Expression of the Gria1-targeting miRNA miR-501-3p is increased locally in dendrites after NMDAR activation and is required for NMDAR-dependent inhibition of GluA1 expression and long-lasting spine shrinkage and elimination.
Abstract
The number of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptors (AMPARs) in synapses determines synaptic strength. AMPAR expression can be regulated locally in dendrites by synaptic activity. The mechanisms of activity-dependent local regulation of AMPAR expression, however, remain unclear. Here, we tested whether microRNAs (miRNAs) are involved in N-methyl-d-aspartate (NMDA) receptor (NMDAR)–dependent AMPAR expression. We used the 3′ untranslated region of Gria1, which encodes the AMPA receptor subunit GluA1, to pull down miRNAs binding to it and analyzed these miRNAs using next-generation deep sequencing. Among the identified miRNAs, miR-501-3p is also a computationally predicted Gria1-targeting miRNA. We confirmed that miR-501-3p targets Gria1 and regulates its expression under physiological conditions. The expression of miR-501-3p and GluA1, moreover, is inversely correlated during postnatal brain development. miR-501-3p expression is up-regulated locally in dendrites through the NMDAR subunit GluN2A, and this regulation is required for NMDA-induced suppression of GluA1 expression and long-lasting remodeling of dendritic spines. These findings elucidate a miRNA-mediated mechanism for activity-dependent, local regulation of AMPAR expression in dendrites.
Introduction
α-Amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptors (AMPARs) are ionotropic glutamate receptors that mediate fast excitatory synaptic transmission in the central nervous system. AMPARs are tetramers composed of four possible subunits (GluA1–4; Shepherd and Huganir, 2007). The number of AMPARs in synapses determines the strength of synaptic transmission, and their abnormal expression has been implicated in cognitive impairments associated with such neurological and neuropsychiatric diseases as Alzheimer’s disease, ischemia, schizophrenia, and depression (Chang et al., 2012).
AMPAR expression is regulated by synaptic activity (Grooms et al., 2006). During activity-dependent synaptic plasticity, for instance, the activation of N-methyl-d-aspartate (NMDA) or metabotropic glutamate receptors affects the abundance of synaptic AMPAR through both posttranslational mechanisms (including phosphorylation, palmitoylation, and ubiquitination) and local translation of dendritic mRNAs encoding AMPAR subunits (Snyder et al., 2001; Ju et al., 2004; Grooms et al., 2006; Sutton et al., 2006; Lu and Roche, 2012). Activity-dependent modulation of AMPAR is an important mechanism that tunes synaptic strength to refine synaptic connectivity during brain development and to store information in the brain during learning and memory (Shepherd and Huganir, 2007). Despite the broad recognition that AMPARs play a pivotal role in brain functions, however, molecular mechanisms underlying their regulation, especially activity-dependent local translation of AMPARs in dendrites, are only incompletely understood.
miRNAs are small noncoding RNAs that regulate gene expression by repressing translation and/or degrading mRNAs. Through imperfect base pairing, miRNAs bind to mRNAs, usually at the 3′ UTR. miRNAs are key regulators of gene expression in neurons and, as such, are essential for various neuronal functions including neuronal differentiation, neurite outgrowth, morphogenesis of dendritic spines, and synaptic plasticity (McNeill and Van Vactor, 2012). miRNAs are transcribed as pri-mRNAs, which are cleaved by Drosha to ∼60–70-nt precursor miRNAs, and then exported to the cytoplasm by exportin 5 (Bartel, 2004). Precursor miRNAs are processed by Dicer into mature miRNAs and incorporated into the RNA-induced silencing complex (RISC; Bartel, 2004).
In neurons, many miRNAs localize to dendrites, where they can be regulated by synaptic activity (Schratt, 2009; Hu et al., 2014). For instance, NMDA receptor (NMDAR) activation inhibits miR-191 expression locally in dendrites, resulting in an elevation of its target tropomodulin-2, which promotes actin depolymerization, shrinkage, and elimination of dendritic spines. In view of this finding, we hypothesize that miRNAs also contribute to activity-dependent local synthesis of AMPARs in dendrites.
To test this hypothesis, we combined miRNA pull-down and computational prediction to search for miRNAs that target mRNAs encoding the AMPAR subunit GluA1. This approach leads to the identification of miR-501-3p as a Gria1-targeting miRNA. Our further analysis of miR-501-3p shows that it is increased locally in dendrites after NMDAR activation and that this up-regulation of miR-501-3p is required for NMDAR-dependent inhibition of GluA1 expression, long-lasting spine shrinkage, and elimination. These findings reveal that miRNAs are important regulators of activity-dependent local synthesis of dendritic AMPARs.
Results
Identification of miRNAs regulating GluA1 expression in a NMDAR-dependent manner
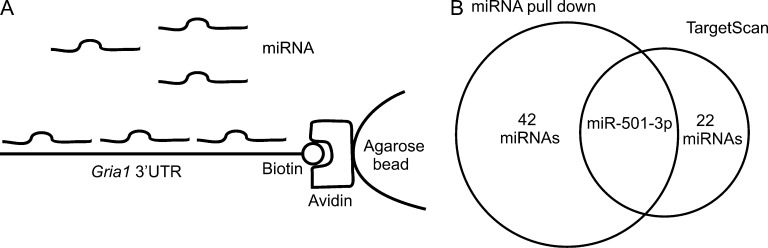
The level of GluA1 protein is reduced upon NMDAR activation (Grooms et al., 2006). To determine whether miRNAs are involved in this process, we first screened for miRNAs that target the GluA1 encoding gene Gria1. The mouse Gria1 mRNA can be targeted by ∼200 miRNAs through both conserved and nonconserved binding sites as predicted by miRNA target prediction tools (such as TargetScan). Computational prediction of miRNA targets, however, has a high false positive error rate (Liu et al., 2014). To avert this problem, we experimentally identified miRNAs targeting Gria1 by using the 3′ UTR of Gria1 mRNA to pull down miRNAs that bind to it (Fig. 1 A). Mouse Gria1 3′ UTR were transcribed in vitro, and the resulting mRNAs were biotinylated at their 3′ ends and immobilized to avidin-agarose beads. The RNA-avidin beads were used to pull down miRNAs isolated from mouse brains. Both eluted and input RNAs from the pull-down assay were analyzed by next-generation deep sequencing. 43 miRNAs that were enriched >10-fold by pull-down were considered candidate Gria1-targeting miRNAs (Fig. 1 B). We then searched the mouse Gria1’s 3′ UTR for miRNA binding sites using TargetScan 6.2. To reduce the false discovery rate, only conserved binding sites were selected. Among the 43 miRNAs identified by the pull-down assay, only miR-501-3p is predicted by TargetScan to bind to Gria1 3′ UTR at a conserved binding site (Fig. 1 B). miR-501-3p, therefore, is our experimentally and computationally identified miRNA-targeting Gria1.
Figure 1.
Identification of Gria1 targeting miRNAs. Small RNAs isolated from the hippocampus of mice (17 d old) were incubated with Gria1 3′ UTR-bound beads for pull-down of Gria1 binding miRNAs. (A) Schematic illustration of the pull-down assay. (B) Overlap of miRNAs identified by pull-down and those predicted by TargetScan to bind to Gria1 3′ UTR at conserved binding sites.
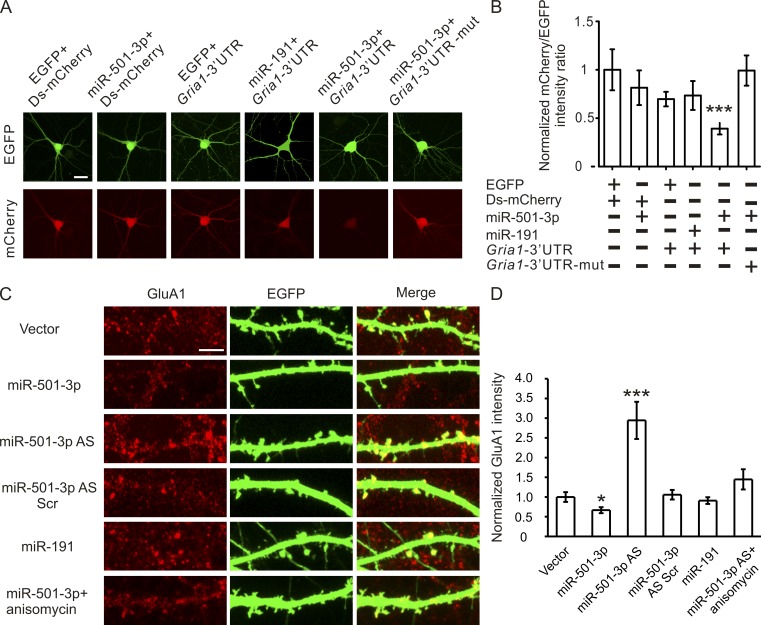
To confirm that miR-501-3p controls GluA1 expression, we generated a reporter construct by inserting the predicted miR-501-3p binding site into the 3′ UTR of destabilized mCherry. We cotransfected this construct, plus a plasmid that expresses both EGFP and miR-501-3p, into cultured hippocampal neurons (14 d in vitro [DIV]). At 3 d after transfection, the effect of miR-501-3p on mCherry protein expression was assessed by measuring the fluorescence intensity ratio between mCherry and EGFP proteins. Our reporter assay showed that cotransfection with the miR-501-3p construct—but not with a construct expressing miR-191 (which is not predicted to target Gria1) or EGFP—inhibited mCherry protein expression (Fig. 2, A and B). Moreover, when the miR-501-3p binding site in the mCherry reporter construct was mutated, this inhibition was abolished (Fig. 2, A and B). These results indicate that the miR-501-3p binding site in the Gria1 gene confers regulation by miR-501-3p.
Figure 2.
Gria1 is a physiological target of miR-501-3p. Cultured hippocampal neurons were transfected with designated constructs at 14 DIV and imaged at 17 DIV. (A) Representative images of neurons cotransfected with the miRNA and the reporter construct. (B) Quantification of A; n = 12–15 neurons for each group. (C) Representative images of dendrites from transfected neurons stained with the GluA1 antibody. (D) Quantification of C; n = 14–20 neurons for each condition. Data are presented as mean ± SEM; one-way ANOVA was used for statistical analysis among different groups; P < 0.05. Mann-Whitney U test is used for statistical analysis; *, P < 0.05; ***, P < 0.005. Bars: (A) 20 µm; (C) 5 µm.
To test whether miR-501-3p regulates GluA1 protein expression under physiological conditions, we transfected cultured hippocampal neurons (14 DIV) with an EGFP construct (for visualization of transfected neurons) along with constructs expressing miR-501-3p or miR-191, antisense oligonucleotides against miR-501-3p, or scrambled oligonucleotides. As the efficiency of lipofectamine-mediated transfection of primary hippocampal neurons is low (<0.05%), dendrites of transfected neurons can be identified by EGFP expression and separated from those of untransfected neurons for immunostaining analysis of GluA1 proteins. GluA1 protein was decreased in miR-501-3p construct transfected, increased in antisense oligonucleotide transfected, and intact in miR-191 construct or scrambled oligonucleotide transfected neurons at 3 d after transfection (Fig. 2, C and D). miR-501-3p knockdown-induced increase in GluA1 protein expression was inhibited by treatment with the translation inhibitor anisomycin (20 µM for 2 h; Fig. 2, C and D), confirming that miR-501-3p represses Gria1 translation. These results indicate that the expression of endogenous GluA1 protein is controlled by miR-501-3p.
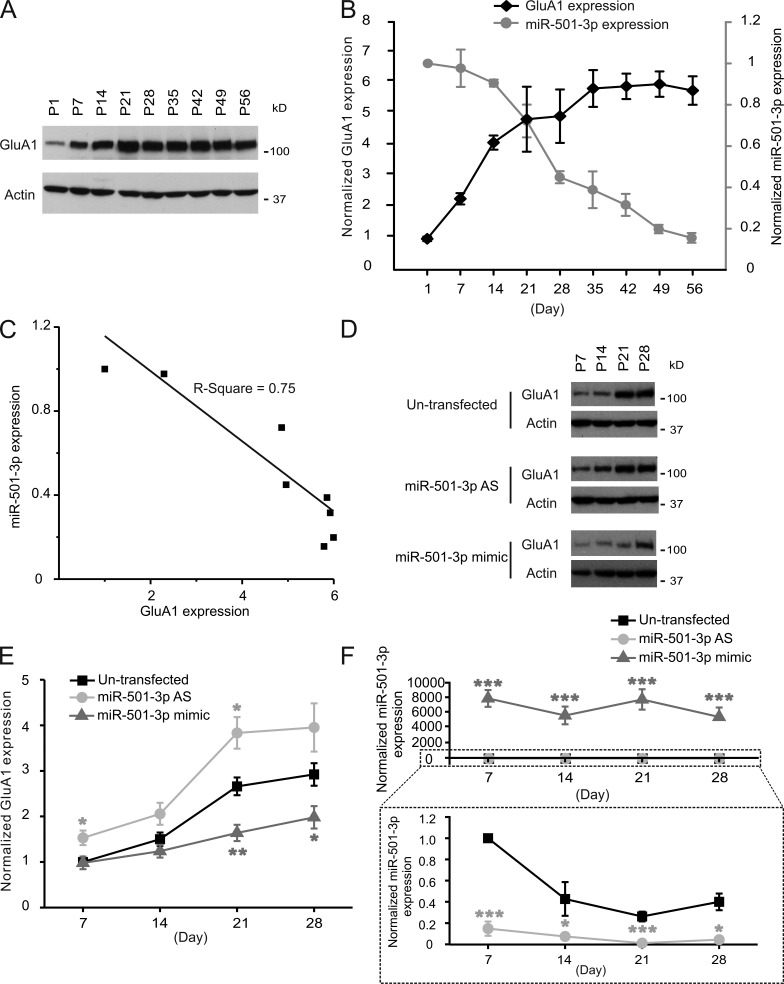
GluA1 protein expression increases during the postnatal development of neurons (Zhu et al., 2000). Because miR-501-3p targets Gria1, it might contribute to GluA1’s developmental change. To test this possibility, we analyzed miR-501-3p expression in developing hippocampal neurons. Small RNAs were isolated from the hippocampus of rats (1–56 d old) and cultured hippocampal neurons (7–28 DIV), and analyzed by quantitative PCR. In the rat hippocampus, the level of GluA1 protein increased between postnatal days 1–35 and then remained stable thereafter (Fig. 3, A and B). Likewise, cultured hippocampal neurons also progressively increased their GluA1 protein expression between 7 and 28 DIV (Fig. 3, D and E). The level of miR-501-3p, in contrast, decreased while that of GluA1 protein became higher (Fig. 3, B, C, and F). The inverse correlation between miR-501-3p and GluA1 protein expression suggests that miR-501-3p might be involved in the developmental increase in GluA1 protein expression. To test this possibility, we transfected cultured hippocampal neurons with miR-501-3p mimic (double-stranded RNAs functionally mimicking endogenous miR-501-3p) at 4, 11, 18, and 25 DIV and analyzed GluA1 proteins at 3 d after each transfection. The developmental increase in GluA1 protein expression was mitigated by miR-501-3p overexpression (Fig. 3, D and E). Transfection of antisense oligonucleotides against miR-501-3p, in contrast, augmented the developmental increase in GluA1 protein expression (Fig. 3, D and E). Hence, the developmental change in GluA1 protein expression is, at least in part, a result of diminishing miR-501-3p expression. Collectively, these results demonstrate that Gria1 is a physiological target of miR-501-3p.
Figure 3.
The expression of miR-501-3p and GluA1 is inversely correlated during development. Proteins and total RNAs were isolated from the hippocampus of rats and cultured hippocampal neurons at indicated ages for immunoblotting (A, B, D, and E) or qRT-PCR (B and F). (A and D) Representative blots. (B and E) Quantification of A and D; n = 3–8 rats or cultures for each age. (B and F) qRT-PCR analysis of miR-501-3p expression in the rat hippocampus (B) and cultured hippocampal neurons (F); n = 3–8 rats or cultures for each age group. (C) Correlation between expression levels of miR-501-3p and GluA1 at different ages in the rat hippocampus. Data are presented as mean ± SEM; one-way ANOVA was used for statistical analysis among different groups; Mann-Whitney U test was used for statistical analysis; *, P < 0.05; **, P < 0.01; ***, P < 0.005.
miR-501-3p is required for NMDA-induced down-regulation of GluA1 expression
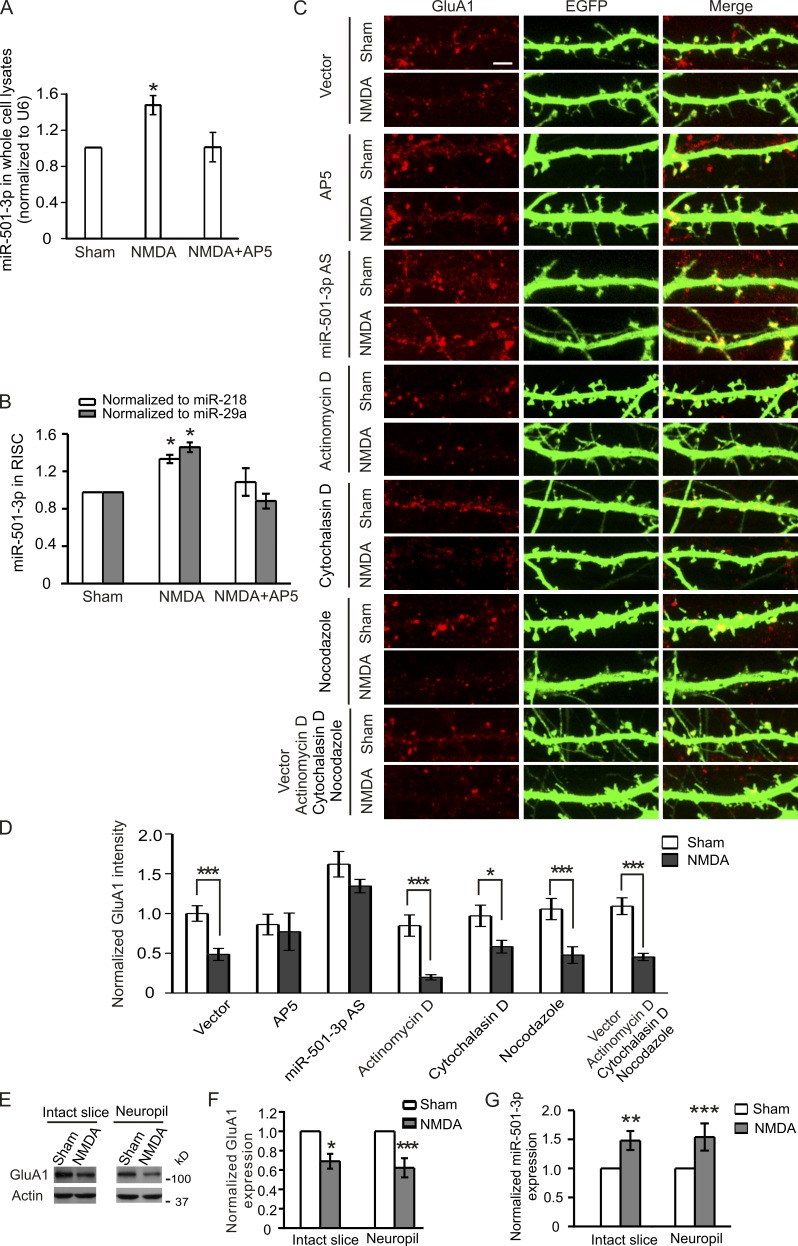
Having validated that Gria1 is a target gene of miR-501-3p, we next examined whether miR-501-3p contributes to the expression change in GluA1 protein induced by NMDAR activation (Grooms et al., 2006). We first tested whether NMDAR activation also affects miR-501-3p expression. Primary hippocampal neurons (17 DIV) were treated with NMDA (30 µM for 5 min) and harvested 90 min after stimulation for RNA isolation and quantitative RT-PCR (qRT-PCR). We detected more miR-501-3p in NMDA-treated than in control cells, and the increase in miR-501-3p was blocked by the NMDAR antagonist (2R)-amino-5-phosphonovaleric acid (AP5; 100 µM; added 10 min before NMDA treatment and present during and after NMDA treatment until the collection of cells; Fig. 4 A). Hence, NMDA treatment enhances miR-501-3p expression.
Figure 4.
Local regulation of dendritic GluA1 by miR-501-3p. In A and B, primary neurons were treated with NMDA (30 µM for 5 min) and collected at 90 min after treatment to test for miR-501-3p. (A) The level of miR-501-3p in whole cell lysates; n = 3–5 experiments. (B) miR-501-3p associated with RISC; n = 3–4 experiments. In C and D, transfected hippocampal neurons (17 DIV; 3 d after transfection) were treated with NMDA, and then stained for GluA1. (C) Representative images of transfected neurons. (D) Quantification of C; n = 14–29 cells for each group; AP5 was added 10 min before treatment and present during and after NMDA treatment. In E–G, hippocampal slices in which cell bodies of CA1 pyramidal neurons were removed or intact hippocampal slices were treated with NMDA (30 µM for 5 min). (E) Representative immunoblots. (F) Quantification of E; n = 4 rats for the intact slice group and 6 rats for the neuropil group. (G) miR-501-3p expression normalized to U6; n = 5 rats for the intact slice group and 6 rats for the neuropil group. Data are presented as mean ± SEM; Mann-Whitney U test is used for statistical analysis; *, P < 0.05; **, P < 0.01; ***, P < 0.005. Bar, 5 µm.
Moreover, we assessed miR-501-3p incorporated into RISC using the cross-linking and immunoprecipitation assay. Primary neurons were transduced with lentivirus expressing Flag-tagged argonoute 2 (Ago2; a protein component of RISC) and then treated with NMDA (30 µM for 5 min). 90 min after stimulation, cells were irradiated with UV light to induce cross-linking of proteins to their bound RNAs. RISC in cross-linked cells were precipitated with an antibody against Flag and analyzed by qRT-PCR for miRNAs, using miR-218 and miR-29a (which are not changed by NMDA stimulation) for normalization (Hu et al., 2014). Our qRT-PCR test showed that miR-501-3p associated with RISC was also increased by NMDA stimulation, and this increase was blocked by AP5 (100 µM; pretreated for 10 min and present during the NMDA treatment and the post-treatment period; Fig. 4 B).
To test whether or not miR-501-3p contributes to NMDA-induced GluA1 protein changes, we transfected cultured hippocampal neurons (14 DIV) with antisense oligonucleotides against miR-501-3p along with an EGFP construct (for visualization of transfected neurons). At 3 d after transfection, neurons were treated with NMDA (30 µM for 5 min) and fixed at 90 min after stimulation for immunostaining against GluA1 proteins. NMDA treatment, as reported earlier (Grooms et al., 2006), decreased the level of dendritic GluA1 proteins in cells transfected with the empty vector (Fig. 4, C and D). This decrease was obliterated by miR-501-3p knockdown, AP5 (100 µM; added 10 min before NMDA stimulation and present during and after NMDA treatment) treatment, or inhibition of protein degradation (by cotreatment with MG132 and leupeptin), but not by the translation inhibitor anisomycin (Fig. 4, C and D; and Fig. S1). These findings indicate that NMDA-induced miR-501-3p up-regulation causes a suppression of GluA1 translation, thereby disturbing the balance between GluA1 protein synthesis and degradation and resulting in a decrease in GluA1 proteins.
Because dendritic GluA1 protein expression is inhibited by NMDA stimulation (Fig. 4, C and D), we next tested whether miR-501-3p regulates GluA1 proteins locally in dendrites. To analyze local GluA1 protein synthesis, we blocked transcription and intracellular trafficking of miRNAs in primary hippocampal neurons (17 DIV; 3 d after transfection with the EGFP construct) with the transcription inhibitor actinomycin D (10 µM), the actin polymerization inhibitor cytochalasin D (10 µM), and the microtubule polymerization inhibitor nocodazole (10 µM) during and after NMDA treatment (30 µM for 5 min). 90 min after treatment, neurons were fixed and stained with an anti-GluA1 antibody. The inhibition of transcription and active transport of miRNAs had no effect on NMDA-induced reduction of dendritic GluA1 (Fig. 4, C and D), suggesting that GluA1 can be regulated locally in dendrites by NMDA. We further confirmed this local regulation in hippocampal slices (prepared form rats 18–19 d old). The cell bodies of CA1 pyramidal neurons in the slices were surgically removed before NMDA treatment (30 µM for 5 min). At 90 min after NMDA stimulation, the CA1 neuropil was collected for immunoblotting against GluA1 and qRT-PCR for miR-501-3p. In intact hippocampal slices, NMDA treatment caused an increase in miR-501-3p and a reduction of GluA1 protein in the CA1 area (Fig. 4, E–G). These NMDA-induced changes were preserved in the CA1 neuropil of the slice in which cell bodies were removed before NMDA stimulation (Fig. 4, E–G), suggesting that miR-501-3p can repress GluA1 protein expression locally in dendrites. Collectively, these results indicate that NMDA-induced changes in GluA1 expression are caused, at least in part, by local suppression of dendritic GluA1 protein expression by miR-501-3p.
miR-501-3p–mediated repression of GluA1 expression is required for long-lasting spine remodeling induced by NMDAR activation
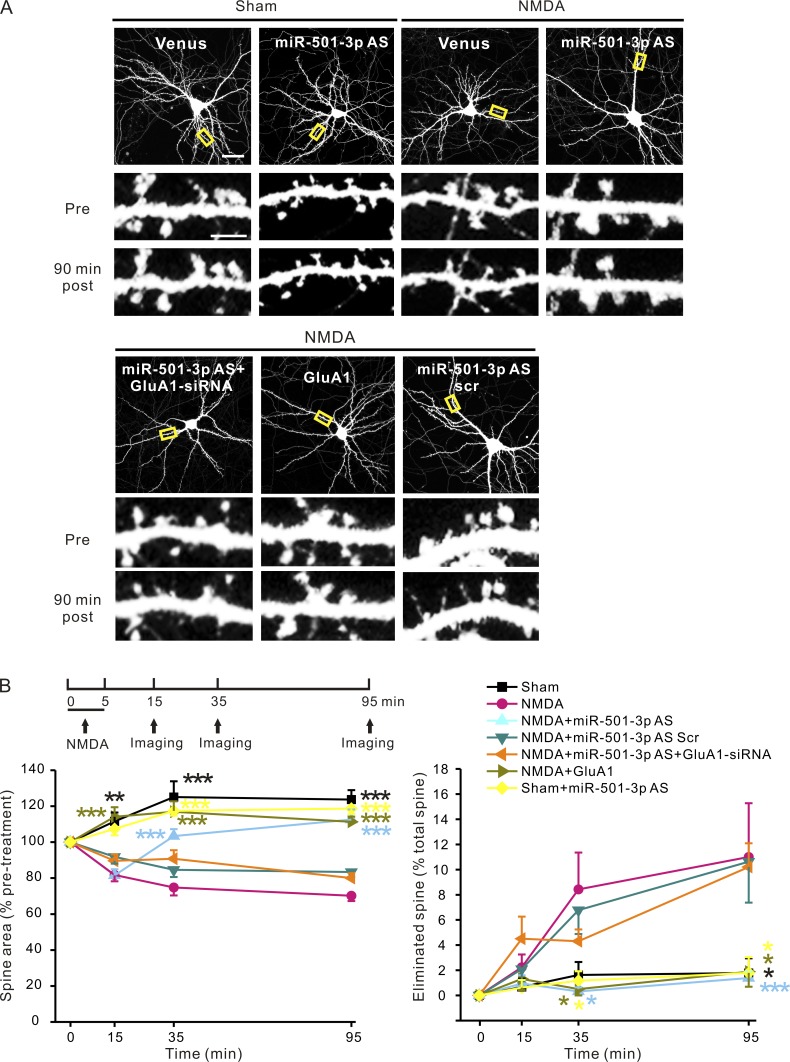
The number of AMPAR in synapses positively correlates with the size of dendritic spines (Baude et al., 1995; Matsuzaki et al., 2001, 2004; Passafaro et al., 2003). Having found that miR-501-3p contributes to down-regulation of GluA1 protein by NMDAR activation, we tested whether this regulation is also involved in NMDAR-mediated spine remodeling. To analyze the morphological changes of spines induced by NMDAR activation, neurons (17 DIV; 3–5 d after transfection with the venus, a YFP mutant construct for visualization of dendritic spines) were treated with NMDA (30 µM for 5 min), and the same spines were imaged before and at 10, 30, and 90 min after NMDA stimulation. Consistent with our previous findings (Hu et al., 2014), NMDA treatment caused retraction and rapid, long-lasting shrinkage of spines (Fig. 5). To test whether miR-501-3p is involved in this process, we transfected neurons (14 DIV) with the venus construct along with miR-501-3p antisense or scrambled oligonucleotides. Although transfection of miR-501-3p antisense oligonucleotides caused an increase in dendritic GluA1 protein (Fig. 2), it left the size and density of spines intact and had no effect on spine size and spine elimination in unstimulated neurons during the live imaging experiment (Fig. 5 and Fig. S2). After NMDA treatment, spines in neurons transfected with miR-501-3p antisense oligonucleotides shrunk at 10 min, but they recovered thereafter, reaching the prestimulation size by 90 min after stimulation (Fig. 5). NMDA-induced spine elimination and increase in GluA1 protein expression were also inhibited by transfection of miR-501-3p antisense oligonucleotides (Fig. 5 and Fig. S3). Transfection of scrambled oligonucleotides had no effect on NMDA-induced spine shrinkage, spine retraction, or GluA1 protein increase (Fig. 5 and Fig. S3). These results indicate that miR-501-3p is required for long-lasting spine restructuring induced by NMDAR activation.
Figure 5.
Suppression of GluA1 expression by miR-501-3p is required for long-lasting spine remodeling induced by NMDAR activation. Cultured hippocampus neurons (14 DIV) were transfected with designated plasmids or oligonucleotides, treated with NMDA (30 µM for 5 min) at 3–5 d after transfection, and imaged before and at 10, 30, and 90 min after treatment. (A) Representative images; yellow boxes indicate the dendrites in the high magnification images. (B) Quantification of A; n = 6–8 neurons for each group. Data are presented as mean ± SEM; one-way ANOVA was used for comparing spine size and elimination among different groups and P < 0.005 for all time points except for spine elimination at 10 min after NMDA treatment. Mann-Whitney U test was used for comparison between the NMDA treated, venus transfected group versus all other groups at the same time point. The asterisks are color-coded and indicate that the conditions labeled with the same color are significantly different from the NMDA treated, venus transfected group at the same time point. *, P < 0.05; **, P < 0.01; ***, P < 0.005. Bars: (A, top) 20 µm; (A, high magnification) 5 µm.
To test whether miR-501-3p contributes to NMDA-induced spine changes by regulating GluA1 expression, we transfected neurons with miR-501-3p antisense oligonucleotides along with a construct expressing a specific and efficient siRNA against mRNAs encoding GluA1 (Fig. S4). At 3–5 d after transfection, neurons were treated with NMDA (30 µM for 5 min) and imaged before and after stimulation. We found that cotransfection of the GluA1 siRNA construct obliterated the effect of miR-501-3p antisense oligonucleotides on NMDA-induced spine remodeling and GluA1 protein expression (Fig. 5 and Fig. S3). Transfection of a construct expressing GluA1, moreover, also blocked NMDA-induced spine shrinkage and retraction (Fig. 5). Hence, GluA1 mediates the effect of miR-501-3p on spine remodeling after NMDAR activation. LTD and the basal levels of spine size and density, however, were left intact in GluA1 overexpressing neurons (Fig. S2 and Fig. S5), indicating that the inhibition of NMDA-induced spine plasticity by GluA1 overexpression is not a result of its effects on LTD or the baseline of spine structure. Collectively, these results indicate that the suppression of GluA1 expression by miR-501-3p is required for prolonged spine shrinkage and elimination induced by NMDAR activation.
miR-501-3p is up-regulated by NMDAR activation at both transcriptional and posttranscriptional levels through GluN2A
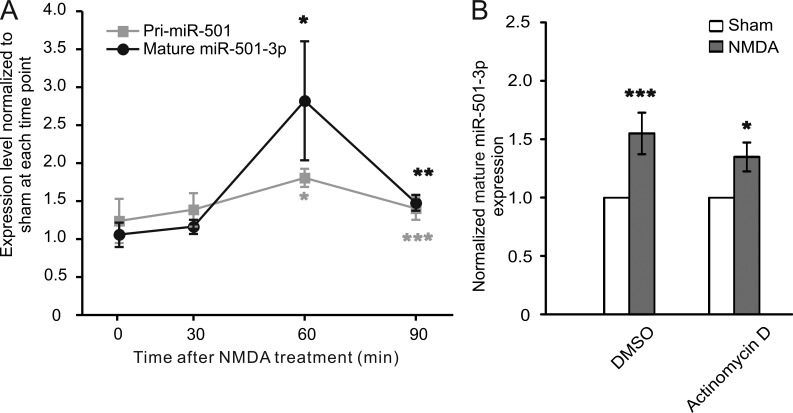
Having found that miR-501-3p is essential for NMDAR-mediated regulation of GluA1 expression, we went on to investigate how miR-501-3p is regulated by NMDAR. We analyzed the temporal profile of miR-501-3p changes in primary hippocampal neurons (17 DIV) after NMDA stimulation (30 µM for 5 min). We found that miR-501-3p trended upward and peaked at 60 min after treatment (Fig. 6 A).
Figure 6.
miR-501-3p expression is regulated both transcriptionally and posttranscriptionally. Cultured hippocampal neurons (17 DIV) were treated with NMDA (30 µM for 5 min) alone or along with designated inhibitors and harvested for qRT-PCR. (A) The temporal profile of pri- and mature miR-501-3p expression after NMDA treatment; the levels of pri- and mature miR-501-3p in NMDA-treated cells are normalized to and compared with those in sham-treated cells at the same time point for statistical analysis; n = 4–6 experiments for each time point. (B) The effect of actinomycin D on NMDA-induced changes in mature miR-501-3p; n = 7 experiments for each condition. Data are shown as mean ± SEM; Mann-Whitney U test was used for statistical analysis. *, P < 0.05; **, P < 0.01; ***, P < 0.005.
To test whether miR-501-3p is regulated at the transcriptional level, we analyzed pri-miR-501-3p by qRT-PCR and found that it was increased at 60 and 90 min after NMDA stimulation (Fig. 6 A). Hence, NMDA treatment enhances miR-501-3p transcription. In the presence of the transcription inhibitor actinomycin D, however, mature miR-501-3p still increased after NMDA stimulation (Fig. 6 B), suggesting that miR-501-3p is also regulated at posttranscriptional levels.
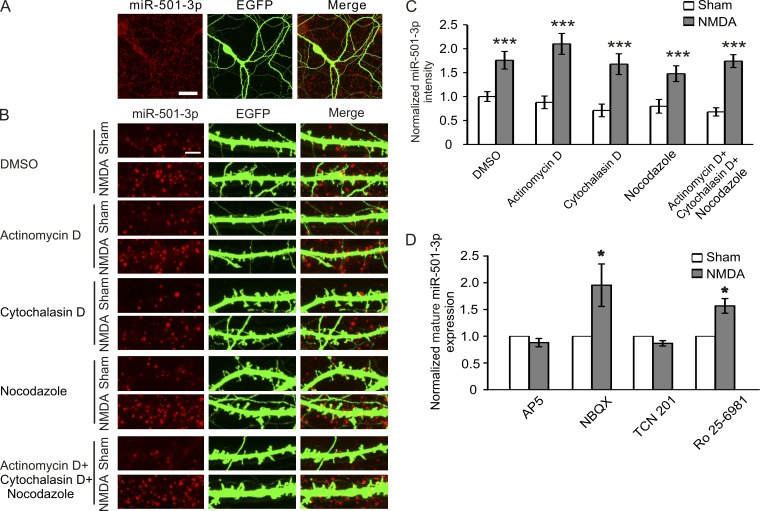
Because posttranscriptional processing of miRNAs can take place in dendrites where the components of RISC are found (Lugli et al., 2005; Bicker et al., 2013), we next tested whether NMDA stimulation induces regulation of miR-501-3p locally. To this end, we first tested whether miR-501-3p is localized in dendrites by using in situ hybridization. Signals from probes for miR-501-3p were detected in both cell bodies and dendrites (Fig. 7 A), indicating that miR-501-3p is indeed localized in dendrites. To test whether miR-501-3p is regulated locally, we treated hippocampal neurons (17 DIV; 3 d after cotransfection with the miR-501-3p and the EGFP construct) with NMDA (30 µM for 5 min), along with actinomycin D, cytochalasin D, and nocodazole (all at 10 µM) to block transcription and the intracellular transport of miRNAs. NMDA-induced increases in dendritic miR-501-3p were not affected by the transcription or transport inhibitors (Fig. 7, B and C), indicating a local regulation of miR-501-3p in dendrites. The local regulation of miR-501-3p is further supported by our finding that NMDA stimulation increases miR-501-3p in CA1 neurons whose cell bodies have been removed (Fig. 4 G).
Figure 7.
miR-501-3p expression is regulated locally in dendrites. Cultured hippocampal neurons (17 DIV) were treated with NMDA (30 µM for 5 min) alone or along with designated inhibitors and fixed at 90 min after treatment for in situ hybridization. (A) Subcellular localization of miR-501-3p. (B) Representative images of dendrites from transfected neurons. (C) Quantification of B; n = 15–23 neurons for each condition. (D) Effects of AMPAR and NMDAR blockers on NMDA-induced changes in mature miR-501-3p; n = 4–9 experiments for each condition. Bars: (A) 20 µm; (B) 5 µm. Data are shown as mean ± SEM; Mann-Whitney U test was used for statistical analysis. *, P < 0.05; ***, P < 0.005.
We also examined which signaling pathways regulate miR-501-3p expression and found that the NMDAR antagonist AP5 (100 µM; pretreated for 10 min and present during and after NMDA treatment) and the GluN2A blocker TCN 201 (10 µM) inhibited, whereas the GluN2B blocker Ro 25–6891 (3 µM) and the AMPAR blocker NBQX (2,3-Dioxo-6-nitro-1,2,3,4-tetrahydrobenzo[f]quinoxaline-7-sulfonamide; 10 µM) had no effect on, NMDA-induced up-regulation of miR-501-3p (Fig. 7 D). These results indicate that miR-501-3p is regulated by GluN2A-dependent mechanisms after NMDAR activation.
Collectively, these findings indicate that NMDAR activation regulates miR-501-3p at both transcriptional and posttranscriptional levels through GluN2A-mediated signaling pathways.
Discussion
This study examines the role of miRNAs in activity-dependent regulation of GluA1 expression. Using a miRNA pull-down assay, we identify miRNAs that bind to the 3′ UTR of mRNAs encoding GluA1. We selected miR-501-3p as a candidate activity-dependent regulator of GluA1 because its targeting to Gria1 is confirmed both experimentally and computationally, and it is up-regulated by NMDA stimulation. Our subsequent knockdown experiment shows that miR-501-3p is indeed required for NMDA-induced decreases in GluA1 expression. In addition, NMDA-induced long-lasting spine remodeling requires miR-501-3p–mediated suppression of GluA1 expression. This regulation of GluA1 by miR-501-3p, moreover, is found to occur locally in dendrites.
The synaptic expression of AMPAR is dependent on their regulated trafficking to/away from synapses and protein synthesis. AMPAR synthesis can be regulated by synaptic activity. The mechanism of how activity controls AMPA receptor translation, however, remains poorly understood. To throw light on this important question, this study investigates the role of miRNAs in NMDAR-dependent regulation of GluA1 expression. We chose to focus on the GluA1 subunit because it is Ca2+ permeable and Ca2+ influx through Ca2+-permeable AMPARs can trigger long-term synaptic plasticity (Liu and Savtchouk, 2012). Also, GluA1 expression is suppressed by NMDAR activation and changes during synaptic scaling, a form of homeostatic plasticity induced by long-term changes in network activity (Turrigiano, 2008; Huganir and Nicoll, 2013). We show that GluA1 expression is regulated by miR-501-3p. This is demonstrated by both a reporter assay and a test for endogenous GluA1. The reporter assay is used because it allows us to analyze individual intact neurons for protein expression and neuronal morphology simultaneously. Results from our reporter assay indicate that the 3′ UTR of Gria1 that includes a computationally predicted miR-501-3p binding site confers repression of reporter expression by miR-501-3p. We further confirmed that this regulation is mediated by miR-501-3p as it is obliterated by mutating the seed region of miR-501-3p binding site. Although the GluR1 mRNA is likely targeted by many other miRNAs, miR-501-3p knockdown causes an elevation of endogenous GluA1, suggesting that miR-501-3p is an essential regulator of GluA1 expression under physiological conditions.
The level of miR-501-3p is increased by NMDAR activation, suggesting that miR-501-3p mediates the effect of NMDAR on GluA1 expression. This is indeed supported by our finding that miR-501-3p knockdown blocks NMDA-induced down-regulation of GluA1 protein expression. In addition, we found that miR-501-3p localizes to dendrites, and inhibitors of transcription and intracellular transport have no effect on NMDA-induced suppression of dendritic GluA1 expression. miRNAs in mammalian cells primarily exist in messenger ribonucleoprotein complexes and processing bodies (ribonucleoprotein complexes containing RNA-processing enzymes; Pitchiaya et al., 2012). Only few ribonucleoprotein complexes and processing bodies move in a directional manner by passive diffusion, and the majority of them are confined to an area of a few micrometers squared if without cytoskeleton motor protein-mediated active transport (Fusco et al., 2003; Leung and Sharp, 2013). Hence, our finding that NMDA still induces changes to dendritic GluA1 expression when active intracellular transport is inhibited suggests that miR-501-3p can regulate GluA1 expression locally in dendrites.
Investigating how NMDARs regulate miR-501-3p expression, we find that NMDAR subunit GluN2A, but not GluN2B or AMPAR, mediates the effect of NMDA on miR-501-3p. In addition to mature miR-501-3p, pri-miR-501-3p is also increased by NMDA treatment, suggesting that transcription contributes to the expression change in miR-501-3p. Because transcription inhibition reduces but does not abolish NMDA-induced increases in mature miR-501-3p, it appears that miR-501-3p is also regulated posttranscriptionally.
NMDA stimulation induces shrinkage of dendritic spines, and the modified spines can either stay small for prolonged periods of time or be eliminated (Zhou et al., 2004). This long-lasting spine change is thought to be a structural basis of information storage in the brain for cognitive functions such as learning and memory. Our results indicate that long-lasting maintenance of NMDA-induced spine modification needs suppression of GluA1 expression and that this regulation is mediated, at least in part, by miR-501-3p. Because Gria1 is targeted by many miRNAs and miR-501-3p also has multiple target mRNAs, our findings do not exclude the possibility that other Gria1-targeting miRNAs or miR-501-3p targets also contribute to NMDAR-dependent spine remodeling. We noted that although GluA1 overexpression inhibits NMDA-induced spine remodeling, it has no effect on LTD induction. NMDA-induced spine shrinkage can be caused by a reduction of synaptic AMPA receptors as a consequence of GluA1 down-regulation, as the size of dendritic spines correlates with AMPAR content in synapses (Baude et al., 1995; Matsuzaki et al., 2001, 2004; Passafaro et al., 2003).
Our earlier studies found that overexpression of miR-191 in hippocampal neurons obliterates NMDA-induced spine elimination and long-lasting spine shrinkage by inhibiting actin depolymerization (Hu et al., 2014). Our results in this study show that miR-501-3p knockdown inhibits NMDA-induced spine elimination and initial spine shrinkage, but only partially blocks persistent spine shrinkage. miR-501-3p regulates spines by repressing GluA1 expression. Hence, both miR-191 and miR-501-3p are essential for spine elimination and prolonged spine shrinkage. The fact that miR-191 and miR-501-3p regulate dendritic spines by acting on different target genes and cellular processes indicates that their effects on spines are specific. The partial blockage of NMDA-induced long-lasting spine shrinkage by miR-501-3p knockdown suggests that other mechanisms also contribute to persistent GluA1 down-regulation and prolonged spine shrinkage, such as phosphorylation and ubiquitination of GluA1 (Ehlers, 2000; Oh et al., 2006; Man et al., 2007; Kessels et al., 2009; Schwarz et al., 2010).
In sum, our study elucidates important roles of miRNAs, exemplified by miR-501-3p, in NMDAR-dependent GluA1 expression and spine remodeling.
Materials and methods
Animals, DNA constructs, and reagents
All animal procedures followed the National Institutes of Health guideline Using Animals in Intramural Research and were approved by the National Institutes of Mental Health Animal Care and Use Committee. To generate constructs expressing miRNAs, rat pre-miR-501 was amplified from genomic DNAs by PCR and cloned into the BglII–HindIII site of the pGSuper (promoter: H1) vector. The siRNA construct was made by inserting annealed oligos containing the siRNA sequence against GluR1 (5′-gcagacggaaattgcttat-3′) into the pSuper (promoter: H1) vector. The cDNA of Gria1 was obtained by RT-PCR and cloned into the GW1 (promoter: CMV) vector. To generate the Gria1 reporter construct, the 3′ UTR of Gria1 was amplified from mouse brain cDNA by PCR and cloned into the pCMV-mCherry-Ds vector behind the mCherry sequence. The mutant Gria1 reporter was generated by PCR using KOD DNA Polymerase (EMD Millipore) with primers (5′-gcatgtggcactcactgaagcgtattggtgggagtggctgcataagaagtgg-3′ and 5′-ccacttcttatgcagccactcccaccaatacgcttcagtgagtgccacatgc-3′) followed by DpnI digestion. The miR-501-3p precursor and Ago2 cDNA were cloned into the pRRLsin.CMV.GFPpre vector for lentivirus production. The following reagents were obtained commercially: miR-501-3p antisense oligonucleotide (5′-CCAAAUCCUUGCCCGGGUGCAUU-3′; Integrated DNA Technologies), miR-501-3p scrambled antisense oligonucleotide (5′-GCCAGTTGTCCTACGCGCTCTAA-3′; Integrated DNA Technologies), NMDA (Sigma-Aldrich), actinomycin D (Sigma-Aldrich), AP5 (Sigma-Aldrich), Ro 25–6891(Sigma-Aldrich), TCN 201 (Tocris Bioscience), Cytochalasin (Sigma-Aldrich), Nocodazole (Sigma-Aldrich), NBQX (Sigma-Aldrich), anti-GluA1 (1:7 for Immunocytochemistry; EMD Millipore; 1:1,000 for Western blot; EMD Millipore), anti-HA antibody (1:1,000; Covance), anti-myc antibody (1:1,000; EMD Millipore), anti–digoxigenin-POD antibody (1:100; Roche), and anti-actin antibody (1:5,000; Sigma-Aldrich).
miRNA pull-down
The 3′ UTR of mouse Gria1 was amplified by PCR and cloned into the pCR4-TOPO vector (promoter: T3; Invitrogen). RNAs were synthesized by in vitro transcription using T3 transcriptase (Roche) and RNA 3′ end biotinylation kit (Thermo Fisher Scientific), and then immobilized to beads (Thermo Fisher Scientific). Total RNAs were extracted from the hippocampus of mice (17 d old) using mirVana miRNA Isolation kit (Ambion), denatured, and incubated with 3′ UTR-bound streptavidin beads in the hybridization buffer (50% formamide, 10% dextran sulfate, 5× SSC, 500 mg/ml yeast tRNA, and 1× Denhardt’s solution) for 60 min at 35°C with gentle rotation. The beads were washed with 1× SSC/0.1% SDS (three times) at room temperature, and then with 0.5× SSC (three times at room temperature and once at 42°C). Bound RNAs on the beads were eluted with water (65°C for 2 min).
Construction of miRNA sequencing libraries and next generation deep sequencing
Approximately 17–27-nt-long RNA fragments (enriched for miRNAs) were isolated from the input and eluted RNAs of the pull-down assay using denaturing PAGE (15%). Deep-sequencing libraries were constructed by using Small RNA Sample Prep kit (Illumina). 3′ and 5′ adaptors were ligated to purified miRNAs sequentially. Adaptor-tagged miRNAs were reverse transcribed and amplified by low-cycle PCR. The PCR products were purified by PAGE (8%) and sequenced using an Genome Analyzer II (Illumina).
Deep-sequencing data analysis
Raw sequence reads were first consolidated by clustering identical sequence reads. Only reads containing complete 3′ and 5′ adaptor sequences were subjected to downstream analysis. After trimming of 3′ and 5′ adaptor sequences, the remaining sequences were aligned to miRNA hairpin sequences downloaded from the miRBase database (http://www.mirbase.org/) using Bowtie v. 0.2.1. Mapped reads were further filtered to remove those that do not map to mature miRNAs. Low-abundance miRNAs (read counts ≤5 in input or ≤50 in eluted RNAs) were excluded from analysis. The expression count of each miRNA was normalized to the total number of mapped reads of the corresponding sample, log2 transformed, and normalized across all samples. miRNAs with ≥10-fold enrichment by pull-down were identified as candidate Gria1-targeting miRNAs.
qRT-PCR
miRNAs were purified using mirVana miRNA Isolation kit (Ambion), and then transcribed into cDNAs and amplified by PCR with the Taqman primers for miRNAs with miRBase ID rno-miR-501-3p, mo-miR-218a-5p, and mo-miR-29a-3p or primers (pri-miR-501: 5′-ctgctctgctcgtcctctct-3′ and 5′-ctcctgtcctcacatgcaga-3′) specific for miRNAs of interest with the 7900 Fast Real-Time PCR System (Applied Biosystems). U6 (NCBI accession no. NR_004394) was analyzed (using Taqman primers, 5′-gcttcggcagcacatatactaa-3′ and 5′-aaaatatggaacgcttcacga-3′) as the endogenous control gene for normalization of input RNAs.
Neuronal culture and transfection
Hippocampal and cortical neuron cultures were prepared from embryonic day 18–19 rat embryos. The hippocampus was removed from the brain and digested with trypsin. Dissociated hippocampal neurons were seeded on coverslips or plates coated with poly-d-lysine (30 µg/ml) and laminin (2 µg/ml) at a density of 750 cells/mm2 (for spine morphology analysis) or 450 cells/mm2 (for immunocytochemistry). Cortical neurons were seeded on a plate coated with poly-d-lysine (30 µg/ml) at a density of 1,000 cells/mm2. Neurons were grown in Neurobasal medium supplemented with 2% B27 and 2 mM glutamax, and transfected with Lipofectamine 2000. All reagents for neuronal cultures were purchased from Invitrogen.
Time-lapse imaging
Primary hippocampal neurons were placed in an imaging chamber and perfused with ACSF (124 mM NaCl, 3 mM KCl, 26 mM NaHCO3, 1.25 mM NaH2PO4, 2.5 mM CaCl2, 1.3 mM MgSO4, and 10 mM d-glucose; bubbled with 95% O2/5% CO2; 30°C] at 2 ml/min. Images were acquired using a BX61WI confocal microscope (Olympus) with a 60× water immersion objective (NA 1.35) and the Olympus Fluoview software. Z-stack images were collapsed to make 2D projections for image analysis. Spine area and density were analyzed with MetaMorph software (Molecular Devices). Images were thresholded to remove background. Spines in secondary dendrites were then manually selected and analyzed for area and number using the Integrated Morphometry Analysis function of MetaMorph. The length of dendrites was measured using the Region Measurement function of MetaMorph. Spine density was calculated by dividing the total number of spines on a given dendrite by the length of the dendrite. One-way analysis of variance (ANOVA) was used to test for differences among groups, and two-tailed Mann-Whitney U test was used for post hoc analysis.
Immunocytochemistry
Hippocampal neurons were fixed in PBS containing 4% formaldehyde and 4% sucrose. After rinse with PBS, neurons were incubated with primary antibodies overnight at 4°C, followed by incubation with fluorophore-conjugated (Alexa Fluor 647) secondary antibodies. Z-stack Images were acquired by using a confocal microscope (LSM510; Carl Zeiss) with a 63× (NA 1.4) or 40× (NA 1.2) oil immersion objective at room temperature and collapsed to make 2D projections with the LSM software (Carl Zeiss). MetaMorph software was used to measure integrated fluorescence intensity of GluA1 on the dendrite. All image acquisition and image analysis were done blindly to treatment. Data were analyzed with the Kolmogorov-Smirnov and χ2 test for distribution. As the image data are not normally distributed, two-tailed Mann-Whitney U test was used to calculate p-values.
In situ hybridization
In situ hybridization of miRNAs was performed as described previously (Hu et al., 2014). Hippocampal neurons were fixed in PBS containing 4% formaldehyde and 4% sucrose. After permeabilization with 0.5% Triton X-100 (in PBS), neurons were hybridized with 5′ and 3′ digoxigenin-labeled, locked nucleic acid–modified oligonucleotide probes (Exiqon) in hybridization buffer at 59°C for 1 h. After wash with 0.1× SSC (three times at 65°C) and then with 2× SSC (two times at room temperature), hybridized probes were detected by incubation with horseradish peroxidase–conjugated anti-digoxigenin antibody (Roche) followed by amplification with Cy5-conjugated tyramide (PerkinElmer).
Cross-linking and immunoprecipitation
Cultured cortical neurons grown on 10-cm plates were transduced with lentivirus expressing Flag-tagged Ago2 at 7 DIV and treated with NMDA at 17 DIV. Cross-linking and immunoprecipitation of Ago2-associated RNAs was performed as described previously (Hu et al., 2014). At 90 min after NMDA treatment, neurons were cross-linked by UV irradiation (480,000 µJ/cm2), and then lysed in 1× RIPA buffer containing 0.1 U/µl RNase inhibitor followed by sonication. After centrifugation at 13,000 rpm for 30 min at 4°C, supernatant was incubated (overnight at 4°C, with rotation) with anti-Flag antibody–conjugated beads (Sigma-Aldrich) in the immunoprecipitation buffer (25 mM Hepes, pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 1 mM EGTA, 10% glycerol, 0.1% NP-40, 1 mM NaF, 1 mM 2-glycerophosphate, and 1 mM Na3VO4). The beads were washed five times with the immunoprecipitation buffer. Bound RNAs were extracted with phenol/chloroform, precipitated with ethanol, and analyzed by qRT-PCR.
Hippocampal slice culture and transfection
Hippocampal slices were cultured as described previously (Jiao and Li, 2011). Brains were removed from Sprague-Dawley rats (postnatal day 6–8) and cut into slices with a vibratome (Leica) in icy cutting solution (238 mM sucrose, 2.5 mM KCl, 26 mM NaHCO3, 1 mM NaH2PO4, 5 mM MgCl2, 11 mM d-glucose, and 1 mM CaCl2). Hippocampal slices (350 µm) were incubated on semipermeable membrane inserts (EMD Millipore) in a culture medium (78.8% minimum essential medium, 20% heat-inactivated horse serum, 25 mm Hepes, 10 mM d-glucose, 26 mM NaHCO3, 2 mM CaCl2, 2 mM MgSO4, 0.0012% ascorbic acid, and 1 µg/ml insulin; pH 7.3; 320–330 mOsm). Neurons were biolistically transfected using the gene gun (Helios Gene-gun system; Bio-Rad Laboratories) at 3 DIV. Electrophysiological recordings were performed at 4–7 d after transfection.
Electrophysiology
Hippocampal slices were recorded as described previously (Han et al., 2013). Slices were kept under 25°C in ACSF (bubbled with 95% O2/5% CO2, supplemented with 25 µM picrotoxin and 2 µM 2-chloroadenosine) at the rate of 2 ml/min. EPSCs of CA1 pyramidal cells evoked by stimulating the Schaffer collateral pathway were recorded in the whole-cell voltage-clamp mode at the holding potential of −70 mV. The patch pipette (4–7 MΩ) solution contained 130 mM cesium methanesulfonate, 8 mM NaCl, 4 mM Mg-ATP, 0.3 mM Na-GTP, 0.5 mM EGTA, 10 mM Hepes, and 5 mM QX-314 at pH 7.3. LTD was induced by pairing low-frequency stimulations (1 Hz) with a postsynaptic depolarization to −45 mV for 300 s.
Online supplemental material
Fig. S1 shows that NMDA-induced GluA1 reduction is caused by miR-501-3p–mediated suppression (related to Fig. 4). Fig. S2 shows that miR-501-3p antisense oligonucleotides and GluA1 overexpression have no effect on basal spine number and size (related to Fig. 5). Fig. S3 shows GluA1 protein expression in neurons used for live imaging experiments (related to Fig. 5). Fig. S4 shows that GluA1 siRNAs effectively and specifically knock down GluA1 expression (related to Fig. 5). Fig. S5 shows that GluA1 overexpression has no effect on LTD (related to Fig. 5). Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201404092/DC1.
Supplementary Material
Acknowledgments
We thank Dr. Heinz Arnheiter for intense discussion of this study and Dr. Elizabeth Sherman (National Institute of Mental Health) for editing of the manuscript.
This work was supported by the National Institute of Mental Health and the National Heart, Lung, and Blood Institute Intramural Programs.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- AMPAR
- α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor
- ANOVA
- analysis of variance
- AP5
- (2R)-amino-5-phosphonovaleric acid
- DIV
- days in vitro
- NMDA
- N-methyl-d-aspartate
- NMDAR
- NMDA receptor
- qRT-PCR
- quantitative RT-PCR
- RISC
- RNA-induced silencing complex
References
- Bartel D.P.2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 116:281–297 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- Baude A., Nusser Z., Molnár E., McIlhinney R.A., and Somogyi P.. 1995. High-resolution immunogold localization of AMPA type glutamate receptor subunits at synaptic and non-synaptic sites in rat hippocampus. Neuroscience. 69:1031–1055 10.1016/0306-4522(95)00350-R [DOI] [PubMed] [Google Scholar]
- Bicker S., Khudayberdiev S., Weiß K., Zocher K., Baumeister S., and Schratt G.. 2013. The DEAH-box helicase DHX36 mediates dendritic localization of the neuronal precursor-microRNA-134. Genes Dev. 27:991–996 10.1101/gad.211243.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang P.K., Verbich D., and McKinney R.A.. 2012. AMPA receptors as drug targets in neurological disease—advantages, caveats, and future outlook. Eur. J. Neurosci. 35:1908–1916 10.1111/j.1460-9568.2012.08165.x [DOI] [PubMed] [Google Scholar]
- Ehlers M.D.2000. Reinsertion or degradation of AMPA receptors determined by activity-dependent endocytic sorting. Neuron. 28:511–525 10.1016/S0896-6273(00)00129-X [DOI] [PubMed] [Google Scholar]
- Fusco D., Accornero N., Lavoie B., Shenoy S.M., Blanchard J.M., Singer R.H., and Bertrand E.. 2003. Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Curr. Biol. 13:161–167 10.1016/S0960-9822(02)01436-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grooms S.Y., Noh K.M., Regis R., Bassell G.J., Bryan M.K., Carroll R.C., and Zukin R.S.. 2006. Activity bidirectionally regulates AMPA receptor mRNA abundance in dendrites of hippocampal neurons. J. Neurosci. 26:8339–8351 10.1523/JNEUROSCI.0472-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M.H., Jiao S., Jia J.M., Chen Y., Chen C.Y., Gucek M., Markey S.P., and Li Z.. 2013. The novel caspase-3 substrate Gap43 is involved in AMPA receptor endocytosis and long-term depression. Mol. Cell. Proteomics. 12:3719–3731 10.1074/mcp.M113.030676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z., Yu D., Gu Q.H., Yang Y., Tu K., Zhu J., and Li Z.. 2014. miR-191 and miR-135 are required for long-lasting spine remodelling associated with synaptic long-term depression. Nat. Commun. 5:3263 10.1038/ncomms4263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huganir R.L., and Nicoll R.A.. 2013. AMPARs and synaptic plasticity: the last 25 years. Neuron. 80:704–717 10.1016/j.neuron.2013.10.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao S., and Li Z.. 2011. Nonapoptotic function of BAD and BAX in long-term depression of synaptic transmission. Neuron. 70:758–772 10.1016/j.neuron.2011.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju W., Morishita W., Tsui J., Gaietta G., Deerinck T.J., Adams S.R., Garner C.C., Tsien R.Y., Ellisman M.H., and Malenka R.C.. 2004. Activity-dependent regulation of dendritic synthesis and trafficking of AMPA receptors. Nat. Neurosci. 7:244–253 10.1038/nn1189 [DOI] [PubMed] [Google Scholar]
- Kessels H.W., Kopec C.D., Klein M.E., and Malinow R.. 2009. Roles of stargazin and phosphorylation in the control of AMPA receptor subcellular distribution. Nat. Neurosci. 12:888–896 10.1038/nn.2340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung A.K., and Sharp P.A.. 2013. Quantifying Argonaute proteins in and out of GW/P-bodies: implications in microRNA activities. Adv. Exp. Med. Biol. 768:165–182 10.1007/978-1-4614-5107-5_10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Li J., and Cairns M.J.. 2014. Identifying miRNAs, targets and functions. Brief. Bioinform. 15:1–19 10.1093/bib/bbs075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.J., and Savtchouk I.. 2012. Ca2+ permeable AMPA receptors switch allegiances: mechanisms and consequences. J. Physiol. 590:13–20 10.1113/jphysiol.2011.213926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W., and Roche K.W.. 2012. Posttranslational regulation of AMPA receptor trafficking and function. Curr. Opin. Neurobiol. 22:470–479 10.1016/j.conb.2011.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugli G., Larson J., Martone M.E., Jones Y., and Smalheiser N.R.. 2005. Dicer and eIF2c are enriched at postsynaptic densities in adult mouse brain and are modified by neuronal activity in a calpain-dependent manner. J. Neurochem. 94:896–905 10.1111/j.1471-4159.2005.03224.x [DOI] [PubMed] [Google Scholar]
- Man H.Y., Sekine-Aizawa Y., and Huganir R.L.. 2007. Regulation of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor trafficking through PKA phosphorylation of the Glu receptor 1 subunit. Proc. Natl. Acad. Sci. USA. 104:3579–3584 10.1073/pnas.0611698104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki M., Ellis-Davies G.C., Nemoto T., Miyashita Y., Iino M., and Kasai H.. 2001. Dendritic spine geometry is critical for AMPA receptor expression in hippocampal CA1 pyramidal neurons. Nat. Neurosci. 4:1086–1092 10.1038/nn736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki M., Honkura N., Ellis-Davies G.C., and Kasai H.. 2004. Structural basis of long-term potentiation in single dendritic spines. Nature. 429:761–766 10.1038/nature02617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeill E., and Van Vactor D.. 2012. MicroRNAs shape the neuronal landscape. Neuron. 75:363–379 10.1016/j.neuron.2012.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh M.C., Derkach V.A., Guire E.S., and Soderling T.R.. 2006. Extrasynaptic membrane trafficking regulated by GluR1 serine 845 phosphorylation primes AMPA receptors for long-term potentiation. J. Biol. Chem. 281:752–758 10.1074/jbc.M509677200 [DOI] [PubMed] [Google Scholar]
- Passafaro M., Nakagawa T., Sala C., and Sheng M.. 2003. Induction of dendritic spines by an extracellular domain of AMPA receptor subunit GluR2. Nature. 424:677–681 10.1038/nature01781 [DOI] [PubMed] [Google Scholar]
- Pitchiaya S., Androsavich J.R., and Walter N.G.. 2012. Intracellular single molecule microscopy reveals two kinetically distinct pathways for microRNA assembly. EMBO Rep. 13:709–715 10.1038/embor.2012.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt G.2009. microRNAs at the synapse. Nat. Rev. Neurosci. 10:842–849 10.1038/nrn2763 [DOI] [PubMed] [Google Scholar]
- Schwarz L.A., Hall B.J., and Patrick G.N.. 2010. Activity-dependent ubiquitination of GluA1 mediates a distinct AMPA receptor endocytosis and sorting pathway. J. Neurosci. 30:16718–16729 10.1523/JNEUROSCI.3686-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd J.D., and Huganir R.L.. 2007. The cell biology of synaptic plasticity: AMPA receptor trafficking. Annu. Rev. Cell Dev. Biol. 23:613–643 10.1146/annurev.cellbio.23.090506.123516 [DOI] [PubMed] [Google Scholar]
- Snyder E.M., Philpot B.D., Huber K.M., Dong X., Fallon J.R., and Bear M.F.. 2001. Internalization of ionotropic glutamate receptors in response to mGluR activation. Nat. Neurosci. 4:1079–1085 10.1038/nn746 [DOI] [PubMed] [Google Scholar]
- Sutton M.A., Ito H.T., Cressy P., Kempf C., Woo J.C., and Schuman E.M.. 2006. Miniature neurotransmission stabilizes synaptic function via tonic suppression of local dendritic protein synthesis. Cell. 125:785–799 10.1016/j.cell.2006.03.040 [DOI] [PubMed] [Google Scholar]
- Turrigiano G.G.2008. The self-tuning neuron: synaptic scaling of excitatory synapses. Cell. 135:422–435 10.1016/j.cell.2008.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Homma K.J., and Poo M.M.. 2004. Shrinkage of dendritic spines associated with long-term depression of hippocampal synapses. Neuron. 44:749–757 10.1016/j.neuron.2004.11.011 [DOI] [PubMed] [Google Scholar]
- Zhu J.J., Esteban J.A., Hayashi Y., and Malinow R.. 2000. Postnatal synaptic potentiation: delivery of GluR4-containing AMPA receptors by spontaneous activity. Nat. Neurosci. 3:1098–1106 10.1038/80614 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.