Abstract
Members of the endosomal sorting complex required for transport (ESCRT) machinery function in membrane remodeling processes during multivesicular endosome biogenesis, cytokinesis, retroviral budding, and plasma membrane repair. During lumenal vesicle formation at endosomes, the ESCRT-II complex and the ESCRT-III subunit VPS-20 play a specific role in regulating assembly of ESCRT-III filaments, which promote vesicle scission. Previous work suggests that Vps20 isoforms, like other ESCRT-III subunits, exhibits an autoinhibited, closed conformation in solution, and its activation depends on an association with ESCRT-II specifically at membranes. However, we show here that C. elegans ESCRT-II and VPS-20 interact directly in solution, both in cytosolic cell extracts and using recombinant proteins in vitro. Moreover, we demonstrate that purified VPS-20 exhibits an open, extended conformation, irrespective of ESCRT-II binding, in contrast with the closed, autoinhibited architecture of another ESCRT-III subunit, VPS-24. Our data argue that individual ESCRT-III subunits adopt distinct conformations, which are tailored for their specific functions during ESCRT-mediated membrane reorganization events.
Keywords: conformational change, membrane curvature, molecular modeling, multivesicular body, small angle x-ray scattering (SAXS)
INTRODUCTION
Active cell surface receptors and other transmembrane proteins undergo downregulation through the generation of multivesicular endosomes (MVEs), which sequester cargoes within the lumen of the endosome and block interactions with cytoplasmic effectors [1–3]. Biogenesis of MVEs is dependent upon a collection of proteins known as the ESCRT machinery, which functions in the identification of cargoes for internalization, membrane deformation to generate nascent vesicles that bud away from the cytoplasm, and the scission event that releases these vesicles into the endosome lumen [4,5]. Transmembrane proteins destined for degradation are modified by ubiquitin conjugation, which acts as a sorting signal recognized by early-acting components of the ESCRT machinery [6,7]. In addition to MVE biogenesis, subsets of the ESCRT machinery also play roles in cytokinesis, the budding of several enveloped viruses, and plasma membrane repair [8–10]. These processes are topologically equivalent, suggesting a specialized function for the ESCRT machinery in promoting scission events from the cytoplasmic surface of membranes.
The ESCRT machinery is made of five complexes (ESCRT-0, ESCRT-I, ESCRT-II, ESCRT-III, and Vps4-Vta1), and a set of accessory proteins. ESCRT-0, ESCRT-I, and ESCRT-II form stable, heteropolymeric complexes in the cytoplasm, which contain ubiquitin binding domains that directly interact with cargo [1,3,4]. In vitro studies suggest that ESCRT-I and ESCRT-II can contribute to membrane deformation on the surface of giant unilamellar vesicles (GUVs), and upon addition of the ESCRT-III subunits, Vps20 and Vps32, release nascent vesicles into the lumen of GUVs [11]. This suggests that ESCRT-III has a direct role in membrane scission. Unlike upstream ESCRT complexes, ESCRT-III polymerizes only on lipid bilayers in vivo and requires the Vps4-Vta1 complex to disassemble [1,3,12].
Based on genetic and biochemical studies in yeast, the core ESCRT-III subunits are recruited in a sequential fashion to the site of MVE formation and include Vps20, Vps32, Vps24 and Vps2, respectively [13]. Vps20 binds to each of the two Vps25 subunits of ESCRT-II [14]. In vitro experiments suggest that conformational changes may occur in Vps20 when mixed with ESCRT-II and liposomes, and these structural changes are associated with the recruitment and subsequent activation of Vps32 [15]. Vps32 is the most abundant ESCRT-III subunit and polymerizes to form filaments, which play a direct role in membrane scission [13,15]. Additionally, a ‘supercomplex’ of ESCRT-II and Vps20 preferentially binds to regions of high curvature, spatially regulating Vps32 filament formation [16]. Vps24 may act as a cap for the forming filament and also binds to Vps2, which plays an important role in the Vps4-dependent disassembly of the ESCRT-III complex [13,17,18].
The ESCRT-III proteins possess a similar overall architecture, with a basic amino-terminus and an acidic carboxyl-terminus, both of which are composed of a series of alpha helices. Crystallographic data on the helical core of human Vps24 (CHMP3) revealed a 70 Å helical hairpin (composed of helices 1 and 2) that forms an asymmetric four-helix bundle with two short helices (helices 3 and 4) [19]. The carboxyl-terminal portions (including helix 5) of the ESCRT-III proteins have been proposed to act as autoinhibitory domains, which prevent interaction between individual ESCRT-III subunits within the cytoplasm [20–24]. Two structures of human Vps24 have placed helix 5 in different positions. One shows helix 5 in a closed conformation, where it is packed against the helical hairpin loop [23], while the other structure represents an open conformation where helix 5 lies perpendicular to the four helix bundle [19]. Based on a crystal structure of the binding interface between Vps20 and ESCRT-II, the position of helix 5 should not be impacted by their association [25], suggesting that ESCRT-II binding to Vps20 would not promote its transition to a more open, active conformation. Here, we take advantage of small-angle X-ray scattering (SAXS) to demonstrate that Vps20, unlike Vps24, adopts an extended, open conformation, independently of ESCRT-II. Our findings indicate that free ESCRT-III subunits exhibit distinct conformations in solution, and additionally suggest that autoinhibition does not represent a general mechanism to prevent cytosolic assembly of ESCRT-III polymers in vivo.
EXPERIMENTAL
Immunoprecipitation and Mass Spectrometry
Adult hermaphrodites were grown synchronously in liquid culture following a previously described protocol [26]. Immunoprecipitations were also conducted as described previously [26], with the exception that detergent was eliminated from buffers when generating a high speed cytosolic extract. C. elegans ESCRT-II antibodies were raised in rabbits by immunization (Covance) with a strep-tagged form of the intact ESCRT-II complex (VPS-22, VPS-25, and VPS-36) produced in E. coli. Antibodies were subsequently affinity purified from serum by binding to a column harboring the same recombinant ESCRT-II complex. Antibodies directed against VPS-20 and STAM-1 have been described previously [16]. For mass spectrometry analysis, proteins were precipitated by using TCA and were subsequently processed for MudPIT analysis [26,27]. The spectra were searched with either the SEQUEST or ProLuCID [28] algorithm against the WormBase C. elegans database.
Protein purification, gel filtration studies, and circular dichroism
Recombinant protein expression was performed using BL21-T1R (DE3) E. coli. All proteins utilized in this study were generated using C. elegans cDNA clones. For ESCRT-II, all subunits were cloned into the polycistronic expression vector pST39 [29] with a single tag appended to VPS-25 to enable purification. VPS-24, VPS-20ΔC (amino acids 1–170), and VPS-25110–183 (amino acids 110–183) were cloned into pGEX6P-1, which encodes a cleavable, amino-terminal GST tag. VPS-20 was cloned into His-Sumo-pET28d with a cleavable, amino-terminal His-Sumo tag. Protein purifications were conducted using glutathione agarose beads (for GST-VPS-24, GST-VPS-20ΔC, and GST-VPS-25110–183) or nickel affinity resin (for intact ESCRT-II and His-Sumo-VPS-20). The GST moiety was removed from VPS-24, VPS-20ΔC, and VPS-25110–183 using Prescission protease. The His-Sumo tag was removed with Sumo Protease and after cleavage the sample was mixed with fresh nickel resin to isolate VPS-20. Following affinity purification, all proteins (1 mL) were applied to a S200 size exclusion column (GE Healthcare) equilibrated in either SAXS buffer (50 mM Tris (pH 8.0), 10 mM DTT, 100 mM NaCl) or ESCRT-II/VPS-20 interaction buffer (50 mM Hepes (pH 7.6), 1 mM DTT, 100 mM NaCl). Samples for SAXS were dialyzed overnight into the SAXS buffer to ensure proper buffer matching (VPS-20 sample without salt was dialyzed into SAXS buffer without NaCl). For ESCRT-II/VPS-20 interaction studies, both proteins were independently re-run over the S200 size exclusion column along with a 1:2 molar ratio (ESCRT-II:VPS-20) that had been mixed for 1 hour at 4°C. 1 mL fractions were collected for each run and analyzed by SDS-PAGE analysis. Densitometry of each band was used to calculate peak elution volumes. C. elegans extracts generated in the absence of detergent were applied onto a Superose 6 gel filtration column, and eluted fractions were separated by SDS-PAGE and immunoblotted using antibodies directed against ESCRT-II and VPS-20. Light-scattering data was collected using a Wyatt mini-DAWN TREOS three-angle light scattering detector coupled to a high-resolution size-exclusion column. Data was collected at a flow rate of 0.5 mL/min and analyzed with the ASTRA software to determine molecular weight of protein [30].
For circular dichroism studies, proteins were analyzed using a 0.1 cm path length quartz cell. Proteins were dialyzed overnight into 25 mM sodium phosphate (pH 7.2), and spectra were collected using a Model 202SF Circular Dichroism Spectrophotometer at 25°C.
Small angle X-ray scattering data collection
SAXS data was collected at the National Magnetic Resonance Facility at Madison (NMRFAM) using a Bruker Nanostar. The Bruker Nanostar had a wavelength of 1.54 Å with a sample-detector distance set to 67.7 cm resulting in a momentum transfer range of 0.01 < q < 0.40 Å−1. Data for SAXS buffer and protein were collected at 25°C for 2–4 hours. To account for excluded volume of the protein, buffer was subtracted. For optimal data, merged data sets were generated from two different concentrations using the lowest angles from one dataset and the highest angles from another. An overlapping region of at least 60 points was utilized for the merging process. This was conducted in accordance with the Primus analysis guide. Guinier analysis was conducted to determine the Rg value through the use of the Primus software (ATSAS) [31,39]. A comparison of concentrations was conducted to detect overall quality of data.
SAXS analysis and modeling
Processing of SAXS data was performed with Gnom software (ATSAS) [32] to determine the Dmax of the protein and the Porod volume. An approximate molecular weight (MW) can be determined by using the Porod volume in the following equation: MW = (Porod volume × 1.2)/2. Specifically, the Gnom software generates the pair distance distribution function (PDDF) for each sample, which describes the probability of finding two atoms a select distance apart. Dmax refers to the distance where the probability is zero to find two atoms said distance apart. This distance defines the maximum dimension and governs the overall shape of the SAXS envelope. The Dmax was adjusted (1–2 Å increments) until the following criteria were met: there was a smooth drop to zero probability at Dmax, agreement between the Rg from the Guiner transform and the Rg from the PDDF was achieved, and agreement between the experimental scatter data and the regularized scattering from the PDDF was reached. To avoid over-smoothing the regularization parameter, alpha, was kept below 5. The scattering amplitudes of the PDDF were used to calculate 15 replicate ab initio dummy atom models using DAMM~F [33]. The DAMAVER program was used to determine an averaged model reporting mean NSD value to describe the uniqueness of the overall model [34]. Models generated from DAMAVER were superimposed with crystal structures (2GD5 and 3FRT) using the Supcomb20 algorithm [34,35].
Production of Liposomes and Co-floatation Assays
Liposomes (36.5% phosphatidylcholine, 30% phosphatidylethanolamine (PE), 30% phosphatidylserine, 3% phosphatidylinositol 3-phosphate, and 0.5% rhodamine-labeled PE) were prepared by extrusion through polycarbonate filters with pore sizes of 50 and 400 nm (Avanti Polar Lipids). Dynamic light scattering measurements were conducted to determine the actual size distribution of liposomes generated. For co-flotation assays, liposomes were incubated with protein in buffer (50 mM Hepes (pH 7.6), 100 mM NaCl, and 1 mM DTT) prior to mixing with Accudenz density medium. Mixtures were overlaid with decreasing concentrations of Accudenz (0–40%) and centrifuged for 2 h at 280,000 × g. During this period, liposomes and associated proteins floated to the buffer/Accudenz interface and were harvested by hand. Recovery of liposomes was normalized based on the fluorescence intensity of the sample, and equivalent fractions (when comparing flotation experiments that used liposomes of differing sizes) were separated by SDS-PAGE and subjected to immunoblot analysis to determine the relative amount of protein that bound [16].
RESULTS
ESCRT-II interacts directly with VPS-20 in solution
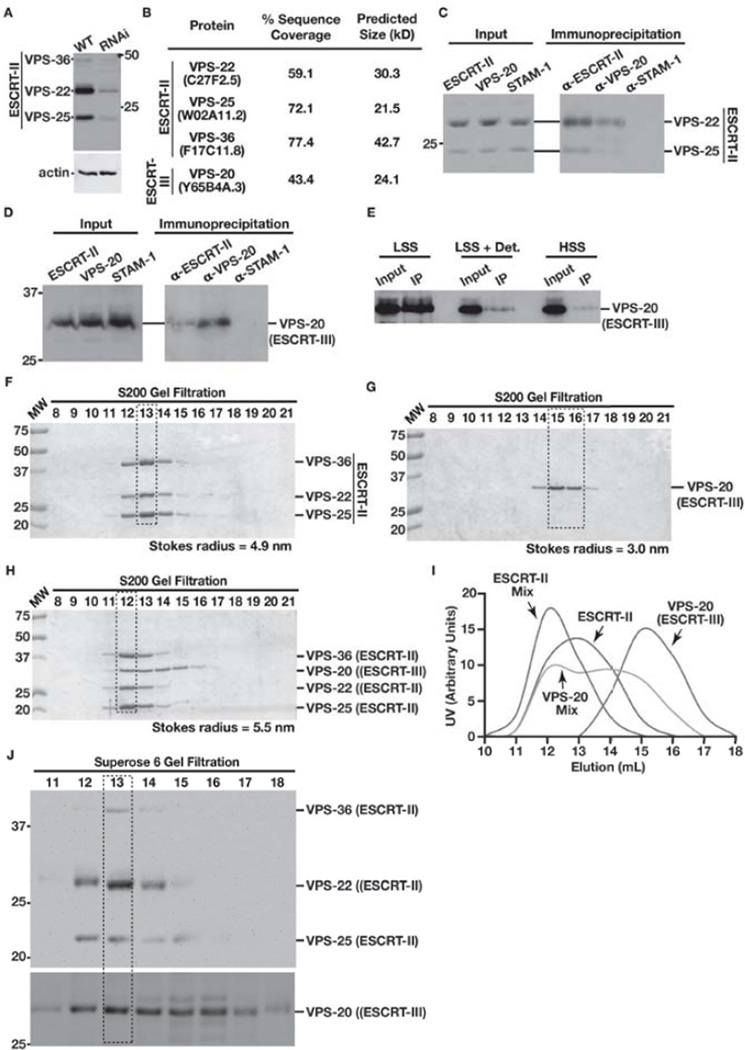
ESCRT-II and Vps20 have been shown previously in yeast to associate only at endosomal membranes [15]. A mechanism underlying this specificity is lacking, and structural studies fail to support the idea that these factors would be unable to bind in solution [25]. We used affinity-purified antibodies directed against ESCRT-II to conduct a series of immunoprecipitations from C. elegans embryo extracts. The specificity of our antibodies was confirmed using C. elegans extracts generated from wild-type animals and animals depleted of all three ESCRT-II subunits (Figure 1A). Multidimensional protein identification technology (MudPIT) was used to identify ESCRT-II interacting partners [36]. A directed search for all components of the ESCRT machinery that are expressed in worms revealed the presence of only the ESCRT-II subunits (VPS-22, VPS-25, and VPS-36) and the ESCRT-III protein VPS-20 (Figure 1B). To confirm this finding, we immunoprecipitated ESCRT-II and VPS-20 separately from C. elegans extracts, and conducted a set of immunoblot analyses. Consistent with our unbiased mass spectrometry-based approach, we found that ESCRT-II and VPS-20 bind to one another in solution, but neither interacts with upstream components of the ESCRT machinery (Figures 1C and 1D).
Figure 1. ESCRT-II binds directly to VPS-20 in solution.
(A) The specificity of antibodies directed against ESCRT-II was tested using C. elegans extracts generated from wild type animals and animals depleted of all three ESCRT-II subunits. The three bands corresponding to VPS-36, VPS-22, and VPS-25 are substantially diminished following RNA interference. Molecular mass markers are shown on the right, and an immunoblot directly against actin was conducted as a loading control. (B) Antibodies directed against all three subunits of the ESCRT-II complex were used to isolate ESCRT-II and any interacting partners from C. elegans embryo extracts and subjected to solution mass spectrometry analysis. Identified components of the ESCRT machinery, their percent sequence coverage, and their molecular masses are shown. (C and D) Antibodies directed against ESCRT-II, VPS-20, or STAM-1 were used to conduct immunoprecipitations from C. elegans embryo extracts. Recovered proteins were separated by SDS-PAGE and immunobloted using antibodies directed against ESCRT-II (panel C) or VPS-20 (panel D). Inputs for each immunoprecipitation are shown on the left. (E) Immunoprecipitations using ESCRT-II antibodies were conducted using a low speed supernatant (LSS) in the presence (middle) or absence (left) of detergent (0.05% NP40 and 1% Triton X-100) or a high speed supernatant (HSS) generated in the absence of detergent (right). Equivalent amounts of starting material (input) were used for each immunoprecipitation, and the presence of VPS-20 was detected using VPS-20 specific antibodies by immunoblot analysis. (F–H) ESCRT-II alone (panel D), VPS-20 alone (panel E), or a mixture of ESCRT-II and VPS-20 (F; at a 1:2 molar ratio) were separated by size exclusion chromatography. Stokes radii were calculated based on the elution profile of characterized standards. Peak fractions are highlighted (boxes with dashed lines). (I) A graphical representation of the densitometry measurements conducted on each gel shown in panels D, E, and F. Based on this graph, peak elution volumes were determined to be the following: 12.9 mL for ESCRT-II alone, 15.1 mL for VPS-20 alone, 12.1 mL for ESCRT-II within the mixture of it and VPS-20, and two peaks of 12.2 mL (associated with ESCRT-II) and 14 mL (free of ESCRT-II) for VPS-20 within mixture. (J) A C. elegans extract was separated by size exclusion chromatography, and fractions eluted were immunoblotted using antibodies directed against ESCRT-II (top) and VPS-20 (bottom). The peak fraction in which ESCRT-II and VPS-20 are both found is highlighted (boxes with dashed lines).
Since extracts used for immunoprecipitation were generated in the presence of detergent, the interaction between ESCRT-II and Vps20 observed may represent a remnant of an endosome-associated complex that was released upon membrane dissolution. We therefore conducted a series of additional immunoprecipitations using ESCRT-II antibodies and extracts that either contained membranes (i.e., a low speed supernatant that was centrifuged at 1000 RPM to clear cell debris following lysis) or were subjected to a high speed centrifugation step (50,000 RPM) in the absence of detergent (i.e., a cytosolic extract devoid of membranes). We found that a significant amount of VPS-20 was recovered from a membrane-containing extract, consistent with the idea that the ESCRT-II/VPS-20 complex exists on endosomal membranes. In contrast, we observed significantly less VPS-20 following ESCRT-II immunoprecipitation from a cytosolic extract (Figure 1E). Treatment of the low speed supernatant with detergent prior to ESCRT-II immunoprecipitation similarly reduced the interaction between ESCRT-II and VPS-20, suggesting that the presence of membranes stabilizes their association. These findings are consistent with previous work demonstrating that the ESCRT-II/VPS-20 complex binds avidly to membranes [15,16]. However, our data do not rule out the possibility that ESCRT-II initially binds to VPS-20 in the cytoplasm and is subsequently recruited rapidly onto endosomal membranes.
To explore this question further and determine whether the interaction between ESCRT-II and VPS-20 was direct, we purified recombinant forms of the proteins and measured their association using size exclusion chromatography. Independently, ESCRT-II and VPS-20 exhibit Stokes radii of approximately 4.9 nm and 3.0 nm, respectively (Figures 1F and 1G). However, when combined at a molar ratio of 1:2 (to account for the presence of two VPS-20 binding sites in ESCRT-II), a new complex containing both ESCRT-II and VPS-20 forms in solution, with an enlarged Stokes radius of approximately 5.5 nm (Figures 1H and 1I). Since the concentrations of recombinant ESCRT-II and VPS-20 used in these experiments are likely to be substantially higher than that of their endogenous counterparts in the cytoplasm, we also investigated whether native ESCRT-II and VPS-20 associate. To do so, we subjected a C. elegans cytosolic extract generated in the absence of detergent to size exclusion chromatography. Strikingly, following immunoblot analysis, we found that a large pool of VPS-20 co-eluted with ESCRT-II (Figure 1J). Collectively, these data suggest that ESCRT-II and VPS-20 interact directly in solution, but are subject to rapid recruitment onto membranes following their co-assembly in cells.
Small angle X-ray scattering defines unique conformations for two ESCRT-III subunits
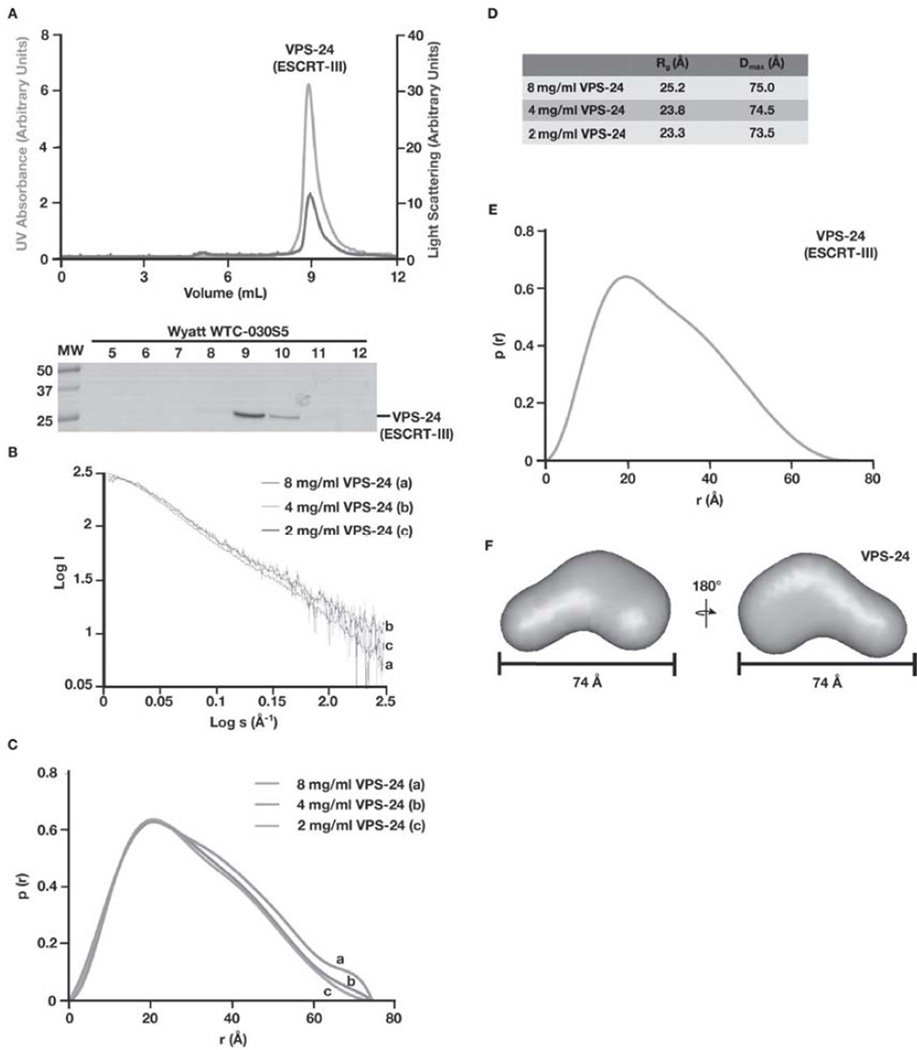
The interface between ESCRT-II and ESCRT-III has been determined crystallographically and identifies an amino-terminal helix in Vps20 (helix 1, part of an asymmetric 4-helix bundle), which mediates its association with ESCRT-II [25]. This interaction is not predicted to alter the autoinhibitory conformation that is observed in other ESCRT-III subunits, such as human Vps24 and Ist1 [19,23]. Combined with our findings that ESCRT-II and VPS-20 interact in solution, we questioned the proposed role for ESCRT-II in relieving VPS-20 from an autoinhibited state to nucleate ESCRT-III polymerization. To explore the structures of full-length ESCRT-III subunits, none of which are amenable to x-ray crystallography, we took advantage of small angle X-ray scattering (SAXS), a low-resolution structural approach that permits ab initio modeling to determine the conformations of proteins in solution. Previous studies using SAXS demonstrated that full-length human Vps24 exhibits a closed conformation under low salt conditions [22,37]. To validate this approach using C. elegans ESCRT-III proteins, we prepared purified, recombinant C. elegans VPS-24 for small angle x-ray scattering (SAXS) analysis. Based on size exclusion chromatography combined with multi-angle light scattering (SEC-MALS), untagged VPS-24 exhibits a molecular mass of 24.5 kD (Figure 2A), consistent with it existing as a monomer in solution, even at a concentration as high as 340 µM. SAXS analysis of VPS-24 produced consistent and interpretable scattering profiles across three protein concentrations (Figure 2B). From these data, we determined the radius of gyration (Rg) and maximal inter-atomic distance (Dmax) to range from 23.3–25.2 Å and 73.5–75.0 Å, respectively (Figures 2C and 2D). We generated a merged data set between two concentrations, which produced an Rg of 23.3 Å and a Dmax of 74.0 Å (Figure 2E). The Porod volume was used to estimate the molecular weight to be ~ 21 kD, consistent with its predicted molecular weight of 23.6 kD, based on amino acid composition. These data confirm that C. elegans VPS-24 is a monodispersed monomer in solution.
Figure 2. Analysis of C. elegans VPS-24 by SAXS suggests that it exhibits a compact, auto-inhibited conformation in solution.
(A) Purified, recombinant VPS-24 was separated over a gel filtration column that was coupled to a multi-angle light scattering device. Both the UV absorbance (green) and light scattering (blue) profiles are plotted (top) and eluted fractions were separated by SDS-PAGE and stained using Coomassie to highlight the elution profile of VPS-24 (bottom). (B) Log of scattered intensity vs. log of s is shown across the three concentrations of VPS-24 used in SAXS experiments (8 mg/mL, 4 mg/mL, and 2 mg/mL). S is equal to 4πsin(θ)/λ (λ = wavelength and 20 = scattering angle). Plots for each concentration of protein are labeled alphabetically (8 mg/mL is denoted ‘a’, 4 mg/mL is denoted ‘b’, and 2 mg/mL is denoted with ‘c’). (C) Pair distance distribution function plots comparing the three concentrations of VPS-24 used in SAXS experiments. Plots for each concentration of protein are labeled alphabetically (8 mg/mL is denoted ‘a’, 4 mg/mL is denoted ‘b’, and 2 mg/mL is denoted with ‘c’). (D) A summary of the Rg and Dmax values for the three concentrations of C. elegans VPS-24 tested. (E) Pair distance distribution function plot of the merged data set generated from two different SAXS data sets (8 mg/mL and 2 mg/mL) determined a Dmax of 74 Å for VPS-24. (F) A low resolution, ab initio model of VPS-24 was determined based on fifteen structures that were generated using the program DAMMIF and averaged with DAMAVER, resulting in a NSD value of 0.67. Two views, rotated 180° related to one another, are shown.
An overall protein envelope was determined ab initio from the scattering profile [33]. 15 independent models of VPS-24 were aligned and averaged [34], with a nominal spatial discrepancy (NSD) of 0.67 (Figure 2F). The overall envelope of VPS-24 reveals a compact structure in agreement with the previously published SAXS envelope of human Vps24, which exhibited a Dmax of 75 Å [22]. Together, our findings demonstrate that C. elegans VPS-24 possesses a similar conformation as compared to human Vps24 and validates our use of SAXS to study the structures of full-length ESCRT-III subunits.
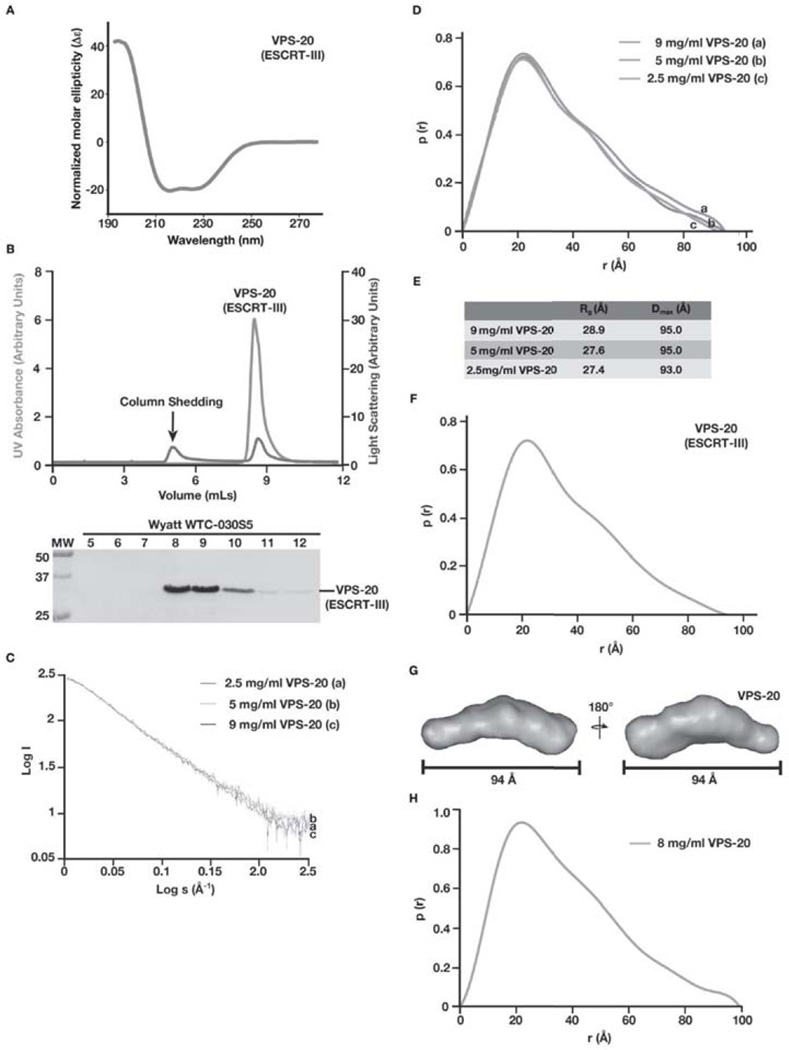
To determine the conformation of VPS-20 in solution, we purified a recombinant form of the full-length, untagged protein to homogeneity. Based on circular dichroism spectroscopy, the protein appeared folded and exhibited a characteristic spectrum for an alpha-helical protein, as is expected for ESCRT-III subunits (Figure 3A). Additionally, our previous work highlighted the functionality of recombinant VPS-20 in facilitating ESCRT-III filament assembly in the presence of ESCRT-II [16]. SEC-MALS analysis demonstrated that VPS-20 elutes in a single peak with a molecular mass of 25.5 kD (Figure 3B), similar to its predicted molecular weight of 24.1 kD, based on amino acid composition. These data indicate that VPS-20 is monomeric in solution. SAXS analysis of VPS-20 produced consistent and interpretable scattering profiles across three unique protein concentrations (Figure 3C). In contrast to our findings using VPS-24, VPS-20 exhibits Rg and Dmax values ranging from 27.4–28.9 Å and 93.0–95.0 Å, respectively (Figures 3D and 3E). We generated a merged data set between two concentrations, producing an overall Rg of 27.3 Å and a Dmax of 94.0 Å (Figure 3F). The Porod volume estimated the molecular weight to be ~25 kD, consistent with the predicted molecular weight of a VPS-20 monomer.
Figure 3. C. elegans VPS-20 exhibits an extended, open conformation in solution.
(A) Circular dichroism (CD) spectroscopy was used to characterize VPS-20. Samples were analyzed at a concentration of 1 µM, and the data were normalized. CD spectra were collected at 25°C in 25 mM sodium phosphate (pH 7.2) using a 1 mm path length quartz cell. (B) Purified, untagged VPS-20 was separated over a gel filtration column that was coupled to a multi-angle light scattering device. Both the UV absorbance (green) and light scattering (blue) profiles are plotted (top) and eluted fractions were separated by SDS-PAGE and stained using Coomassie to highlight the elution profile of VPS-20 (bottom). A small light scattering peak, which contains no protein, is indicated (arrow), and is a result of non-specific column shedding. (C) Log of scattered intensity vs. log of s is shown across the three concentrations of VPS-20 used in SAXS experiments (9 mg/mL, 5 mg/mL, and 2.5 mg/mL). Plots for each concentration of protein are labeled alphabetically (2.5 mg/mL is denoted ‘a’, 5 mg/mL is denoted ‘b’, and 9 mg/mL is denoted with ‘c’). (D) Pair distance distribution function plots comparing the three concentrations of VPS-20 used in SAXS experiments. Plots for each concentration of protein are labeled alphabetically (9 mg/mL is denoted ‘a’, 5 mg/mL is denoted ‘b’, and 2.5 mg/mL is denoted with ‘c’). (E) A summary of the Rg and Dmax values for the three concentrations of C. elegans VPS-20 tested. (F) Pair distance distribution function plot of the merged data set generated from two different SAXS data sets (5 mg/mL and 2.5 mg/mL) determined a Dmax of 94 Å for VPS-20. (G) A low resolution, ab initio model of VPS-20 was determined based on fifteen structures that were generated using the program DAMMIF and averaged with DAMAVER, resulting in a NSD value of 0.69. Two views, rotated 180° related to one another, are shown. (H) Pair distance distribution function plot of VPS-20 (8 mg/mL), as measured by SAXS in the absence of salt.
An overall protein envelope for VPS-20 was obtained using ab initio modeling. 15 independent models produced an average envelope with a NSD value of 0.69 (Figure 3G). In contrast to the conformation of VPS-24, the overall shape of VPS-20 in solution revealed an elongated conformation. To verify that the limited concentration of salt (100 mM) used in our experiments was not affecting the shape of VPS-20, we conducted SAXS analysis on VPS-20 in the absence of salt, which yielded similar Dmax and Rg values of 99.5 Å and 29.6 Å, respectively (Figure 3H). Together, these data indicate that VPS-20 and VPS-24 do not share a similar conformation in solution.
VPS-20 exhibits an open conformation independent of ESCRT-II in solution
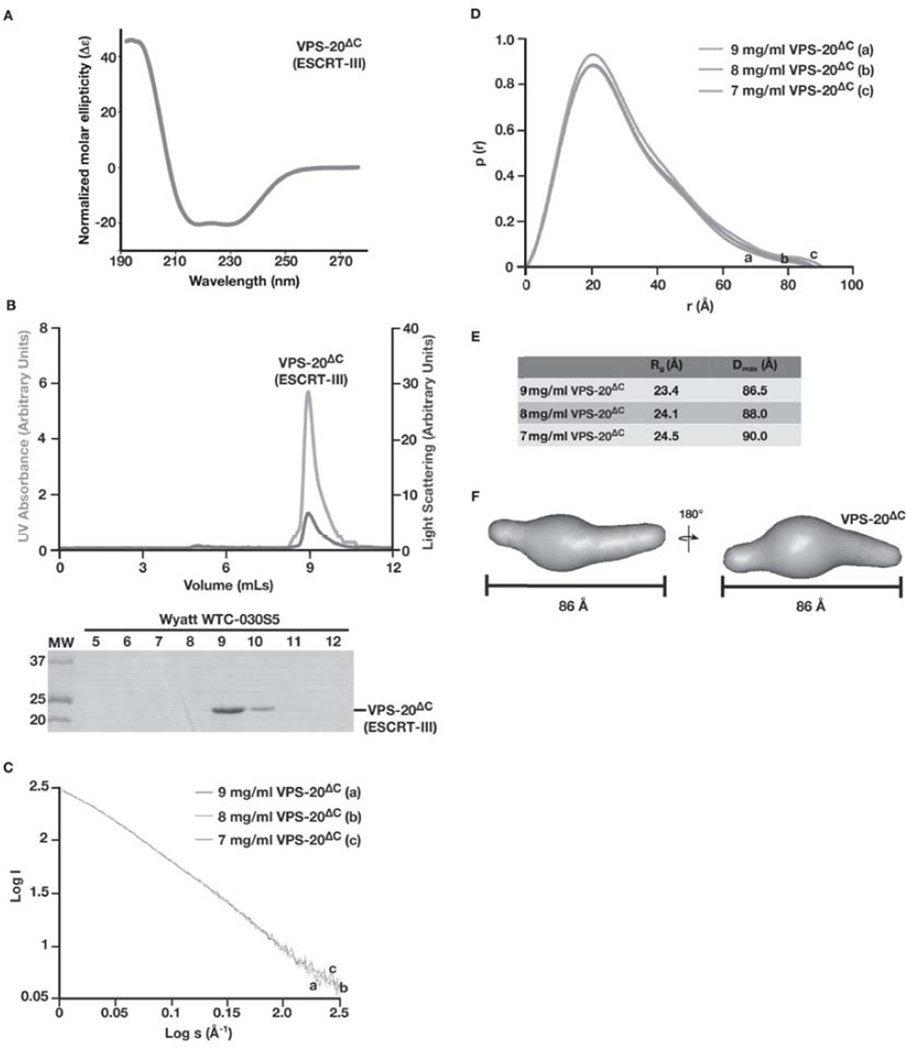
Although the SAXS envelope for VPS-20 indicates that it exhibits an extended conformation in solution, the orientation of the protein is difficult to assign unambiguously. We therefore generated a truncated form of C. elegans VPS-20 containing its asymmetric helical core (VPS-20ΔC; helices 1–5 encoded by amino acids 1–170). Circular dichroism spectroscopy indicated that the protein was folded and exhibited a spectrum characteristic of an alpha-helical protein, similar to that observed using full length VPS-20 (Figure 4A). Based on SEC-MALS, VPS-20ΔC exhibited a molecular mass of 20.3 kD (Figure 4B), nearly identical to that predicted based on its amino acid composition (19.7 kD). These data strongly suggest it is monomeric in solution. We therefore analyzed VPS-20ΔC using SAXS, which produced interpretable scattering profiles (Figure 4C) and revealed that VPS-20ΔC possesses Rg and Dmax values ranging from 24.1–24.5 Å and 86.5–90.0 Å, respectively (Figures 4D and 4E). The Porod volume determined the molecular weight to be 18.6 kD, similar to that of a monomeric form of VPS-20ΔC. 15 independent models produced an envelope with a NSD value of 0.65 (Figures 4F). The overall shape of VPS-20ΔC was similar to the elongated conformation observed following SAXS analysis of full length VPS-20.
Figure 4. Analysis of C. elegans VPS-20Δc by SAXS suggests that it exhibits an extended conformation, similar to the full length protein in solution.
(A) CD spectroscopy was used to characterize VPS-20ΔC. Samples were analyzed at a concentration of 1 µM, and the data were normalized. CD spectra were collected at 25°C in 25 mM sodium phosphate (pH 7.2) using a 1 mm path length quartz cell. (B) Purified, untagged VPS-20ΔC (amino acids 1–170) was separated over a gel filtration column that was coupled to a multi-angle light scattering device. Both the UV absorbance (green) and light scattering (blue) profiles are plotted (top) and eluted fractions were separated by SDS-PAGE and stained using Coomassie to highlight the elution profile of VPS-20ΔC (bottom). (C) Log of scattered intensity vs. log of s is shown across the three concentrations of VPS-20ΔC used in SAXS experiments (9 mg/mL, 8 mg/mL, and 7 mg/mL). Plots for each concentration of protein are labeled alphabetically (9 mg/mL is denoted ‘a’, 8 mg/mL is denoted ‘b’, and 7 mg/mL is denoted with ‘c’). (D) Pair distance distribution function plots comparing the three concentrations of VPS-20ΔC used in SAXS experiments. Plots for each concentration of protein are labeled alphabetically (9 mg/mL is denoted ‘a’, 8 mg/mL is denoted ‘b’, and 7 mg/mL is denoted with ‘c’). (E) A summary of the Rg and Dmax values for the three concentrations of C. elegans VPS-20ΔC tested. (F) A low resolution, ab initio model of VPS-20ΔC was determined based on fifteen structures that were generated using the program DAMMIF and averaged with DAMAVER, resulting in a NSD value of 0.65. Two views, rotated 180° related to one another, are shown. VPS-20ΔC exhibits a Dmax of 86 Å.
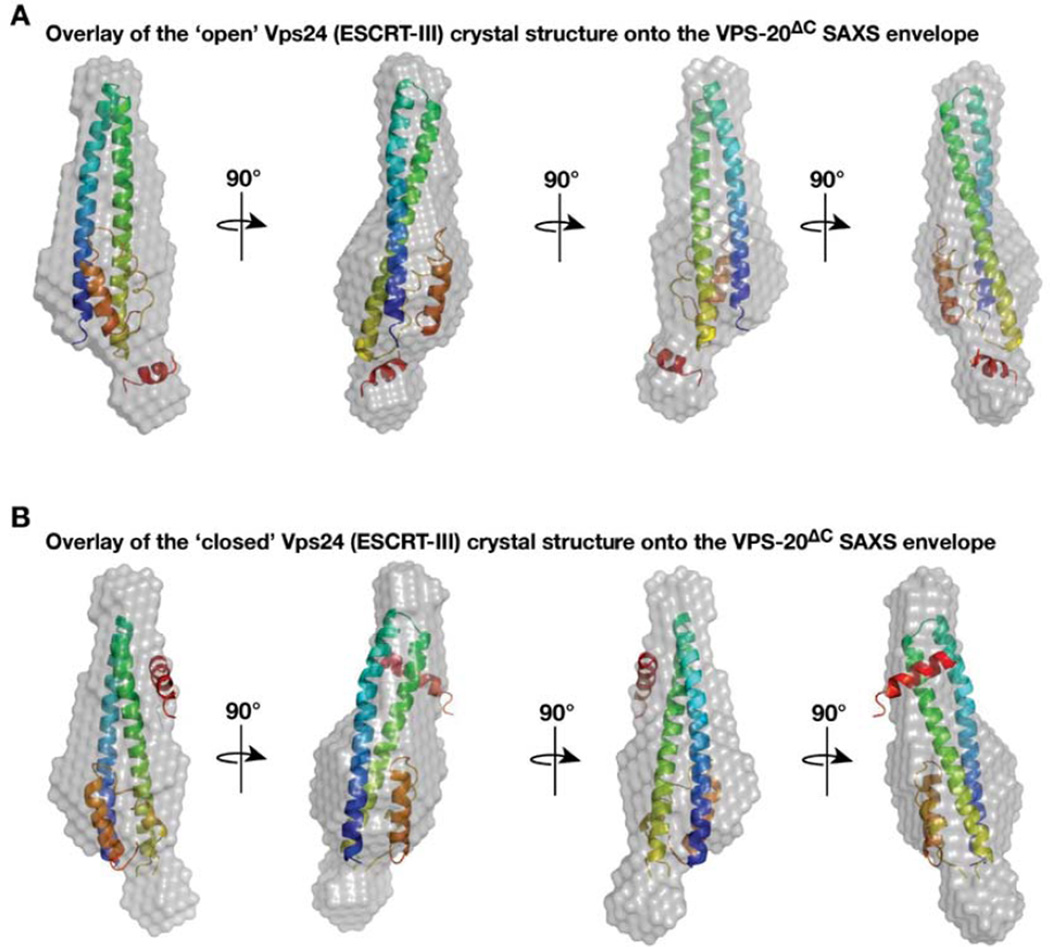
Although the positioning of carboxyl-terminal helices within ESCRT-III subunits varies, depending on whether they exhibit an open or a closed conformation, the helical core domain is structurally stable [19,23]. For example, two crystallographic structures for Vps24 have been determined. One of these structures utilizes a carboxyl-terminal truncation, similar to the truncation we produced in VPS-20ΔC, which displaces helix 5 away from the helical hairpin and represents an open conformation [19]. The alternative structure of Vps24 represents a closed conformation, in which helix 5 is packed against the helical hairpin loop [23]. Unbiased in silico methods were used to fit both crystal structures within the SAXS envelope of VPS-20ΔC [35]. While the open form of human Vps24 fits well into the SAXS envelope of VPS-20ΔC in all orientations (Figure 5A), the closed form could not be modeled into the envelope without helix 5 extending beyond its boundaries (Figure 5B). These data strongly suggest that VPS-20ΔC represents the open conformation of VPS-20.
Figure 5. Overlays of two human Vps24 crystal structures onto the VPS-20ΔC SAXS envelope.
(A) The program Supcomb20 was used to align one of the human Vps24 crystal structures (2GD5, representing its open conformation) with the SAXS model for VPS-20ΔC. (B) Alignment of another human Vps24 crystal structure (3FRT, representing its closed conformation) with the SAXS model for VPS-20ΔC reveals a poor fit.
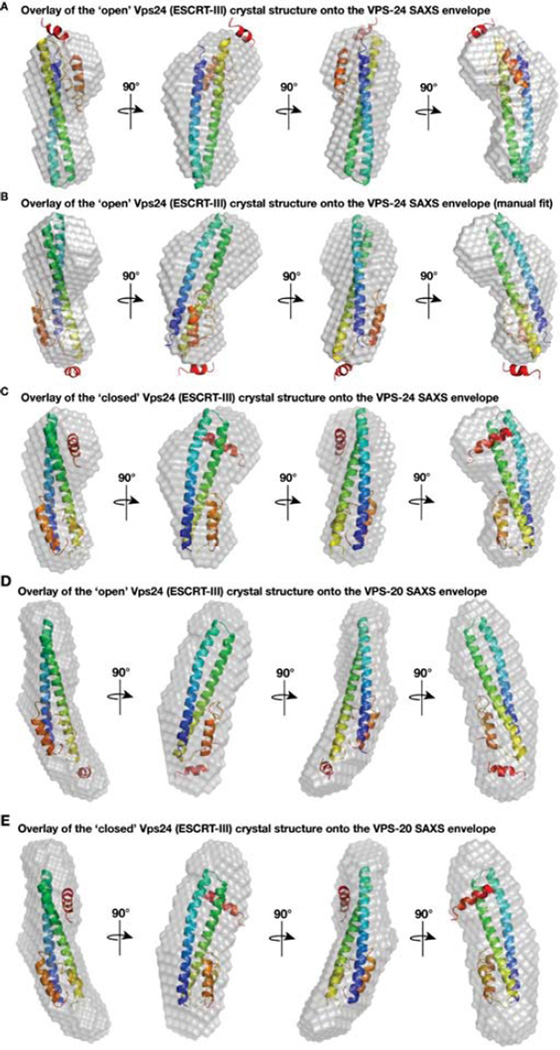
We also overlaid the closed and open human Vps24 structures onto our full-length C. elegans VPS-24 SAXS envelope. In the open conformation, we found that helix 5 extends beyond the SAXS model using an unbiased modeling program (Figure 6A). We additionally attempted to manually fit the open structure, but still encountered difficulty in accommodating helix 5 within the boundaries of the SAXS envelope (Figure 6B). In contrast, we were able to easily fit the closed Vps24 conformation onto the SAXS structure in an unbiased manner, confirming that in solution, VPS-24 exhibits an autoinhibited, closed state in which helix 5 is folded onto the helical hairpin loop (Figure 6C).
Figure 6. The closed human Vps24 crystal structure overlays well with the full length C. elegans VPS-24 SAXS envelope but not with the full length VPS-20 envelope.
(A) The program Supcomb20 was used to align one of the human Vps24 crystal structures (2GD5, representing its open conformation) with the SAXS model for C. elegans VPS-24, yielding a poor fit. (B) Manual refinement of the human Vps24 structure (2GD5) was performed in an attempt to optimize alignment with the SAXS model for C. elegans VPS-24. (C) The program Supcomb20 was used to align another human Vps24 structure (3FRT, representing the closed conformation) with the SAXS model for C. elegans VPS-24, which yields an excellent fit. (D) The program Supcomb20 was used to align one of the human Vps24 crystal structures (2GD5, representing its open conformation) with the SAXS model for full length VPS-20. (E) Alignment of another human Vps24 crystal structure (3FRT, representing its closed conformation) with the SAXS model for VPS-20 reveals a poor fit, with multiple regions of unaccounted density in the SAXS envelope.
Informed by the orientation of the human Vps24 crystal structure on the SAXS envelope generated by truncated VPS-20, we performed similar docking studies using our SAXS model for full-length VPS-20. Analogous to VPS-20ΔC, we found that the open configuration of Vps24 fit nicely into the envelope (Figure 6D). Additionally, we identified a density in our SAXS envelope around helix 5, which likely accounts for the remaining carboxyl-terminal residues of VPS-20, which were too unstructured to be modeled using crystallographic data obtained for human Vps24 [19]. In contrast, when the structure representing the closed conformation of Vps24 was docked, we failed to identify a satisfactory conformation that did not leave substantial density both above and below the crystal structure (Figure 6E). Moreover, since VPS-20 adopts an elongated conformation in solution, with a Dmax of 94 Å, the crystal structure in which helix 5 is displaced away from the helical hairpin loop helps to account for this increase in diameter that extends beyond the 70 Å helical hairpin. Collectively, our data strongly suggest that VPS-20 adopts an open conformation in solution unlike the downstream ESCRT-III subunit VPS-24, which remains in an autoinhibited, closed state until activation.
Association with ESCRT-II promotes VPS-20 curvature sensing
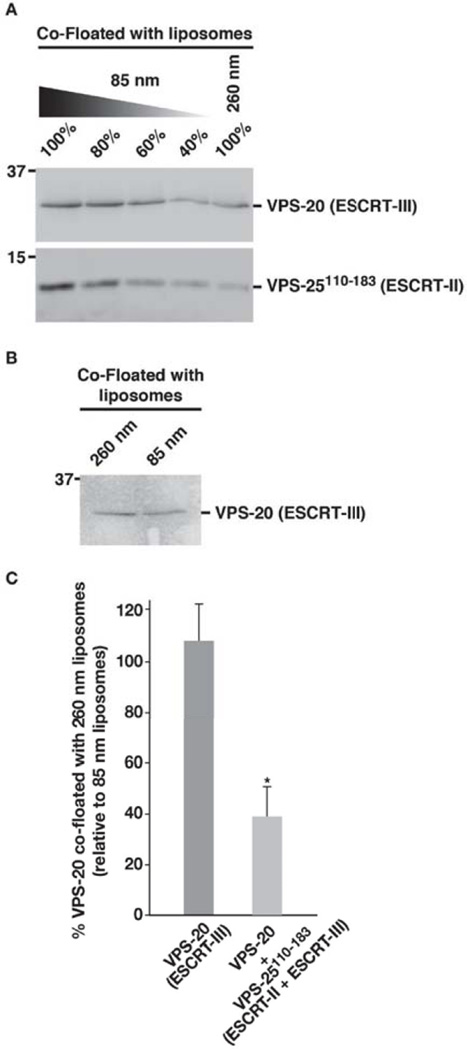
Since our findings suggest that VPS-20 does not require an association with another factor to transition into an open state, we considered alternative roles of its interaction with ESCRT-II. We and others previously demonstrated that a combination of ESCRT-II and VPS-20 binds to membranes with tighter affinity as compared to either alone [16,38]. Additionally, we found that a mixture of these proteins is capable of sensing elevated membrane curvature, while the individual factors cannot [16]. To determine whether either of these properties depends on a conformational change in VPS-20 that is triggered by ESCRT-II binding, we examined the ability of VPS-20 to bind liposomes of varying curvatures in the presence of an ESCRT-II fragment (amino acids 110–183 of VPS-25), which does not associate with lipid bilayers, but was shown previously to interact directly with VPS-20 [25]. Although a mixture of VPS-25110–183 and VPS-20 did not alter the affinity of VPS-20 for liposomes, we found that the presence of the VPS-25 polypeptide was sufficient to induce curvature sensing by VPS-20 (Figure 7A). Specifically, in contrast to VPS-20 alone, which does not exhibit a preference for more highly curved liposomes (Figure 7B), we found that the addition of VPS-25110–183 enabled VPS-20 to bind 85 nm liposomes more than 2-fold better than 260 nm liposomes (Figure 7C). These data suggest that ESCRT-II binding to VPS-20 acts to alter the ability of VPS-20 to sense curvature and highlights a new form of ESCRT-III activation that does not involve a transition from a closed to open conformation.
Figure 7. A fragment of VPS-25 is sufficient to enable VPS-20 to sense membrane curvature.
(A) A co-flotation assay was used to analyze the binding of a mixture of VPS-20 and VPS-25110–183 (amino acids 110–183) to liposomes of different diameters (85 and 260 nm). Fractions were resolved by SDS-PAGE and immunoblotted using VPS-20 or ESCRT-II antibodies. A dilution series of the protein mixture that co-floated with 85 nm liposomes was loaded to quantify the relative amount that co-floated with 260 nm vesicles. Data shown are representative of at least three independent experiments. On the basis of densitometry measurements, we found that less than 50% of the protein mixture co-floated with 260 nm vesicles, relative to 85 nm vesicles. (B) A co-flotation assay performed similarly as described in panel A demonstrates that VPS-20 alone fails to sense the difference in curvature between 85 and 260 nm liposomes. (C) Quantification of the co-flotation assays performed in panels A and B. The percentage of VPS-20 co-floated with 260 nm vesicles relative to that co-floated with 85 nm vesicles is plotted for each condition shown. Error bars represent mean +/− S.E.M. (n=3 each; p < 0.05, t-test).
DISCUSSION
ESCRT-mediated membrane fission is dependent upon the ESCRT-III complex. However, the mechanisms by which ESCRT-III subunits function cooperatively to mediate membrane scission are unknown. Recently, several models have been postulated to describe this process. In the “dome model,” Vps32 forms flat, inward-spiraling filaments that bind and initiate polymerization of Vps24 and Vps2, which generate a tube-like structure with a narrowing diameter that ultimately facilitates membrane cleavage [39]. Another scenario commonly referred to as the “purse string model” asserts that Vps32 filaments are capped by Vps24 and its binding partner Vps2 to promote Vps4-Vta1 complex recruitment [15]. In this case, Vps4-mediated disassembly of ESCRT-III filaments alters its structure to promote membrane scission. Despite mechanistic differences within these models, all require individual ESCRT-III subunits to possess discrete functions during the membrane fission process. However, it has been difficult to structurally distinguish components of the ESCRT-III complex, even though each acts in a non-redundant manner.
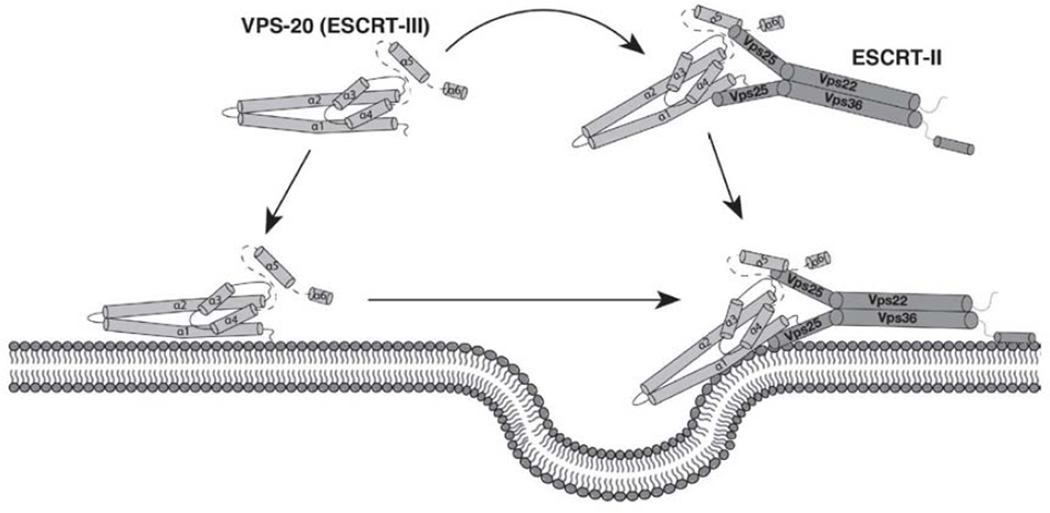
To define potential structural differences among the ESCRT-III subunits, we focused our attention on Vps20, a key factor linking the actions of ESCRT-II and ESCRT-III in MVE biogenesis [13,25]. Although mechanisms underlying Vps20-mediated initiation of Vps32 polymerization remain unclear, the binding interface between Vps20 and the Vps25 subunit of ESCRT-II has been crystallized, revealing a specific association with the amino-terminal portion of helix 1 within Vps20 [25]. This binding surface is not precluded in either the open or closed conformations of other ESCRT-III subunits [19,23], challenging the idea that ESCRT-II activates Vps20 by relieving an autoinhibited conformation [15]. Although ESCRT-II modulates the fluorescence properties of Vps20, interpretation of these data is challenging in the absence of structural information [15]. Moreover, our data demonstrate that ESCRT-II and Vps20 bind avidly in solution, a finding that is inconsistent with the model that ESCRT-II mediates Vps20 activation specifically on the endosomal membrane. Instead, Vps20 may interact directly with ESCRT-II in the cytoplasm, generating a curvature-sensing supercomplex that then binds rapidly to curved membranes and stimulates Vps32 polymerization at nascent vesicle bud necks [16]. In this revised model, alteration of a Vps20 closed conformation by ESCRT-II becomes unnecessary, as curvature sensing is sufficient to spatially regulate assembly of ESCRT-III polymers (Figure 8).
Figure 8. A model describing the role of ESCRT-II in activating VPS-20 during intralumenal vesicle formation at multivesicular endosomes.
The model highlights that VPS-20 exhibits an open conformation independently of ESCRT-II. However, upon binding ESCRT-II, VPS-20 is capable of sensing regions of elevated membrane curvature (e.g., at the forming neck of inward budding vesicles on endosomes), and restricting ESCRT-III distribution to these bent lipid bilayers.
Although high-resolution structures have been determined for the majority of the early-acting ESCRT machinery, the architectures of individual ESCRT-III subunits have been challenging to define using crystallographic methods. SAXS represents an attractive, alternative approach that provides complimentary information of the conformation and overall shape of a protein, albeit at low resolution. An additional advantage of SAXS is that experiments are performed in solution without the need to incorporate artificial fluorescent dyes or tags, which can introduce structural artifacts [31]. Indeed, previous SAXS analysis of human Vps24 showed it to exist in a closed conformation in solution, consistent with models in which ESCRT-III subunits remain autoinhibited in the cytoplasm [19,22]. In the presence of non-physiological concentrations of salt, Vps24 can undergo a conformational change to an elongated state [22], suggesting that SAXS analysis of ESCRT-III subunits is capable of determining their conformation. Our studies on C. elegans VPS-24 confirmed previous results. However, analysis of full-length VPS-20 indicated it to be in an extended conformation, irrespective of salt concentration or the presence of ESCRT-II. These findings again argue against a role for ESCRT-II in promoting a dramatic conformational change in Vps20. In vivo, Vps20 has been suggested to undergo myristoylation [12]. Unfortunately, we were unable to purify in vitro myristoylated VPS-20 at a sufficiently high concentration in a buffer compatible with SAXS. However, it is important to point out that previous NVIR studies focused on the small GTPase Arf1 demonstrated the myristoylation fails to alter the global fold of the protein [40]. Similarly, myristoylation may not have a substantial impact on the extended conformation of VPS-20, although further studies are necessary to demonstrate this directly and determine whether myristoylation of VPS-20 alters its ability to sense membrane curvature in the presence or absence of ESCRT-II. Nevertheless, based on our data, we propose that the conformations of individual ESCRT-III subunits vary, potentially tailoring each to a specific function during membrane scission. Additionally, our findings demonstrate that current models, which suggest all ESCRT-III proteins undergo a structural rearrangement from a closed to an open state to enable complex assembly, are overly simplistic. Only through a systematic structural characterization of each ESCRT-III subunit will the full complexity of ESCRT-III complex assembly be ultimately revealed.
ACKNOWLEDGEMENTS
We thank S. Butcher, J. Burke, B. Bhattacharyya for advice and helpful discussions, M. Anderson and J. Burke for technical assistance with the Bruker Nanostar, E. Chapman for access to a dynamic light scattering instrument, and members of the Audhya lab for critically reading this manuscript.
FUNDING
This work was supported by grants from the NIH (GM088151 to AA, MH067880 to JRY, and GM103533 to JRY). This study made use of the National Magnetic Resonance Facility at Madison, which is supported by NIH grants P41RR02301, P41GM66326, and P41GM10399. Additional equipment was purchased with funds from the University of Wisconsin, the NIH (RR02781, RR08438, and S100RR027000 to S.E.B.), the NSF (DMB-8415048, OIA-9977486, BIR-9214394), and the USDA. Circular dichroism data were obtained at the UW-Madison Biophysics Instrumentation Facility, which was established with support from the UW-Madison and grants BIR-9512577 (NSF) and S10 RR13790 (NIH).
Abbreviations used
- CD
circular dichroism
- ESCRT
endosomal sorting complex required for transport
- GUV
giant unilamellar vesicle
- MudPIT
multidimensional protein identification technology
- MVE
multivesicular endosome
- MW
molecular weight
- NSD
nominal spatial discrepancy
- PDDF
pair distance distribution function
- PE
phosphatidylethanolamine
- SAXS
small-angle X-ray scattering
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- SEC-MALS
size exclusion chromatography-multiangle light scattering
Footnotes
AUTHOR CONTRIBUTION
Amber Schuh, Michael Hanna, John Yates, and Anjon Audhya conceived and designed experiments. Amber Schuh, Michael Hanna, Kyle Quinney, Lei Wang, Ali Sarkeshik, and Anjon Audhya performed experiments and analyzed data. All authors discussed the results, and Amber Schuh and Anjon Audhya wrote the final version of the manuscript.
REFERENCES
- 1.Henne WM, Buchkovich NJ, Emr SD. The ESCRT pathway. Dev. Cell. 2011;21:77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 2.Rusten TE, Vaccari T, Stenmark H. Shaping development with ESCRTs. Nat. Cell Biol. 2011;14:38–45. doi: 10.1038/ncb2381. [DOI] [PubMed] [Google Scholar]
- 3.Schuh AL, Audhya A. The ESCRT machinery: from the plasma membrane to endosomes and back again. Crit. Rev. Biochem. Mol. Biol. 2014;49:242–261. doi: 10.3109/10409238.2014.881777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hurley JH. The ESCRT complexes. Crit. Rev. Biochem. Mol. Biol. 2010;45:463–487. doi: 10.3109/10409238.2010.502516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raiborg C, Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 2009;458:445–452. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- 6.Raiborg C, Rusten TE, Stenmark H. Protein sorting into multivesicular endosomes. Curr. Opin. Cell Biol. 2003;15:446–455. doi: 10.1016/s0955-0674(03)00080-2. [DOI] [PubMed] [Google Scholar]
- 7.Katzmann DJ, Odorizzi G, Emr SD. Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell Biol. 2002;3:893–905. doi: 10.1038/nrm973. [DOI] [PubMed] [Google Scholar]
- 8.von Schwedler UK, Stuchell M, Muller B, Ward DM, Chung HY, Morita E, Wang HE, Davis T, He GP, Cimbora DM, et al. The protein network of HIV budding. Cell. 2003;114:701–713. doi: 10.1016/s0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- 9.Carlton JG, Martin-Serrano J. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science. 2007;316:1908–1912. doi: 10.1126/science.1143422. [DOI] [PubMed] [Google Scholar]
- 10.Jimenez AJ, Maiuri P, Lafaurie-Janvore J, Divoux S, Piel M, Perez F. ESCRT machinery is required for plasma membrane repair. Science. 2014;343:1247136. doi: 10.1126/science.1247136. [DOI] [PubMed] [Google Scholar]
- 11.Wollert T, Hurley JH. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature. 2010;464:864–869. doi: 10.1038/nature08849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Babst M, Katzmann DJ, Estepa-Sabal EJ, Meerloo T, Emr SD. ESCRT-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev. Cell. 2002;3:271–282. doi: 10.1016/s1534-5807(02)00220-4. [DOI] [PubMed] [Google Scholar]
- 13.Teis D, Saksena S, Emr SD. Ordered assembly of the ESCRT-III complex on endosomes is required to sequester cargo during MVB formation. Dev. Cell. 2008;15:578–589. doi: 10.1016/j.devcel.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 14.Teis D, Saksena S, Judson BL, Emr SD. ESCRT-II coordinates the assembly of ESCRT-III filaments for cargo sorting and multivesicular body vesicle formation. EMBO J. 2010;29:871–883. doi: 10.1038/emboj.2009.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saksena S, Wahlman J, Teis D, Johnson AE, Emr SD. Functional reconstitution of ESCRT-III assembly and disassembly. Cell. 2009;136:97–109. doi: 10.1016/j.cell.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fyfe I, Schuh AL, Edwardson JM, Audhya A. Association of the endosomal sorting complex ESCRT-II with the Vps20 subunit of ESCRT-III generates a curvature-sensitive complex capable of nucleating ESCRT-III filaments. J. Biol. Chem. 2011;286:34262–34270. doi: 10.1074/jbc.M111.266411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lata S, Schoehn G, Jain A, Pires R, Piehler J, Gottlinger HG, Weissenhorn W. Helical structures of ESCRT-III are disassembled by VPS4. Science. 2008;321:1354–1357. doi: 10.1126/science.1161070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adell MA, Vogel GF, Pakdel M, Muller M, Lindner H, Hess MW, Teis D. Coordinated binding of Vps4 to ESCRT-III drives membrane neck constriction during MVB vesicle formation. J. Cell Biol. 2014;205:33–49. doi: 10.1083/jcb.201310114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muziol T, Pineda-Molina E, Ravelli RB, Zamborlini A, Usami Y, Gottlinger H, Weissenhorn W. Structural basis for budding by the ESCRT-III factor CHMP3. Dev. Cell. 2006;10:821–830. doi: 10.1016/j.devcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 20.Lin Y, Kimpler LA, Naismith TV, Lauer JM, Hanson PI. Interaction of the mammalian endosomal sorting complex required for transport (ESCRT) III protein hSnf7-1 with itself, membranes, and the AAA+ ATPase SKD1. J. Biol. Chem. 2005;280:12799–12809. doi: 10.1074/jbc.M413968200. [DOI] [PubMed] [Google Scholar]
- 21.Zamborlini A, Usami Y, Radoshitzky SR, Popova E, Palu G, Göttlinger H. Release of autoinhibition converts ESCRT-III components into potent inhibitors of HIV-1 budding. Proc. Natl. Acad. Sci. USA. 2006;103:19140–19145. doi: 10.1073/pnas.0603788103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lata S, Roessle M, Solomons J, Jamin M, Gottlinger HG, Svergun DI, Weissenhorn W. Structural basis for autoinhibition of ESCRT-III CHMP3. J. Mol. Biol. 2008;378:818–827. doi: 10.1016/j.jmb.2008.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bajorek M, Schubert HL, McCullough J, Langelier C, Eckert DM, Stubblefield WM, Uter NT, Myszka DG, Hill CP, Sundquist WI. Structural basis for ESCRT-III protein autoinhibition. Nat. Struct. Mol. Biol. 2009;16:754–762. doi: 10.1038/nsmb.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henne WM, Buchkovich NJ, Zhao Y, Emr SD. The endosomal sorting complex ESCRT-II mediates the assembly and architecture of ESCRT-III helices. Cell. 2012;151:356–371. doi: 10.1016/j.cell.2012.08.039. [DOI] [PubMed] [Google Scholar]
- 25.Im YJ, Wollert T, Boura E, Hurley JH. Structure and function of the ESCRT-II–III interface in multivesicular body biogenesis. Dev. Cell. 2009;17:234–243. doi: 10.1016/j.devcel.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Audhya A, Hyndman F, McLeod IX, Maddox AS, Yates JR, Desai A, Oegema K. A complex containing the Sm protein CAR-1 and the RNA helicase CGH-1 is required for embryonic cytokinesis in Caenorhabditis elegans. J. Cell Biol. 2005;171:267–279. doi: 10.1083/jcb.200506124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheeseman IM, Niessen S, Anderson S, Hyndman F, Yates JR, Oegema K, Desai A. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 2004;18:2255–2268. doi: 10.1101/gad.1234104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu T, Venable JD, Park S, Cociorva D, Lu B, Liao L, Wohlschlegel J, Hewel J, Yates JR. ProLuCID, a Fast and Sensitive Tandem Mass Spectra-based Protein Identification Program. Mol. Cell Proteomics. 2006;5:S174. [Google Scholar]
- 29.Tan S. A modular polycistronic expression system for overexpressing protein complexes in Escherichia coli. Protein Expr. Purif. 2001;21:224–234. doi: 10.1006/prep.2000.1363. [DOI] [PubMed] [Google Scholar]
- 30.Wyatt PJ. Light-scattering and the absolute characterization of macromolecules. Analytica Chimica Acta. 1993;272:1–40. [Google Scholar]
- 31.Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q. Rev. Biophys. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- 32.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Cryst. 2003;36:1277–1282. [Google Scholar]
- 33.Franke D, Svergun DI. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Cryst. 2009;42:342–346. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Cryst. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kozin MB, Svergun DI. Automated matching of high- and low-resolution structural models. J. Appl. Cryst. 2001;34:33–41. [Google Scholar]
- 36.Washburn MP, Wolters D, Yates JR. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 37.Rozycki B, Kim YC, Hummer G. SAXS ensemble refinement of ESCRT-III CHMP3 conformational transitions. Structure. 2011;19:109–116. doi: 10.1016/j.str.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Teo H, Perisic O, Gonzalez B, Williams RL. ESCRT-II, an endosome-associated complex required for protein sorting: crystal structure and interactions with ESCRT-III and membranes. Dev. Cell. 2004;7:559–569. doi: 10.1016/j.devcel.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 39.Fabrikant G, Lata S, Riches JD, Briggs JA, Weissenhorn W, Kozlov MM. Computational model of membrane fission catalyzed by ESCRT-III. PLoS Comput. Biol. 2009;5:e1000575. doi: 10.1371/journal.pcbi.1000575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu T, Kahn RA, Prestegard JH. Structure and membrane interaction of myristoylated ARF1. Structure. 2009;17:79–87. doi: 10.1016/j.str.2008.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]