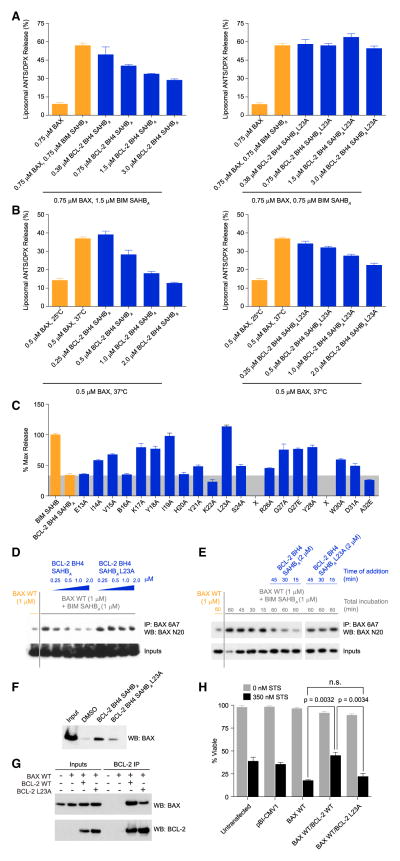
Figure 2. Direct and Sequence-Specific Inhibition of Pro-Apoptotic BAX by the BCL-2 BH4 Domain.
(A and B) BCL-2 BH4 SAHBA dose-responsively impaired BIM SAHBA-triggered (A) and heat-induced (B) BAX-mediated liposomal release. L23A mutagenesis of BCL-2 BH4 SAHBA disrupted the BAX-inhibitory activity. Data are mean ± SD for assays performed in triplicate.
(C) Alanine scanning was used to identify amino acid residues most critical to functional inhibition of BAX-mediated liposomal release by BCL-2 BH4 SAHBA. Data are mean ± SD for assays performed in triplicate.
(D) BCL-2 BH4 SAHBA, but not the L23A mutant, dose-responsively blocked BIM SAHBA-induced exposure of the N-terminal activation epitope on BAX in the presence of liposomes, as assessed by immunoprecipitation with the conformation-specific 6A7 antibody. The data are representative of three independent experiments.
(E) Induction of 6A7 epitope exposure by BIM SAHBA in the presence of liposomes was reversed by subsequent treatment (15, 30, and 45 min after BIM SAHBA exposure) with BCL-2 BH4 SAHBA, but not BCL-2 BH4 SAHBA L23A. The data are representative of three independent experiments.
(F) L23A mutagenesis of the hydrophobic surface of BCL-2 BH4 SAHBA impaired its interaction with native BAX, as demonstrated by streptavidin pull-down of biotinylated BCL-2 BH4 SAHBs from HeLa cell lysates and anti-BAX western analysis.
(G) Co-immunoprecipitation of full-length BCL-2 and BAX co-expressed in HeLa cells was impaired by L23A mutagenesis of the BCL-2 BH4 domain.
(H) Correspondingly, L23A mutagenesis reduced the capacity of BCL-2 to protect against staurosporine-induced apoptosis of HeLa cells co-expressing full-length BAX and BCL-2. Data are mean ± SD for experiments performed in triplicate and repeated three times with similar results. See also Figure S2.
To identify the sequence determinants for this inhibitory activity, we employed alanine scanning mutagenesis and screened for mutants with reduced capacity to suppress BIM SAHBA-triggered BAX activation. We found that mutation of select residues markedly impaired functional activity, including a series of amino acids at the hydrophobic surface of BCL-2 BH4 SAHBA, such as V15, I19, L23, and G27 (Figure 1B), with L23A reflecting the most deleterious mutation (Figures 2A–2C and S2D). L23A mutagenesis of BCL-2 BH4 SAHBC also impaired inhibitory activity (Figures S2B and S2C). Select charged and hydrophilic residues located outside of the core hydrophobic surface were also functionally important (e.g., K17 and Y18), as observed for BH3 interactions, where such residues participate in complementary charge-charge or hydrogen-bonding interactions at the target protein surface (Gavathiotis et al., 2008; Sattler et al., 1997). Taken together, these data indicate that BCL-2 BH4 SAHBs A and C are direct and peptide sequence-specific inhibitors of pro-apoptotic BAX, and that the hydrophobic face of the BH4 helix is a functional interaction surface for targeting BAX directly (Figures 1B and 2C).
We previously demonstrated that direct BH3 engagement and functional activation of BAX correlates with exposure of an N-terminal epitope recognized by the conformation-specific 6A7 antibody (Gavathiotis et al., 2008; Hsu and Youle, 1997). To determine if BCL-2 BH4 SAHBA suppression of BAX-mediated liposomal poration blocks exposure of the 6A7 epitope, we incubated BAX with liposomes, BIM SAHBA, and increasing doses of BCL-2 BH4 SAHBA, and monitored the N-terminal conformational activation of BAX by 6A7 immunoprecipitation. BCL-2 BH4 SAHBA, but not BCL-2 BH4 SAHBA L23A, dose-responsively inhibited 6A7 immunoprecipitation of BAX (Figures 2D and S2E). BCL-2 BH4 SAHBC likewise inhibited 6A7-mediated immunoprecipitation of BAX (Figure S2F). We further observed that BIM SAHBA-triggered exposure of the 6A7 epitope could be reversed by the subsequent addition of BCL-2 BH4 SAHBA, but not BCL-2 BH4 SAHBA L23A (Figure 2E). These data suggest that BCL-2 BH4 engagement blocks and even reverses the N-terminal conformational activation of BAX in the membrane context.
Correlation between BCL-2 BH4 SAHB and BCL-2 Protein Activities
To further validate the sequence- and interface-specific binding activity of BCL-2 BH4 SAHBA as it relates to native BAX, we incubated HeLa cell lysates with biotinylated BCL-2 BH4 SAHBA or its L23A mutant, and performed a streptavidin pull-down to monitor differential peptide engagement of BAX. Whereas BCL-2 BH4 SAHBA co-precipitated native BAX, L23A mutagenesis markedly suppressed this binding activity (Figure 2F). To extend these findings to full-length BCL-2 protein, we co-expressed BAX and the wild-type or L23A mutant form of BCL-2, and found that BCL-2 L23A was less effective at co-immunoprecipitating BAX relative to wild-type BCL-2 (Figure 2G). Importantly, we confirmed that recombinant BCL-2 and BCL-2 L23A proteins (Figure S2G) exhibit a similar overall fold, as assessed by circular dichroism (Figure S2H), and retain nanomolar canonical BH3-binding activity, as documented by direct FP assay using FITC-BIM SAHBA (Figure S2I). Thus, L23A serves as an informative mutant for probing BH4-dependent binding and biological activity in the context of full-length BCL-2 protein. To interrogate the functional consequences of the immunoprecipitation results (Figure 2G), we tested the effect of L23A mutagenesis on the anti-apoptotic activity of BCL-2, co-expressed with BAX, in staurosporine-treated HeLa cells. Whereas co-expression of BCL-2 reversed the apoptotic response observed with expression of BAX alone, L23A mutagenesis impaired the ability of BCL-2 to protect HeLa cells from staurosporine-induced cell death (Figures 2H and S2J). Taken together, we find that mutagenesis of L23 at the hydrophobic face of the BCL-2 BH4 domain helix impairs both the interaction between BCL-2 and BAX and the anti-apoptotic activity of BCL-2 in a cellular context.
BCL-2 BH4 SAHBA Blocks the Conformational Activation of BAX
To investigate the effect of BCL-2 BH4 SAHBA on the structure of full-length BAX, we used HXMS to first determine the baseline deuterium exchange pattern of BAX in solution, which highlighted differences in exchange kinetics between regions of the protein that are unfolded or exposed (e.g., N terminus, α1–α2 loop) and those that are highly structured or buried (e.g., α2, α5, α6, α9) (Figure S3A). We then tracked BAX conformational status by monitoring the kinetics of deuterium exchange in the presence of BCL-2 BH4 SAHBA. Following deuterium exposure, BAX was digested with pepsin, and deuterium incorporation into individual BAX peptic fragments was compared for the BAX-alone versus BAX/BCL-2 BH4 SAHBA samples. Interestingly, the interaction between BCL-2 BH4 SAHBA and BAX induced protection of N-terminal peptides that correspond to (1) the very region recognized by the 6A7 antibody that selectively binds to conformationally activated BAX (Hsu and Youle, 1997) and (2) the α1–α2 loop, whose displacement has been implicated as the initiating conformational change of BAX activation (Gavathiotis et al., 2010) (Figures 3A and 3B). Importantly, we found that the observed changes in deuterium incorporation upon incubating BAX with BCL-2 BH4 SAHBA were peptide sequence-specific, as the L23A mutant had little to no effect on the deuterium exchange pattern of BAX (Figure 3C). Thus, we find that the binding interaction between BAX and BCL-2 BH4 SAHBA in solution causes localized conformational protection of monomeric BAX and, in particular, restrains the N-terminal region.
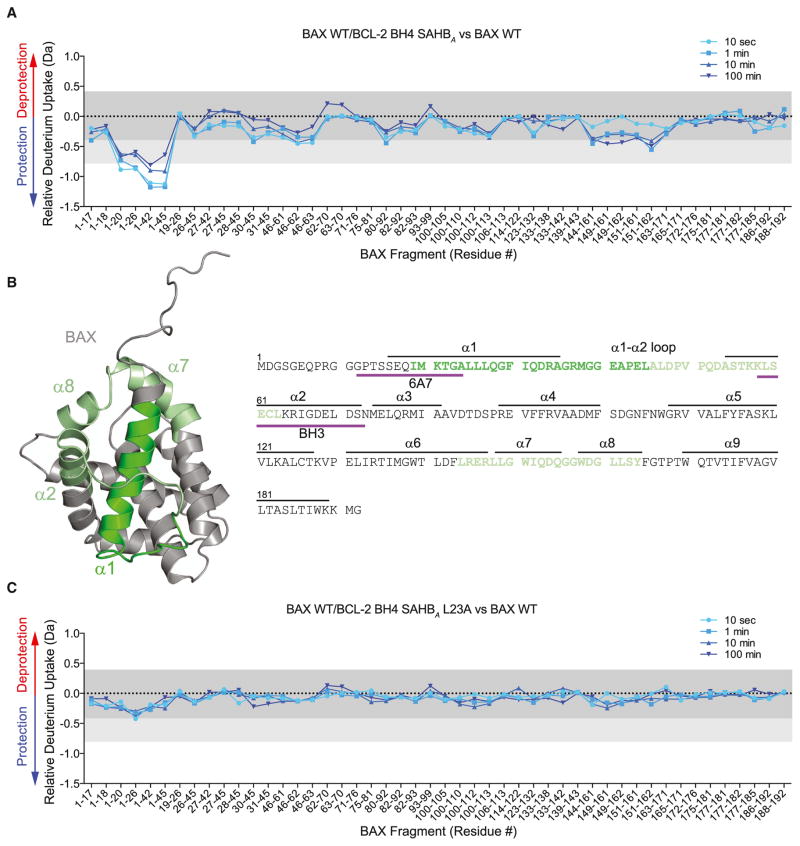
Figure 3. Conformational Stabilization of Monomeric BAX by BCL-2 BH4 SAHBA.
(A and B) BCL-2 BH4 SAHBA-bound BAX manifests a decrease in deuterium incorporation over time compared to unliganded BAX, as measured by HXMS. The most notable localized protection involves N-terminal fragments that comprise portions of the 6A7 epitope and α1–α2 loop (dark green). Significant, but less prominent, protection was also observed in regions that surround the α1–α2 loop and N terminus (light green). The relative difference plot reflects the relative deuterium incorporation of BCL-2 BH4 SAHBA/BAX WT minus the relative deuterium incorporation of BAX WT. Dark gray shading represents changes in the plot that are below the significance threshold of 0.4 Da, whereas light gray shading and the white region highlight changes above the baseline significance threshold of 0.4 Da and the more stringent threshold of 0.8 Da, respectively. The experiments were repeated twice with similar results.
(C) The sequence dependence of BCL-2 BH4 SAHBA-induced conformational stabilization of BAX was confirmed by L23A mutagenesis. BCL-2 BH4 SAHBA L23A had little to no effect on deuterium incorporation by BAX over time. The relative difference plot reflects the relative deuterium incorporation of BCL-2 BH4 SAHBA L23A/BAX WT minus the relative deuterium incorporation of BAX WT. See also Figure S3.
Although a wealth of information has been gleaned from studying BCL-2 family protein interactions in solution, including the blueprint that led to the development of an anti-cancer drug (Sattler et al., 1997; Souers et al., 2013), the major physiologic context for BCL-2 family biology is the mitochondrial outer membrane. The importance of conducting mechanistic studies of BCL-2 family protein interactions in the membrane environment, despite the formidable challenges, has become increasingly apparent (Leber et al., 2010). Thus, we undertook a series of HXMS studies designed to explore the impact of BIM BH3 and BCL-2 BH4 helices on the conformational activation of BAX in a membrane environment. First, we demonstrated that liposomal membranes have little to no effect on the relative deuterium uptake of BAX (Figure S3B). However, upon addition of the BAX-activating BIM BH3 helix, BIM SAHBA (aa 145–164), we observed dose- and time-responsive increases in deuterium exchange in discrete regions of the BAX protein, reflective of induced structural changes (Figures 4A–4C). The most prominent region of protein exposure occurred at the N-terminal face, which harbors both a trigger site for BIM BH3-induced direct BAX activation (Gavathiotis et al., 2008) and an activation epitope recognized by the conformation-specific 6A7 antibody (Hsu and Youle, 1997) (Figures 4A–4C). In addition, increased deuterium incorporation was detected in a portion of BAX α3 that comprises a subregion of the C-terminal binding pocket. At higher BIM SAHBA doses, increased deuterium levels were also observed in the BAX BH3 domain (Figures 4A and 4B), whose exposure is required for the propagation and oligomerization of BAX (Wang et al., 1998). These data are consistent with our prior NMR studies that documented chemical shift changes in these three functional regions of BAX upon BIM BH3 helix treatment (Gavathiotis et al., 2010).
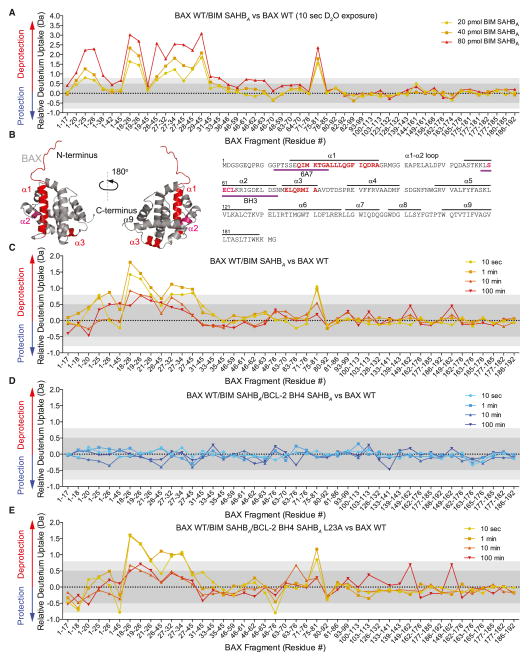
Figure 4. BCL-2 BH4 SAHBA Blocks the Conformational Changes of BAX Induced by BIM SAHBA in a Membrane Environment.
(A) The addition of BIM SAHBA to BAX (40 pmol) in a liposomal environment triggered dose-responsive and regiospecific increases in deuterium incorporation compared to unliganded BAX, as measured by HXMS. The most notable region of deprotection involved N-terminal peptides that comprise portions of the 6A7 activation epitope. Significant deprotection was also observed in BAX α3, which comprises a subregion of the C-terminal hydrophobic groove, and in the BH3 domain (BAX α2). The relative difference plot reflects the relative deuterium incorporation of BIM BH3 SAHBA/BAX WT minus the relative deuterium incorporation of BAX WT. Dark gray shading represents changes in the plot that are below the significance threshold of 0.5 Da, whereas light gray shading and the white region highlight changes above the baseline significance threshold of 0.5 Da and the more stringent threshold of 0.8 Da, respectively. Data represent the average of at least two independent experiments.
(B) The regions of increased deuterium uptake are highlighted in red (> 0.8 Da significance threshold) and pink (> 0.5 Da significance threshold) on the sequence and structure of monomeric BAX (PDB: 1F16).
(C) Incubation of BIM SAHBA (40 pmol) and BAX (40 pmol) in a liposomal environment triggered time-responsive changes in deuterium incorporation that mirrored the regiospecific changes observed upon BIM SAHBA dose escalation. The relative difference plot reflects the relative deuterium incorporation of BIM BH3 SAHBA/BAX WT minus the relative deuterium incorporation of BAX WT.
(D) The addition of BCL-2 BH4 SAHBA completely eliminated the observed increases in BAX deuterium exchange triggered by BIM SAHBA in the membrane environment. The relative difference plot reflects the relative deuterium incorporation of BIM BH3 SAHBA/BCL-2 BH4 SAHBA/BAX WT minus the relative deuterium incorporation of BAX WT.
(E) The mutant BCL-2 BH4 SAHBA L23A construct had no inhibitory effect, highlighting the peptide sequence specificity of BCL-2 BH4 SAHBA activity. The relative difference plot reflects the relative deuterium incorporation of BIM BH3 SAHBA/BCL-2 BH4 SAHBA L23A/BAX WT minus the relative deuterium incorporation of BAX WT. See also Figure S3.
We next tested the effect of adding BCL-2 BH4 SAHBA to the mixture of BIM SAHBA, BAX, and liposomes. Remarkably, BCL-2 BH4 SAHBA completely blocked the observed deuterium exchange upon BIM SAHBA-induced BAX activation (Figure 4D). As important measures of specificity, we confirmed that L23A mutagenesis abrogated the inhibitory effect (Figure 4E), and that BCL-2 BH4 SAHBA had no direct effect on BIM SAHBA under these conditions (Figure S2A). Thus, we linked the inhibitory activity of BCL-2 BH4 SAHBA to peptide sequence-specific conformational restraint of BAX at the very regions previously implicated in the direct activation of BAX by activator BH3 domains.
BH4-Mediated Inhibition of BAX at a Novel Interaction Site
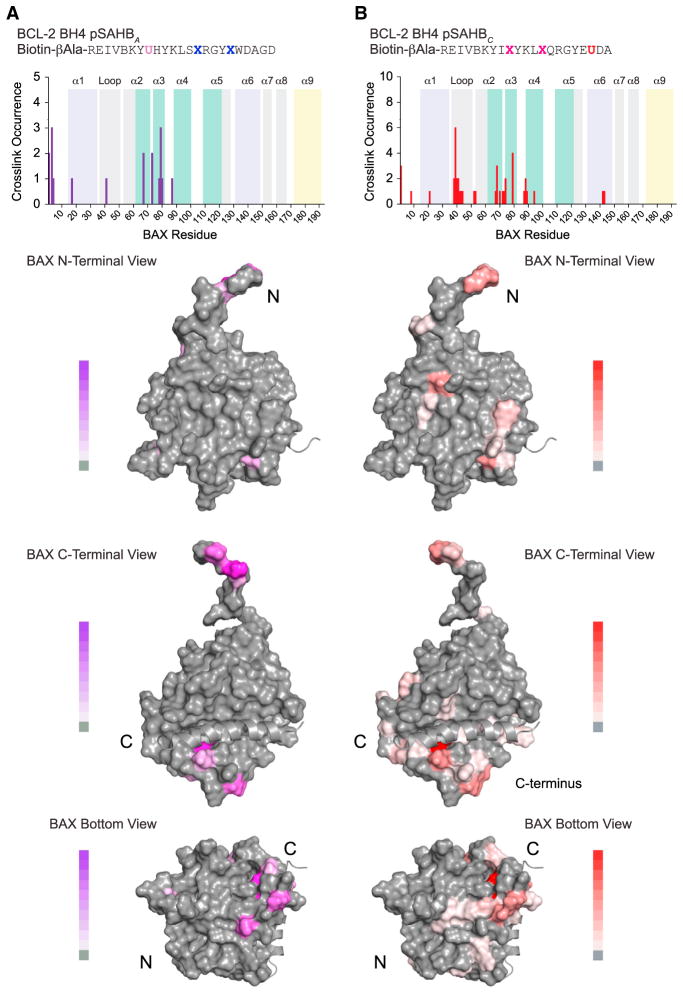
HXMS provides a rigorous measurement of solvent-exposed amide hydrogens to probe secondary structure changes and suggested a suppressive BH4 interaction involving the α1 and α1–α2 loop region of BAX. However, the method cannot always be applied to conclusively localize ligand binding sites, especially when considering that structured hydrophobic interaction surfaces are likely to be protected in both the unbound and bound states (Engen, 2003). Therefore, to locate the BCL-2 BH4 SAHB binding region on BAX, we applied our photoaffinity labeling and mass spectrometry approach (Braun et al., 2010). We synthesized biotinylated BCL-2 BH4 photoreactive SAHBs (pSAHBs) that incorporate a benzophenone moiety at discrete locations. We confirmed that such pSAHBs, containing the A or C staple, retained the capacity to bind BAX, as demonstrated by co-precipitation of native BAX from HeLa cell lysates (Figure S4A) and suppression of BIM SAHBA-induced 6A7 epitope exposure (Figure S4B). Each pSAHB was incubated with full-length recombinant BAX under UV light, and the peptide fragments that covalently linked to the pSAHB via the benzophenone were identified by LC-MS/MS (Braun et al., 2010). The most frequently crosslinked residues for both BCL-2 BH4 pSAHBs A and C localized to a focal surface below and underneath the C-terminal pocket involving BAX α3, and adjacent C-terminal residues of α2 and N-terminal residues of α4; additional cross-linking to the α1–α2 loop and select α1 and α6 residues was also observed (Figures 5A and 5B). These crosslinking results support the intriguing concept that the BCL-2 BH4 domain helix could mediate its inhibitory function by influencing those regions involved in the direct activation of BAX by activator BH3 domain helices (Czabotar et al., 2013; Edwards et al., 2013; Gavathiotis et al., 2010; Leshchiner et al., 2013). Indeed, the α1–α2, α2, and α3 portions of BAX demonstrated by HXMS to undergo conformational change upon BIM SAHBA exposure (Figures 4A–4C) emerge here as the very sites that are crosslinked most frequently by BCL-2 BH4 pSAHBs (Figure 5).
Figure 5. Localization of the BCL-2 BH4 Binding Region on BAX by Photoaffinity Labeling and Mass Spectrometry.
(A and B) Biotinylated BCL-2 BH4 photoreactive SAHBs (pSAHBs) were generated by incorporation of a UV-active benzophenone residue (U) at position 19 (purple) in pSAHBA or position 30 (red) in pSAHBC. pSAHBs were mixed with full-length BAX and subjected to UV irradiation, electrophoresis, excision of the crosslinked protein, trypsin proteolysis, and LC-MS/MS analysis. The plots (above) depict the frequency of crosslinked sites identified across the BAX polypeptide sequence. As a reference, the regions of the BH3-activation sites at the N- and C-terminal faces of BAX are shaded in lavender and teal, respectively, and the C-terminal α9 helix is colored in yellow. The crosslinked residues are mapped onto the solution structure of monomeric BAX (PDB: 1F16) (below) and colored according to the frequency of occurrence. B, norleucine; U, 4-benzoyl-phenylalanine (Bpa). See also Figure S4.
To explore the mechanistic possibility that the BCL-2 BH4 helix directly competes with activator BH3 helices at the same binding sites, we repeated the crosslinking analyses using two BAX constructs that alternatively block or expose the BH3 interaction surface at the C-terminal face of BAX. We previously demonstrated that, upon deletion of the BAX C-terminal helix, an activator BH3 helix preferentially engages the canonical pocket of BAX (as opposed to the α1/α6 trigger site at the N-terminal face), making the BAXΔC construct especially useful for interrogating binding at the C-terminal surface (Edwards et al., 2013). Although BCL-2 BH4 SAHBA directly binds to BAXΔC (Figure S4C), and competes with BIM SAHBA for interaction (Figure S4D), elimination of the C-terminal helix did not significantly increase the frequency or distribution of crosslinking within the canonical pocket, as assessed using our most efficient pSAHB crosslinker, BCL-2 BH4 pSAHBC (Figure S4E). To determine if occlusion of the C-terminal binding pocket impacted the crosslinking pattern of BCL-2 BH4 pSAHBC, we employed our previously reported BAX A112C/V177C construct, which upon oxidation forms an α5–α9 disulfide tether that blocks BH3 binding at the canonical site (Edwards et al., 2013) and prevents mitochondrial translocation of BAX (Gavathiotis et al., 2010). As for BAXΔC, here we find that occlusion of the BAX C-terminal pocket has little to no effect on the crosslinking pattern of BCL-2 BH4 pSAHBC (Figure S4F). Taken together, these data suggest that although the BH4 and BH3 helices compete for interaction with BAX (Figure S4D), the BH4-binding site may be different. What’s more, the consistent pattern of BCL-2 pSAHB crosslinking to inactive, monomeric forms of BAX, including the conformationally restricted, α9-tethered form, suggests that BH4-mediated BAX inhibition may occur at a mechanistically distinct step from the established mode for BCL-2 inhibition of conformationally activated BAX.
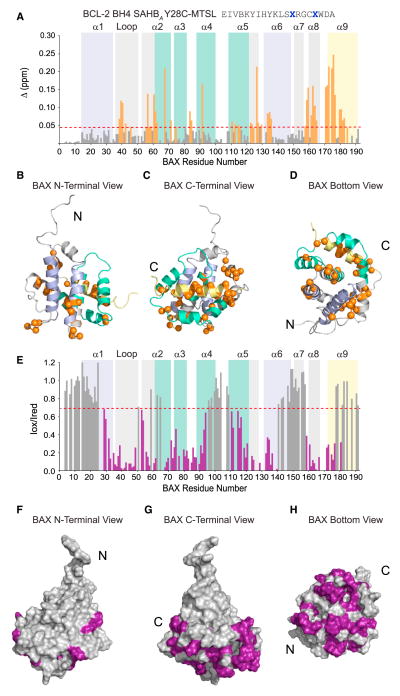
To examine the structural effects of the BH4 interaction and better localize the inhibitory site on BAX, we conducted NMR analyses of 15N-BAX upon titration with a series of BCL-2 BH4 SAHBs. Using S-(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl methanesulfonothioate (MTSL)-derivatized BCL-2 BH4 SAHBA Y28C, we observed discrete chemical shift changes consistent with a fast exchange regime involving surface residues of the α1–α2 loop, α2, and the α2–α3, α3–α4, and α5–α6 hairpins of BAX (Figures 6A–6D). Prominent changes were also detected throughout α8 and α9. We repeated the experiment with a second peptide construct, BCL-2 BH4 SAHBA B16C-MTSL (B, norleu), and observed a similar pattern of chemical shift changes, although of lower magnitude, consistent with a weaker binding interaction and/or decreased solubility of this peptide construct (Figures S5A–S5D). We also performed the HSQC analysis using BCL-2 BH4 SAHBC, which bears an alternate staple position and lacks the Cys-MTSL modification, and again observed a similar pattern of chemical shift changes (Figure S5E).
Figure 6. NMR Analysis of the BCL-2 BH4 SAHBA/BAX Interaction.
(A–D) NMR analysis of 15N-BAX upon titration with BCL-2 BH4 SAHBA Y28C-MTSLred up to a ratio of 1:1.1 BAX:BCL-2 BH4 SAHB revealed chemical shift changes that localized to the α1–α2 loop, α2, the α2–α3, α3–α4, and α5–α6 hairpins, α8, and α9. Cα atoms of affected residues are represented as orange bars in the plot (A) and orange spheres in the ribbon diagrams (B)–(D). The significance threshold of > 0.043 ppm for backbone amide chemical shift changes was calculated based on the average chemical shift across all residues plus the SD, as previously reported (Gavathiotis et al., 2008) and in accordance with standard methods (Marintchev et al., 2007). As a reference, the α1/α6 region that comprises the N-terminal trigger site is colored lavender, those portions of α-helices 2, 3, 4, and 5 that contain residues forming the C-terminal canonical groove are colored teal, and the C-terminal α9 helix is colored yellow.
(E–H) Ratios of BAX cross-peak intensities in the presence of oxidized or reduced (Iox/Ired) BCL-2 BH4 SAHBA Y28C-MTSL plotted versus BAX residue number. BAX residue intensities reduced below a ratio of 0.7 are colored purple in the plot (E) and mapped onto surface views of the BAX structure (F)–(H). BAX residues affected by the MTSL label colocalize to the bottom face of BAX in a region spanning from the base of the C-terminal pocket to the α1–α2 loop of the N-terminal face. See also Figures S5 and S6.
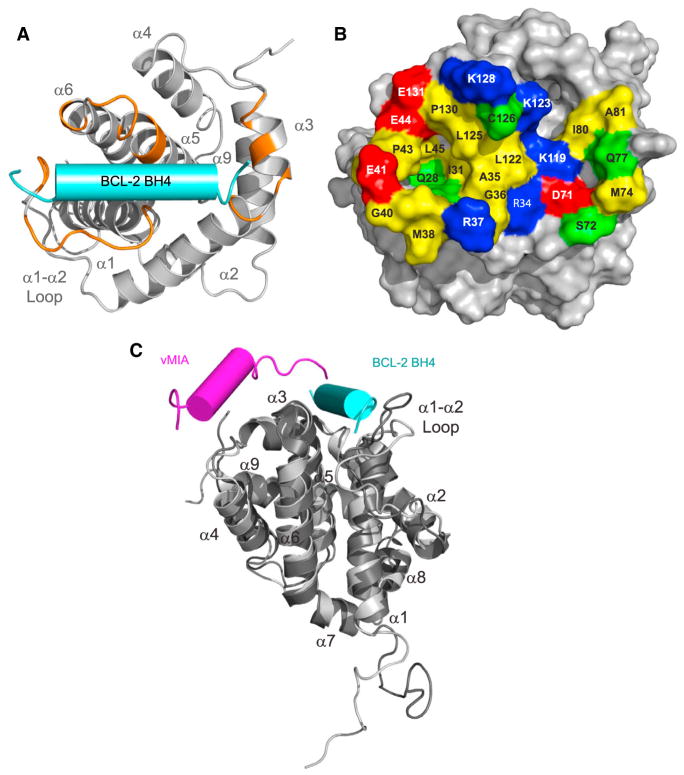
We next compared the 1H-15N HSQC spectra of BAX upon titration with BCL-2 BH4 SAHBA Y28C-MTSL in the oxidized versus reduced state. Residues displaying the greatest reduction in cross-peak intensity between the two states are those in the immediate vicinity of the nitroxide spin label. The ratios of BAX cross-peak intensities (Iox/Ired) revealed striking reductions in cross-peak intensities of select residues in BAX α1, α1–α2 loop, α2, α3, α4, α5, α6, α8, and α9 that colocalize to a region underneath the C-terminal pocket and traverse the bottom face of BAX to the α1–α2 loop (Figures 6E–6H). A similar pattern was observed for BCL-2 BH4 SAHBA M16C-MTSL (Figures S6A–S6D), although the magnitude of changes was relatively lower, as observed in the above-described HSQC study (Figure S6A). Because the degree of paramagnetic relaxation enhancement (PRE) correlates with the distance between the nitroxide label and the affected BAX residues, structure calculations using restraints derived from the NMR analyses were performed and positioned the α-helical BH4 peptide at a cleft predominantly formed by select residues of the α1–α2 loop, α2, α3, α5, and α6 (Figure 7A). Based on these results, the BH4-binding site on BAX comprises a groove lined by a series of hydrophobic residues (e.g., A35, G36, I31, M38, G39, G40, P43, L45, M74, I80, A81, L122, L125, P130) and surrounded by charged and hydrophilic residues (e.g., R34, R37, E41, E44, D71, S72, Q77, K119, K123, C126, K128, E131) (Figure 7B). Taken together, the structural analyses suggest that the BH4 inhibitory site lies at the intersection of key regulatory regions of BAX, namely the α1–α2 loop, the BAX BH3 domain, and a crossroad that links α-helices of the N- and C-terminal faces of BAX.
Figure 7. A Distinct Binding Site for BCL-2 BH4 Inhibition of BAX.
(A) Docking calculations based on the PRE NMR data placed the BCL-2 BH4 helix at an interaction site formed by the confluence of surface residues from the α1 C terminus, α1–α2 loop, and α2–α3 and α5–α6 hairpins. Those residues within 2 Å of the docked BCL-2 BH4 helix (cyan cylinder) are colored in orange on the ribbon structure of BAX.
(B) Surface view of the BH4-interaction site demonstrates a groove comprised of hydrophobic residues (yellow), which are surrounded by a series of positively charged (blue), negatively charged (red), and polar (green) residues.
(C) A structural overlay demonstrates that vMIA (aa 131–150) (PDB: 2LR1) and BCL-2 BH4 (aa 13–32) peptides engage BAX at inhibitory sites at the bottom face of BAX.
DISCUSSION
How the anti- and pro-apoptotic members of the BCL-2 family regulate one another to arrive at a life or death decision for the cell remains a vigorous area of scientific inquiry. The stakes are high because the biochemical, structural, and mechanistic insights can yield new pharmacologic approaches to eliminate pathologic cell survival such as in cancer, or preserve cells from unwanted demise such as in stroke and heart attack. The established mechanism for BCL-2 inhibition of BAX involves capture of the essential BH3 killer domain of BAX in the C-terminal binding groove of BCL-2. This paradigm emerged from the structure of a C-terminally deleted form of BCL-XL in complex with a BH3 domain peptide of BAK (Sattler et al., 1997). The analogous BCL-2 binding pocket is now drugged by ABT-199, a selective BCL-2 inhibitor that reactivates the death pathway in certain BCL-2-dependent human cancers (Souers et al., 2013). Recently, this long-established interaction paradigm was expanded based on site-specific crosslinking of BCL-XL/BAX heterodimers, which revealed that the canonical binding mode is composed of both the BH3-in-groove interface and additional, simultaneous contacts between the α1 helices of BCL-XL and BAX (Ding et al., 2014). Here, we report the unexpected finding that a synthetic BCL-2 BH4 domain helix can bind to full-length, inactive BAX in a completely different interaction mode that directly blocks the initiating conformational changes of BAX activation. These biochemical and structural findings suggest that a distinct regulatory step may exist upstream of the established inhibitory mechanism of BCL-2 C-terminal groove entrapment of the fully activated, BH3-exposed form of BAX.
Importantly, we find that this noncanonical, inhibitory BCL-2 BH4-BAX interaction is both BH4 sequence specific and BH4 helical surface specific. Alanine scanning mutagenesis and functional analyses of the inactivating L23A mutation established the hydrophobic face of the BH4 helix as the critical interaction surface. Therefore, in order for this putative BH4 interaction to occur, the C terminus-inserted form of BCL-2 at the mitochondrial outer membrane must differ from the solution-phase structure, such that the hydrophobic face of the BCL-2 BH4 domain can become exposed for BAX engagement. Indeed, BCL-2 is believed to undergo a conformational change at the mitochondrial membrane in order to target BAX (Dlugosz et al., 2006; Kim et al., 2004). With the BCL-2 BH4 domain tethered to a 65-amino-acid-long unstructured loop, the potential mobility and reach of this α-helix is indeed substantial.
To investigate the structure-function implications of BH4 engagement of BAX, and localize the site of BH4 interaction, we used a diversity of experimental approaches, including HXMS, photoaffinity labeling and mass spectrometry, and NMR analyses. The HXMS studies demonstrated that BH4 binding to BAX in solution notably slowed hydrogen-deuterium exchange in the α1–α2 loop and 6A7 activation epitope, key regulatory regions implicated in BH3-mediated direct activation of BAX at its N-terminal face. HXMS analyses performed in the presence of liposomes further demonstrated that the BCL-2 BH4 helix could suppress BIM BH3-induced conformational changes at α1 (6A7 epitope), α2 (BAX BH3), and α3 (base of C-terminal groove adjacent to α9). The pSAHB crosslinking results (Figures 5, S4E, and S4F) and the NMR analyses both implicate residues in this exact same region as participants in BH4 helix interactions (Figures 6, 7, S5, and S6), suggesting that BH4 binding locks down the very regions essential to conformational activation of BAX. Indeed, allosteric communication between the N- and C-terminal faces of BAX is a key feature of its regulation (Gavathiotis et al., 2010). Here, we define an inhibitory BH4-interaction site that literally bridges these two regulatory faces of BAX.
In further support of this potentially distinct and afferent mode of BCL-2 inhibition, the cytomegalovirus protein vMIA has recently been shown to inhibit the membrane insertion and oligomerization of BAX through an interaction adjacent to the C terminus, restraining BAX’s α3–α4 and α5–α6 hairpins (Ma et al., 2012). Although the vMIA sequence and its binding site on BAX are different from that of BCL-2 BH4 SAHBs, both peptide classes engage BAX on the same “side” of the protein (Figure 7C). Such inhibitory interactions may reflect a common mechanistic control point for stabilizing inactive forms of monomeric BAX from progressing to the more perilous fully activated, BH3-exposed, and mitochondrial membrane-embedded conformation.
Our findings potentially resolve inconsistencies in the literature regarding the functional relevance of the BH4 domain of BCL-2. For example, BCL-2 protein lacking the BH4 domain has been shown to lose anti-apoptotic activity, yet retain the capacity to bind pro-apoptotic BCL-2 family members including BAX (Huang et al., 1998). In contrast, others have found that upon BCL-2 BH4 truncation, heterodimerization with BAX is lost (Hirotani et al., 1999). We find that mutagenesis of L23 in BCL-2 BH4 disrupts BCL-2 BH4 SAHBA activity in vitro and partially impairs BCL-2 binding and inhibitory activity in a cellular context, results that are largely consistent with prior BH4 mutagenesis studies of BCL-2 in cells (Huang et al., 1998; Lee et al., 1996). We speculate that these observations could be reconciled if BCL-2 and BAX interact in two distinct modes at different steps along the BAX activation pathway: (1) a noncanonical anti-apoptotic BH4 in BAX groove interaction to arrest (and perhaps even reverse) the initiation of BAX activation and (2) a canonical pro-apoptotic BH3 in anti-apoptotic groove interaction to block downstream propagation of BAX activation and oligomerization within the mitochondrial membrane. Our observation that a BCL-2 BH4 helix is sufficient for inhibiting the membrane poration activity of BAX suggests that BH4-mediated restraint of BAX can occur independently of the canonical BH3-in-groove interaction. Correspondingly, our data provide a mechanistic explanation for how transduction of an anti-apoptotic BH4 peptide can suppress apoptosis in vitro and in vivo, namely by directly binding to and inhibiting BAX. Finally, the observed engagement of BAX by a BCL-2 BH4 helix and the resultant conformational suppression—and even reversal—of 6A7 activation epitope exposure could also contribute to the reported capacity of anti-apoptotic proteins to deactivate and then retrotranslocate BAX from mitochondria to cytosol (Edlich et al., 2011).
In summary, we find that a synthetic BH4 domain helix can engage a novel inhibitory site on BAX to reinforce its inactive, monomeric state. This inhibitory BH4 interaction could function to restrain monomeric BAX and allosterically block BH3-activating interactions and conformational changes at the mitochondrial membrane. Thus, anti-apoptotic BH4 engagement of BAX could reflect a first line of defense against the propagation of BAX activation at the mitochondria. Just as the “BH3-in-groove” mechanism for apoptotic modulation led to the development of ABT-199 to reactivate apoptosis in BCL-2-dependent cancer (Sattler et al., 1997; Souers et al., 2013), the “BH4-in-groove” interaction paradigm reported here also reveals new pharmacologic opportunities. Compounds that neutralize the BCL-2 BH4 domain could represent a new class of apoptosis sensitizers to combat pathologic cell survival, whereas BH4 mimetics like BCL-2 BH4 SAHBs that target the novel interaction site on BAX may serve as next-generation inhibitors of unwanted or premature cell death.
EXPERIMENTAL PROCEDURES
SAHB Synthesis
Stapled peptides corresponding to the BH4 domain of BCL-2 were synthesized, derivatized, and purified using our established methods (Bird et al., 2008) (Table S1). Stapled peptides were purified by LC-MS to > 95% purity and quantified by amino acid analysis. Lyophilized SAHBs were reconstituted in 100% DMSO and diluted into aqueous buffers for experimentation.
Recombinant BAX Expression and Purification
Recombinant BAX, BAX A112C/V177C, BAXΔC (aa 1–166), and 15N-BAX were generated in bacteria and then purified by affinity and size exclusion chromatography as described (Edwards et al., 2013; Gavathiotis et al., 2008).
Liposomal Release Assay
Large unilamellar vesicles (LUVs) with lipid composition resembling the mitochondrial outer membrane were generated and entrapped with ANTS and DPX as described (Leshchiner et al., 2013; Lovell et al., 2008). For measurement of BIM BH3 SAHBA-induced activation of BAX, recombinant, full-length BAX protein, BIM BH3 SAHBA (aa 145–164), and BCL-2 BH4 SAHBs were added to liposomes (5 μl) at the indicated concentrations to a final volume of 30 μl in 384-well plate format. ANTS release and dequenching due to DPX dissociation (F) was measured over a period of 40–120 min with a Tecan M1000 plate reader (excitation and emission wavelengths of 355 and 520 nm, respectively). For measurement of heat-induced activation of BAX, recombinant BAX and BCL-2 BH4 SAHBs were added to liposomes in 384-well plate format and heated to 37°C during fluorescence measurement by a Tecan M1000 plate reader. Plates were re-read following lysis with 1% Triton X-100 to determine maximal release (F100). Percent ANTS/DPX release was calculated as [(F − F0)/(F100 − F0)] × 100. To compare the relative inhibitory activity of the BCL-2 BH4 SAHBA alanine mutants, the maximal BIM SAHBA-induced BAX-mediated release was used for F100.
6A7 Immunoprecipitation
Recombinant BAX (1 μM) was mixed with 8 μl liposomes,1 μM BIM SAHBA (aa 145–164), and the indicated doses of BCL-2 BH4 SAHBA or BCL-2 BH4 SAHBA L23A in 20 μl liposomal buffer (10 mM HEPES [pH 7.0], 200 mM KCl, 1 mM MgCl2). Following the indicated incubation period at room temperature, the mixture was added to 280 μl 3% BSA in PBS, and 30 μl of the resulting mixture (10%) was reserved for input analysis. Activated BAX was immunoprecipitated with BAX 6A7 antibody (Santa Cruz Biotechnology) conjugated to pre-washed protein A/G beads (Thermo Scientific) or beads alone, and resolved by electrophoresis. Immunoblotting of the inputs and immunoprecipitated samples was performed with the BAX N20 antibody (Santa Cruz).
SAHB Pull-Down Assay
HeLa cells (ATCC) were cultured in DMEM with 10% FBS and penicillin/streptomycin. To generate whole-cell lysates, HeLa cells (6.0 × 106) were trypsinized, washed with cold PBS, and lysed by incubation with 0.5% CHAPS in lysis buffer (150 mM NaCl, 50 mM Tris [pH 7.4]). The total protein concentration of the soluble fraction was measured using a BCA kit according to the manufacturer’s instructions (Thermo Scientific) and diluted to 1 mg/ml. Biotinylated SAHB (20 μM) was added to 500 μl of pre-cleared lysate and incubated for 16 hr at 4°C. The samples were subjected to affinity pull-down by incubation with streptavidin agarose beads (Thermo Scientific), which were successively washed with ice-cold PBS. Following elution into LDS sample buffer, samples were analyzed by western blotting using the BAX N20 antibody (Santa Cruz).
BAX/BCL-2 Co-Immunoprecipitation Assay
Bax and Bcl-2 cDNAs were subcloned into the pBI-CMV1 bidirectional expression vector (Clontech Laboratories), and the L23A Bcl-2 point mutation was generated by site-directed mutagenesis and confirmed by DNA sequencing. The indicated plasmids were transfected with Xtreme-Gene (Roche) according to the manufacturer’s instructions into HeLa cells (2.5 × 105) that were plated in 6-well format 24 hr prior to transfection. The cells were lysed 24 hr post-transfection in buffer containing 0.5% NP-40 and pre-cleared with protein A/G agarose beads (Thermo Scientific). Pre-clarified lysates were immunoprecipitated with BCL-2 antibody (100) (Santa Cruz). Following three successive washes with cold lysis buffer, immunoprecipitated protein was eluted into LDS buffer and analyzed by western blotting using the BAX N20 antibody (Santa Cruz).
Cell Viability Assay
HeLa cells (8.5 × 103) were plated in 96-well format and transfected with the indicated plasmids 24 hr later. Transfected and control wells were treated the next day with the indicated amount of staurosporine (Cell Signaling Technology) for 6 or 24 hr. Cell viability was measured by Cell-TiterGlo assay (Promega), with luminescence detected by Spectromax M5 microplate reader (Molecular Devices).
Lipid-Phase HXMS
Deuterium labeling was initiated with a 12-fold dilution into D2O buffer (10 mM HEPES, pD 6.6, 200 mM KCl, 1 mM MgCl2) of a pre-equilibrated (15 min, RT) aliquot of each protein and protein:ligand stock solution containing 4 μl of liposomes, generated as described above. At the indicated time points, the labeling reaction was quenched with the addition of an equal volume of quench buffer (0.8 M GdmCl, 0.8% FA [pH 2.5]). Proteolysis was performed by incubation on ice with 40 μg porcine pepsin and 20 μg protease isolated from Aspergillus saitoi (Sigma-Aldrich) for 5 min. Digested samples were then processed and analyzed as described for solution-phase HXMS analyses in the Supplemental Experimental Procedures.
pSAHB Interaction Site Analysis
Recombinant BAX, BAXΔC, and BAX A112C/V177C (10 μM) were mixed with BCL-2 BH4 pSAHBs (100 μM) and irradiated (365 nm, Spectroline Handheld UC Lamp Model En280L, Spectronics Corporation) for 2 hr on ice. BAX A112C/V177C crosslinking was performed in buffer containing 10 mM GSSG. Crosslinked and unmodified protein was resolved by electrophoresis, excised, digested with trypsin, and analyzed by LC-MS/MS on a Thermo Orbi-trap Discovery as described (Braun et al., 2010). MS/MS spectra were searched using the SEQUEST algorithm (Eng et al., 1994) in Proteome Discoverer (Thermo Scientific) against a targeted database consisting of BAX or BAXΔC, trypsin, and common keratin contaminants. Benzophenone-cross-linked peptides were identified according to the mass added by the benzophenone-containing pSAHB fragment (BCL-2 BH4 SAHBA, 860.3857 Da; BCL-2 BH4 SAHBC, 803.8143 Da). Proteome Discoverer data were loaded into Scaffold 3 (Proteome Software) and filtered with protein identification probabilities of 95% or greater. pSAHB-modified amino acid residues were plotted as frequency of occurrence versus the protein sequence. To depict the crosslinked sites on the protein structure, a light-to-dark color scale was assigned by converting crosslinking frequencies to percent maximal occurrence and then coloring individual amino acids by groups of ten percentiles with PYMOL (Schrödinger).
Paramagnetic Relaxation Enhancement NMR
Samples for PRE NMR contained uniformly 15N-labeled human BAX at 50 μM prepared in 20 mM potassium phosphate solution at pH 6.0 with 25 mM NaCl and 10% DMSO-d6. Spectra were acquired at 25°C on Bruker 800 MHz and 600 MHz NMR spectrometers, processed using Topspin 3.0, and analyzed with CCPNMR 2.3.1 (Vranken et al., 2005). Two 1H-15N HSQC spectra for each PRE NMR were acquired using a 1:1.1 ratio of BAX to MTSL-labeled BH4 SAHB in the oxidized and reduced state. The reduced compound was generated by exposure to 5 M excess ascorbic acid for 6 hr. PRE effects were measured from the ratio of integrated peak intensities of the oxidized and reduced HSQC spectra. Two BCL-2 BH4 SAHBA peptides were tested with MTSL attached to cysteine residues installed at positions 16 or 28. To evaluate the paramagnetic broadening effects of each MTSL-derivatized BCL2 BH4 SAHB, the calculated ratios from the peak intensities of the oxidized and reduced spectra (Iox/Ired) were determined. BAX residues closest to a paramagnetic label have calculated values of Iox/Ired < 0.7. Measurements for proline or overlapped residues are absent from the intensity ratio plot. Additional NMR and structure calculation methods are described in the Supplemental Experimental Procedures.
Supplementary Material
Highlights.
Stapled BCL-2 BH4 domain helices directly bind to and inhibit pro-apoptotic BAX
BCL-2 BH4 SAHBs block the conformational activation of BH3-triggered BAX
BCL-2 BH4 domain helices inhibit BAX at a novel interaction site
The BH4-in-groove interaction represents a distinct mechanism for BAX blockade
Acknowledgments
We thank E. Smith for editorial and graphics assistance, and S. Cahill for NMR technical support (Albert Einstein College of Medicine Structural NMR Resource). This work was supported by NIH grants 5R01CA050239 and 5R01GM090299 to L.D.W., NIH grants R01GM086507 and R01GM101135 to J.R.E., an NSF Graduate Research Fellowship to L.A.B., additional support to L.A.B. from T32GM007306, and NIH grant R00HL095929, a Kimmel Scholar Award, and a Sinsheimer Scholar Award to E.G. Additionally, E.G is a member of the New York Structural Biology Center (NYSBC), and the data collected at NYSBC was made possible by a grant from NYSTAR. L.D.W. is a scientific advisory board member and consultant for Aileron Therapeutics. J.R.E. is a consultant for the Waters Corporation.
Footnotes
Supplemental Information includes six figures, one table, and Supplemental Experimental Procedures and can be found with this article online at http://dx.doi.org/10.1016/j.molcel.2015.01.014.
AUTHOR CONTRIBUTIONS
L.D.W., M.L.S., and L.A.B. conceived of and designed the research study. L.D.W., M.L.S., L.A.B., and G.H.B. designed and synthesized the SAHBs. L.A.B. and T.E.W. performed the HXMS analyses, which were supervised by and performed in the J.R.E. laboratory. L.A.B., M.L.S., F.W., and R.G. conducted the SAHB binding and biochemical analyses, and L.A.B. executed the cellular studies. L.A.B., C.R.B., and S.L. performed the pSAHB interaction site analyses. T.P.G. and E.G. conducted the NMR studies. L.D.W. and L.A.B. wrote the manuscript, which was edited and reviewed by all co-authors.
References
- Bird GH, Bernal F, Pitter K, Walensky LD. Synthesis and biophysical characterization of stabilized alpha-helices of BCL-2 domains. Methods Enzymol. 2008;446:369–386. doi: 10.1016/S0076-6879(08)01622-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun CR, Mintseris J, Gavathiotis E, Bird GH, Gygi SP, Walensky LD. Photoreactive stapled BH3 peptides to dissect the BCL-2 family interactome. Chem Biol. 2010;17:1325–1333. doi: 10.1016/j.chembiol.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czabotar PE, Westphal D, Dewson G, Ma S, Hockings C, Fairlie WD, Lee EF, Yao S, Robin AY, Smith BJ, et al. Bax crystal structures reveal how BH3 domains activate Bax and nucleate its oligomerization to induce apoptosis. Cell. 2013;152:519–531. doi: 10.1016/j.cell.2012.12.031. [DOI] [PubMed] [Google Scholar]
- Ding J, Mooers BH, Zhang Z, Kale J, Falcone D, McNichol J, Huang B, Zhang XC, Xing C, Andrews DW, Lin J. After embedding in membranes antiapoptotic Bcl-XL protein binds both Bcl-2 homology region 3 and helix 1 of proapoptotic Bax protein to inhibit apoptotic mitochondrial permeabilization. J Biol Chem. 2014;289:11873–11896. doi: 10.1074/jbc.M114.552562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dlugosz PJ, Billen LP, Annis MG, Zhu W, Zhang Z, Lin J, Leber B, Andrews DW. Bcl-2 changes conformation to inhibit Bax oligomerization. EMBO J. 2006;25:2287–2296. doi: 10.1038/sj.emboj.7601126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnini S, Solito R, Monti M, Balduini W, Carloni S, Cimino M, Bampton ET, Pinon LG, Nicotera P, Thorpe PE, Ziche M. Prevention of ischemic brain injury by treatment with the membrane penetrating apoptosis inhibitor, TAT-BH4. Cell Cycle. 2009;8:1271–1278. doi: 10.4161/cc.8.8.8301. [DOI] [PubMed] [Google Scholar]
- Edlich F, Banerjee S, Suzuki M, Cleland MM, Arnoult D, Wang C, Neutzner A, Tjandra N, Youle RJ. Bcl-x(L) retrotranslocates Bax from the mitochondria into the cytosol. Cell. 2011;145:104–116. doi: 10.1016/j.cell.2011.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards AL, Gavathiotis E, LaBelle JL, Braun CR, Opoku-Nsiah KA, Bird GH, Walensky LD. Multimodal interaction with BCL-2 family proteins underlies the proapoptotic activity of PUMA BH3. Chem Biol. 2013;20:888–902. doi: 10.1016/j.chembiol.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Engen JR. Analysis of protein complexes with hydrogen exchange and mass spectrometry. Analyst (Lond) 2003;128:623–628. doi: 10.1039/b212800b. [DOI] [PubMed] [Google Scholar]
- Engen JR. Analysis of protein conformation and dynamics by hydrogen/deuterium exchange MS. Anal Chem. 2009;81:7870–7875. doi: 10.1021/ac901154s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavathiotis E, Suzuki M, Davis ML, Pitter K, Bird GH, Katz SG, Tu HC, Kim H, Cheng EHY, Tjandra N, Walensky LD. BAX activation is initiated at a novel interaction site. Nature. 2008;455:1076–1081. doi: 10.1038/nature07396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavathiotis E, Reyna DE, Davis ML, Bird GH, Walensky LD. BH3-triggered structural reorganization drives the activation of proapoptotic BAX. Mol Cell. 2010;40:481–492. doi: 10.1016/j.molcel.2010.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirotani M, Zhang Y, Fujita N, Naito M, Tsuruo T. NH2-terminal BH4 domain of Bcl-2 is functional for heterodimerization with Bax and inhibition of apoptosis. J Biol Chem. 1999;274:20415–20420. doi: 10.1074/jbc.274.29.20415. [DOI] [PubMed] [Google Scholar]
- Hotchkiss RS, McConnell KW, Bullok K, Davis CG, Chang KC, Schwulst SJ, Dunne JC, Dietz GPH, Bähr M, McDunn JE, et al. TAT-BH4 and TAT-Bcl-xL peptides protect against sepsis-induced lymphocyte apoptosis in vivo. J Immunol. 2006;176:5471–5477. doi: 10.4049/jimmunol.176.9.5471. [DOI] [PubMed] [Google Scholar]
- Hsu YT, Youle RJ. Nonionic detergents induce dimerization among members of the Bcl-2 family. J Biol Chem. 1997;272:13829–13834. doi: 10.1074/jbc.272.21.13829. [DOI] [PubMed] [Google Scholar]
- Huang DC, Adams JM, Cory S. The conserved N-terminal BH4 domain of Bcl-2 homologues is essential for inhibition of apoptosis and interaction with CED-4. EMBO J. 1998;17:1029–1039. doi: 10.1093/emboj/17.4.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim PK, Annis MG, Dlugosz PJ, Leber B, Andrews DW. During apoptosis bcl-2 changes membrane topology at both the endoplasmic reticulum and mitochondria. Mol Cell. 2004;14:523–529. doi: 10.1016/s1097-2765(04)00263-1. [DOI] [PubMed] [Google Scholar]
- Leber B, Lin J, Andrews DW. Still embedded together binding to membranes regulates Bcl-2 protein interactions. Oncogene. 2010;29:5221–5230. doi: 10.1038/onc.2010.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee LC, Hunter JJ, Mujeeb A, Turck C, Parslow TG. Evidence for α-helical conformation of an essential N-terminal region in the human Bcl2 protein. J Biol Chem. 1996;271:23284–23288. doi: 10.1074/jbc.271.38.23284. [DOI] [PubMed] [Google Scholar]
- Leshchiner ES, Braun CR, Bird GH, Walensky LD. Direct activation of full-length proapoptotic BAK. Proc Natl Acad Sci USA. 2013;110:E986–E995. doi: 10.1073/pnas.1214313110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell JF, Billen LP, Bindner S, Shamas-Din A, Fradin C, Leber B, Andrews DW. Membrane binding by tBid initiates an ordered series of events culminating in membrane permeabilization by Bax. Cell. 2008;135:1074–1084. doi: 10.1016/j.cell.2008.11.010. [DOI] [PubMed] [Google Scholar]
- Ma J, Edlich F, Bermejo GA, Norris KL, Youle RJ, Tjandra N. Structural mechanism of Bax inhibition by cytomegalovirus protein vMIA. Proc Natl Acad Sci USA. 2012;109:20901–20906. doi: 10.1073/pnas.1217094110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma P, Schwarten M, Schneider L, Boeske A, Henke N, Lisak D, Weber S, Mohrlüder J, Stoldt M, Strodel B, et al. Interaction of Bcl-2 with the autophagy-related GABAA receptor-associated protein (GABARAP): biophysical characterization and functional implications. J Biol Chem. 2013;288:37204–37215. doi: 10.1074/jbc.M113.528067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marintchev A, Frueh D, Wagner G. NMR methods for studying protein-protein interactions involved in translation initiation. Methods Enzymol. 2007;430:283–331. doi: 10.1016/S0076-6879(07)30012-8. [DOI] [PubMed] [Google Scholar]
- Monaco G, Decrock E, Nuyts K, Wagner LE, 2nd, Luyten T, Strelkov SV, Missiaen L, De Borggraeve WM, Leybaert L, Yule DI, et al. α-helical destabilization of the Bcl-2-BH4-domain peptide abolishes its ability to inhibit the IP3 receptor. PLoS ONE. 2013;8:e73386. doi: 10.1371/journal.pone.0073386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliari LJ, Kuwana T, Bonzon C, Newmeyer DD, Tu S, Beere HM, Green DR. The multidomain proapoptotic molecules Bax and Bak are directly activated by heat. Proc Natl Acad Sci USA. 2005;102:17975–17980. doi: 10.1073/pnas.0506712102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petros AM, Medek A, Nettesheim DG, Kim DH, Yoon HS, Swift K, Matayoshi ED, Oltersdorf T, Fesik SW. Solution structure of the antiapoptotic protein bcl-2. Proc Natl Acad Sci USA. 2001;98:3012–3017. doi: 10.1073/pnas.041619798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler M, Liang H, Nettesheim D, Meadows RP, Harlan JE, Eberstadt M, Yoon HS, Shuker SB, Chang BS, Minn AJ, et al. Structure of Bcl-xL-Bak peptide complex: recognition between regulators of apoptosis. Science. 1997;275:983–986. doi: 10.1126/science.275.5302.983. [DOI] [PubMed] [Google Scholar]
- Shi XE, Wales TE, Elkin C, Kawahata N, Engen JR, Annis DA. Hydrogen exchange-mass spectrometry measures stapled peptide conformational dynamics and predicts pharmacokinetic properties. Anal Chem. 2013;85:11185–11188. doi: 10.1021/ac403173p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souers AJ, Leverson JD, Boghaert ER, Ackler SL, Catron ND, Chen J, Dayton BD, Ding H, Enschede SH, Fairbrother WJ, et al. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat Med. 2013;19:202–208. doi: 10.1038/nm.3048. [DOI] [PubMed] [Google Scholar]
- Sugioka R, Shimizu S, Funatsu T, Tamagawa H, Sawa Y, Kawakami T, Tsujimoto Y. BH4-domain peptide from Bcl-xL exerts anti-apoptotic activity in vivo. Oncogene. 2003;22:8432–8440. doi: 10.1038/sj.onc.1207180. [DOI] [PubMed] [Google Scholar]
- Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- Walensky LD, Bird GH. Hydrocarbon-stapled peptides: principles, practice, and progress. J Med Chem. 2014;57:6275–6288. doi: 10.1021/jm4011675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walensky LD, Pitter K, Morash J, Oh KJ, Barbuto S, Fisher J, Smith E, Verdine GL, Korsmeyer SJ. A stapled BID BH3 helix directly binds and activates BAX. Mol Cell. 2006;24:199–210. doi: 10.1016/j.molcel.2006.08.020. [DOI] [PubMed] [Google Scholar]
- Wang K, Gross A, Waksman G, Korsmeyer SJ. Mutagenesis of the BH3 domain of BAX identifies residues critical for dimerization and killing. Mol Cell Biol. 1998;18:6083–6089. doi: 10.1128/mcb.18.10.6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.