WISp39 associates with Hsp90, Coronin 1B, and Slingshot phosphatase to regulate Cofilin activation and Arp2/3 complex localization at the leading edge of migrating cells.
Abstract
We previously identified Waf1 Cip1 stabilizing protein 39 (WISp39) as a binding partner for heat shock protein 90 (Hsp90). We now report that WISp39 has an essential function in the control of directed cell migration, which requires WISp39 interaction with Hsp90. WISp39 knockdown (KD) resulted in the loss of directional motility of mammalian cells and profound changes in cell morphology, including the loss of a single leading edge. WISp39 binds Coronin 1B, known to regulate the Arp2/3 complex and Cofilin at the leading edge. WISp39 preferentially interacts with phosphorylated Coronin 1B, allowing it to complex with Slingshot phosphatase (SSH) to dephosphorylate and activate Cofilin. WISp39 also regulates Arp2/3 complex localization at the leading edge. WISp39 KD-induced morphological changes could be rescued by overexpression of Coronin 1B together with a constitutively active Cofilin mutant. We conclude that WISp39 associates with Hsp90, Coronin 1B, and SSH to regulate Cofilin activation and Arp2/3 complex localization at the leading edge.
Introduction
Cell motility in mammalian cells requires tightly regulated actin dynamics. Production of branched actin networks driven by the Arp2/3 complex regulates cell shape and production of lamellipodia (Pollard and Cooper, 2009). In addition to the actin-nucleating activity of the Arp2/3 complex, actin dynamics at the leading edge require actin-depolymerizing factor Cofilin to maintain actin turnover by severing and depolymerizing actin filaments (Bamburg et al., 1999; Pollard and Borisy, 2003). Cofilin is inactivated by phosphorylation and is activated by Slingshot phosphatase (SSH; Agnew et al., 1995; Niwa et al., 2002; Nishita et al., 2005; Delorme et al., 2007). Actin treadmilling is thus driven by Arp2/3 nucleation at the leading edge balanced with Cofilin-dependent severing at the other end (Svitkina and Borisy, 1999).
The Arp2/3 complex and Cofilin are coordinately regulated at the leading edge by Coronin 1B (Mishima and Nishida, 1999; Cai et al., 2005, 2007; Chan et al., 2011). Depletion of Coronin 1B increases the phosphorylation of Cofilin, resulting in its inactivation (Cai et al., 2007). In addition, Coronin 1B binds the Arp2/3 complex in a phosphorylation-dependent manner (Cai et al., 2005). When Coronin 1B is dephosphorylated on Ser2 it binds and inhibits Arp2/3 complex function, whereas phosphorylation by PKC at Ser2 reduces this association (Cai et al., 2005). The phosphorylation status of Coronin 1B Ser2 impacts cell migration, as overexpression of the Coronin 1B(S2A) mutant increases cell speed (Cai et al., 2005). Coronin 1B also disassembles actin filament branches by causing dissociation of the Arp2/3 complex (Cai et al., 2008). Thus, the regulation of phosphorylated Coronin 1B is essential to control Arp2/3 complex activity and ultimately the rate of actin nucleation and branching at the leading edge.
Coronin 1B interacts with and is dephosphorylated by SSH, and this interaction promotes the dephosphorylation and activation of Cofilin (Cai et al., 2007). One crucial missing piece of information is how the Coronin 1B and SSH interaction is regulated and how this interaction controls the dephosphorylation of Cofilin. Here, we report that Waf1 Cip1 stabilizing protein 39 (WISp39), a heat shock protein 90 (Hsp90) binding protein we have previously described (Jascur et al., 2005), is a key element in the regulation of Coronin 1B phosphorylation. WISp39 increases the binding of SSH to phosphorylated Coronin 1B in a complex that excludes Arp2/3. The association of phosphorylated Coronin 1B with SSH leads to the dephosphorylation of Coronin 1B and Cofilin. Loss of WISp39 thus reduces Coronin 1B association with SSH, decreases Cofilin activation, and reduces the localization of the Arp2/3 complex at the leading edge, causing a decrease in directed cell motility. Furthermore, WISp39 function in directional migration depends on Hsp90 binding. We conclude that WISp39, acting in a complex with SSH, regulates Coronin 1B function and is essential to the control of cell polarity and directed motility.
Results
WISp39 knockdown (KD) decreases directional motility
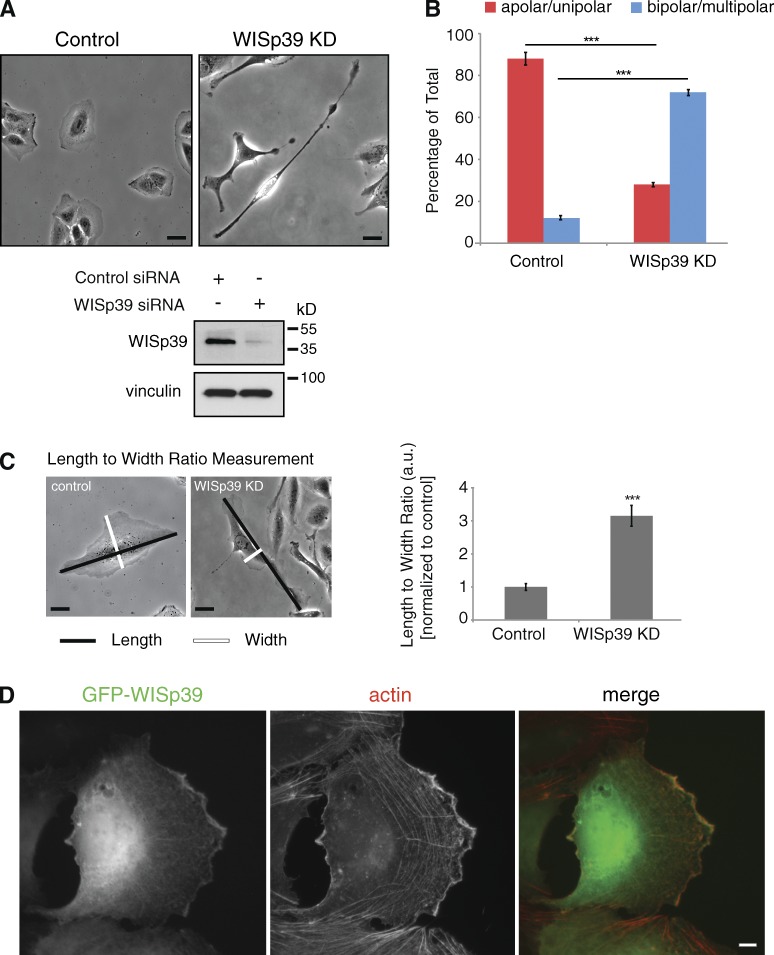
We suppressed WISp39 expression using a siRNA that we have previously demonstrated specifically targets human WISp39 (Jascur et al., 2005). WISp39 siRNA-transfected (referred to hereafter as WISp39 KD) U2OS cells frequently became elongated and irregular in morphology compared with controls (Fig. 1 A). Quantitation of cell shape parameters demonstrated a substantial increase in the number of lamellipodia, recorded as the percentage of cells with two or more lamellipodia when not bordered by other cells (Fig. 1 B). The length to width ratio of the WISp39 KD cells also changed substantially compared with controls (Fig. 1 C and Table 1). Immunofluorescence showed cytoplasmic WISp39 colocalized with actin at the leading edge (Fig. 1 D) and could impact cell migration.
Figure 1.
Loss of WISp39 results in elongated cell morphology. (A) Subconfluent U2OS cells were transfected with either control (left) or human WISp39 siRNA (WISp39 KD; right) for 48 h and representative 20× phase images were taken. Western blot shows KD of WISp39 at 48 h. Vinculin is a loading control. (B) Depletion of WISp39 changes cell polarity. Cells were scored as either apolar (round)/unipolar (one lamellipodium) or bipolar/multipolar (two or more lamellipodia) and are presented as a percentage of total cells scored from more than five independent experiments. n = 1,165 control cells; n = 1,483 WISp39 KD cells. Data represent the means ± SD. Student’s t test; ***, P ≤ 0.001. (C) Quantitation of the maximum length to maximum width ratio of control cells versus WISp39 KD cells from an experiment performed as in A. Number of cells scored per group is 50 from more than three independent experiments. Data represent the means ± SD. Student’s t test; ***, P ≤ 0.001. a.u., arbitrary unit. (D) Cytoplasmic WISp39 localizes to actin filaments at the leading edge. U2OS cells were transfected with GFP-WISp39, fixed with paraformaldehyde, and stained with rhodamine-phalloidin (actin). Bars, 10 µm.
Table 1.
Quantitation of cell shape in different KD and rescue conditions
| Condition | Maximum length | Maximum width | Perimeter |
| µm | µm | µm | |
| Control | 33.09 ± 5.12 | 22.26 ± 3.35 | 88.44 ± 12.02 |
| WISp39 KD | 91.69 ± 21.34a | 19.58 ± 11.95a | 220.26 ± 59.39a |
| WISp39 siRNA + GFP-mWISp39WT | 38.58 ± 8.76b | 19.72 ± 4.87b | 96.30 ± 21.53b |
| WISp39 siRNA + GFP-mWISp39ΔTPR | 85.13 ± 6.9a | 18.09 ± 7.55a | 265.96 ± 60.37a |
| Coronin 1B KD | 93.25 ± 11.36a | 14.56 ± 5.79a | 209.98 ± 25.22a |
| p21 KD | 36.60 ± 4.08b | 23.56 ± 2.41b | 96.18 ± 8.95b |
Quantitation of cell size and shape parameters of the following cells: control, WISp39 KD, Coronin 1B KD, p21 KD, and WISp39 KD reconstituted with WT WISp39 or with WISp39ΔTPR.
P ≤ 0.001 compared to control cells; Student’s t test.
Not significant compared to control cells; Student’s t test.
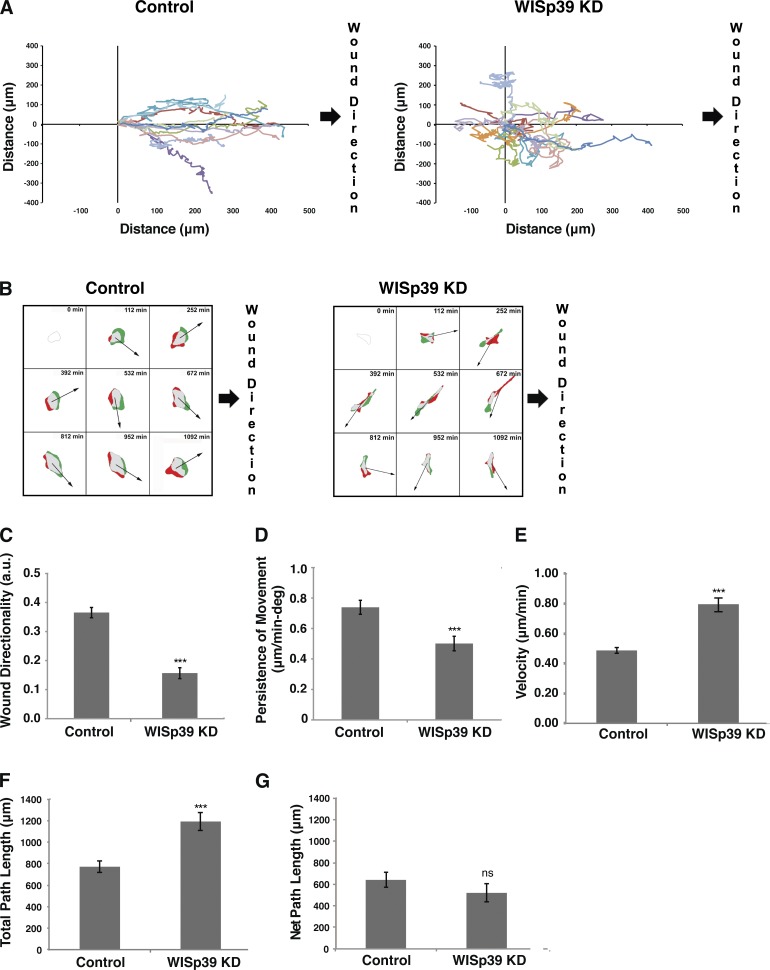
To quantitate the effect of WISp39 KD on directional motility, confluent cultures of both control and WISp39 KD were wounded and then recorded for 24 h using phase-contrast time-lapse microscopy. The movements of individual representative cells were plotted relative to the direction of the wound. Tracking plots show that WISp39 KD cells lost the uniform directional wound closing motility exhibited by control cells (Fig. 2 A and Videos 1 and 2). Whereas control cells at the wound edge mostly moved in concert with the confluent sheet to close the wound, WISp39 KD cells migrated singly and exhibited chaotic and undirected motility at the leading edge. The chaotic migration of WISp39 KD cells correlated with the existence of bipolar or multipolar lamellipodia (Video 2). The presence of more than one lamellipodium sometimes caused a single cell to migrate in opposing directions at once and leave severed fragments. Although cell–cell contact was reduced after WISp39 KD, tracking plots of control cells that separated from the sheet showed that loss of cell–cell contact was not sufficient to cause them to migrate in a chaotic manner or show profound changes in cell morphology (unpublished data). We also observed that some WISp39 KD cells undergo cell death (Video 2). To ensure that the morphological changes observed in WISp39 KD cells are not caused by the stress of impending death, we performed additional experiments with the apoptosis inhibitor ZVAD (benzyloxycarbonyl-valine-alanine-aspartic acid). Treatment with ZVAD suppressed apoptotic cell death in WISp39 KD cells but had no effect on the morphological changes (Fig. S1) or chaotic migration characteristics (Videos 3–6) in these cells. Therefore, the phenotypes observed in WISp39 KD cells cannot simply be attributed to cells undergoing apoptosis.
Figure 2.
WISp39 KD cells exhibit decreased directional motility. (A) WISp39 KD leads to chaotic migration. U2OS cells plated on glass coverslips were transfected with either control or WISp39 siRNA. After 48 h, the cells were confluent, and migration was initiated by creating a wound with a pipette tip. 20× phase images were taken over after a 24-h period as described in the Materials and methods. The movements of individual representative cells, as shown, were obtained using MetaMorph nuclear tracking. WISp39 KD cells lacked directional movement toward the wound, which is on the right side of the frame. The data shown are from a single representative experiment (n = 10), out of three independent experiments. (B) Time-lapse videos were used to calculate protrusion differences as described in Materials and methods. Green areas are the protruded areas, and red areas are the retracted areas calculated between four successive frames of 7-min intervals each (i.e., the 28 min preceding the tile image) beginning at the 112-min mark. Arrows indicate the direction of cell movement averaged from four successive frames. Time between tiles is 140 min. Expected direction of migration toward wound closure is on the right side of each frame. Note that WISp39 KD cells did not consistently move in a defined direction relative to the protrusion. (C) Quantitation of directionality in actively migrating cells collected using DIAS software. Directionality is a measure of path variance from the central axis of the wound. Number of control and WISp39 KD cells scored is 25 each from three independent experiments. a.u., arbitrary units. (D) Quantitation of persistence in actively migrating cells. Persistence is the ratio of net translocation distance to the total length of migration. Number of control and WISp39 KD cells scored is 25 each from three independent experiments. deg, degree. (E) Quantitation of velocity in actively migrating cells. Velocity was measured using the central difference method as described in the DIAS manual. The number of control and WISp39 KD cells scored is 25 each from three independent experiments. (F) Quantitation of total path length, the distance the cell traveled from the first frame to the last frame taken as a straight line approximation every four frames. The number of control and WISp39 KD cells scored is 25 each from three independent experiments. (G) Quantitation of net path, the distance from the starting frame to the last frame. The number of control and WISp39 KD cells scored is 25 each from three independent experiments. Data shown in C–G represent the means ± SD. Student’s t test; ***, P ≤ 0.001.
Areas of cell protrusion (Fig. 2 B, green) and retraction (Fig. 2 B, red) were also recorded (Fig. 2 B). Whereas the control migrated uniformly toward the wound edge and the trajectory matched closely to the area of cell protrusion, WISp39 KD cells moved randomly relative to the direction of wound closure, and the trajectory of migration did not match with regions of recent cell protrusions (Fig. 2 B). Quantitative analysis of the cell population showed that wound directionality, defined as direction of migration relative to the central axis of the wound, was substantially reduced in WISp39 KD cells (Fig. 2 C). In addition, persistence of movement, the net displacement relative to total distance traveled, was also decreased relative to controls (Fig. 2 D). The increased velocity of WISp39 KD cells (Fig. 2 E) resulted in an increase in total path length (Fig. 2 F) but decreased the net path length as a result of increased turning frequency compared with control cells (Fig. 2 G).
WISp39 effect on motility requires Hsp90 binding
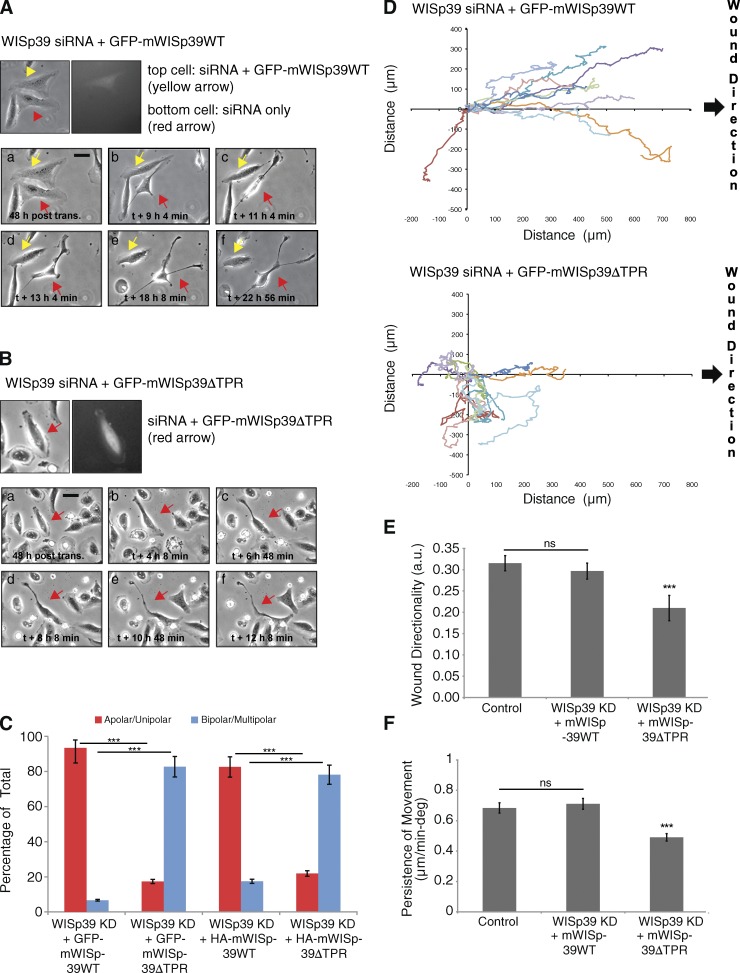
To confirm that these results represented the specific effect of WISp39 KD, we rescued WISp39 KD cells with a plasmid expressing GFP-tagged wild-type (WT) mouse WISp39 that was resistant to the human sequence-specific siRNA (see Materials and methods). WISp39 KD cells expressing GFP-mWISp39 WT remained more uniform in shape compared with WISp39 KD cells, which were multipolar in appearance (Fig. 3 A).
Figure 3.
WISp39 KD effect on motility is rescued by WT WISp39 and requires Hsp90. (A) Images of WISp39 KD rescue by WT WISp39. U2OS cells were cotransfected with human WISp39 siRNA and GFP-mouse WISp39 WT plasmid. 20× phase-contrast images were taken after 48 h of transfection. Top images show a cell transfected with rescue GFP plasmid (yellow arrows), whereas the neighboring cell is not. This cell (yellow arrows) retains normal apolar/unipolar morphology throughout the experiment unlike its neighbor (red arrows), which becomes elongated and multipolar. post trans., posttransfection. Bar, 10 µm. (B) Images of failure of WISp39 KD rescue by WISp39ΔTPR. U2OS cells were cotransfected with human WISp39 siRNA and mouse GFP-WISp39ΔTPR (which does not bind Hsp90) for 48 h and imaged as in A. The cell containing GFP-WISp39ΔTPR (top, red arrows) gradually loses its normal appearance and becomes elongated. Bar, 10 µm. (C) Quantitation of WISp39 KD rescue experiments. U2OS cells were cotransfected with human WISp39 siRNA and a plasmid expressing GFP-tagged mouse WISp39 WT or WISp39ΔTPR. In additional experiments, cells were transfected with WISp39 siRNA, HA-tagged mWISp39-WT or -ΔTPR, and GFP-H2B to mark transfected cells. Abnormal cell polarity of WISp39 KD cells is rescued by WT WISp39 but not by WISp39ΔTPR. Cells were scored as either apolar/unipolar or bipolar/multipolar and presented as a percentage of total cells scored from more than three independent experiments. Number of rescued cells scored: GFP-mouse WISp39 WT (30); GFP-mouse WISp39ΔTPR (75); HA-mouse WISp39 WT (172); HA-mouse WISp39ΔTPR (105). Note that only fluorescent cells were scored. (D) Migration assay of WISp39 KD rescue with WT WISp39 versus WISp39ΔTPR. Migration was initiated by wounding and the experiment was conducted as in Fig. 2 A. GFP-WISp39 WT rescues the WISp39 KD phenotype, whereas WISp39ΔTPR does not. The data shown are from a single representative experiment (n = 10), out of three independent experiments. (E and F) Quantitation of cell motility parameters in U2OS cells cotransfected with human WISp39 siRNA and either GFP-mouse WISp39 WT or GFP-mouse WISp39ΔTPR plasmid. Controls were transfected with control siRNA. Measurements of wound directionality and persistence were performed as described in Fig. 2. Number of control and WISp39 KD cells scored is 25 each from three independent experiments. Data shown in C, E, and F represent the means ± SD. ANOVA followed by Tukey’s test; ***, P ≤ 0.001. a.u., arbitrary unit; deg, degree.
We used the rescue assay to determine whether Hsp90 binding was important for the restoration of WISp39 function. For this, we used a mouse GFP-WISp39 mutant in which Lys285 and Arg289 in the tetratricopeptide repeat (TPR) binding motif, required for association with Hsp90, were mutated to alanine (mWISp39ΔTPR; Jascur et al., 2005). WISp39 KD cells expressing GFP-mWISp39ΔTPR were elongated with the irregular shape and multipolarity typical of WISp39 KD cells (Fig. 3, B and C). To rule out nonspecific effects caused by the GFP tag on WISp39, we performed additional rescue experiments using HA-tagged mouse WISp39 WT or WISp39ΔTPR combined with GFP-H2B to mark transfected cells. Whereas mWISp39 WT rescued cells were either apolar or unipolar in appearance and appeared similar to control cells, mWISp39ΔTPR-rescued cells uniformly showed multipolarity (Fig. 3 C), thus exhibiting the same distortion of cell shape apparent in WISp39 KD cells (Fig. 1 A and Table 1). Tracking analysis of representative GFP-expressing cells showed that mutant WISp39 lacking the Hsp90 binding domain did not rescue directionality of motility (Fig. 3 D). Consistent with these result, quantitative analysis showed that both directionality of cell migration and persistence of movement were restored to control levels by expression of mWISp39 WT but not by mWISp39ΔTPR (Fig. 3, E and F). We conclude that the WISp39 siRNA effects can be attributed to specific KD of WISp39 and that the directed cell motility function of WISp39 requires binding to Hsp90.
Coronin 1B binds to WISp39
To identify WISp39 interactors involved in migration, we performed a yeast two-hybrid screen on an embryonic mouse library using full-length mouse WISp39 as bait. We recovered a partial clone of mouse Coronin 1B, which has 100% sequence homology to the human orthologue and contains the fourth and fifth “blade” of Coronin 1B (mouse aa 170–279; UniProt accession no. Q9WUM3). Yeast two-hybrid analysis showed a specific interaction between the two proteins, as indicated by a positive signal with multiple probes (unpublished data).
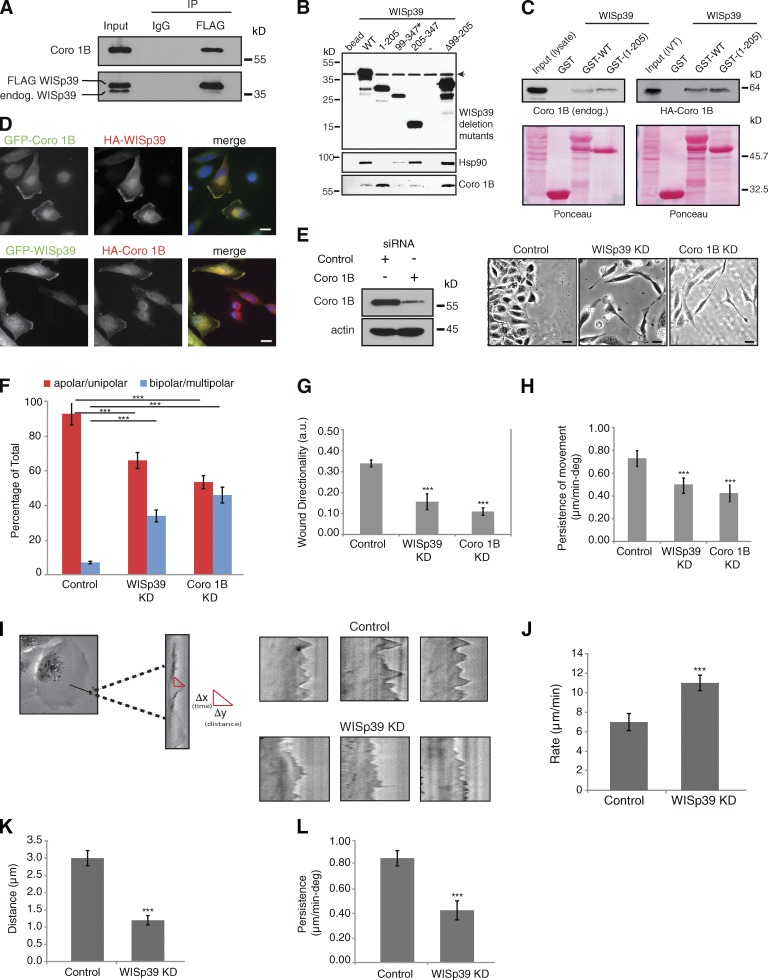
We confirmed the interaction between human WISp39 and Coronin 1B by expressing FLAG-tagged human WISp39 in cells and showing that it coimmunoprecipitated endogenous Coronin 1B from human cell lysates (Fig. 4 A). To determine the binding site on WISp39 for Coronin 1B, we expressed truncation mutants of WISp39 in HEK293 cells and performed pull-down assays from cell lysates. The results indicate that Coronin 1B binds to WISp39 in its N-terminal region between residues 1–98. In contrast, Hsp90 binds the C-terminal region (residues 205–347) of WISp39 (Fig. 4 B), supporting our previous results showing that WISp39 binds to Hsp90 through two conserved residues R285 and K289 in the C-terminal TPR region (Jascur et al., 2005). These results are consistent with the possibility that the three proteins could exist together in a multiprotein complex. The binding of WISp39 with Coronin 1B was further confirmed using purified bacterially expressed GST-tagged WISp39 WT to pull down Coronin 1B from cell lysates (Fig. 4 C). Furthermore, in vitro translated Coronin 1B binds to bacterially purified GST-tagged WISp39 WT or WISp39(1–205) (Fig. 4 C). Finally, cytoplasmic WISp39 strongly colocalizes with Coronin 1B at the actin-rich leading edge (Fig. 4 D).
Figure 4.
Coronin 1B binds to WISp39 and colocalizes with WISp39. (A) Coronin 1B (Coro 1B) binds WISp39. Extracts from U2OS cells transfected with FLAG-WISp39 were immunoprecipitated (IP) with either anti-FLAG or mouse IgG and subsequently Western blotted for associated endogenous Coronin 1B (top) and WISp39 (bottom). The experiment was repeated three times with similar results. endog., endogenous; IP, immunoprecipitation. (B) Mapping of the Coronin 1B binding region in WISp39. HEK293 cells were transfected with HA-tagged WISp39 WT or WISp39 truncation mutants. WISp39 and its associated proteins were isolated as described in the Materials and methods. The bound proteins were detected by Western blotting. Coronin 1B binds residues 1–98 of WISp39. Mutant 99–347 (asterisk) is only present in the nucleus, whereas the other WISp39 mutants and the WT WISp39 are both nuclear and cytoplasmic. Mutant Δ99–205 lacks residues 99–205 of WISp39. Arrow indicates a nonspecific band. Minus sign indicates another control lane. (C) In vitro binding of WISp39 to Coronin 1B. (left top) HeLa cell lysates were incubated with bacterially expressed GST-WISp39 WT or GST-WISp39(1–205) bound to glutathione-Sepharose, and the associated Coronin 1B was detected by Western blotting with Coronin 1B–specific antibodies. (right top) In vitro transcribed and translated (IVT) HA–Coronin 1B was incubated with GST-WISp39 WT or GST-WISp39(1–205) bound to glutathione-Sepharose, and the associated HA–Coronin 1B was detected by Western blotting with HA-specific antibodies. Ponceau staining of the nitrocellulose membranes is shown. The association of IVT HA–Coronin 1B with GST-WISp39 WT or GST-WISp39(1–205) is not notably different (right), perhaps because cellular proteins that associate with WT WISp39–Hsp90 reduce the fraction of WISp90–Hsp90 complex available for binding Coronin 1B. The experiments were repeated more than three times with similar results. (D) WISp39 colocalizes with Coronin 1B at the leading edge. HeLa cells were transfected with GFP–Coronin 1B and HA-WISp39 or HA–Coronin 1B and GFP-WISp39. Cells were fixed and stained with the anti-HA antibody. Bars, 10 µm. (E) Loss of Coronin 1B yields a morphology similar to WISp39 KD. U2OS cells transfected with either control siRNA or Coronin 1B siRNA or WISp39 siRNA for 48 h were wounded as in Fig. 2 A and then imaged. Phase-contrast images (20×) of Coronin 1B KD and WISp39 KD show elongated and multipolar morphology. Western blot shows KD of Coronin 1B at 48 h. Actin is a loading control. Bars, 10 µm. (F) Quantitation of Coronin 1B KD morphology. U2OS cells transfected with either control siRNA or Coronin 1B siRNA or WISp39 siRNA for 48 h were wounded as in Fig. 2 A. Cells were scored from phase-contrast images (20×) as either apolar/unipolar or bipolar/multipolar and are presented as a percentage of total cells scored from more than three independent experiments (control = 200, Coronin 1B KD = 404, and WISp39 KD = 258). Note that the cells were scored after wounding; therefore, they are somewhat constrained from producing multipolar morphology by the surrounding cells, unlike sparsely plated cells in Fig. 1 B. Thus, there are fewer multipolar cells in comparison to Fig. 1 B. (G and H) Decrease in directionality and persistence of movement in Coronin 1B KD cells. Quantitation of motility parameters was performed as in Fig. 2. Number of cells scored from three independent experiments: control (20); Coronin 1B KD (20); WISp39 KD (20). a.u., arbitrary unit. (I) Kymograph analysis. (left) A magnified region of the cell is outlined in the rectangular box. Sample kymographs for control and WISp39 KD cells are shown on the right. Experimental parameters are detailed in J–L. (J–L) Protrusion parameters of control and WISp39 KD cells. Kymographs were constructed along a 1-pixel-wide axis oriented in the direction of the protrusion and perpendicular to the edge of the lamellipodia using MetaMorph software. Rate, distance, and persistence of protrusions were calculated from the distance of protrusion (y axis) and time (x axis). Δy = distance of lamellipodia protrusions; Δx = persistence (time) of lamellipodia protrusions; Δy/Δx = rate of lamellipodia protrusions. 10 cells from three independent experiments (8–10 protrusions/cell) were analyzed. deg, degree. Data shown in F–H and J–L represent the means ± SD. (F–H) ANOVA followed by Tukey’s test; ***, P ≤ 0.001. (J–L) Student’s t test; ***, P ≤ 0.001.
Consistent with the importance of WISp39 to Coronin 1B function, WISp39 KD yielded a phenotype similar to the Coronin 1B KD phenotype (Fig. 4 E). After transfection of U2OS cells with Coronin 1B siRNA, the cells became elongated and multipolar in appearance, similar to WISp39 KD (Fig. 4, E and F). Time-lapse imaging of Coronin 1B KD cells (Video 7) showed the cells migrated in wound-healing assays in a chaotic fashion, similar to WISp39 KD. Quantitation of the motility parameters showed that WISp39 and Coronin 1B KD both decreased cell directionality and persistence in wound-healing assays (Fig. 4, G and H).
Kymograph recordings of the leading edge showed a substantial difference in lamellipodial appearance in WISp39 KD cells. Similar to Coronin 1B KD cells (Cai et al., 2007), WISp39 KD cells consistently extended protrusions at a higher rate than controls (Fig. 4, I and J). However, both the distance and persistence of the leading edge protrusions were decreased (Fig. 4, K and L).
We have previously demonstrated that WISp39 interacts with cyclin-dependent kinase inhibitor p21 and Hsp90 in a trimeric complex that stabilizes p21 against degradation during the cellular response to ionizing radiation (Jascur et al., 2005). As cytoplasmic p21 has been found to play a role in migration (Tanaka et al., 2002; Lee and Helfman, 2004), we asked whether p21 was involved in WISp39-directed migration in these cells. However, we found that suppression of p21 with siRNA did not influence any parameters of motility, including directionality of migration toward wound closure and persistence of migration (Fig. S2). Therefore, we conclude that WISp39 has two discrete functions, one involving the stabilization of p21, and another function interacting with Coronin 1B to regulate cell migration.
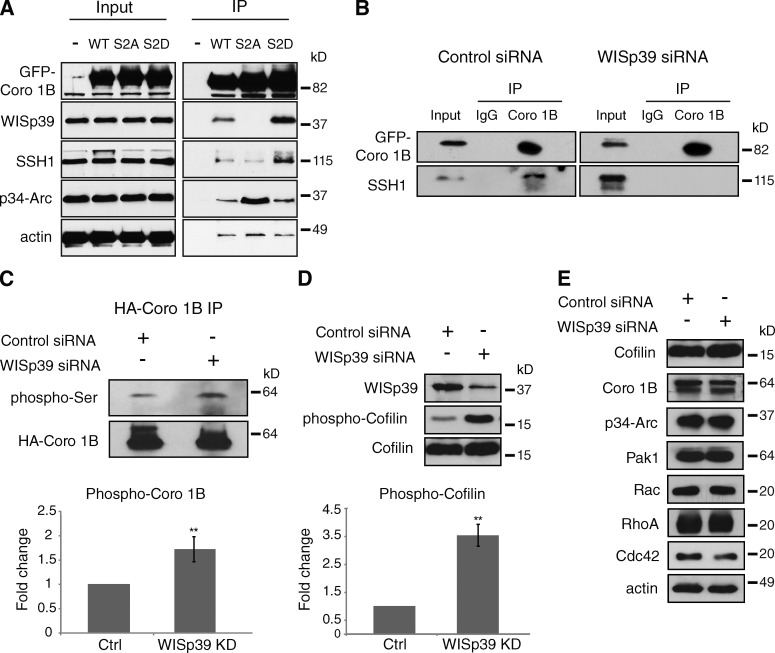
Phosphorylated Coronin 1B binds WISp39 and SSH
SSH binds and dephosphorylates Coronin 1B (Cai et al., 2007), and dephosphorylated Coronin 1B binds the Arp2/3 complex (Cai et al., 2005). Phosphorylation of Coronin 1B creates an inactive pool that can be activated by SSH. However, how an inactive pool of phosphorylated Coronin 1B becomes activated by SSH has remained unclear. We therefore addressed the role of WISp39 in regulating the dephosphorylation of Coronin 1B by SSH. For this, we expressed the phosphomimetic mutant of Coronin 1B(S2D) or the nonphosphorylatable mutant of Coronin 1B(S2A) in cells, and we found that only Coronin 1B(S2D) coimmunoprecipitated with endogenous WISp39 and SSH from cell extracts (Fig. 5 A). In contrast, Coronin 1B(S2A) did not coimmunoprecipitate with either protein. WT Coronin 1B, a mixture of phosphorylated and unphosphorylated forms, bound the two proteins weakly (Fig. 5 A). Furthermore, endogenous Arp2/3 complex (as indicated by p34-Arc subunit) coimmunoprecipitates with Coronin 1B(S2A), but not Coronin 1B(S2D), as has been shown previously (Cai et al., 2005). Collectively, these results are consistent with the existence of a complex between WISp39, SSH, and phosphorylated Coronin 1B that excludes Arp2/3.
Figure 5.
Phosphorylated Coronin 1B associates with WISp39 and SSH. (A) Coronin 1B (Coro 1B) associates with WISp39 and SSH when it is phosphorylated. HEK293 cells were transfected with GFP-tagged WT, S2A, or S2D Coronin 1B. Lysates were immunoprecipitated with anti-GFP antibody and analyzed by Western blotting with indicated antibodies. The experiments were repeated three times with similar results. (B) Coronin 1B associates with SSH only in the presence of WISp39. U2OS cells were cotransfected with control or WISp39 siRNA and GFP-tagged WT Coronin 1B. Lysates were immunoprecipitated with anti-GFP antibody and analyzed for association with SSH by Western blotting. The experiments were repeated more than three times with similar results. (C) Coronin 1B phosphorylation increases in the absence of WISp39. HeLa cells were cotransfected with control or WISp39 siRNA and HA-tagged WT Coronin 1B. Lysates were immunoprecipitated with the anti-HA antibody (HA–Coronin 1B) and analyzed for phosphorylation of Coronin 1B on Ser2 by Western blotting with phospho-PKC substrate-specific antibody (phospho-Ser). Quantitation of the phospho-Ser Coronin 1B in control and WISp39 KD cells from three experiments is shown. Student’s t test; **, P ≤ 0.01. (D) WISp39 KD increases phosphorylated Cofilin. U2OS cells were transfected with control or WISp39 siRNA, and the cell lysates were analyzed for phosphorylation by Western blotting with antibodies recognizing phospho-Cofilin. Quantitation of the phospho-Cofilin in control and WISp39 KD cells from three experiments is shown. Student’s t test; **, P ≤ 0.01. (E) WISp39 KD does not influence the levels of other motility-associated proteins. U2OS cells were transfected with control or WISp39 siRNA, and the cell lysates were analyzed by Western blotting with the indicated antibodies. Ctrl, control; IP, immunoprecipitation.
Further evidence for a complex of Coronin 1B, WISp39, and SSH comes from determining the interaction between Coronin 1B and SSH in the presence and absence of WISp39. In extracts from GFP–Coronin 1B-expressing cells transfected with control siRNA, we observed endogenous SSH coimmunoprecipitated with GFP–Coronin 1B, consistent with Cai et al. (2007). However, when cells were depleted of WISp39, SSH failed to coimmunoprecipitate with GFP–Coronin 1B (Fig. 5 B). Loss of WISp39 resulted in increased phosphorylation of Coronin 1B (Fig. 5 C), presumably as a result of the inability of SSH to associate with Coronin 1B and dephosphorylate it in the absence of WISp39. We also saw a dramatic increase in the phosphorylation of Cofilin in lysates from WISp39 KD cells compared with controls (Fig. 5 D). These results are consistent with the requirement of the Coronin 1B–SSH complex to activate Cofilin, as Coronin 1B KD or the expression of catalytically inactive SSH increases the levels of inactive phosphorylated Cofilin (Cai et al., 2007). Together, our data show that WISp39 promotes the association of phosphorylated Coronin 1B with SSH and that WISp39 controls the phosphorylation of both Coronin 1B and Cofilin by regulating the Coronin 1B–SSH association.
As we previously found that WISp39 increases the stability of p21 (Jascur et al., 2005), we checked whether the loss of WISp39 affected the stability of the proteins we were examining. We did not detect a difference in the protein levels of Coronin 1B, p34-Arc, or Cofilin after WISp39 KD (Fig. 5 E). We also did not detect any difference in the protein levels of the Rho GTPases Rac, RhoA, Cdc42, nor of the major regulating kinase Pak1, which could influence Cofilin phosphorylation (Fig. 5 E).
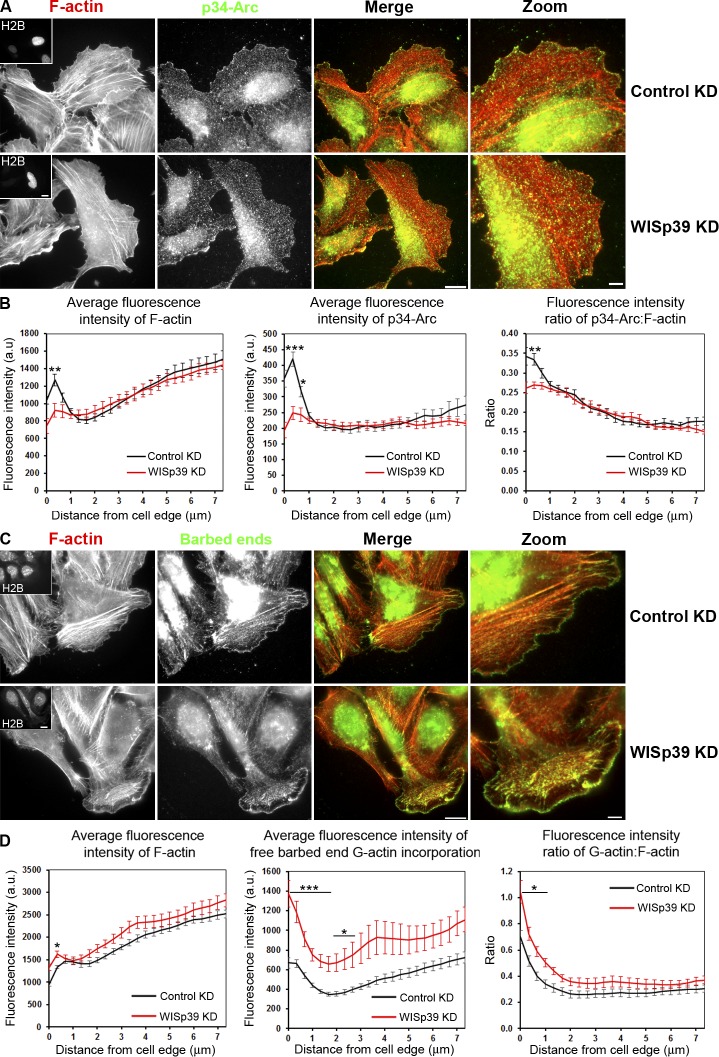
WISp39 KD decreases the localization of the Arp2/3 complex and increases the concentration of barbed ends at the leading edge
The phosphorylation state of Coronin 1B determines its binding to the Arp2/3 complex (Fig. 5 A; Cai et al., 2005), and WISp39 influences the phosphorylation of Coronin 1B. We therefore investigated the effect of WISp39 on Arp2/3 complex distribution at the leading edge. WISp39 colocalizes with the p34-Arc subunit of the Arp2/3 complex at the leading edge, as indicated by the robust correlation coefficients obtained in linear regression analysis (Fig. S3). We next determined whether the loss of WISp39 would have an effect on Arp2/3 distribution at the leading edge. We consistently found less p34-Arc subunit at the leading edge of WISp39 KD cells compared with controls, irrespective of the cell morphology (Fig. 6, A and B; and Fig. S4). This result was unexpected, as increased phosphorylation of Coronin 1B in WISp39 KD cells should reduce the ability of Coronin 1B to bind and inhibit the Arp2/3 complex, resulting in increased localization of active Arp2/3 complex to the leading edge. As WISp39 affects Arp2/3 complex localization at the leading edge, we next checked whether the barbed ends were affected by the loss of WISp39. Despite the decreased Arp2/3 complex localization, WISp39 depletion increased the concentration of barbed ends relative to total actin at the leading edge (Fig. 6, C and D). Our data are consistent with a previous study showing increased free barbed ends in Coronin 1B KD cells (Cai et al., 2007).
Figure 6.
WISp39 KD decreases the localization of Arp2/3 complex and increases the generation of free barbed ends at the cell leading edge. (A) WISp39 KD decreases Arp2/3 complex localization at the cell leading edge. Immunolocalization of the Arp2/3 subunit p34-Arc and F-actin phalloidin staining in cells transfected for 48 h with control or WISp39 siRNA. U2OS cells were transfected with WISp39 siRNA in combination with a GFP plasmid expressing histone H2B to visualize transfected cells. After 48 h, the cells were fixed, and immunofluorescence images were acquired as described in Materials and methods. Insets in the first image show a cell expressing GFP-H2B, cotransfected with the siRNA. Magnified regions are shown in the right-most image. (B) Quantification of fluorescence intensity (±SEM) of F-actin, p34-Arc, and ratio of p34-Arc/F-actin in control and WISp39-depleted cells, measured from the cell edge (0) to the cell center (7 µm). Bands of constant distance to the cell edge were constructed, and individual fluorescence intensities were accumulated and averaged in each band to produce graphs of fluorescence intensities versus distance to the cell edge (Fig. S5). The data shown represent one experiment and are averaged from n ≥ 11 cells for each condition. Only cells with a spread morphology have been included. Student’s t test; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001 compared with control siRNA cells. The experiment was repeated three times, with similar results. (C) G-actin incorporation marking free barbed ends and F-actin phalloidin staining in cells transfected for 48 h with control or WISp39 siRNA. U2OS cells were transfected with WISp39 siRNA in combination with GFP plasmid expressing histone H2B for 48 h, and immunofluorescence localization of barbed ends was performed by live-cell microscopy. Insets in the first image show the expression of H2B, cotransfected with the siRNA. Magnified regions are shown in the right-most image. WISp39 depletion induces the formation of free barbed filament ends. (D) Quantification of fluorescence intensity of F-actin, free barbed ends, and the ratio of free barbed ends: F-actin in control and WISp39 KD cells, measured from the cell edge (distance = 0) to the cell center (7 µm). Quantitation was performed as in B. The data shown represent one experiment and are averaged from n ≥ 15 cells for each condition (±SEM). Only cells with a spread morphology have been included. Student’s t test; *, P ≤ 0.05; ***, P ≤ 0.001 compared with control siRNA cells. The experiment was repeated four times, with similar results. The mean fluorescent F-actin intensity is elevated slightly relative to B, and this variation may arise because the analysis in B was performed on fixed cells. a.u., arbitrary unit. Bars: (left images and insets) 15 µm; (zoom images) 5 µm.
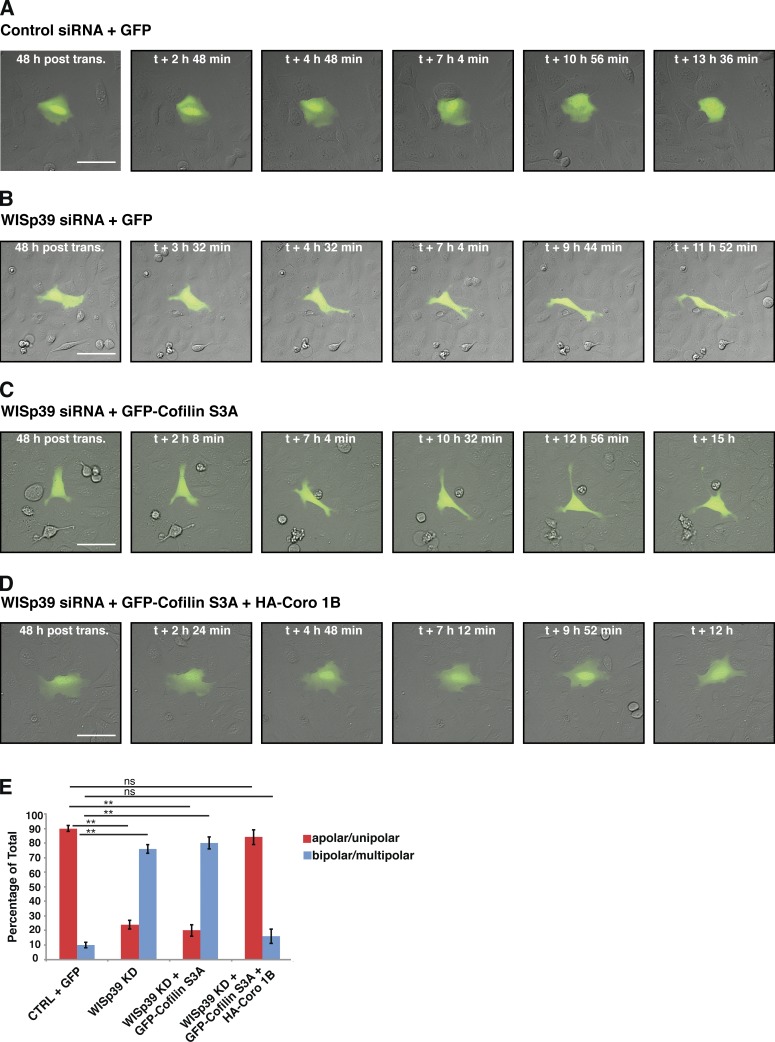
WISp39 KD phenotype is rescued by overexpression of Coronin 1B and constitutively active Cofilin
Because WISp39 promotes the dephosphorylation of Coronin 1B and Cofilin, the profound changes in cell morphology and directional migration observed in WISp39 KD cells may ultimately be a result of the loss of proper Coronin 1B and Cofilin activation and function, which in turn would affect Arp2/3 complex function. To test this, we attempted to rescue the phenotype of WISp39 KD by expressing constitutively active Cofilin(S3A) alone or in combination with Coronin 1B WT (Fig. 7). Cofilin(S3A) alone was unable to rescue the phenotype created by depletion of WISp39, as measured by morphology changes from apolar to bi- and multipolar cells (Fig. 7, C and E). In contrast, Cofilin(S3A) expressed together with Coronin 1B WT was able to rescue the elongated, irregular shape and multipolarity typical of WISp39 KD cells to the extent that they appeared similar to control cells (Fig. 7, D and E). Collectively, these results suggest that WISp39 plays an important role in regulating actin-dependent lamellipodial dynamics at the leading edge, by binding phosphorylated Coronin 1B in a complex with SSH and coordinating the activation of Cofilin and the regulation of the Arp2/3 complex.
Figure 7.
WISp39 KD can be rescued by expression of Cofilin(S3A) and WT Coronin 1B. (A and B) Images of control and WISp39 KD cells. U2OS cells were transfected with control (A) or WISp39 (B) siRNA and with GFP plasmid to visualize transfected cells. 20× DIC images were taken at 48 h after transfection over a period of 16 h. A control cell (A) retains normal apolar/unipolar morphology throughout the experiment, unlike a WISp39 KD cell (B), which becomes elongated and multipolar. (C) Images of failure of Cofilin(S3A) to rescue WISp39 KD. U2OS cells were cotransfected with human WISp39 siRNA and GFP-Cofilin(S3A) for 48 h, and one cell was imaged as in A. WISp39 KD cell transfected with GFP-Cofilin(S3A) shows abnormal cell polarity similar to that of WISp39 KD cells. (D) Images of rescue of WISp39 KD by Cofilin(S3A) and Coronin 1B. U2OS cells were cotransfected with human WISp39 siRNA and GFP-Cofilin(S3A) and HA–Coronin 1B WT for 48 h, and 20× DIC images were taken. WISp39 KD cell transfected with GFP-Cofilin(S3A) and HA–Coronin 1B (Coro 1B) retains normal apolar/unipolar morphology throughout the experiment. (E) Quantitation of WISp39 KD rescue by Cofilin(S3A) alone or Cofilin(S3A) and Coronin 1B WT. U2OS cells were cotransfected for 48 h. Cells were scored as either apolar/unipolar or bipolar/multipolar and presented as a percentage of total cells scored. Number of rescued cells scored: control siRNA + GFP (22); WISp39 KD + GFP (21); WISp39KD + GFP-Cofilin(S3A) (25); WISp39KD + GFP-Cofilin(S3A) + HA–Coronin 1B WT (25). Note that only fluorescent cells were scored. Data represent the means ± SD. Student’s t test; **, P ≤ 0.01; ns, not significant. CTRL, control; post trans., posttransfection. Bars, 50 µm.
Discussion
We demonstrate that WISp39 is important for controlling directed cell motility in mammalian cells. We show that WISp39 preferentially binds phosphorylated Coronin 1B and establishes a complex with SSH. Our data show that WISp39, in a multiprotein complex with Coronin 1B and SSH, plays an important role in the dephosphorylation of Cofilin and localization of the Arp2/3 complex at the leading edge. As a result, ablation of WISp39 decreases directed cell motility. Thus, by controlling Coronin 1B function as a regulator of the Arp2/3 complex and Cofilin, WISp39 is essential to the control of cell polarity and directed motility.
WISp39 interacts with phosphorylated Coronin 1B
We find that WISp39 interacts with Coronin 1B, a protein required for normal leading edge dynamics (Chan et al., 2011). Endogenous Coronin 1B coimmunoprecipitates with WISp39 in cell extracts, and the two proteins bind in vitro. WISp39 and Coronin 1B also colocalize at the leading edge. WISp39 KD cells show an increased rate of protrusion and a decrease in both protrusion distance and persistence at the leading edge, identical to the effect of Coronin 1B KD in cells (Cai et al., 2007).
WISp39 specifically binds to Coronin 1B that is phosphorylated on Ser2, and this complex contains negligible Arp2/3 complex. Coronin 1B binds to the Arp2/3 complex and inhibits actin nucleation by the complex (Mishima and Nishida, 1999; Cai et al., 2005, 2007; Chan et al., 2011). This binding is regulated by PKC phosphorylation of Coronin 1B on Ser2. Phosphorylation reduces its association with the Arp2/3 complex, and the nonphosphorylatable mutant S2A shows stronger interaction with the Arp2/3 complex than WT Coronin 1B (Cai et al., 2005). Coronin 1B also interacts with SSH and has been reported to associate in an apparent ternary complex with the Arp2/3 complex (Cai et al., 2007), but the phosphorylation state of Coronin 1B in this complex was not assessed. Our data show that Coronin 1B exists in two distinct complexes; unphosphorylated Coronin 1B bound to the Arp2/3 complex without SSH or WISp39 and phosphorylated Coronin 1B bound to WISp39 and SSH but not the Arp2/3 complex. Immunoprecipitation results confirm that two complexes exist. The phosphomimetic Coronin 1B(S2D) mutant associates robustly with SSH and WISp39 but negligibly with the Arp2/3 complex, whereas nonphosphorylatable Coronin 1B(S2A) associates robustly with the Arp2/3 complex but negligibly with SSH and WISp39. Furthermore, loss of WISp39 decreases the association of SSH with Coronin 1B, suggesting that SSH requires WISp39 to associate with Coronin 1B. Thus, we conclude that WISp39 is an integral component of the phosphorylated Coronin 1B–SSH complex.
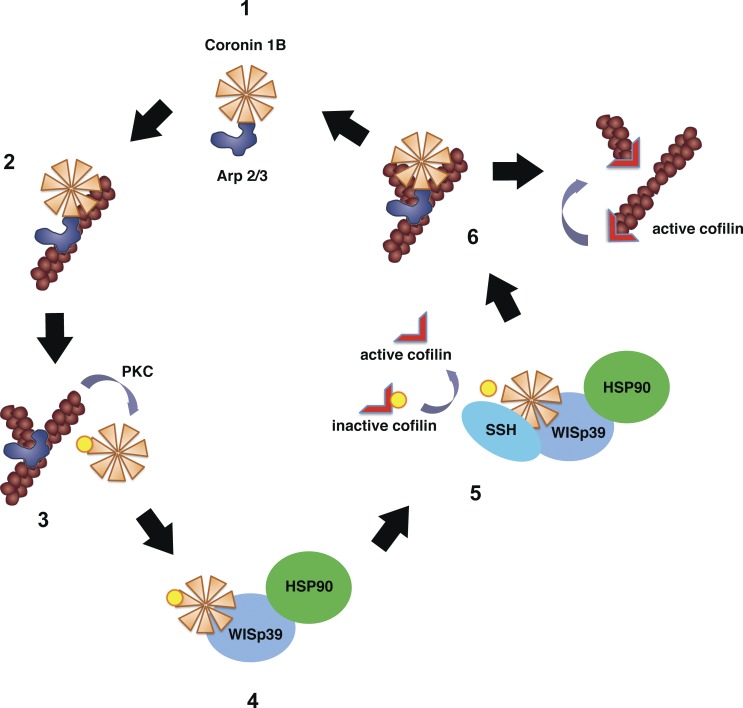
Increased phosphorylation of Coronin 1B in WISp39 KD cells reduces the ability of Coronin 1B to bind the Arp2/3 complex and consequently inhibit it. Although this should result in more active Arp2/3 complex and increased localization at the leading edge, instead we observe a decreased localization of the Arp2/3 complex at the leading edge in WISp39 KD cells. It is possible that a decrease in dephosphorylated Coronin 1B in WISp39 KD cells reduces the efficiency of Arp2/3 complex localization to the side of the actin filament (Fig. 8, step 2; Humphries et al., 2002) and therefore decreases the abundance of Arp2/3 complex at the leading edge.
Figure 8.
Model of WISp39 function. Unphosphorylated Coronin 1B binds Arp2/3 complex (step 1; Cai et al., 2005) and localizes it to the side of the actin filament (step 2) but prevents branching by Arp2/3 complex (Humphries et al., 2002). Coronin 1B phosphorylated by PKC on Ser2 loses its affinity for Arp2/3 complex (Cai et al., 2005; this paper), allowing Arp2/3 complex-mediated branching (step 3). Phosphorylated Coronin 1B binds WISp39 in a complex with SSH and Hsp90 (step 4; this paper). SSH dephosphorylates Coronin 1B and Cofilin (step 5; Cai et al., 2007; this paper). Dephosphorylated Coronin 1B binds Arp2/3 complex and removes it from the branch (step 6; Cai et al., 2008). The destabilized branch is then severed by active Cofilin (Pollard and Borisy, 2003).
The effect of WISp39 on cell motility
We show that WISp39 is essential to the regulation of cell polarity and directed motility. Consistent with a regulatory role, siRNA-mediated depletion of WISp39 increases the lamellipodial protrusion rate but decreases both protrusion persistence and protrusion distance of migrating cells. Expression of the WISp39 plasmid in siRNA-treated cells restores directionality to control levels. In addition, Hsp90 is an important component of the regulatory complex, as WISp39 mutants that do not bind Hsp90 do not rescue the directionality defect or the decreased persistence of movement. Hsp90 has been shown to bind actin (Koyasu et al., 1986) and to increase the stability of proteins that control actin dynamics (Li et al., 2006; Park et al., 2007). Our observations suggest a novel mechanism by which Hsp90 participates in actin-mediated cell motility, through WISp39.
The role of WISp39 in cell motility can be understood in the context of its interaction with Coronin 1B. Coronin 1B is known to control the phosphorylation of Cofilin (Cai et al., 2007), and we find that WISp39 also promotes the phosphorylation of Cofilin. We observed that the inactive phosphorylated form of Cofilin is strongly increased in the absence of WISp39 and that the morphological defects we observed in WISp39-depleted cells could be rescued by coexpression of Coronin 1B and constitutively active Cofilin(S3A). The overexpressed Coronin 1B likely provides a pool of dephosphorylated Coronin 1B, which is not only necessary to localize Arp2/3 complex to the side of the actin filament but also to remove the Arp2/3 complex from the actin branch to facilitate Cofilin function (Fig. 8, steps 2 and 6; Cai et al., 2008). These results show that WISp39 is important for the regulation of cell motility and that WISp39 KD does indeed disrupt the function of Cofilin, which requires the Coronin 1B–SSH complex to dephosphorylate and activate it. We observe one difference between the effect of WISp39 KD and Coronin 1B KD on cells, namely that WISp39 KD cells migrate faster than controls, whereas Coronin 1B KD cells migrate slower (Cai et al., 2007). Importantly, Cofilin KD cells also migrate faster and show increase polarity similar to WISp39 KD cells (Sidani et al., 2007). Therefore, the WISp39 KD phenotype most likely represents a phenotype with attributes of both Coronin 1B KD and Cofilin KD.
We find that WISp39 colocalizes with Arp2/3 and although WISp39 down-regulation results in the reduction of the Arp2/3 complex at the leading edge, we observed a dramatic increase in the abundance of F-actin barbed ends. An increase in free barbed ends was also shown to occur after Coronin 1B KD (Cai et al., 2007). However, Arp2/3 complex localization was not directly tested in Coronin 1B KD cells in those studies. Increased barbed ends can result from an increase in actin filament severing activity when active Cofilin is at lower concentrations with respect to F-actin (Andrianantoandro and Pollard, 2006) through a mechanism involving Aip1 (Chen et al., 2015). Decreasing the active pool of Cofilin through increased phosphorylation in WISp39 KD cells (Fig. 5) may enhance severing and generate more barbed ends through a similar mechanism. Further work is necessary to determine the cause of the increase in concentration of barbed ends at the leading edge, both in WISp39 KD and in Coronin 1B KD cells.
Our data support a key role for WISp39 in regulating actin dynamics, thus sustaining directed cell motility (Fig. 8). Our evidence suggests that WISp39 acts as a crucial scaffold that integrates Coronin 1B, SSH, and Arp2/3 complex at the leading edge. In the absence of WISp39, the complex is disrupted, which leads to the loss of Cofilin activation, with consequences for the dynamics of the lamellipodia. Future studies will focus on elucidating the detailed mechanism by which WISp39 coordinates Coronin 1B, Arp2/3 complex, and Cofilin activity in the lamellipodia to achieve directed cell motility.
Materials and methods
Cell culture and extracts
U2OS, HeLa, and HEK293 cell lines were purchased from ATCC. Cells were trypsinized, washed, and lysed with buffer containing 50 mM Tris, 0.5% NP-40, 150 mM NaCl, 50 mM sodium fluoride, 1 mM sodium orthovanadate, 1 mM phenylmethylsulfonyl fluoride, 10 µg/ml aprotinin, and 5 µg/ml leupeptin. After incubation for 30 min on ice, the extracts were centrifuged at 14,000 rpm in a microfuge (Beckman Coulter) for 20 min at 4°C, and the supernatant was collected.
Antibodies
The following antibodies were used: rabbit polyclonal actin (A 2066) and mouse monoclonal Flag M2 antibody (F3165) obtained from Sigma-Aldrich; Hsp90 A/B (N-17; sc-1055) and rabbit polyclonal Vinculin (H-300) from Santa Cruz Biotechnology, Inc.; mouse monoclonal phospho-Cofilin (#3313), rabbit polyclonal Pak1 (#2602), rabbit polyclonal Cdc42 (#2462), rabbit polyclonal Rac1/2/3 (#2467), rabbit polyclonal phospho-(Ser)PKC substrate antibody (#2261), and rabbit polyclonal RhoA (#2117) from Cell Signaling Technology; and rabbit polyclonal Coronin 1B (A301-316A) and rabbit polyclonal SSH (A301-307A) from Bethyl Laboratories, Inc. Other antibodies were rabbit polyclonal p34-Arc (07–227; EMD Millipore), rabbit polyclonal Cofilin (ACFL02; Cytoskeleton, Inc.), and HA mouse monoclonal antibody (clone 16B12; Covance). Anti-WISp39 antibodies were raised by immunizing rabbits with purified recombinant full-length GST-WISp39 protein (Jascur et al., 2005). The sera were preadsorbed on a GST column before purification on an agarose matrix with covalently attached GST-WISp39.
Expression vectors
HA-tagged mouse WISp39 WT and WISp39ΔTPR (Lys285 and Arg289 mutated to Ala) in pJF-HA vectors have been previously described (Jascur et al., 2005). In brief, pJF-HA plasmid (with SRa promoter) was derived from pJFE14-SRa plasmid by introducing a HA epitope tag upstream of the Eco RI–Bam HI cloning site. HA-tagged WISp39ΔTPR (Lys285Ala and Arg289Ala) was generated by site-directed mutagenesis using QuikChange II (Agilent Technologies) in the pJF-HA vector. We also generated GFP-tagged mouse and human plasmids (mouse EGFPC2-WISp39 WT and EGFPC2-WISp39ΔTPR), human EGFPC2-WISp39 (all EGFPC2 plasmids contain the cytomegalovirus [CMV] promoter) as well as pFLAG-CMV2-WISp39 and human CMV-HA–Coronin 1B. Other plasmids used were GFP–Coronin 1B, GFP–Coronin 1B(S2A), and GFP–Coronin 1B(S2D) in pMSCV (J. Bear, University of North Carolina, Chapel Hill, Chapel Hill, NC), pCDNA-GFP-Cofilin(S3A) (G. Bokoch, Scripps Institute, La Jolla, CA), and myc-His–tagged SSH in pCDNA 3.1 with C-terminal myc-His6 tag (cloning site Hind III–Xho I; K. Mizuno, Tohoku University, Miyagi, Japan). Additional plasmids are described in the next section.
Immunoprecipitation and pull-down assays
Most assays are as described previously (Jascur et al., 2005). In brief, lysates were cleared at 14,000 g for 10 min and incubated with primary antibody for 1 h at 4°C followed by the addition of 20 µl of 50% slurry of ImmunoPure-immobilized protein A/G beads and further incubation at 4°C for an additional 45 min. The immune complexes were collected, washed with buffer three times, separated by SDS-PAGE, and transferred to nitrocellulose membrane for Western blotting. For Fig. 4 B, we generated pKLO.1 plasmids (with a CMV promoter) expressing C-terminal HA-tagged WISp39 (WT or truncation mutants), with the IgG-binding site of ProtA (protein A) from Staphylococcus aureus upstream of a cleavage site for human rhinovirus 3C protease (also known as the PreScission [Prp] site) to enable proteolytic release of HA-WISp39 proteins from beads. HEK293 cells were transfected with ProtA-Prp-WISp39 fusion constructs, and the cell lysates were incubated with IgG beads to recover the ProtA-Prp-WISp39 fusion protein and its associated proteins. After washing, protease was added to release the bound material, which was loaded on gels and Western blotted. For Fig. 4 C (left), we generated pGEX-4T1 plasmids (containing a tac promoter) expressing GST-tagged WISp39 (WT or truncation mutant) in bacteria. HeLa cell lysates were incubated with glutathione-Sepharose–bound GST-WISp39 to recover WISp39-associated proteins. After washing, the proteins were eluted with Laemmli buffer, and proteins were separated by SDS-PAGE and then transferred to nitrocellulose membrane for Western blotting. For Fig. 4 C (right), we generated HA-tagged pCDNA3.1–Coronin 1B plasmid to express in vitro translated Coronin 1B. In brief, Coronin 1B was in vitro transcribed and translated using the quick coupled transcription/translation system (TNT; Promega). The reaction was then incubated with glutathione-Sepharose–bound GST-WISp39, and the bound proteins were analyzed by Western blotting.
siRNA transfection
Cells were transfected with RNA oligomers using Oligofectamine (Life Technologies) for 4–6 h in serum-free medium, and then, 20% of serum was added (Jascur et al., 2005). WISp39 siRNA (5′-AAUUCUAGAGCAUACUCAAdTdT-3′ and 5′-ACUGGUUGCUGAACUUGAAdTdT-3′) and Luciferase GL3 siRNA (5′-CUUACGCUGAGUACUUCGAdTdT-3′) used as a negative control were synthesized by Thermo Fisher Scientific (Jascur et al., 2005). The siRNA designed to deplete human WISp39 differs by seven nucleotides from the mouse sequence and does not affect the levels of coexpressed mouse WISp39. Coronin 1B siRNA (5′-GACGAAGUCAUAGCCAGCdTdT-3′ and 5′-GGGUGGGCUCUAUAUUUUCdTdT-3′; Cai et al., 2007) was synthesized by Ambion (Life Technologies).
Live-cell imaging
Confluent U2OS monolayers, grown in each chamber of a LabTek 8-chamber glass slide in DMEM (Mediatech) supplemented with 10% fetal bovine serum, were wounded with a pipette tip, and filming was initiated 1 h after wounding. Low magnification phase-contrast videos were taken at 37°C using an inverted microscope (TE-200; Nikon) equipped with a 20× Plan Fluor phase lens (NA 0.5), a 12-bit chilled charge-coupled device (CCD) camera (C4742-95 12G04; Hamamatsu Photonics), and a robotic XYZ microscope stage (MS-2000; Applied Scientific Instruments, Inc.) equipped with linear positional feedback controllers (Haidenhain) on all three axes. Images (200-ms exposure) were collected at 7-min intervals for 24 h. Up to 30 fields were simultaneously counted in each experiment. Image stacks were generated in MetaMorph (Universal Imaging Corp.) and then converted to 14-s QuickTime (Apple) videos at 17 frames/s. For Fig. 7 and Fig. S1, cells were grown in Liebovitz’s L-15 medium (Mediatech) supplemented with 10% fetal bovine serum, and live-cell images were captured in differential interference contrast (DIC) at 37°C with an inverted microscope (IX81; Olympus) using an LC Plan FIuor 20×/NA 0.40 objective, within a DeltaVision Deconvolution system (Applied Precision) equipped with a CCD camera (CoolSNAP; Photometrics). The images were acquired with softWoRx software (Applied Precision).
DIAS image analysis
Cell shape and direction were calculated using DIAS (Dynamic Image Analysis System; Soll Technologies, Inc.) software. Directionality is a measure of path linearity and was derived by dividing net by total path length. Total path length is the distance the cell traveled from the first frame to the last frame taken as a straight line approximation every four frames; net path length is the distance of the cell from the starting frame to the last frame. Maximum length to maximum width ratio was quantitated as follows. Maximum length is defined as the major axis, which is the longest distance between any two points along the cell boundary; maximum width is defined as the longest chord that is perpendicular to the major axis. Persistence, a measurement of how far a cell moves in the same direction, was measured as the frame-to-frame velocity divided by a function of direction change in the same set of image frames.
Statistical analysis
Unpaired two-tailed Student’s t test was performed using Excel (Microsoft) and Prism version 6 (GraphPad Software). One-way analysis of variance (ANOVA) followed by Tukey’s test was performed using StatPlus software version 5 (AnalystSoft).
Kymography analysis
To analyze the leading edge of WISp39 KD and control cells, we captured 300 frames from each cell at intervals of 1 s using phase-contrast microscopy. For kymograph analysis, a single pixel line was drawn in MetaMorph perpendicular to the cell edge (4–8 lines drawn per cell, at least five cells per group). The region was then extracted from each image of the time lapse to generate a montage of the region over time. Lamellipodia were identified as regions where the cell edge extends (y axis = distance) with respect to time (x axis). A bounding rectangle drawn for each lamellipodium was used to calculate the protrusion time or persistence (x axis) and the protrusion distance (y axis), and the protrusion velocity was calculated by dividing the distance by time.
Immunofluorescence microscopy and analysis of Arp2/3 complex and barbed ends
U2OS cells were plated on glass coverslips 2 d before the experiment. Cells were transfected with control or WISp39 siRNA, in combination with 20 ng DNA encoding GFP-tagged H2B. 48 h after transfection, cell layers were wounded. Cells were allowed to migrate into the wound for 5 h before being processed for immunofluorescence microscopy as previously described (Delorme-Walker et al., 2011). In brief, cells were fixed in cytoskeletal buffer (CB; 10 mM MES, 3 mM MgCl2, 138 mM KCl, and 2 mM EGTA, pH 6.9) containing 4% paraformaldehyde, permeabilized in CB containing 0.5% Triton X-100, and blocked with 2% BSA in CB. Cells were then immunolabeled for p34-Arc (1:300; EMD Millipore) using Alexa Fluor 647–conjugated anti–rabbit secondary antibodies (1:250; Life Technologies). Immunofluorescence localization of free barbed filament ends was performed in live cells permeabilized for 2 min with 0.25 mg/ml saponin in rinsing buffer (20 mM Hepes, pH 7.5, 138 mM KCl, 4 mM MgCl2, 3 mM EGTA, and 1 mM ATP) in the presence of 0.5 mM X-rhodamine actin. Cells were then immediately fixed in CB containing 0.5% glutaraldehyde and processed as previously described (Symons and Mitchison, 1991). F-actin was visualized using Alexa Fluor 350–conjugated phalloidin at 1:10. Cells were mounted on slides with ProLong Gold antifade reagent (Life Technologies). Immunofluorescence images of fixed cells were acquired on an inverted microscope (Eclipse TE 2000-U; Nikon) equipped with an electronically controlled shutter, filter wheels, and a 14-bit cooled CCD camera (CoolSNAP HQ) controlled by MetaMorph software using a 60×/1.4 NA Plan Apochromat DIC objective lens (Nikon).
Quantification of the fluorescence intensity of F-actin, p34-Arc, and free barbed filament ends as a function of a distance from the cell leading edge was obtained with custom software, QFSM (quantitative fluorescent speckle microscopy), written in MATLAB (MathWorks; Mendoza et al., 2011). This software is freely available at http://lccb.hms.harvard.edu/software.html. The quantification represents fluorescence intensity along the entire leading edge as illustrated in Fig. S5. Bands of constant distance to the cell edge were constructed, and individual fluorescence intensities were accumulated and averaged in each band to produce fluorescence intensities versus distance to the cell edge graphs. Only cells expressing GFP-H2B, used as a control for transfection efficiency with siRNA, were analyzed.
Stained cells in Fig. 4 D and Fig. S3 were imaged with an inverted microscope (IX81) using a Plan Apochromat 60×/1.35 NA oil objective, within a DeltaVision Deconvolution system equipped with a CoolSNAP CCD camera. Secondary antibodies used were Alexa Fluor 488 and 594 (Life Technologies). The images were acquired with softWoRx software. For Fig. S3, images were analyzed using the plugin Coloc2 included in the Fiji software package (version 1.49g; National Institutes of Health). The different colocalization algorithms of the plugin were analyzed with a 2.5 point spread function value for these images.
Yeast two hybrid
The screen was performed as described previously (Jascur et al., 2005) except that cDNA encoding full-length mouse WISp39 cloned in frame to the Gal4 DNA binding domain in pGBKT7 (Takara Bio Inc.) was used as the bait. A mouse whole embryo (stage embryonic day 17.5 [E17.5]) cDNA library, cloned into pACT2 (Takara Bio Inc.), was used for screening (gift from T. Maynard and P. Barnes, University of North Carolina, Chapel Hill). AH-109 yeast strain was sequentially transformed with pGBKT7-WISp39 and then with pACT mouse E17.5 cDNA library. Double transformants were allowed to grow on synthetic dextrose medium deficient in leucine (Leu), tryptophan (Trp), and histidine (His). Colonies were then restreaked on plates lacking Leu, Trp, His, and adenine and containing X-α-Gal. Clones that were phenotypically His+, Ade+, and lacZ+ were rescued and sequenced. Approximately 106 transformants were screened.
Online supplemental material
Fig. S1 shows that the change in cell morphology and the chaotic migration observed in WISp39 KD cells is not caused by induction of apoptosis. Fig. S2 shows that p21 KD does not promote abnormal cellular morphology or directional migration defects characteristic of WISp39 KD. Fig. S3 shows colocalization and quantitation of WISp39 and Arp2/3 complex at the cell leading edge. Fig. S4 shows additional images of decreased Arp2/3 complex localization at the cell leading edge in WISp39 KD cells and quantification of cells with different lamellipodium sizes. Fig. S5 shows how quantification of the fluorescence intensity in Fig. 6 was performed. Videos 1 and 2 show live-cell imaging of control siRNA (Video 1) and WISp39 siRNA (Video 2)–transfected cells. Video 3 shows live-cell imaging of control cells without ZVAD. Video 4 shows live-cell imaging of control cells treated with ZVAD. Video 5 shows live-cell imaging of WISp39 KD cells without ZVAD. Video 6 shows live-cell imaging of WISp39 KD cells treated with ZVAD Video 7 shows that changed cell morphology and chaotic migration of Coronin 1B KD cells is similar to WISp39 KD. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201410095/DC1.
Supplementary Material
Acknowledgments
We dedicate this paper to the fond memory of Arun Fotedar.
We thank Clare Waterman for the use of microscopes and James Bamburg for helpful discussions. We thank Leslie Boyd from the Sanford-Burnham Cellular Imaging facility for helpful discussions. We are grateful to Tom Maynard, Paul Barnes, James Bear, Gary Bokoch, and Kensaku Mizuno for the kind gift of reagents. We thank Mourad Bendjennat for generating some of the data presented in the manuscript. We thank Tiffany Dzou for assistance with the yeast two-hybrid screen.
This work was supported by National Institutes of Health grants to R. Fotedar (CA108947 and CA101810) and R.L. Margolis (GM088716).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- ANOVA
- analysis of variance
- CB
- cytoskeletal buffer
- CCD
- charge-coupled device
- CMV
- cytomegalovirus
- DIC
- differential interference contrast
- Hsp90
- heat shock protein 90
- KD
- knockdown
- Prp
- PreScission
- SSH
- Slingshot phosphatase
- TPR
- tetratricopeptide repeat
- WISp39
- Waf1 Cip1 stabilizing protein 39
- WT
- wild type
References
- Agnew B.J., Minamide L.S., and Bamburg J.R.. 1995. Reactivation of phosphorylated actin depolymerizing factor and identification of the regulatory site. J. Biol. Chem. 270:17582–17587 10.1074/jbc.270.29.17582 [DOI] [PubMed] [Google Scholar]
- Andrianantoandro E., and Pollard T.D.. 2006. Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol. Cell. 24:13–23 10.1016/j.molcel.2006.08.006 [DOI] [PubMed] [Google Scholar]
- Bamburg J.R., McGough A., and Ono S.. 1999. Putting a new twist on actin: ADF/cofilins modulate actin dynamics. Trends Cell Biol. 9:364–370 10.1016/S0962-8924(99)01619-0 [DOI] [PubMed] [Google Scholar]
- Cai L., Holoweckyj N., Schaller M.D., and Bear J.E.. 2005. Phosphorylation of coronin 1B by protein kinase C regulates interaction with Arp2/3 and cell motility. J. Biol. Chem. 280:31913–31923 10.1074/jbc.M504146200 [DOI] [PubMed] [Google Scholar]
- Cai L., Marshall T.W., Uetrecht A.C., Schafer D.A., and Bear J.E.. 2007. Coronin 1B coordinates Arp2/3 complex and cofilin activities at the leading edge. Cell. 128:915–929 10.1016/j.cell.2007.01.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L., Makhov A.M., Schafer D.A., and Bear J.E.. 2008. Coronin 1B antagonizes cortactin and remodels Arp2/3-containing actin branches in lamellipodia. Cell. 134:828–842 10.1016/j.cell.2008.06.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K.T., Creed S.J., and Bear J.E.. 2011. Unraveling the enigma: progress towards understanding the coronin family of actin regulators. Trends Cell Biol. 21:481–488 10.1016/j.tcb.2011.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Courtemanche N., and Pollard T.D.. 2015. Aip1 promotes actin filament severing by cofilin and regulates constriction of the cytokinetic contractile ring. J. Biol. Chem. 290:2289–2300 10.1074/jbc.M114.612978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delorme V., Machacek M., DerMardirossian C., Anderson K.L., Wittmann T., Hanein D., Waterman-Storer C., Danuser G., and Bokoch G.M.. 2007. Cofilin activity downstream of Pak1 regulates cell protrusion efficiency by organizing lamellipodium and lamella actin networks. Dev. Cell. 13:646–662 10.1016/j.devcel.2007.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delorme-Walker V.D., Peterson J.R., Chernoff J., Waterman C.M., Danuser G., DerMardirossian C., and Bokoch G.M.. 2011. Pak1 regulates focal adhesion strength, myosin IIA distribution, and actin dynamics to optimize cell migration. J. Cell Biol. 193:1289–1303 10.1083/jcb.201010059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries C.L., Balcer H.I., D’Agostino J.L., Winsor B., Drubin D.G., Barnes G., Andrews B.J., and Goode B.L.. 2002. Direct regulation of Arp2/3 complex activity and function by the actin binding protein coronin. J. Cell Biol. 159:993–1004 10.1083/jcb.200206113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jascur T., Brickner H., Salles-Passador I., Barbier V., El Khissiin A., Smith B., Fotedar R., and Fotedar A.. 2005. Regulation of p21(WAF1/CIP1) stability by WISp39, a Hsp90 binding TPR protein. Mol. Cell. 17:237–249 10.1016/j.molcel.2004.11.049 [DOI] [PubMed] [Google Scholar]
- Koyasu S., Nishida E., Kadowaki T., Matsuzaki F., Iida K., Harada F., Kasuga M., Sakai H., and Yahara I.. 1986. Two mammalian heat shock proteins, HSP90 and HSP100, are actin-binding proteins. Proc. Natl. Acad. Sci. USA. 83:8054–8058 10.1073/pnas.83.21.8054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., and Helfman D.M.. 2004. Cytoplasmic p21Cip1 is involved in Ras-induced inhibition of the ROCK/LIMK/cofilin pathway. J. Biol. Chem. 279:1885–1891 10.1074/jbc.M306968200 [DOI] [PubMed] [Google Scholar]
- Li R., Soosairajah J., Harari D., Citri A., Price J., Ng H.L., Morton C.J., Parker M.W., Yarden Y., and Bernard O.. 2006. Hsp90 increases LIM kinase activity by promoting its homo-dimerization. FASEB J. 20:1218–1220 10.1096/fj.05-5258fje [DOI] [PubMed] [Google Scholar]
- Mendoza M.C., Er E.E., Zhang W., Ballif B.A., Elliott H.L., Danuser G., and Blenis J.. 2011. ERK-MAPK drives lamellipodia protrusion by activating the WAVE2 regulatory complex. Mol. Cell. 41:661–671 10.1016/j.molcel.2011.02.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima M., and Nishida E.. 1999. Coronin localizes to leading edges and is involved in cell spreading and lamellipodium extension in vertebrate cells. J. Cell Sci. 112:2833–2842. [DOI] [PubMed] [Google Scholar]
- Nishita M., Tomizawa C., Yamamoto M., Horita Y., Ohashi K., and Mizuno K.. 2005. Spatial and temporal regulation of cofilin activity by LIM kinase and Slingshot is critical for directional cell migration. J. Cell Biol. 171:349–359 10.1083/jcb.200504029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa R., Nagata-Ohashi K., Takeichi M., Mizuno K., and Uemura T.. 2002. Control of actin reorganization by Slingshot, a family of phosphatases that dephosphorylate ADF/cofilin. Cell. 108:233–246 10.1016/S0092-8674(01)00638-9 [DOI] [PubMed] [Google Scholar]
- Park S.J., Suetsugu S., Sagara H., and Takenawa T.. 2007. HSP90 cross-links branched actin filaments induced by N-WASP and the Arp2/3 complex. Genes Cells. 12:611–622 10.1111/j.1365-2443.2007.01081.x [DOI] [PubMed] [Google Scholar]
- Pollard T.D., and Borisy G.G.. 2003. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 112:453–465 10.1016/S0092-8674(03)00120-X [DOI] [PubMed] [Google Scholar]
- Pollard T.D., and Cooper J.A.. 2009. Actin, a central player in cell shape and movement. Science. 326:1208–1212 10.1126/science.1175862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidani M., Wessels D., Mouneimne G., Ghosh M., Goswami S., Sarmiento C., Wang W., Kuhl S., El-Sibai M., Backer J.M., et al. . 2007. Cofilin determines the migration behavior and turning frequency of metastatic cancer cells. J. Cell Biol. 179:777–791 10.1083/jcb.200707009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina T.M., and Borisy G.G.. 1999. Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J. Cell Biol. 145:1009–1026 10.1083/jcb.145.5.1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons M.H., and Mitchison T.J.. 1991. Control of actin polymerization in live and permeabilized fibroblasts. J. Cell Biol. 114:503–513 10.1083/jcb.114.3.503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H., Yamashita T., Asada M., Mizutani S., Yoshikawa H., and Tohyama M.. 2002. Cytoplasmic p21Cip1/WAF1 regulates neurite remodeling by inhibiting Rho-kinase activity. J. Cell Biol. 158:321–329 10.1083/jcb.200202071 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.