YB-1, which is upregulated in human sarcomas, controls the availability of the stress granule nucleator G3BP1 and thereby controls stress granule assembly.
Abstract
Under cell stress, global protein synthesis is inhibited to preserve energy. One mechanism is to sequester and silence mRNAs in ribonucleoprotein complexes known as stress granules (SGs), which contain translationally silent mRNAs, preinitiation factors, and RNA-binding proteins. Y-box binding protein 1 (YB-1) localizes to SGs, but its role in SG biology is unknown. We now report that YB-1 directly binds to and translationally activates the 5′ untranslated region (UTR) of G3BP1 mRNAs, thereby controlling the availability of the G3BP1 SG nucleator for SG assembly. YB-1 inactivation in human sarcoma cells dramatically reduces G3BP1 and SG formation in vitro. YB-1 and G3BP1 expression are highly correlated in human sarcomas, and elevated G3BP1 expression correlates with poor survival. Finally, G3BP1 down-regulation in sarcoma xenografts prevents in vivo SG formation and tumor invasion, and completely blocks lung metastasis in mouse models. Together, these findings demonstrate a critical role for YB-1 in SG formation through translational activation of G3BP1, and highlight novel functions for SGs in tumor progression.
Introduction
Messenger RNA translation is tightly controlled in response to cellular stress, primarily at the initiation step (Sonenberg and Hinnebusch, 2009). Under diverse forms of cell stress such as oxidative stress, hypoxia, or nutrient deprivation, translation initiation is rapidly blocked, likely as a means to limit energy-demanding protein synthesis (Anderson and Kedersha, 2006). Under these and other stress conditions, stalled mRNA preinitiation complexes form mRNA–protein complexes (mRNPs) that are first stored in the cytoplasm as translationally inactive small granules called P-bodies (PBs) containing translation initiation factors such as eIF4E. PBs then transition into highly specialized cytoplasmic structures known as stress granules (SGs) by secondary and tertiary aggregation (Anderson and Kedersha, 2008). While PBs are associated with mRNA decay, SGs, which contain additional proteins such as eIF3, eIF4G, and PABP, are thought to represent sites of mRNA storage and triage to facilitate translational reprogramming. SG aggregation requires specific RNA-binding proteins including TIA-1 and G3BP1 (Anderson and Kedersha, 2009). TIA1 possess Gln or Asn (Q/N)-rich prion-like interaction domains, whereas G3BP1 contains low-complexity (LC) or intrinsically disordered (ID) regions necessary for dimerization (Tourrière et al., 2003; Gilks et al., 2004; Kedersha et al., 2013). These domains underlie the ability of these proteins to function as nucleating factors for SG assembly. Knockdown (kd) of TIA1 or G3BP1 severely impairs SG assembly under arsenite-induced oxidative stress (Tourrière et al., 2003; Gilks et al., 2004). Moreover, overexpression of TIA1 or G3BP1 alone is sufficient to induce SG nucleation even in the absence of stress (Kedersha et al., 1999; Tourrière et al., 2003).
In addition to nucleating proteins, SG assembly requires non-polysome-bound mRNAs (Kedersha and Anderson, 2002, 2007; Buchan and Parker, 2009). Although incompletely characterized, some mRNA species stored in SGs are postulated to be highly expressed transcripts (Stöhr et al., 2006; Anderson and Kedersha, 2008), thus limiting ATP-demanding translation of these and other mRNAs during cell stress to preserve energy, and to prevent misfolded protein accumulation (Kimball et al., 2003). Moreover, storage of mRNAs in SGs blocks their degradation and allows cells to rapidly restore synthesis of vital proteins during recovery from cell stress (Lavut and Raveh, 2012). In contrast, some mRNAs are excluded from SGs, such as chaperones and repair enzymes, possibly to ensure their continued translation during acute stress (Arimoto et al., 2008; Yamasaki and Anderson, 2008). Therefore, SG formation appears to be critical to reprogram mRNA translation under adverse conditions to facilitate cellular adaptive responses. Although SGs are involved in many normal processes including cellular signaling (Kedersha et al., 2013), mutations of SG proteins are found in degenerative disorders including amyotrophic lateral sclerosis (ALS), frontotemporal lobar degeneration (FTLD), and inclusion body myopathy (IMD; Li et al., 2013; Vanderweyde et al., 2013; Toyoshima and Takahashi, 2014). Whether SGs play specific functions in tumor cells is very poorly understood.
In addition to nucleating factors, other RNA-binding proteins also localize to SGs, although with largely unknown consequences (Anderson and Kedersha, 2009). One example is YB-1, a highly conserved cold shock domain (CSD) family protein that binds DNA and RNA (Kohno et al., 2003). YB-1 associates with both PBs (Yang and Bloch, 2007) and SGs (Kedersha and Anderson, 2007), but how YB-1 functions in the biology of these structures is unknown. YB-1 is a multifunctional protein with roles in transcription (Ohga et al., 1998), oxidative phosphorylation (Matsumoto et al., 2012), and cytosolic mRNA translational control (Eliseeva et al., 2011). Cytosolic YB-1 functions as a component of translationally inactive mRNPs to directly block translation initiation and to inhibit mRNA degradation (Evdokimova et al., 2001). In addition to being a global translational repressor, YB-1 selectively activates translation of specific transcripts via its ability to bind UTRs of target mRNAs. For example, we showed that YB-1 directly binds to 5′ UTRs and activates translation of mRNAs encoding SNAIL1, TWIST, and other transcription factors to mediate an epithelial-to-mesenchymal transition (EMT) and metastasis in breast cancer cells (Evdokimova et al., 2009). YB-1 is also reported as a downstream translational effector of mTORC1 (Hsieh et al., 2012; Thoreen et al., 2012), with potential roles in cancer initiation and metastasis (Hsieh et al., 2012). Indeed, elevated expression of YB-1 correlates with poor patient survival and drug resistance in diverse tumor types, and YB-1 levels are elevated in metastatic tumors (Lovett et al., 2010; Eliseeva et al., 2011; Wu et al., 2012).
Human sarcomas are aggressive bone and soft tissue tumors with high YB-1 expression (Oda et al., 1998, 2008; Fujiwara-Okada et al., 2013). We recently used a novel Click-SILAC mass spectrometry approach to show that in human sarcomas, YB-1 regulates acute synthesis of proteins involved in diverse biological pathways including metabolism, cell stress responses, mitochondrial functions, protein folding, and mRNA translation (Somasekharan et al., 2012). We wondered if this might be linked to a function for YB-1 in SGs, which could potentially affect translation of broad classes of mRNAs. We therefore investigated whether YB-1 is involved in stress-induced SGs in sarcoma cells. We report that YB-1 is important for the formation of SGs, but not PBs, in human sarcoma cell lines, both in vitro or in vivo when xenografted into immunocompromised mice. YB-1 regulates SG formation by directly binding to the G3BP1 5′ UTR and activating G3BP1 mRNA translation. YB-1 and G3BP1 protein expression are strongly correlated in human sarcoma specimens, and G3BP1 expression is associated with poor outcome in sarcomas. Finally, G3BP1 kd dramatically reduces sarcoma metastasis in mouse models, highlighting a novel link between YB-1–mediated SG formation and sarcoma progression.
Results
YB-1 is required for SG formation in stressed sarcoma cells
YB-1 is known to associate with both PBs and SGs (Kedersha and Anderson, 2007; Yang and Bloch, 2007; Onishi et al., 2008). To confirm this in sarcoma cells, we assessed localization of YB-1 to these structures in sarcoma cell lines. In U2OS osteosarcoma cells grown under ambient conditions, a minority of YB-1 colocalizes with the PB marker DDX6 in cytosolic granules, confirming that YB-1 localizes to PBs in these cells (Fig. S1 A, left). However, under arsenite-mediated oxidative stress, YB-1 instead localized in close proximity to, but not overlapping with, PBs, as indicated by immunofluorescence (IF) for DDX6 (see the enlarged views in Fig. S1 A, right). To determine if the latter represent SGs, we extended these studies to include other stress forms known to induce SGs, including H2O2, ER stress (thapsigargin), heat shock (42°C), UV irradiation, and hypoxia (1% O2; Moeller et al., 2004; Anderson and Kedersha, 2008). YB-1 colocalized with TIA-1 and G3BP1 in SGs in response to each stress (Fig. S1 B), confirming the association of YB-1 with SGs. Live cell imaging confirmed that YB-1 and G3BP1 colocalize in SGs with similar kinetics under arsenite stress, as shown in Videos 1–3.
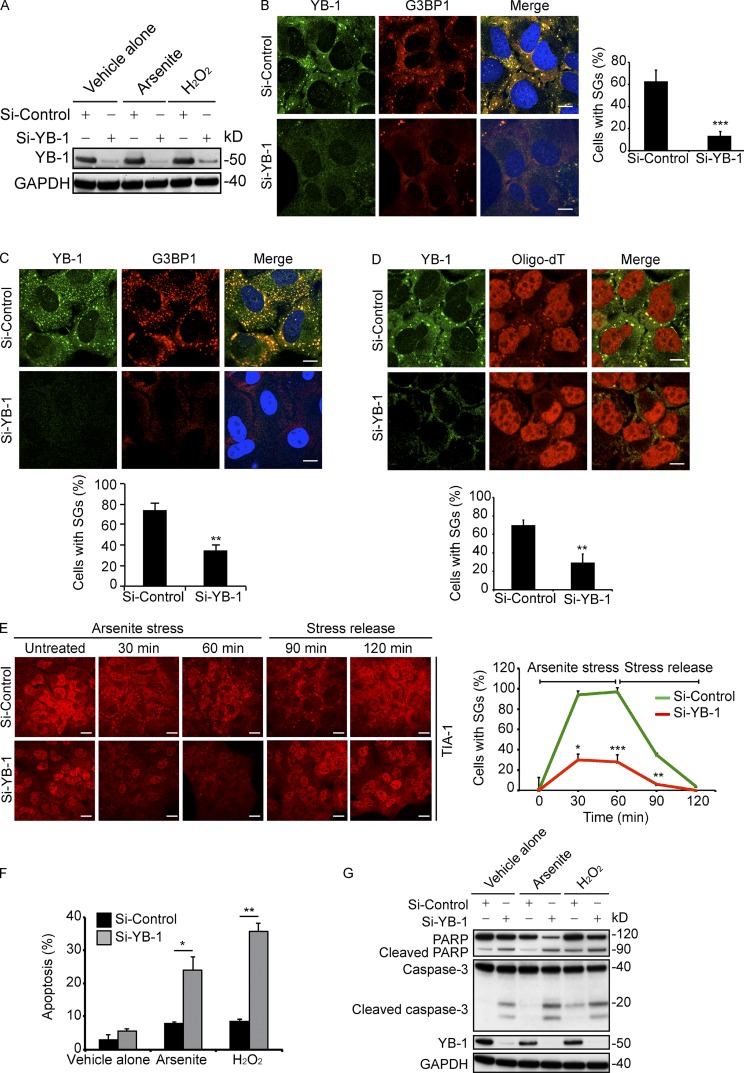
We next tested whether YB-1 is required for PB or SG formation, which to our knowledge has not been established. We performed siRNA-mediated YB-1 kd in U2OS cells (Fig. 1 A), and monitored PB and SG formation under ambient conditions or arsenite and H2O2 treatment. Compared with siRNA controls, YB-1 kd did not appreciably alter PB formation in U2OS cells under ambient conditions (Fig. S1 C). Although this does not absolutely rule out the possibility, this argues against a major role for YB-1 in PB formation. In contrast, YB-1 kd markedly attenuated SG formation under both arsenite (Fig. 1 B) and H2O2 treatment (Fig. 1 C). A second independent YB-1 siRNA showed identical results (Fig. S2, A and B). Moreover, YB-1 kd significantly reduced SG-associated poly(A)+ mRNAs in arsenite-treated cells (Fig. 1 D), in spite of comparable levels of nuclear poly(A)+ mRNA pools in control cells. Similar results were obtained using RH-30 rhabdomyosarcoma (Fig. S2 C) and MNNG osteosarcoma cells (Fig. S2 D), as well as for DU-145 prostate carcinoma cells (Fig. S2 E), which points to a broader mechanism in transformed cells. Ectopic reexpression of Myc-tagged YB-1 in U2OS cells with YB-1 kd efficiently rescued SG formation under arsenite treatment but not under ambient conditions (Fig. S2 F; see also Fig. S3 A). A time-course of SG formation in U2OS cells in response to arsenite showed that YB-1 kd cells were reduced more than threefold in SG formation compared with control cells (Fig. 1 E). This trend was observed throughout the 60-min time-course of arsenite treatment as well as during the recovery period, in which SGs almost completely disappeared in YB-1 kd cells by 30 min after arsenite removal. Similar results were obtained for H2O2 and the plant phytogen piperlongumine (unpublished data). These results indicate that YB-1 is critical for oxidative stress-induced SG formation in sarcoma cells.
Figure 1.
YB-1 kd impairs SG assembly and sensitizes cells to oxidative stress. (A–C) U2OS cells transfected with siControl or siYB-1 siRNAs were treated with vehicle alone, arsenite (0.5 mM), or H2O2 (0.5 mM) for 1 h and immunoblotted using anti–YB-1 or anti-GAPDH antibodies (A). SGs were detected by IF in arsenite- (B) or H2O2-treated (C) siControl or siYB-1 U2OS cells using the indicated antibodies. Slides were counterstained with DRAQ5 to detect nuclei. SGs were quantified using ImageJ software by counting cells containing SGs divided by total cells, and represented by bar graphs. (D) Arsenite (0.5 mM)-treated siControl or siYB-1 U2OS cells were subjected to in situ hybridization using 5-FAM-oligodT and counterstained with anti–YB-1 antibodies. SGs were quantified as in B. (E) U2OS cells transfected with siControl or siYB-1 siRNAs were treated with arsenite (0.5 mM) for 60 min, then placed in full media without arsenite for another 60 min. Cells were fixed at the indicated time points and subjected to IF using anti–TIA-1 antibodies. SGs were quantified as in B. (F) U2OS cells transfected with siControl or siYB-1 siRNAs were treated with arsenite (0.5 mM) or H2O2 (0.5 mM) for 5 h, and apoptosis was measured by Annexin V-FITC flow cytometry. (G) Cell lysates from F were subjected to immunoblotting using the indicated antibodies. Mean values ± SD (error bars) are shown for three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Bars, 10 µm.
Given that SGs provide protection from cell stress (Arimoto et al., 2008; Lavut and Raveh, 2012), we next tested whether YB-1 loss affects cell viability under oxidative stress. Indeed, using Annexin V-FITC–based flow cytometry, the percentage of Annexin V-FITC–positive apoptotic cells in U2OS cells with YB-1 kd treated with arsenite or H2O2 was two to fourfold higher than in control siRNA cells (Fig. 1 F). This was confirmed by immunoblotting for PARP or caspase-3 cleavage, both of which were markedly increased in YB-1 kd compared with the control cells treated (Fig. 1 G). Similar results were obtained using RH-30, MNNG, and DU-145 cells treated with arsenite and H2O2 (Fig. S2 G) or hypoxia (Fig. S2 H), although stress treatment effects varied among cell lines. Therefore, blocking YB-1 expression results in reduced SG formation and enhanced oxidative stress or hypoxia-induced apoptosis.
YB-1 posttranscriptionally regulates synthesis of the G3BP1-nucleating factor
Next, we wished to determine the mechanism by which YB-1 regulates SG formation. First, we examined if YB-1 functions as an SG-nucleating factor as described for major SG proteins such as TIA-1 and G3BP1 (Tourrière et al., 2003; Gilks et al., 2004). A characteristic of such factors is that their overexpression is sufficient to induce SGs even in the absence of cell stress (Kedersha et al., 2013). To test this, we overexpressed YB-1 in U2OS cells by transiently transfecting cells with Myc-tagged YB-1, and then analyzed SG formation. Using G3BP1, eIF3η, and TIA-1 as SG markers, YB-1 overexpression did not induce SG formation under ambient conditions and Myc-YB-1 remained diffusely cytosolic, even though these cells were competent to form SGs under arsenite (Fig. S3 A; see also Fig. S2 F). Therefore YB-1 overexpression does not by itself induce SG formation upon under basal conditions, which argues against YB-1 as an SG nucleating factor.
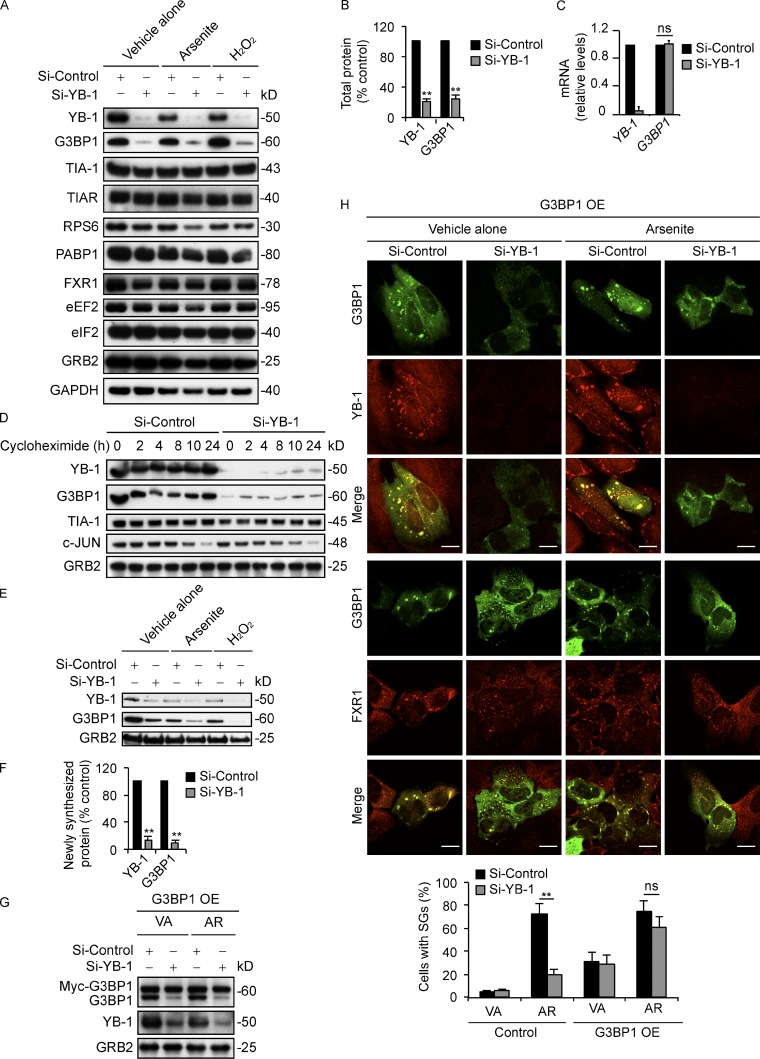
The data in the previous paragraph pointed to other potential mechanisms for regulation of SGs by YB-1. Because YB-1 is known to regulate mRNA translation (Evdokimova et al., 2006, 2009), we tested whether YB-1 might influence the expression of known SG-associated proteins. Levels of SG resident proteins, including TIA-1, TIAR, the 40S ribosomal protein RPS6, PABP1, and FXR1 or translational components such as eEF2 and eIF2, were not altered by YB-1 kd in U2OS cells under either ambient conditions or oxidative stress (Fig. 2 A). Unexpectedly, however, YB-1 kd led to markedly reduced levels of G3BP1 compared with siControl cells, both under ambient conditions or with arsenite or H2O2 treatment (Fig. 2, A and B). Similar results were obtained in RH-30, MNNG, and DU-145 cell lines (Fig. S3 B). This was not due to reduced global protein synthesis, as overall rates of protein synthesis in YB-1 kd cells were actually higher than in siControl cells (Fig. S3 C), in agreement with our previously published results (Somasekharan et al., 2012). As shown by qRT-PCR, G3BP1 mRNA expression levels were unaffected by YB-1 kd (Fig. 2 C), which indicates that YB-1 regulates G3BP1 protein abundance posttranscriptionally, either at the level of protein stability or synthesis. We first tested the former possibility by monitoring protein levels in YB-1 kd and control cells by immunoblotting after a time course of treatment with the translation inhibitor cycloheximide (CHX; Fig. 2 D). Similar to TIA-1 and GRB2, treatment with CHX for the indicated time points failed to significantly alter G3BP1 degradation rates in siYB-1 compared with control cells. We used c-Jun, a protein with a known short half-life, as a positive control to confirm that its expression decreases with CHX treatment. These results confirm that G3BP1 is a relatively stable protein and that YB-1 does not appreciably affect degradation of G3BP1.
Figure 2.
YB-1 regulates G3BP1 mRNA translation. (A) U2OS cells transfected with siControl or siYB-1 siRNAs were treated with vehicle alone, arsenite (0.5 mM), or H2O2 (0.5 mM) for 1 h. Lysates were analyzed by immunoblotting using the indicated antibodies. (B) Band intensities from A were quantified by densitometry. Mean values ± SD (error bars) are shown for three independent experiments. **, P < 0.01. (C) Total RNA isolated from siControl and siYB-1 U2OS cells under ambient conditions was subjected to RT-PCR using primers for YB-1 and G3BP1. Mean values ± SD (error bars) are shown for three independent experiments. ns, nonsignificant. (D) U2OS cells transfected with siControl or siYB-1 siRNAs were incubated with CHX for the indicated times, and lysates were immunoblotted using the indicated antibodies. (E) U2OS cells transfected with siControl or siYB-1 siRNAs were treated with vehicle alone, arsenite (0.5 mM), or H2O2 (0.5 mM) for 1 h in the presence of AHA to capture newly synthesized proteins, and immunoblotted using the indicated antibodies. (F) Band intensities from E were quantified using densitometry. Mean values ± SD (error bars) are shown for three independent experiments. **, P < 0.01. (G) siControl or siYB-1 kd U2OS cells were transfected with Myc-G3BP1 (G3BP1 overexpression [OE]), and treated with vehicle alone (VA) or arsenite (AR; 0.5 mM) for 1 h. Lysates were then prepared and immunoblotted using antibodies against G3BP1, YB-1, and GRB2. (H) The same cells were fixed and subjected to IF using the indicated antibodies. SGs were quantified as in Fig. 1 B. Mean values ± SD (error bars) are shown for three independent experiments. **, P < 0.01. ns, nonsignificant. Bars, 10 µm.
Next, we analyzed whether YB-1 instead affects G3BP1 protein synthesis. Because G3BP1 is a stable protein, we first monitored levels of newly synthesized G3BP1 in YB-1 kd and control cells using l-azidohomoalanine (AHA) incorporation. Cells were treated with AHA, and AHA-labeled newly synthesized proteins were biotinylated and subjected to immunoprecipitation with Streptavidin beads as described previously (Somasekharan et al., 2012). Newly synthesized G3BP1 was markedly reduced in YB-1 kd cells under both ambient conditions or after arsenite and H2O2 treatment compared with siControl cells (Fig. 2, E and F), which points to a role for YB-1 in promoting new G3BP1 protein synthesis. We therefore focused on this process to potentially explain the requirement for YB-1 in SG assembly. Indeed, several publications confirm that G3BP1 is both necessary and sufficient to induce SG formation (Tourrière et al., 2003; Reineke et al., 2012). We verified this in U2OS cells, as G3BP1 kd with two independent siRNAs (Fig. S3 D) prevented YB-1–, TIA-1–, FMRP-, and eIF3η-associated SG formation under arsenite stress in these cells (Fig. S3, E and F). Notably, G3BP1 kd did not alter protein levels of YB-1 (Fig. S3 G), which indicates that YB-1 functions upstream of G3BP1, rather than the reverse. Moreover, ectopic overexpression of G3BP1 in YB-1 kd cells (Fig. 2 G) restored SG formation (Fig. 2 H), which supports a model whereby SG formation via YB-1 is due largely to its ability to induce G3BP1 synthesis.
YB-1 promotes G3BP1 translation through the G3BP1 5′ UTR
Because YB-1 binds RNA and is a known translational regulator, we hypothesized that its effects on G3BP1 synthesis are through translational activation of G3BP1 mRNAs. To examine this, we first assessed if YB-1 physically interacts with G3BP1 transcripts in U2OS cells. We immunoprecipitated YB-1–associated mRNAs from cell extracts using YB-1 antibodies or control normal rabbit serum (NRS), and then assayed for the presence of YB-1–bound G3BP1 or control XIAP mRNAs using semiquantitative RT-PCR. As shown in Fig. 3 A, G3BP1 RT-PCR products were strongly enriched in siControl cell lysates after immunoprecipitation with anti–YB-1 but not nonspecific antibodies, whereas this enrichment was lost in siYB-1 cell immunoprecipitates. Next, to show that YB-1 associates with actively translated G3BP1 messages, we performed sucrose gradient centrifugation to isolate polysomal fractionated (ribosome-bound) mRNAs, as described previously (Evdokimova et al., 2009), followed by immunoprecipitation with anti–YB-1 antibodies to pull down YB-1 and its bound transcripts from polysomal fractions. This demonstrated that significant amounts of YB-1 are present in polysomes under both ambient and arsenite stress conditions, using RPS6 as a known polysome-associated control protein (Fig. S4 A). Moreover, semiquantitative RT-PCR showed that YB-1 associates with G3BP1 mRNAs in polysomes from U2OS cells, under both ambient and arsenite-treated conditions (Fig. 3 B). These results indicate that YB-1 interacts with actively translated G3BP1 mRNAs in cells even under ambient conditions, and that a pool of YB-1 remains available in polysomes in association with G3BP1 transcripts under arsenite stress.
Figure 3.
YB-1 regulates G3BP1 translation through the G3BP1 5′ UTR. (A) mRNA transcripts bound to YB-1 were riboimmunoprecipitated (RIPed) using anti–YB-1 antibodies or normal rabbit serum (NRS) from siControl and siYB-1 kd cell lysates. Captured mRNAs were reverse-transcribed and PCR amplified using primers specific for G3BP1 or XIAP as a control. (B) YB-1–bound mRNAs were RIPed using anti–YB-1 or control anti-GRB2 antibodies from polysomes prepared from vehicle alone and arsenite-treated U2OS cells, and subjected to semiquantitative RT-PCR using G3BP1- and XIAP-specific primers. (C) Constructs containing 5′ UTR sequences of G3BP1 (black) or β-Globin (gray) fused in frame to Luciferase were used for in vitro coupled transcription translation. Increasing concentrations of recombinant YB-1 were added to the assay mixture, and luciferase activity was measured. Error bars indicate SD. (D) RNA EMSA analysis to measure direct binding of YB-1 to the full-length G3BP1 5′ UTR. Biotin-labeled full-length G3BP1 5′ UTR mixed with recombinant GST-YB-1 was subjected to EMSA. The arrowhead indicates a probe mobility shift in the presence of 0.4 µg of GST-YB-1, and enhanced intensity at 0.8 µg of GST-YB-1. A 200-fold molar excess concentration of unlabeled full-length G3BP1 5′ UTR was added to demonstrate specificity of 5′ UTR G3BP1/YB-1 complex formation. As a control, recombinant GST was used in place of GST-YB-1. The broken line indicates that intervening lanes have been spliced out. (E) The full-length 5′ UTR G3BP1 (FL, 1–171) or deletion mutants (M1, Δ105–112; M2, Δ141–171; M3, Δ99–171; and M4, Δ141–171) were cloned in frame with Luciferase and used for in vitro coupled transcription/translation assays ±0.5 pmol YB-1 as described in C. Error bars indicate SD. (F) RNA EMSA showing that YB-1 binds to the full-length (FL, 1–171) G3BP1 5′ UTR but not M3 and M4 mutants. (G) Biotin end-tagged full length or the indicated deletion mutants of the G3BP1 5′ UTR were subjected to RNA affinity chromatography from U2OS lysates using Streptavidin beads. Affinity-purified proteins were immunoblotted using anti–YB-1 antibodies. Biotin end-tagged 5′ UTR of β-Globin was used as a control. (H) Full-length G3BP1 5′ UTR or the M4 deletion mutant (Δ48–171) were transfected into siControl or siYB-1 kd U2OS cells. Lysates were prepared and subjected to RNA affinity chromatography and immunoblotted as described in G to detect 5′ UTR–bound YB-1. Untransfected cells served as controls.
Because YB-1 binds 5′ UTRs of SNAIL1, TWIST, c-MYC, and other mRNAs to translationally activate these messages (Cobbold et al., 2008; Evdokimova et al., 2009), we wondered whether YB-1 influences G3BP1 mRNA translation by acting on the G3BP1 5′ UTR (171 nt; NM_005754). Using an in vitro cell-free translation system, increasing amounts of recombinant YB-1 were mixed with a Luciferase reporter linked to the 5′ UTR of G3BP1 mRNA, or of β-Globin as a negative control. YB-1 strongly and selectively induced translation of the 5′-UTR-G3BP1-Luc construct compared with 5′-UTR-β-Globin-Luc (Fig. 3 C). As a control, recombinant SRp55, another RNA-binding protein (Tran and Roesser, 2003), was substituted for YB-1 to determine the specificity of 5′ UTR stimulation, but SRp55 did not activate 5′-UTR-G3BP1-Luc translation (Fig. S4 B). Notably, further increases in YB-1 levels resulted in translational repression of both reporter mRNAs (Fig. 3 C), in agreement with YB-1’s known translation inhibitory activity (Evdokimova et al., 2006, 2009; Skabkin et al., 2006). In keeping with this, increased levels of YB-1 overexpression reduce G3BP1 expression and reverse SG formation in cellulo (Fig. S4, C and D). These results clearly indicate that cellular G3BP1 protein expression is strongly linked to levels of YB-1.
YB-1 directly binds to the 5′ UTR of G3BP1
We next tested if YB-1 directly binds to the G3BP1 5′ UTR by using an RNA electrophoretic mobility shift assay (EMSA). As shown in Fig. 3 D, we observed that recombinant GST-tagged YB-1 formed a specific complex with a biotin-tagged probe consisting of the full-length G3BP1 5′ UTR using EMSA; this was not observed with GST alone. Moreover, complex formation was completely blocked when a 200-fold molar excess concentration of unlabeled probe was added, demonstrating the specificity of 5′ UTR G3BP1/YB-1 complex formation and confirming that YB-1 directly binds to the G3BP1 5′ UTR. We then cloned the 5′ UTR into a bicistronic vector in a promoterless region upstream of Firefly luciferase, in which Renilla luciferase was under separate control by the SV40 promoter (Fig. S4 E; Young et al., 2008), and measured Firefly/Renilla luciferase ratios (Fluc/Rluc). As shown in Fig. S4 E, the G3BP1 5′ UTR strongly induced Fluc activity under both ambient and arsenite stress conditions, which suggests that it may possess internal ribosome entry site (IRES)-like activity.
To further analyze this UTR, we generated G3BP1 5′ UTR deletion mutants and linked them to Luc reporters, including mutant M1 lacking nt 105–112 (Δ105–112) encoding a previously reported putative YB-1–binding motif (Paranjape and Harris, 2007), M2 (Δ141–171), M3 (Δ99–171), and M4 (Δ48–171; Fig. 3 E). Predicted structures were generated using VARNAGUI (http://varna.lri.fr) and melting temperatures were calculated using RNAfold (http://rna.tbi.univie.ac.at); they are shown in (Fig. S4, F–J). Mutants were then compared with the full-length G3BP1 5′-UTR construct for in vitro translation activity using the cell-free translation system plus/minus recombinant YB-1 (see Materials and methods). While translation of the M2 mutant was only modestly reduced compared with the full-length 5′-UTR, translation of M3 and M4 mutant constructs was dramatically decreased (Fig. 3 E). This was validated in cellulo by transfecting full-length and M4 mutant 5′-UTR-Luc construct into siYB-1 and siControl cells and monitoring reporter activity. Again, full-length G3BP1 5′ UTR showed significantly higher activity in siControl compared with siYB-1 cells, whereas the M4 mutant construct was only poorly translated and not affected by YB-1 expression (Fig. S4 K). Consistent with these results, M3 and M4 mutants failed to bind YB-1 in RNA EMSA experiments (Fig. 3 F). Together, these results indicate that YB-1 directly binds to the 5′ UTR of G3BP1 transcripts to translationally activate expression of the G3BP1 nucleating factor. Although these studies implicate distal portion of the 5′ UTR for YB-1 activation, further studies are necessary to determine the specific UTR sequences or structures bound by YB-1.
Finally, we performed RNA affinity chromatography (Baird et al., 2007) from U2OS cell lysates using biotin end-tagged full-length or G3BP1 5′ UTR deletion mutants as bait. YB-1 strongly interacted with the full-length and M2 mutant 5′ UTR, but binding was markedly reduced in M3 and M4 mutants that failed to activate translation in the reporter assays as described in the previous paragraph (Fig. 3 G). The 5′ UTR of β-Globin was used as a control, and showed only background binding of YB-1. To validate this in cellulo, we used a novel in vivo RNA transfection technique to directly transfect biotin end-tagged versions of the G3BP1 5′ UTR into siYB-1 and siControl U2OS cells, followed by RNA affinity chromatography and YB-1 immunoblotting. We observed strong binding of YB-1 to the full-length G3BP1 5′ UTR in siControl U2OS cells, which, as expected, was lost after YB-1 kd. However, the M4 mutant 5′ UTR of G3BP1 failed to bind YB-1 in cellulo (Fig. 3 H). Together, these results provide strong evidence that YB-1 directly binds to the G3BP1 5′ UTR and positively regulates G3BP1 translation.
YB-1 regulates G3BP1 expression in vivo
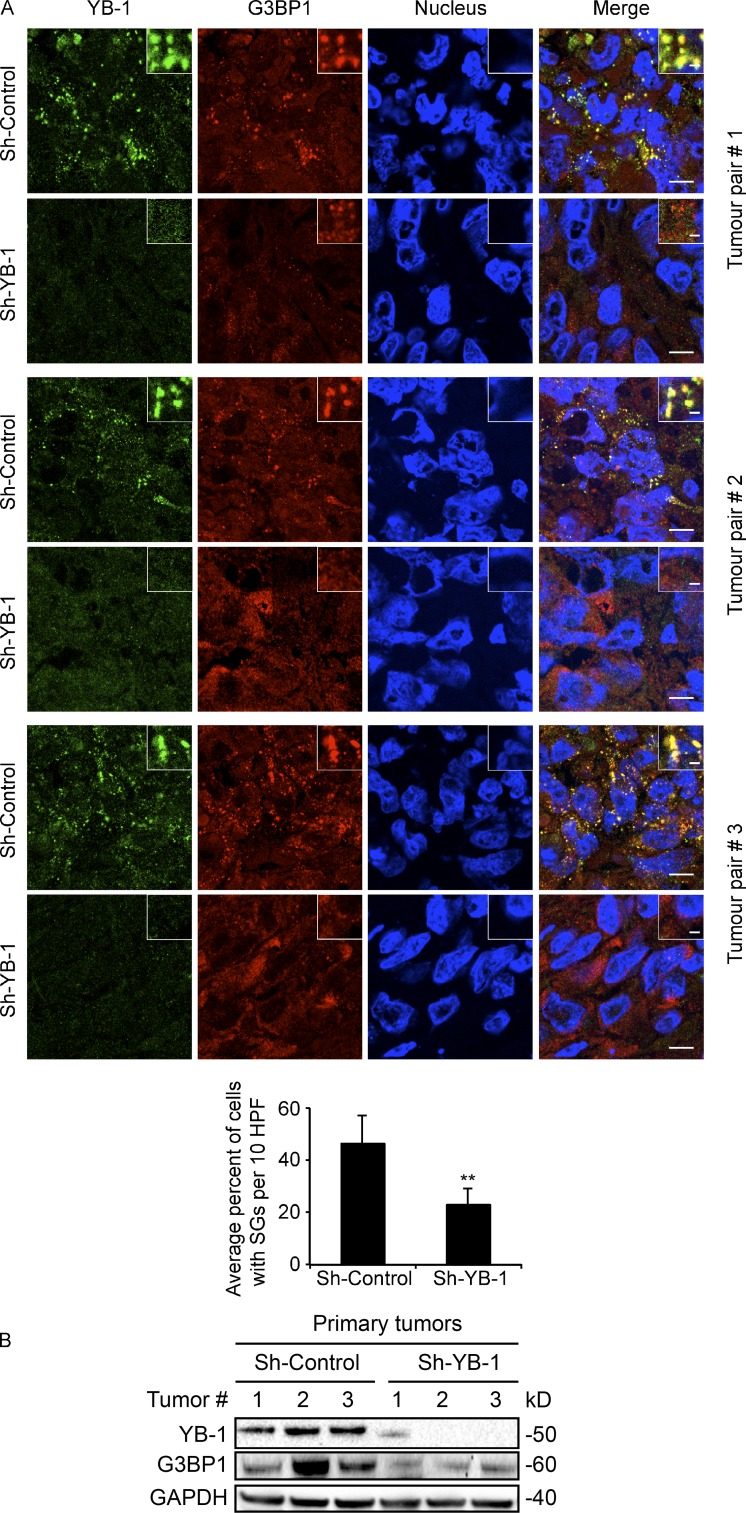
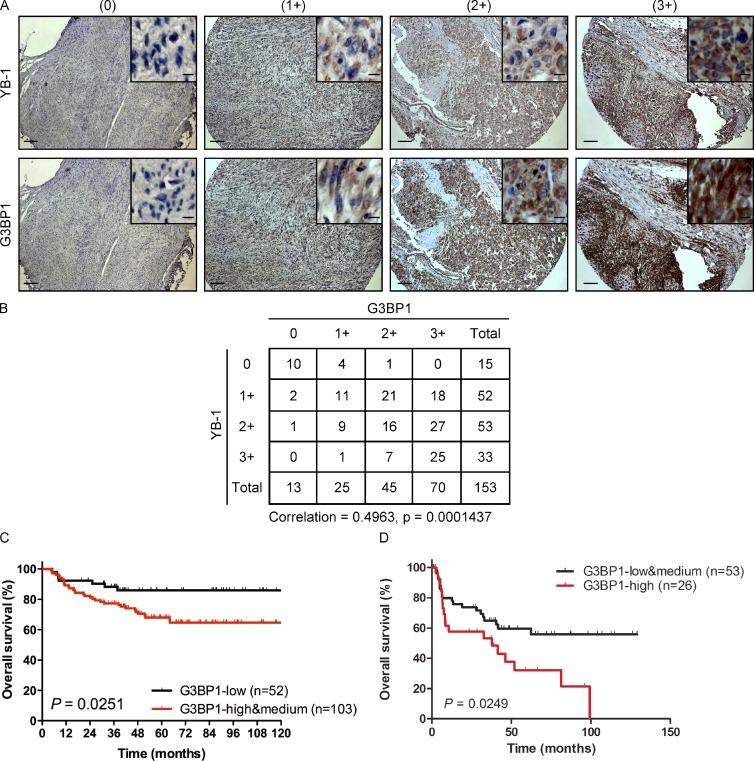
SG formation in vivo has previously been documented in pathological tumor and brain tissues (Moeller et al., 2004; Vanderweyde et al., 2012). Given that YB-1 regulates SG formation in cultured sarcoma cells, we next wished to determine the relevance of YB-1 for SG formation in vivo. We therefore used a murine renal subcapsular implantation model to establish human sarcoma xenografts in mice as described previously (Mendoza-Naranjo et al., 2013). MNNG osteosarcoma cell lines transfected with control or YB-1 shRNAs for stable YB-1 kd were implanted under the renal capsules of immunocompromised mice, and implantation site tumors were surgically removed after 4–5 wk. We confirmed that this model is appropriate for analysis of stress responses, as areas of hypoxia (detected by pimonidazole staining; Raleigh et al., 1999), ER stress (detected by increased expression of the ER stress marker, Bip; Boelens et al., 2007), and oxidative stress (detected by protein carbonylation; Dalle-Donne et al., 2006) could be readily detected in tumors but not in normal mouse kidney (Fig. S5, A–C). Viable regions of the excised tumors were examined by immunoblot analysis, in order to assess SG associated proteins, and by cryosectioning to detect SGs (Figs. 4 A and S5 D). This clearly demonstrated cytosolic SGs in control tumor sections, with colocalization of YB-1 and G3BP1, FMRP, and eIF3η. However, SGs were significantly reduced in MNNG tumors with stable YB-1 kd. Reduced expression of YB-1 in tumor lysates was confirmed by immunoblotting, and, importantly, YB-1 kd strongly correlated with reduced G3BP1 expression in the same lysates (Figs. 4 B and S5 E). Second, we assessed whether YB-1 and G3BP1 protein expression are correlated in human sarcoma specimens using tissue microarrays (TMAs) of 153 different human sarcoma cases (Fig. 5, A and B). Serial sections of TMAs were subjected to sequential immunohistochemistry (IHC) using antibodies to YB-1 and G3BP1. YB-1 expression strongly correlated with G3BP1 in human sarcomas (Spearman correlation of 0.4963 and a p-value = 0.0001437), which is consistent with our in vitro and in vivo results and supports a positive role for YB-1 in regulating G3BP1 expression in human sarcomas. Moreover, G3BP1 expression in sarcoma patient samples strongly correlates with survival (Fig. 5, C and D), highlighting G3BP1 as a potential prognostic marker in sarcomas. Together, these results provide compelling evidence that YB-1 is a critical regulator of G3BP1 levels and SG formation in vivo.
Figure 4.
YB-1 regulates G3BP1 expression in vivo. (A) MNNG shControl or shYB-1 kd cells were implanted under the renal capsules of three independent pairs of NOD/SCID mice, and primary site tumors were collected 4–5 wk after implantation. Viable regions of tumors were cryosectioned and subjected to IF with anti–YB-1 (green) or anti-G3BP1 (red) antibodies, and counterstained with DRAQ5 to detect nuclei. Insets show fivefold higher magnification of indicated areas. 10 high-power fields (HPF) of representative tumor sections from each implantation site tumor (n = 3 per group) were used for quantification of SGs as in Fig. 1 B, as shown below with a bar graph. Mean values ± SD (error bars) are shown for three independent tumors. **, P < 0.01. Bars: (main panels) 10 µm; (enlarged insets) 1 µm. (B) Lysates extracted from tumor tissues from A were subjected to immunoblot analysis using the indicated antibodies.
Figure 5.
Expression of G3BP1 correlates with YB-1 and is linked to poor outcomes. (A) Serial sections of three TMAs containing 153 different human sarcoma cases were subjected to sequential IHC using antibodies to YB-1 or G3BP1 as indicated. Panels show expression of YB-1 and G3BP1 by IHC in four different representative sarcoma cases. Insets show 10-fold higher magnification of representative areas. Immunostaining for each protein was scored as no (0), low (1+), moderate (2+), or high expression (3+) as indicated. Bars: (main panels) 100 µm; (enlarged insets) 20 µm. (B) Tabulated values from IHC scoring of A, with a Spearman’s rank correlation coefficient between expression of each protein of 0.4963 and a p-value of 0.0001437. (C and D) Kaplan-Meier estimates for overall survival (event = deceased) in relation to G3BP1 expression for Ewing sarcoma patients with localized disease (high and medium vs. low G3BP1 expression; C) and metastatic disease (high vs. low and medium G3BP1 expression; D).
G3BP1 regulates in vivo SG formation, invasion, and metastasis
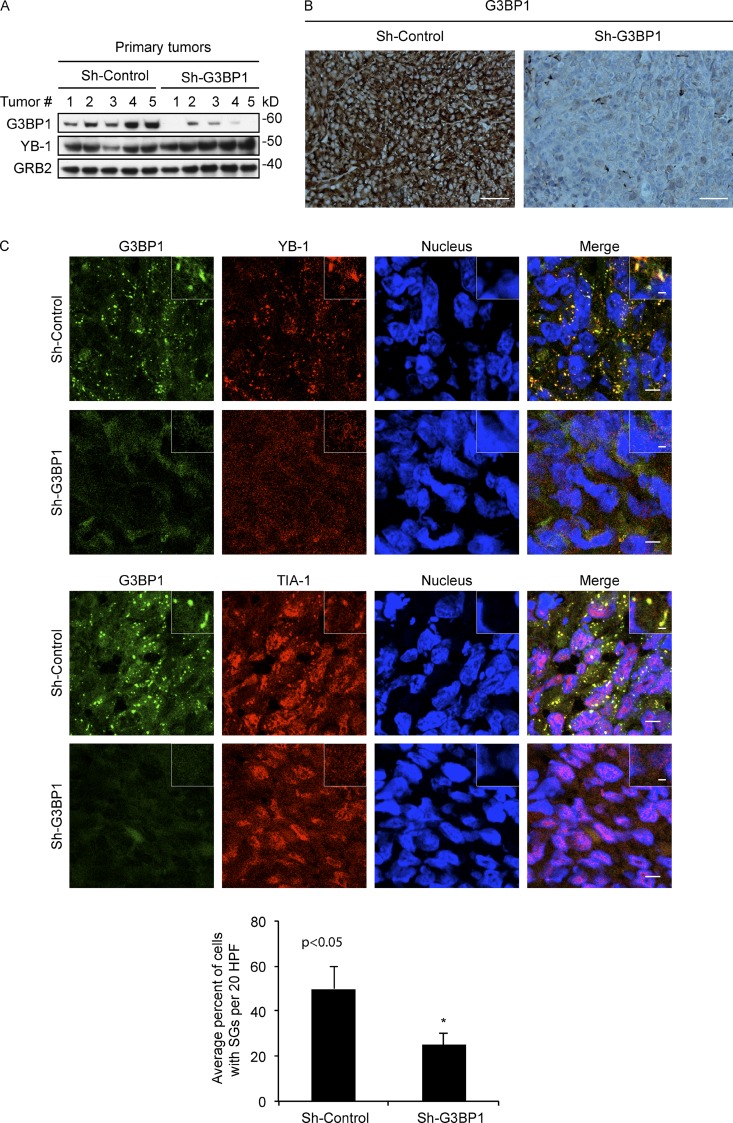
Given the link between G3BP1 expression and poor prognosis in sarcomas (Fig. 5, C and D), we wondered whether SG assembly is linked to cancer progression. To test this, we used the renal subcapsular implantation model to implant MNNG cells with or without stable G3BP1 kd under mouse renal capsules, and monitored in vivo SG formation, local invasion, and metastasis to the lungs. We previously used this model to monitor metastatic capacity of human sarcoma cell lines in vivo (Mendoza-Naranjo et al., 2013). By analyzing viable regions of implantation site tumors, we confirmed that expression levels of G3BP1 were significantly reduced in kd as compared with control tumors by immunoblotting (Fig. 6 A) and IHC (Fig. 6 B). In addition, IF of tumor cryosections showed that SGs were significantly reduced in G3BP1 kd tumors compared with controls (Fig. 6 C). Notably, histological analysis showed highly invasive local growth of control tumors into adjacent normal kidney (Fig. 7 A, left), but this feature was dramatically inhibited in G3BP1 kd tumors (Fig. 7 A, right). In fact, the tumor margins of the latter were reverted to noninvasive so-called “pushing” borders abutting but not penetrating neighboring normal kidney, which is characteristic of a more benign growth pattern (Fig. 7 A, right). We did not detect differences in cell migration or invasion between G3BP1 kd and control MNNG cells in vitro (unpublished data), which suggests that the SG effects on cell motility are an in vivo property. Primary tumors with G3BP1 kd also showed significant increases in necrosis, as determined morphologically (Fig. 7 B). Notably, although control MNNG tumors metastasized to lungs within the 4–5 wk timeframe before mice had to be sacrificed, G3BP1 kd completely blocked MNNG metastasis to lungs (Fig. 7 C). Importantly, there were no differences in proliferation rates between control and G3BP1 kd in implantation site tumors, as assessed by expression of the Ki67 proliferation marker (Fig. 7 D); i.e., enhanced metastasis of control tumors was not due to increased proliferation rates. These results show for the first time, to our knowledge, that SG assembly is associated with tumor invasion and metastasis.
Figure 6.
G3BP1 regulates SG formation in vivo. (A) MNNG shControl or shG3BP1 cells were implanted under the renal capsules of NOD/SCID mice (eight mice per group), and primary site tumors were collected 4–5 wk after implantation. Lysates extracted from tumor tissues were immunoblotted using the indicated antibodies. (B) Viable regions of shControl and shG3BP1 tumors were cryosectioned and subjected to G3BP1 IHC. Bars, 100 µm. (C) Viable regions of tumors were cryosectioned and subjected to IF with the indicated antibodies and DRAQ5 to counterstain nuclei. Insets show fivefold higher magnification of representative areas. Quantification of SGs as in Fig. 1 B is shown below as a bar graph, in which 20 high-power fields of representative tumor sections from each tumor group (n = 5) were used for SG quantification. Mean values ± SD (error bars) are shown. *, P < 0.05. Bars: (main panels) 10 µm; (enlarged insets) 1 µm.
Figure 7.
G3BP1 kd inhibits local invasion and lung metastasis. (A) H&E-stained representative sections of implantation site tumors from NOD/SCID mice bearing renal subcapsular tumor xenografts of MNNG cell lines with (shG3BP1) or without (shControl) G3BP1 kd. Arrowheads show highly invasive growth pattern of shControl tumors (left) and noninvasive borders of shG3BP1 tumors (right). (B) H&E-stained sections of MNNG shControl and shG3BP1 tumors, with asterisks showing microscopic areas of necrosis. The latter is quantitated (bar graph, right) by measuring average percent geographical necrosis in 20 high-power fields from each tumor type (n = 5 tumors per group). Mean values ± SD (error bars) are shown. *, P < 0.05. (C) H&E-stained lung sections from mice bearing renal subcapsular tumor xenografts of MNNG cells with (shG3BP1) or without (shControl) G3BP1 kd. The arrowhead (left) shows a metastatic pulmonary lesion. The bar graph shows the total number of mice with at least one lung metastasis (Mets positive, red) in the indicated groups (n = 8 mice/group). Mean values ± SD (error bars) are shown. *, P < 0.05. (D) Primary site tumors with (shG3BP1) or without (shControl) G3BP1 kd were stained with antibodies to the Ki67 proliferation marker. The percentage of Ki67-positive cells in each group is quantitated in the bar graph (right) by counting cells in 20 high-power fields from each tumor type (n = 5 tumors per group). ns, nonsignificant. Error bars indicate SD. Bars, 100 µm.
Discussion
Translation of cellular mRNAs is tightly regulated to allow rapid adaptation to environmental changes. In response to adverse conditions such as oxidative and metabolic stress or hypoxia, overall mRNA translation is inhibited to preserve energy balance and prevent accumulation of damaged and misfolded proteins. Nevertheless, a subset of these proteins are still synthesized, including stress response proteins that protect and repair cells from damage. Induction of SGs contributes to this process by providing temporary storage for translationally inhibited transcripts, preserving them for eventual translation once the stress is removed (Kedersha et al., 2013). Here we show that the YB-1 translation factor is critical for SG assembly in cultured human sarcoma cells under diverse stress forms, as well as in sarcoma xenografts using in vivo mouse models. Moreover, YB-1 provides cellular protection from oxidative stress and other adverse conditions. Mechanistically, this occurs through direct binding to the 5′ UTR of G3BP1 to stimulate its translation. Moreover, stable G3BP1 kd in human MNNG osteosarcoma cells significantly reduces in vivo SG formation and local invasive capacity in renal subcapsule tumor xenografts, without changing tumor proliferative rates. Importantly, xenograft tumors lose their capacity to metastasize to lungs when G3BP1 is inactivated. To our knowledge, this is the first report to show that SGs are important for tumor progression.
Controlling levels of SG-associated proteins critically impacts SG formation, as many of these proteins provide structural scaffolds for SG assembly. TDP-43, an RNA-binding protein required for SG formation, influences SG dynamics via transcriptional regulation of SG-nucleating proteins such as G3BP1 and TIA-1, and G3BP1 transcripts levels and SG formation are drastically reduced in TDP-43–deficient cells (McDonald et al., 2011). Prolonged infection with poliovirus results in cleavage of G3BP1 by viral 3C proteinase, leading to dispersal of SGs, but expression of a cleavage-resistant G3BP1 restores SG formation during poliovirus infection (White et al., 2007). HIV1 viral infection blocks SGs by recruiting G3BP1 away from preformed SGs (Valiente-Echeverría et al., 2014). Apart from such mechanisms, regulation of SG protein expression is poorly understood, especially at the level of mRNA translation. Direct translational induction of G3BP1 mRNA by YB-1 therefore provides a potential new mechanism to regulate SG formation.
YB-1 binds to the 5′ UTR of G3BP1 transcripts to up-regulate translation, whereas YB-1 down-regulation compromises G3BP1 protein synthesis and SG formation. Cell-free in vitro translation assays demonstrate that the G3BP1 5′ UTR is sufficient for direct YB-1–mediated translational control, and deletion mutations of this region abrogate YB-1–mediated translational activation. Whether this UTR contains IRES-like regions, as suggested by our data (Fig. S4 E), remains to be rigorously determined, as does the specific mechanism of YB-1–mediated activation. Like SNAIL1 and TWIST 5′ UTRs, the G3BP1 5′ UTR is predicted to form complex stem loops, and highly stable stem-loop structures are known to decrease translational efficiency (Davuluri et al., 2000; Vogel et al., 2010; Araujo et al., 2012). Therefore, one feasible explanation is that YB-1 binds to and unwinds these secondary structures to allow for more efficient G3BP1 translation. This is consistent with our observation of YB-1–mediated translational activation of SNAIL1 and TWIST (Evdokimova et al., 2009), particularly given that YB-1 has known helicase activity (Skabkin et al., 2004). However, more studies are necessary to confirm this, and also to determine where YB-1 specifically interacts along the G3BP1 5′ UTR. It is reasonable to predict that this 5′ UTR and those of other mRNA targets such as SNAIL1, TWIST, and c-MYC (Cobbold et al., 2008; Evdokimova et al., 2009) possess specific architectural features or regulatory motifs that mediate binding and translational regulation by YB-1. However, we have to date failed to identify such motifs, other than that nucleotides 99–171 of the G3BP1 5′ UTR appear to be critical for YB-1–mediated translational activation.
A role for YB-1 in SG formation in tumor cells in vivo has potential pathological relevance, given that both YB-1 and its translational target G3BP1 are associated with cancer. YB-1 is an established metastatic marker, and high expression of YB-1 correlates with tumor aggressiveness and poor patient survival in diverse tumor types (Kohno et al., 2003; Lasham et al., 2013). G3BP1 is markedly overexpressed in human tumors, including those of the breast, head, neck, colon, and thyroid (Guitard et al., 2001; French et al., 2002). Expression of G3BP1 is associated with lymph node metastasis and survival in esophageal squamous cell carcinoma (Zhang et al., 2007). This suggests that G3BP1 has pro-oncogenic functions, similar to YB-1 (Evdokimova et al., 2009). Accumulating evidence suggests that SG formation is protective against stress-induced cell damage and death (Arimoto et al., 2008; Tsai and Wei, 2010), but only a few studies implicate SGs in cancer biology (Moeller et al., 2004; Fournier et al., 2010). We found that reducing YB-1 expression in sarcoma cell lines decreases SG formation in vivo, and markedly increases their sensitivity to oxidative stress in vitro. Therefore, YB-1 may exert its protective effects against oxidative stress at least in part by facilitating SG assembly, providing tumor cells with protection during chemotherapy and radiation, therefore supporting their survival and progression.
Exactly where in the cell YB-1 activates G3BP1 translation remains unknown. Although YB-1 localizes to SGs under cell stress, it is likely that such localization occurs independently of its ability to activate G3BP1 translation, as SGs are themselves translationally inactive (Anderson and Kedersha, 2006). Indeed, YB-1 association with SGs is likely due to its general RNA binding activity (Lyabin et al., 2014) and hence its association with SG-resident mRNAs. In fact, although we see interactions between YB-1 and SG proteins such as G3BP1 and TIA-1, these interactions are lost when lysates are treated with RNase A (i.e., such interactions are RNA dependent; unpublished data). Because G3BP1 transcripts were present among YB-1–bound mRNAs immunoprecipitated from polysomes of arsenite-treated cells, at least a fraction of G3BP1 mRNAs are excluded from SGs, in addition to other transcripts such as those encoding chaperones and repair enzymes that ensure cell survival under acute cell stress (Arimoto et al., 2008; Yamasaki and Anderson, 2008). This also implies that a pool of cytosolic YB-1 exists outside of SGs under cell stress that can be accessed for translational activation of stress-related mRNAs. What regulates this distribution is a critical question that is currently under investigation in our laboratory.
Also unexplained is how SG formation promotes invasion and metastasis. One possibility is that SGs might sequester mRNAs encoding factors that inhibit invasion and metastasis. This would sustain these mRNAs in a silenced state to provide SG-proficient cells with an enhanced capacity to invade and metastasize. It is also possible that SGs can selectively release mRNAs encoding matrix-degrading enzymes for translation to facilitate invasive capacity. Supporting this hypothesis, we found that control primary implantation site tumors could efficiently invade neighboring kidney, whereas in vivo G3BP1 kd reverted the growth pattern to noninvasive borders that did not penetrate neighboring normal kidney. Collectively, our study highlights YB-1 as a critical regulator of SG assembly through its ability to enhance G3BP1 translation to facilitate SG nucleation, providing new insights into SG biology and its potential link with cancer progression.
Materials and methods
Reagents
DMEM, MEM, Roswell Park Memorial Institute Medium (RPMI), l-glutamine, FBS, and trypsin were from Sigma-Aldrich. Antibodies against YB-1 (rabbit), biotin HRP-linked antibody (goat), ribosomal protein S6 (RPS6; rabbit), PARP (rabbit), caspase-3 (rabbit), c-Jun (rabbit), Myc-tag (rabbit), PABP1 (rabbit), eIF2α (rabbit), eEF2 (rabbit), and GAPDH (rabbit) were from Cell Signaling Technology. Antibodies against G3BP1 (mouse) and GRB2 (mouse), FACS tubes, Annexin V-FITC, and propidium iodide were from BD. Antibodies against YB-1 (mouse), TIA-1 (goat), TIAR (goat), eIF3η (goat), FMRP (FMR1; goat), and FXR1 (goat); and siRNAs against G3BP1 (sc-75076-A and C) and shRNA lentiviral particles (consisting of a pool of 3–5 lentiviral vector plasmids) against G3BP1 and YB-1 were from Santa Cruz Biotechnology, Inc. Antibodies against DDX6 (rabbit) and G3BP1 (rabbit) were from Novus Biologicals. Fluorescently labeled secondary antibodies (mouse, Alexa Fluor 488/594; rabbit, Alexa Fluor 488/594; and goat, Alexa Fluor 488/594), Click-iT Protein Reaction Buffer kit, biotin alkyne, l-azidohomoalanine (AHA), TRIzol, and Protein A Magnetic Beads were from Life Technologies. TransIT-mRNA transfection reagent was from Mirus. siRNAs against YB-1 (siRNA1, 5′-UGACACCAAGGAAGAUGUA-3′; siRNA 2, 5′-GUGAGAGUGGGGAAAAGAA-3′) were from GE Healthcare. TranscriptAid T7 High Yield Transcription kit, RNA 3′ End Biotinylation kit, LightShift Chemiluminescent RNA EMSA kit, and Zeba Spin desalting columns (7K MWCO, 2 mL) were from Thermo Fisher Scientific. Myc-DDK–tagged G3BP1 (CMV promoter) and Myc-DDK-tagged TIA-1 (CMV promoter) were from OriGene. 5-FAM-oligodT probe was from Integrated DNA Technologies. RIP-Assay Starter kit YBX1 was from MBL International Corporation. QuantiTect Reverse Transcription kit was from QIAGEN. FluorSave was from Merck. TNT Quick Coupled Transcription/Translation system, Bright-Glo Luciferase Assay System, and RNasin (RNA inhibitor) were from Promega. Recombinant YB-1 was from EMD Millipore. The LightSwitch Luciferase Assay kit was from Switchgear Genomics, and the Protein Carbonyl Colorimetric Assay kit was from Cayman Chemical.
Cell culture
U2OS cells were cultured in DMEM, DU-145 and MNNG cells were cultured in MEM, and RH-30 cells were cultured in RPMI. All media were supplemented with 10% FBS and 1× penicillin/streptomycin (Life Technologies).
Stress treatments and apoptosis assays
Stress treatments were performed by treating cells with vehicle alone or different stress inducers such as arsenite (0. 5 mM for 1 h), H2O2 (0.5 mM for 1 h), thapsigargin (10 µM for 1 h), UV rays (90 mJ/cm2 using a UV Stratalinker 2400 apparatus from Stratagene followed by 1 h incubation), heat shock (42°C for 1 h), or hypoxia (1% O2 for 6 h). For measuring apoptosis, cells transfected with siControl or siYB-1 siRNAs were treated with vehicle alone, arsenite (0.5 mM), or H2O2 (0.5 mM) for 5 h or hypoxia (1% O2) for 24 h. Cells were then harvested, washed with PBS, and suspended in Annexin V-FITC binding buffer (0.01 M Hepes/NaOH, pH 7.4, 0.14 M NaCl, and 2.5 mM CaCl2) at a concentration of 106 cells/ml. 100 µl of cell suspensions were transferred into a 5 ml FACS tube, and 5 µl of Annexin V-FITC and 5 µl of propidium iodide (1 mg/ml) were added. Cells were then incubated in the dark at room temperature for 15 min before analyzing apoptosis by fluorescence using FACSCalibur (BD).
Microscopy
Cells seeded at 20–25% confluence in 6-cm culture dishes containing round cover glasses (12CIR-1D; Thermo Fisher Scientific) were transfected with scrambled, YB-1, or G3BP1 siRNAs. 3 d after transfection, cells were treated with vehicle alone or exposed to different stress conditions. IF was performed as described previously (Somasekharan et al., 2013). Cells were fixed in 4% paraformaldehyde (PAF) for 20 min and permeabilized with PBS-T (0.05% Triton X-100 in PBS) for 20 min. The cells were then blocked for 30 min in PBS-T containing 5% BSA and incubated with primary antibodies (1:100) for 1 h in PBS containing 2.5% BSA. Cells were washed in PBS-T for 30 min (3 × 10 min) followed by incubation with secondary antibodies (1:200) in PBS-T containing 2.5% BSA for 1 h. Cells were then washed in PBS-T for 30 min (3 × 10 min). For the staining of in vivo SGs, cryosections were cut at 4 µm thickness and were processed for IF as described earlier in this paragraph. For the localization of mRNA in the SGs, in situ hybridization was performed as described previously (Khong and Jan, 2011). In brief, coverslips containing cells were incubated in hybridization buffer (2× SSC, 20% formamide, 0.2% BSA, and 1 µg/µl yeast tRNA) for 15 min at 37°C. Subsequently, cells were hybridized with 5-FAM-oligodT at 37°C. After 24 h of incubation, cells were washed twice with 2× SSC and 20% formamide for 5 min at 37°C, twice with 2× SSC for 5 min each at 37°C, and once with 1× SSC for 5 min at 37°C. The cells were then counterstained with antibodies as described. All the cells processed as in this paragraph were immersed in DRAQ5 (10 µM; Biostatus) for nuclear staining, mounted with FluorSave, and viewed using an inverted confocal microscope (Eclipse Ti-E; Nikon) with 40× and 100× oil-immersion objective lenses. Images were captured using EZ-C1 software and were further processed using ImageJ software.
For live cell imaging, U2OS cells were cotransfected with GFP-YB-1 and RFP-G3BP1. After 24 h of transfection, cultures were placed in a 37°C chamber equilibrated with humidified air containing 5% CO2. Before live cell imaging, the media was changed to fresh DMEM containing 0.5 mM arsenite. Time-lapse images were taken with a microscope (Axio Observer Z1; Carl Zeiss) using a 40× objective lens and analyzed by AxioVision software. Images were captured at every 2 min for 1 h, and the movies were generated with the time-lapse series using ImageJ software.
For the detection of necrosis, invasion, and metastasis, paraffin-embedded primary tumor and the lung sections were stained with hematoxylin and eosin (H&E). Pimonidazole staining of hypoxic regions in the tumor was performed as described previously (Bennewith and Durand, 2004). Pimonidazole hydrochloride (20 mg/ml was dissolved in double-distilled water) was injected i.p. at a dose of 100 mg/kg mouse body weight. Tumors were then collected, cryosectioned, and stained with DAPI before observing them in a fluorescence microscope. For IHC of patient tumors, three TMAs containing 153 different human sarcoma samples were used, including SFT961 containing a broad variety of low- and high-grade sarcoma types, supplied by US Biomax, and two TMAs predominantly containing myxoid liposarcoma and synovial sarcoma samples, provided by T. Nielsen (Genetic Pathology Evaluation Centre, University of British Columbia), were subjected to IHC using anti–YB-1 (rabbit) or anti-G3BP1 (rabbit) antibodies. Sections were counterstained with hematoxylin and viewed using a microscope (Axio Observer Z1; Carl Zeiss) with 10× and 40× objective lenses. Images were captured using AxioVision software (Carl Zeiss). A board-certified pathologist scored the staining for each antibody as 0 (staining absent), 1+ (staining in <10% of cells), 2+ (staining in 10–50% of cells), and 3+ (staining in >50% of cells). A Spearman’s rank correlation coefficient analysis was performed to assess correlations between expressions of each protein.
Quantification of SGs, lung metastases, necrosis, and Ki67 staining
The number of SG-bearing cells was quantified using the ImageJ software. IF images were randomly taken with a 40× objective lens in 20 different fields. The plug-in “Analyze particles” was used to count the number of SGs. The percentage of cells with SGs was calculated by counting the number of cells displaying SGs and expressing them as 100 × [(number of cells with SGs)/(total number of cells)]. For assessing pulmonary metastases, 4-µm-thick sections of H&E-stained lung tissues were histologically analyzed by a board-certified pathologist, and the number of mice/condition (n = 8) bearing lung metastases was calculated. The percentage of tumor necrosis was measured using ImageJ software. 20 randomly chosen high-power fields (40×) per tumor section (n = 5) were analyzed. Areas of whole and necrotic tumor surfaces were measured and the percentage of necrotic area was calculated as 100 × (necrotic area/total tumor area). Cell proliferation was assessed using proliferation marker protein Ki67. Quantification of proliferating cells positively stained with Ki67 was conducted by analyzing 20 randomly chosen high-power fields (40×) per tumor section (n = 5). The ImageJ plug-in “ImmunoRatio” directly provided the percentage of Ki67-positive cells.
Immunoblot analysis
Cells were gently scraped off from the culture dishes with a cell scraper, washed with PBS, and lysed using lysis buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% Triton X-100, and 1× protease inhibitor). Cell lysates were centrifuged at 14,000 rpm for 10 min and the supernatant was saved. Protein concentration was determined using a Bradford assay (Bio-Rad Laboratories). Protein lysates were mixed with 2× loading dye, and equal amount of proteins were separated in 4–12% gradient SDS-PAGE and immunoblotted into nitrocellulose membrane using wet transfer as described previously (Montessuit et al., 2010). Quantification of newly synthesized proteins was performed as described previously (Somasekharan et al., 2012). In brief, cells were treated with vehicle alone, arsenite, or H2O2 for 1 h in DMEM containing AHA in place of methionine. AHA-labeled cells were suspended in lysis buffer (1% SDS in 50 mM Tris, pH 7.5). The cell lysates were subjected to click reaction using biotin-alkyne to add a biotin tag to AHA. The click reaction products were then passed through Zeba Spin Desalting Columns (7K MWCO, 2 ml) to remove the unincorporated reaction components, and to replace the lysis buffer with PBS. The newly synthesized proteins were then affinity purified on magnetic beads (Streptavidin) and subjected to immunoblot analysis. For the CHX treatment, siControl and siYB-1 cells were treated with vehicle alone or 100 µM CHX for 2, 4, 8, 10, and 24 h. Lysates were extracted from these cells and subjected to immunoblot analysis.
Isolation of RNA, riboimmunoprecipitation (RIP), and RT-PCR
Total RNA was extracted using TRIzol reagent according to the manufacturer’s instructions. The direct interaction between G3BP1 mRNA and YB-1 was analyzed by RIP using a RIP-Assay Starter kit YBX1 according to the manufacturer’s instructions. In brief, U2OS cells (1.5 × 107) transfected with siControl or siYB-1 siRNAs were harvested, washed with PBS, and lysed in 500 µl lysis buffer containing 1× protease inhibitor and 5 mM DTT followed by centrifugation at 12,000 g for 5 min. The supernatant was then mixed with 25 µl protein A magnetic beads and precleared for 1 h. In parallel, protein A magnetic beads (25 µl) suspended in lysis buffer were incubated with either anti–normal rabbit serum (anti-NRS; nonspecific control) or anti–YB-1 with gentle agitation for 1 h. Subsequently, beads were washed once with lysis buffer and incubated with 500 µl of precleared cell lysate with gentle agitation for 3 h. Beads were then washed four times with wash buffer and the RNA was extracted using 400 µl of master solution and precipitated with 600 µl of 2-propanol. The pellet was washed twice with 0.5 ml of 70% ethanol, dried, and dissolved in nuclease-free water. The amount of RNA was quantified using NanoDrop (Thermo Fisher Scientific). cDNAs were synthesized using the QuantiTect Reverse Transcription kit. Equal amounts of cDNAs were subjected to semi-quantitative PCR or RT-PCR in an ABI PRISM 7500 Sequence Detection System (Applied Biosystems) using the following primers. For G3BP1, 5′-ATGCAGTCTACGGACAGAAAGA-3′ (forward) and 5′-GAGCATCAACATGGCGAATCT-3′ (reverse); for XIAP, 5′-GACAGTATGCAAGATGAGTCAAGTCA-3′ (forward) and 5′-GCAAAGCTTCTCCTCTTGCAG-3′ (reverse). For the RIP of YB-1–bound transcripts from polysomes, U2OS cells were treated with vehicle alone or arsenite, cell lysates were prepared, and polysomes were purified as described previously (Evdokimova et al., 2006). In brief, the cell lysates were centrifuged at 10,000 rpm for 15 min to remove the nuclei and mitochondria, and the supernatants were then centrifuged at 100,000 rpm in a TLA-100 centrifuge (Beckman Coulter) for 15 min to separate postpolysomal supernatant and polysomes. Additional purification of polysomes was then performed by centrifuging the resuspended pellet through a 30% sucrose cushion for 15 min at 100,000 rpm. The isolated polysomes were subjected to RIP using anti–YB-1 or anti-GRB2 antibodies as described in this paragraph, and the extracted mRNAs were subjected to semiquantitative PCR using primers for G3BP1 or XIAP (nonspecific control).
Luciferase reporter assays
Reporter constructs for the in vitro luciferase assays were custom-made from GenScript. The 5′ UTR region of full-length G3BP1 (FL, 1–171) or deletion mutants (M1, Δ105–112; M2, Δ141–171; M3, Δ99–171; M4, Δ48–171), linked to T7 promoter sequences at their 5′ ends, were gene synthesized and cloned into the BglII–NcoI site upstream of RenSP (optimized luciferase gene) in pLightSwitch_5′ UTR vector with ACTB promoter (SwitchGear Genomics). Plasmid DNA (1 µg) was used for in vitro transcription/translation assay using a TNT Quick Coupled Transcription/Translation system in the presence of increasing concentrations of recombinant GST-YB-1. As a control, 5′ UTR of β-Globin fused to Firefly Luciferase in pcDNA3 with T7 promoter was used as described previously (Evdokimova et al., 2009). Recombinant SRp55 was added in place of recombinant GST-YB-1 as a nonspecific control. After 90 min of incubation at 30°C, 2.5 µl of each reaction mixture was removed and luciferase activity was measured. For the in vivo experiments, the full-length G3BP1 (FL, 1–171) or deletion mutant (M4 [Δ48–171]), cloned using the same approach described in this paragraph, were transfected into siControl and siYB-1 cells. After 24 h of transfection, the cells were harvested and the luciferase activity was measured using a luciferase assay system. For measuring the IRES activities of 5′ UTR of G3BP1, the full-length (FL, 1–171) 5′ UTR of G3BP1 was PCR amplified and cloned into the SpeI–EcoR1 site in the pRF bicistronic vector with SV40 promoter (Young et al., 2008) upstream of the firefly luciferase gene to generate pRF-G3BP1. pRF-G3BP1 and control pRF plasmids were then transfected into U2OS cells. 24 h after transfection, cells were treated with vehicle alone or 0.5 mM arsenite for 30, 90, and 180 min, and the renilla and firefly luciferase activities were measured using a dual-luciferase reporter assay system.
EMSA
RNA EMSA to detect direct binding of YB-1 to G3BP1 5′ UTR was performed using a LightShift Chemiluminescent RNA EMSA kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. In brief, 6.25 nM biotin tagged full-length (FL, 1–171) or mutants (M3, Δ99–171; and M4, Δ48–171) of G3BP1 5′ UTR’s were incubated with recombinant GST-YB-1 (0.4 and 0.8 µg) in binding buffer (100 mM Hepes, pH 7.3, 200 mM KCl, 10 mM MgCl2, 10 mM DTT, 5% glycerol, and 2 µg tRNA) in a total volume of 20 µl for 30 min. Control reactions were set up using 200-fold molar excess concentration of unlabeled probe in place of labeled probe as well as recombinant GST in place of recombinant GST-YB-1. The reaction products were mixed with 5 µl of 5× loading buffer and subjected to electrophoresis in a 4–6% native polyacrylamide gel in 0.5× TBE buffer (100 V for 8 × 8 × 0.1 cm gel) until the bromophenol blue dye has migrated ∼3/4 down the length of the gel. The RNA–protein complexes were then transferred to a nylon membrane and cross-linked for 60 s at 120 mJ/cm2 using a commercial UV light cross-linker (Agilent Technologies) equipped with 254-nm UV light lamps. The membranes were then immunoblotted using anti-biotin antibodies.
In vivo murine renal subcapsular implantation model
The University of British Columbia Animal Care Committee approved murine renal subcapsular implantation studies. Xenograft cell blocks for implantation were prepared using 106 cells of shControl, shYB-1, or shG3BP1, which were mixed with 50 µl of 5× DMEM-tail collagen gel. They were then seeded in cell culture plates and incubated for 30 min. The cell suspensions were then implanted into NOD-SCID male mice (6–8 wk old, maintained in the animal resource facility at the BC Cancer Research Centre). In brief, a 0.5-cm incision was made to exteriorize the kidney, which was then removed and laid on the body wall. A 2–4-mm incision was made in the kidney capsule and cell suspensions were inserted into the pocket under the capsule. Once the grafting procedure was completed, the kidney was eased back into the body cavity, and the skin edges were sutured. After 4–5 wk, mice were sacrificed, and primary tumors and lungs were surgically removed and processed for microscopy and immunoblotting.
Protein carbonyl content measurement
To measure the carbonyl content in proteins, 2 mg/sample of normal kidney or shControl and shYB-1 tumor xenografts (n = 5 per each tumor group) were subjected to carbonyl content measurement using a Protein Carbonyl Colorimetric Assay kit according to the manufacturer’s instructions.
RNA affinity chromatography and RNA transfection
The RNA affinity chromatography was performed as described previously (Baird et al., 2007). U2OS cells were harvested and homogenized in binding buffer containing 10 mM Tris-HCl, pH 7.4, 1.5 mM MgCl2, 10 mM KCl, 0.05% NP-40, and 1× protease inhibitors using a Dounce homogenizer. The lysates were centrifuged at 10,000 g at 4°C for 10 min, and the supernatant was saved. The lysates (2 mg) were then precleared by incubating with 20 µl of RNasin (RNase inhibitor), 12 µg of yeast tRNA, and 100 µl Streptavidin magnetic beads in a total volume of 1 ml at 4°C for 2 h. In parallel, 100 µg of biotinylated RNA was incubated with 100 µl of Streptavidin magnetic beads at 4°C for 2 h on a rotator. The beads were washed and mixed with the precleared lysate and incubated at 4°C for 30 min in a rotator. The beads were then washed five times in the binding buffer, boiled in loading dye, and subjected to immunoblotting. For RNA transfection, the full-length G3BP1 (FL, 1–171) or deletion mutant (M4, Δ48–171) was in vitro transcribed using a TranscriptAid T7 High Yield Transcription kit and tagged with biotin at its 3′ end using an RNA 3′ End Biotinylation kit (Thermo Fisher Scientific). The biotin-tagged RNA (15 µg) was transfected into siControl or siYB-1 U2OS cells using TransIT-mRNA transfection reagent. Cells were harvested 5 h after transfection, and subjected to RNA affinity chromatography as described in this paragraph.
Survival analyses
Microarray data of 234 primary Ewing sarcoma (GSE12102, GSE17618, and GSE34620; Scotlandi et al., 2009; Savola et al., 2011; Postel-Vinay et al., 2012), for which well-curated clinical annotations were available, were normalized simultaneously by robust multi-array average (RMA) using custom brainarray CDF files (v18, ENTREZG; Irizarry et al., 2003). Thereafter, patients were grouped into a cohort showing no evidence for metastasis at diagnosis (localized disease, n = 155) and another cohort of patients with evidence for metastasis at diagnosis (metastatic disease, n = 79). For subsequent survival analyses, samples were ranked according to their G3BP1 expression intensities (10146_at) within each cohort and stratified according to their G3BP1 expression levels as indicated. Analysis of overall survival was performed using the Kaplan-Meier method within the GraphPad Prism 5 software (GraphPad Software). Statistical significance was assessed with a log-rank test.
Online supplemental material
Fig. S1 shows localization of YB-1 in SGs under different stress conditions. Fig. S2 shows that the down-regulation of YB-1 reduces SG assembly and sensitizes RH-30, MNNG, and DU-145 cells to stress-induced apoptosis. Fig. S3 documents the selective regulation of G3BP1 by YB-1. Fig. S4 shows the translational regulation of G3BP1 by YB-1. Fig. S5 shows the evidence of hypoxia, ER, and oxidative stress in the tumors. For the movies, U2OS cells transfected with GFP-YB-1 (green) and RFP-G3BP1 (red) were treated with 0.5 mM arsenite. Images were analyzed by time-lapse fluorescence microscopy using a fluorescence microscope (Zeiss Axio Observer Z1; Carl Zeiss). Frames were taken every 2 min for 1 h. Video 1 shows localization of GFP-YB-1 in SGs after 0.5 mM arsenite treatment over 1 h. Video 2 shows localization of RFP-G3BP1 in SGs after 0.5 mM arsenite treatment over 1 h. Video 3 shows the merge showing colocalization of GFP-YB-1 and RFP-G3BP1 in SGs under these conditions. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201411047/DC1.
Supplementary Material
Acknowledgments
We thank Torsten Nielsen for sarcoma TMAs, Kevin Bennewith for pimonidazole staining, and Julie Lorette, Tina Yang, Brian Kwok, and Joan Mathers for technical assistance.
This work was supported by funds to P.H. Sorensen from Terry Fox Research Institute Team grant 1021 and Prostate Cancer Canada-Movember Foundation Team grant T2013-1. T.G.P. Grunewald is supported by the German Research Foundation (DFG GR3728/2-1). The work was also funded by the British Columbia Cancer Foundation through generous donations from Team Finn and other riders in the Ride to Conquer Cancer.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- AHA
- l-azidohomoalanine
- CHX
- cycloheximide
- EMSA
- electrophoretic mobility shift assay
- H&E
- hematoxylin and eosin
- IF
- immunofluorescence
- IHC
- immunohistochemistry
- IRES
- internal ribosome entry site
- kd
- knockdown
- PB
- P-body
- RIP
- riboimmunoprecipitation
- SG
- stress granule
- TMA
- tissue microarray
References
- Anderson P., and Kedersha N.. 2006. RNA granules. J. Cell Biol. 172:803–808 10.1083/jcb.200512082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P., and Kedersha N.. 2008. Stress granules: the Tao of RNA triage. Trends Biochem. Sci. 33:141–150 10.1016/j.tibs.2007.12.003 [DOI] [PubMed] [Google Scholar]
- Anderson P., and Kedersha N.. 2009. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat. Rev. Mol. Cell Biol. 10:430–436 10.1038/nrm2694 [DOI] [PubMed] [Google Scholar]
- Araujo P.R., Yoon K., Ko D., Smith A.D., Qiao M., Suresh U., Burns S.C., and Penalva L.O.. 2012. Before it gets started: Regulating translation at the 5′ UTR. Comp. Funct. Genomics. 2012:475731 10.1155/2012/475731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimoto K., Fukuda H., Imajoh-Ohmi S., Saito H., and Takekawa M.. 2008. Formation of stress granules inhibits apoptosis by suppressing stress-responsive MAPK pathways. Nat. Cell Biol. 10:1324–1332 10.1038/ncb1791 [DOI] [PubMed] [Google Scholar]
- Baird S.D., Lewis S.M., Turcotte M., and Holcik M.. 2007. A search for structurally similar cellular internal ribosome entry sites. Nucleic Acids Res. 35:4664–4677 10.1093/nar/gkm483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennewith K.L., and Durand R.E.. 2004. Quantifying transient hypoxia in human tumor xenografts by flow cytometry. Cancer Res. 64:6183–6189 10.1158/0008-5472.CAN-04-0289 [DOI] [PubMed] [Google Scholar]
- Boelens J., Lust S., Offner F., Bracke M.E., and Vanhoecke B.W.. 2007. Review. The endoplasmic reticulum: a target for new anticancer drugs. In Vivo. 21:215–226. [PubMed] [Google Scholar]
- Buchan J.R., and Parker R.. 2009. Eukaryotic stress granules: the ins and outs of translation. Mol. Cell. 36:932–941 10.1016/j.molcel.2009.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobbold L.C., Spriggs K.A., Haines S.J., Dobbyn H.C., Hayes C., de Moor C.H., Lilley K.S., Bushell M., and Willis A.E.. 2008. Identification of internal ribosome entry segment (IRES)-trans-acting factors for the Myc family of IRESs. Mol. Cell. Biol. 28:40–49 10.1128/MCB.01298-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalle-Donne I., Aldini G., Carini M., Colombo R., Rossi R., and Milzani A.. 2006. Protein carbonylation, cellular dysfunction, and disease progression. J. Cell. Mol. Med. 10:389–406 10.1111/j.1582-4934.2006.tb00407.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davuluri R.V., Suzuki Y., Sugano S., and Zhang M.Q.. 2000. CART classification of human 5′ UTR sequences. Genome Res. 10:1807–1816 10.1101/gr.GR-1460R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliseeva I.A., Kim E.R., Guryanov S.G., Ovchinnikov L.P., and Lyabin D.N.. 2011. Y-box-binding protein 1 (YB-1) and its functions. Biochemistry Mosc. 76:1402–1433 10.1134/S0006297911130049 [DOI] [PubMed] [Google Scholar]
- Evdokimova V., Ruzanov P., Imataka H., Raught B., Svitkin Y., Ovchinnikov L.P., and Sonenberg N.. 2001. The major mRNA-associated protein YB-1 is a potent 5′ cap-dependent mRNA stabilizer. EMBO J. 20:5491–5502 10.1093/emboj/20.19.5491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evdokimova V., Ruzanov P., Anglesio M.S., Sorokin A.V., Ovchinnikov L.P., Buckley J., Triche T.J., Sonenberg N., and Sorensen P.H.. 2006. Akt-mediated YB-1 phosphorylation activates translation of silent mRNA species. Mol. Cell. Biol. 26:277–292 10.1128/MCB.26.1.277-292.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evdokimova V., Tognon C., Ng T., Ruzanov P., Melnyk N., Fink D., Sorokin A., Ovchinnikov L.P., Davicioni E., Triche T.J., and Sorensen P.H.. 2009. Translational activation of snail1 and other developmentally regulated transcription factors by YB-1 promotes an epithelial-mesenchymal transition. Cancer Cell. 15:402–415 10.1016/j.ccr.2009.03.017 [DOI] [PubMed] [Google Scholar]
- Fournier M.J., Gareau C., and Mazroui R.. 2010. The chemotherapeutic agent bortezomib induces the formation of stress granules. Cancer Cell Int. 10:12 10.1186/1475-2867-10-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French J., Stirling R., Walsh M., and Kennedy H.D.. 2002. The expression of Ras-GTPase activating protein SH3 domain-binding proteins, G3BPs, in human breast cancers. Histochem. J. 34:223–231 10.1023/A:1021737413055 [DOI] [PubMed] [Google Scholar]
- Fujiwara-Okada Y., Matsumoto Y., Fukushi J., Setsu N., Matsuura S., Kamura S., Fujiwara T., Iida K., Hatano M., Nabeshima A., et al. . 2013. Y-box binding protein-1 regulates cell proliferation and is associated with clinical outcomes of osteosarcoma. Br. J. Cancer. 108:836–847 10.1038/bjc.2012.579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilks N., Kedersha N., Ayodele M., Shen L., Stoecklin G., Dember L.M., and Anderson P.. 2004. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell. 15:5383–5398 10.1091/mbc.E04-08-0715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guitard E., Parker F., Millon R., Abecassis J., and Tocqué B.. 2001. G3BP is overexpressed in human tumors and promotes S phase entry. Cancer Lett. 162:213–221 10.1016/S0304-3835(00)00638-8 [DOI] [PubMed] [Google Scholar]
- Hsieh A.C., Liu Y., Edlind M.P., Ingolia N.T., Janes M.R., Sher A., Shi E.Y., Stumpf C.R., Christensen C., Bonham M.J., et al. . 2012. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 485:55–61 10.1038/nature10912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U., and Speed T.P.. 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 4:249–264 10.1093/biostatistics/4.2.249 [DOI] [PubMed] [Google Scholar]
- Kedersha N., and Anderson P.. 2002. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 30:963–969 10.1042/BST0300963 [DOI] [PubMed] [Google Scholar]
- Kedersha N., and Anderson P.. 2007. Mammalian stress granules and processing bodies. Methods Enzymol. 431:61–81 10.1016/S0076-6879(07)31005-7 [DOI] [PubMed] [Google Scholar]
- Kedersha N.L., Gupta M., Li W., Miller I., and Anderson P.. 1999. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2α to the assembly of mammalian stress granules. J. Cell Biol. 147:1431–1442 10.1083/jcb.147.7.1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Ivanov P., and Anderson P.. 2013. Stress granules and cell signaling: more than just a passing phase? Trends Biochem. Sci. 38:494–506 10.1016/j.tibs.2013.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khong A., and Jan E.. 2011. Modulation of stress granules and P bodies during dicistrovirus infection. J. Virol. 85:1439–1451 10.1128/JVI.02220-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimball S.R., Horetsky R.L., Ron D., Jefferson L.S., and Harding H.P.. 2003. Mammalian stress granules represent sites of accumulation of stalled translation initiation complexes. Am. J. Physiol. Cell Physiol. 284:C273–C284 10.1152/ajpcell.00314.2002 [DOI] [PubMed] [Google Scholar]
- Kohno K., Izumi H., Uchiumi T., Ashizuka M., and Kuwano M.. 2003. The pleiotropic functions of the Y-box-binding protein, YB-1. BioEssays. 25:691–698 10.1002/bies.10300 [DOI] [PubMed] [Google Scholar]
- Lasham A., Print C.G., Woolley A.G., Dunn S.E., and Braithwaite A.W.. 2013. YB-1: oncoprotein, prognostic marker and therapeutic target? Biochem. J. 449:11–23 10.1042/BJ20121323 [DOI] [PubMed] [Google Scholar]
- Lavut A., and Raveh D.. 2012. Sequestration of highly expressed mRNAs in cytoplasmic granules, P-bodies, and stress granules enhances cell viability. PLoS Genet. 8:e1002527 10.1371/journal.pgen.1002527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.R., King O.D., Shorter J., and Gitler A.D.. 2013. Stress granules as crucibles of ALS pathogenesis. J. Cell Biol. 201:361–372 10.1083/jcb.201302044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett D.H., Cheng S., Cape L., Pollock A.S., and Mertens P.R.. 2010. YB-1 alters MT1-MMP trafficking and stimulates MCF-7 breast tumor invasion and metastasis. Biochem. Biophys. Res. Commun. 398:482–488 10.1016/j.bbrc.2010.06.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyabin D.N., Eliseeva I.A., and Ovchinnikov L.P.. 2014. YB-1 protein: functions and regulation. Wiley Interdiscip Rev RNA. 5:95–110 10.1002/wrna.1200 [DOI] [PubMed] [Google Scholar]
- Matsumoto S., Uchiumi T., Tanamachi H., Saito T., Yagi M., Takazaki S., Kanki T., and Kang D.. 2012. Ribonucleoprotein Y-box-binding protein-1 regulates mitochondrial oxidative phosphorylation (OXPHOS) protein expression after serum stimulation through binding to OXPHOS mRNA. Biochem. J. 443:573–584 10.1042/BJ20111728 [DOI] [PubMed] [Google Scholar]
- McDonald K.K., Aulas A., Destroismaisons L., Pickles S., Beleac E., Camu W., Rouleau G.A., and Vande Velde C.. 2011. TAR DNA-binding protein 43 (TDP-43) regulates stress granule dynamics via differential regulation of G3BP and TIA-1. Hum. Mol. Genet. 20:1400–1410 10.1093/hmg/ddr021 [DOI] [PubMed] [Google Scholar]
- Mendoza-Naranjo A., El-Naggar A., Wai D.H., Mistry P., Lazic N., Ayala F.R., da Cunha I.W., Rodriguez-Viciana P., Cheng H., Tavares Guerreiro Fregnani J.H., et al. . 2013. ERBB4 confers metastatic capacity in Ewing sarcoma. EMBO Mol. Med. 5:1019–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller B.J., Cao Y., Li C.Y., and Dewhirst M.W.. 2004. Radiation activates HIF-1 to regulate vascular radiosensitivity in tumors: Role of reoxygenation, free radicals, and stress granules. Cancer Cell. 5:429–441 10.1016/S1535-6108(04)00115-1 [DOI] [PubMed] [Google Scholar]
- Montessuit S., Somasekharan S.P., Terrones O., Lucken-Ardjomande S., Herzig S., Schwarzenbacher R., Manstein D.J., Bossy-Wetzel E., Basañez G., Meda P., and Martinou J.C.. 2010. Membrane remodeling induced by the dynamin-related protein Drp1 stimulates Bax oligomerization. Cell. 142:889–901 10.1016/j.cell.2010.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda Y., Sakamoto A., Shinohara N., Ohga T., Uchiumi T., Kohno K., Tsuneyoshi M., Kuwano M., and Iwamoto Y.. 1998. Nuclear expression of YB-1 protein correlates with P-glycoprotein expression in human osteosarcoma. Clin. Cancer Res. 4:2273–2277. [PubMed] [Google Scholar]
- Oda Y., Kohashi K., Yamamoto H., Tamiya S., Kohno K., Kuwano M., Iwamoto Y., Tajiri T., Taguchi T., and Tsuneyoshi M.. 2008. Different expression profiles of Y-box-binding protein-1 and multidrug resistance-associated proteins between alveolar and embryonal rhabdomyosarcoma. Cancer Sci. 99:726–732 10.1111/j.1349-7006.2008.00748.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohga T., Uchiumi T., Makino Y., Koike K., Wada M., Kuwano M., and Kohno K.. 1998. Direct involvement of the Y-box binding protein YB-1 in genotoxic stress-induced activation of the human multidrug resistance 1 gene. J. Biol. Chem. 273:5997–6000 10.1074/jbc.273.11.5997 [DOI] [PubMed] [Google Scholar]
- Onishi H., Kino Y., Morita T., Futai E., Sasagawa N., and Ishiura S.. 2008. MBNL1 associates with YB-1 in cytoplasmic stress granules. J. Neurosci. Res. 86:1994–2002 10.1002/jnr.21655 [DOI] [PubMed] [Google Scholar]
- Paranjape S.M., and Harris E.. 2007. Y box-binding protein-1 binds to the dengue virus 3′-untranslated region and mediates antiviral effects. J. Biol. Chem. 282:30497–30508 10.1074/jbc.M705755200 [DOI] [PubMed] [Google Scholar]
- Postel-Vinay S., Véron A.S., Tirode F., Pierron G., Reynaud S., Kovar H., Oberlin O., Lapouble E., Ballet S., Lucchesi C., et al. . 2012. Common variants near TARDBP and EGR2 are associated with susceptibility to Ewing sarcoma. Nat. Genet. 44:323–327 10.1038/ng.1085 [DOI] [PubMed] [Google Scholar]
- Raleigh J.A., Chou S.C., Arteel G.E., and Horsman M.R.. 1999. Comparisons among pimonidazole binding, oxygen electrode measurements, and radiation response in C3H mouse tumors. Radiat. Res. 151:580–589 10.2307/3580034 [DOI] [PubMed] [Google Scholar]
- Reineke L.C., Dougherty J.D., Pierre P., and Lloyd R.E.. 2012. Large G3BP-induced granules trigger eIF2α phosphorylation. Mol. Biol. Cell. 23:3499–3510 10.1091/mbc.E12-05-0385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savola S., Klami A., Myllykangas S., Manara C., Scotlandi K., Picci P., Knuutila S., and Vakkila J.. 2011. High expression of complement component 5 (C5) at tumor site associates with superior survival in Ewing’s sarcoma family of tumour patients. ISRN Oncol. 2011:168712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotlandi K., Remondini D., Castellani G., Manara M.C., Nardi F., Cantiani L., Francesconi M., Mercuri M., Caccuri A.M., Serra M., et al. . 2009. Overcoming resistance to conventional drugs in Ewing sarcoma and identification of molecular predictors of outcome. J. Clin. Oncol. 27:2209–2216 10.1200/JCO.2008.19.2542 [DOI] [PubMed] [Google Scholar]
- Skabkin M.A., Kiselyova O.I., Chernov K.G., Sorokin A.V., Dubrovin E.V., Yaminsky I.V., Vasiliev V.D., and Ovchinnikov L.P.. 2004. Structural organization of mRNA complexes with major core mRNP protein YB-1. Nucleic Acids Res. 32:5621–5635 10.1093/nar/gkh889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skabkin M.A., Liabin D.N., and Ovchinnikov L.P.. 2006. Nonspecific and specific interaction of Y-box binding protein 1 (YB-1) with mRNA and posttranscriptional regulation of protein synthesis in animal cells. Mol. Biol. 40:551–563 10.1134/S0026893306040078 [DOI] [PubMed] [Google Scholar]
- Somasekharan S.P., Stoynov N., Rotblat B., Leprivier G., Galpin J.D., Ahern C.A., Foster L.J., and Sorensen P.H.. 2012. Identification and quantification of newly synthesized proteins translationally regulated by YB-1 using a novel Click-SILAC approach. J. Proteomics. 77:e1–e10 10.1016/j.jprot.2012.08.019 [DOI] [PubMed] [Google Scholar]
- Somasekharan S.P., Koc M., Morizot A., Micheau O., Sorensen P.H., Gaide O., Andera L., and Martinou J.C.. 2013. TRAIL promotes membrane blebbing, detachment and migration of cells displaying a dysfunctional intrinsic pathway of apoptosis. Apoptosis. 18:324–336 10.1007/s10495-012-0782-6 [DOI] [PubMed] [Google Scholar]
- Sonenberg N., and Hinnebusch A.G.. 2009. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 136:731–745 10.1016/j.cell.2009.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöhr N., Lederer M., Reinke C., Meyer S., Hatzfeld M., Singer R.H., and Hüttelmaier S.. 2006. ZBP1 regulates mRNA stability during cellular stress. J. Cell Biol. 175:527–534 10.1083/jcb.200608071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen C.C., Chantranupong L., Keys H.R., Wang T., Gray N.S., and Sabatini D.M.. 2012. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 485:109–113 10.1038/nature11083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourrière H., Chebli K., Zekri L., Courselaud B., Blanchard J.M., Bertrand E., and Tazi J.. 2003. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 160:823–831 10.1083/jcb.200212128 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Toyoshima Y., and Takahashi H.. 2014. TDP-43 pathology in polyglutamine diseases: with reference to amyotrphic lateral sclerosis. Neuropathology. 34:77–82 10.1111/neup.12053 [DOI] [PubMed] [Google Scholar]
- Tran Q., and Roesser J.R.. 2003. SRp55 is a regulator of calcitonin/CGRP alternative RNA splicing. Biochemistry. 42:951–957 10.1021/bi026753a [DOI] [PubMed] [Google Scholar]
- Tsai N.P., and Wei L.N.. 2010. RhoA/ROCK1 signaling regulates stress granule formation and apoptosis. Cell. Signal. 22:668–675 10.1016/j.cellsig.2009.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valiente-Echeverría F., Melnychuk L., Vyboh K., Ajamian L., Gallouzi I.E., Bernard N., and Mouland A.J.. 2014. eEF2 and Ras-GAP SH3 domain-binding protein (G3BP1) modulate stress granule assembly during HIV-1 infection. Nat. Commun. 5:4819 10.1038/ncomms5819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderweyde T., Yu H., Varnum M., Liu-Yesucevitz L., Citro A., Ikezu T., Duff K., and Wolozin B.. 2012. Contrasting pathology of the stress granule proteins TIA-1 and G3BP in tauopathies. J. Neurosci. 32:8270–8283 10.1523/JNEUROSCI.1592-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderweyde T., Youmans K., Liu-Yesucevitz L., and Wolozin B.. 2013. Role of stress granules and RNA-binding proteins in neurodegeneration: a mini-review. Gerontology. 59:524–533 10.1159/000354170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel C., Abreu R.S., Ko D., Le S.Y., Shapiro B.A., Burns S.C., Sandhu D., Boutz D.R., Marcotte E.M., and Penalva L.O.. 2010. Sequence signatures and mRNA concentration can explain two-thirds of protein abundance variation in a human cell line. Mol. Syst. Biol. 6:400 10.1038/msb.2010.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J.P., Cardenas A.M., Marissen W.E., and Lloyd R.E.. 2007. Inhibition of cytoplasmic mRNA stress granule formation by a viral proteinase. Cell Host Microbe. 2:295–305 10.1016/j.chom.2007.08.006 [DOI] [PubMed] [Google Scholar]
- Wu Y., Yamada S., Izumi H., Li Z., Shimajiri S., Wang K.Y., Liu Y.P., Kohno K., and Sasaguri Y.. 2012. Strong YB-1 expression is associated with liver metastasis progression and predicts shorter disease-free survival in advanced gastric cancer. J. Surg. Oncol. 105:724–730 10.1002/jso.23030 [DOI] [PubMed] [Google Scholar]
- Yamasaki S., and Anderson P.. 2008. Reprogramming mRNA translation during stress. Curr. Opin. Cell Biol. 20:222–226 10.1016/j.ceb.2008.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W.H., and Bloch D.B.. 2007. Probing the mRNA processing body using protein macroarrays and “autoantigenomics”. RNA. 13:704–712 10.1261/rna.411907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R.M., Wang S.J., Gordan J.D., Ji X., Liebhaber S.A., and Simon M.C.. 2008. Hypoxia-mediated selective mRNA translation by an internal ribosome entry site-independent mechanism. J. Biol. Chem. 283:16309–16319 10.1074/jbc.M710079200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H.Z., Liu J.G., Wei Y.P., Wu C., Cao Y.K., and Wang M.. 2007. Expression of G3BP and RhoC in esophageal squamous carcinoma and their effect on prognosis. World J. Gastroenterol. 13:4126–4130 10.3748/wjg.v13.i30.4126 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.