Abstract
Rationale: Diagnosis of ventilator-associated pneumonia (VAP) is imprecise.
Objectives: To (1) determine whether alternate-day surveillance mini–bronchoalveolar lavage (mini-BAL) in ventilated adults could reduce time to initiation of targeted treatment and (2) evaluate the potential for automated microscopy to reduce analysis time.
Methods: Adult intensive care unit patients who were anticipated to require ventilation for at least a further 48 hours were included. Mini-BALs were processed for identification, quantitation, and antibiotic susceptibility, using (1) clinical culture (50 ± 7 h) and (2) automated microscopy (∼5 h plus offline analysis).
Measurements and Main Results: Seventy-seven mini-BALs were performed in 33 patients. One patient (3%) was clinically diagnosed with VAP. Of 73 paired samples, culture identified 7 containing pneumonia panel bacteria (>104 colony-forming units/ml) from five patients (15%) (4 Staphylococcus aureus [3 methicillin-resistant S. aureus], 2 Stenotrophomonas maltophilia, 1 Klebsiella pneumoniae) and resulted in antimicrobial changes/additions to two of five (40%) of those patients. Microscopy identified 7 of 7 microbiologically positive organisms and 64 of 66 negative samples compared with culture. Antimicrobial responses were concordant in four of five comparisons. Antimicrobial changes/additions would have occurred in three of seven microscopy-positive patients (43%) had those results been clinically available in 5 hours, including one patient diagnosed later with VAP despite negative mini-BAL cultures.
Conclusions: Microbiological surveillance detected infection in patients at risk for VAP independent of clinical signs, resulting in changes to antimicrobial therapy. Automated microscopy was 100% sensitive and 97% specific for high-risk pneumonia organisms compared with clinical culturing. Rapid microscopy-based surveillance may be informative for treatment and antimicrobial stewardship in patients at risk for VAP.
Keywords: nosocomial infections, ventilator-associated pneumonia, microbiological techniques
At a Glance Commentary
Scientific Knowledge on the Subject
Clinical cultures of surveillance mini–bronchoalveolar lavage specimens can detect lung infections independent of clinical signs, and result in earlier antibiotic changes/additions compared with ventilator-associated pneumonia (VAP) diagnosis based on clinical signs alone.
What This Study Adds to the Field
Samples processed for rapid microbiological diagnosis by automated microscopy have the potential to detect VAP before clinical diagnosis by reducing time to identification and antimicrobial susceptibility results to approximately 5 hours compared with 50 ± 7 hours for clinical cultures.
Ventilator-associated pneumonia (VAP) is a common, life-threatening, hospital-acquired infectious complication of prolonged mechanical ventilation in critically ill patients (1–4). Delayed recognition and treatment is associated with increased morbidity, mortality, and health care resource use (5–9). Clinical diagnosis without microbiological testing is imprecise (10, 11), and laboratory cultures needed for improved diagnostic accuracy (12) typically require 2–3 days to produce results. Clinical guidelines (11, 13) advise initiating broad-spectrum therapy at the time a patient presents with symptoms consistent with pneumonia, after a lower respiratory specimen is acquired for microbiological confirmation. Empiric broad-spectrum therapy may fail, with multidrug resistance contributing to risk in approximately 20% of health care–associated infections (9, 14). Switching therapy from inadequate to adequate antibiotics must, however, occur in less than 1 day to improve outcomes (15). Prolonged broad-spectrum empiric therapy can also contribute to the selection and spread of multidrug resistant organisms (MDROs) (16, 17). Rapid deescalation to optimal therapy based on resistance testing (11, 18, 19) is therefore important for antimicrobial stewardship.
A significant proportion of patients may develop a bacterial infection without meeting the criteria for clinical diagnosis despite a substantial microbiological burden (20). By implication, surveillance to detect clinically cryptic infection could have significant clinical value. However, the usefulness of this approach has not been rigorously evaluated (21). We therefore conducted a pilot study to test the hypothesis that intermittent microbiological lung surveillance for a panel of potential MDROs in mechanically ventilated intensive care unit (ICU) patients at risk for but in advance of VAP symptoms would shorten the time to initiation of adequate therapy. We additionally hypothesized that compared with conventional culturing methods, analysis of organisms extracted directly from mini–bronchoalveolar lavage (BAL) specimens using a novel, automated microscopy phenotypic testing technique would further reduce time to diagnosis and initiation of adequate antibiotics and guide deescalation to specific therapy.
Some of the data in this study were presented at the American Thoracic Society 2011 International Conference (22).
Methods
Complete methods may be found in the online supplement.
The Colorado Multiple Institutional Review Board approved the study. Informed consent (from the patient or by surrogate) was obtained.
Patients
Patients were recruited from August 1, 2009 to September 30, 2010 when they were admitted to a 24-bed medical ICU at Denver Health Medical Center (Denver, CO). Patients 18 years of age and older were eligible if the first study procedure could be performed within 72 hours of intubation, and if more than 48 hours of ventilation was anticipated after the first procedure. Patients were excluded for diffuse bronchiectasis, massive hemoptysis, cystic fibrosis, participation in a drug or device trial within 30 days, if pregnant or nursing, had an advanced directive to withhold life support, or were expected to survive less than 14 days.
Baseline demographics were collected including Acute Physiology and Chronic Health Evaluation (APACHE) II and Clinical Pulmonary Infection Score (CPIS). Clinical symptoms of VAP were evaluated daily. Additional process and outcome measures were collected including duration of ventilation, number of mini-BAL procedures, volume of BAL fluid returned, length of ICU stay, adverse events, discharge vital status, and mortality.
Mini-BAL Surveillance
On Days 1, 3, 5, 7, and 10 of ventilation, distal airway specimens were collected by nonbronchoscopic mini-BAL as previously described (23) (see the online supplement). Mini-BAL samples were divided into two aliquots, with one analyzed by routine clinical microbiological culturing and the other with a custom-built automated microscopy instrument.
Clinical Culture Testing and Impact on Prescribing
The hospital laboratory performed semiquantitative microbiological culturing, identification (ID), and antibiotic susceptibility testing (AST) according to standard practices. The hospital laboratory reported data to ICU clinicians to assist medical decision-making. A threshold of at least 104 cfu/ml was used to classify specimen content as microbiologically positive or negative. Antibiotics prescribed or changed as a result of mini-BAL culture were collected for patients with positive clinical microbiology cultures.
Automated Microscopy Testing and Potential Impact on Prescribing
Two research laboratories (Denver Health Infectious Diseases Research [Denver, CO] and Accelerate Diagnostics, Inc. [Tucson, AZ]) separately and prospectively collected microscopy images on various specimens. Automated microscopy tests are summarized in Table 1. Microscopy image analysis for organism identification was performed offline, using manual expert interpretation assisted by computerized image analysis of morphology and growth rates. Automated image analysis was used to perform quantitation, and antibiotic susceptibility testing involved manual expert interpretation assisted by computerized image analysis of organism quantitation and growth rates on exposure to antibiotics listed in Table 1. Microscopy results were not reported to ICU clinicians in real time. The investigators (I.S.D. and C.S.P.) evaluated microscopy results to identify hypothetical drug choices for each case, simulating clinical decision-making had the microscopy results been available at the point of care. The hypothetical drug choices (drug and dose) were compared for concordance with actual therapy prescribed during patient care.
Table 1.
Automated Microscopy Tests
| Test | Antibiotic/Media/Additive |
|---|---|
| MRSA phenotype (cefoxitin) | Cefoxitin |
| Clindamycin resistance | Clindamycin |
| Amikacin resistance | Amikacin |
| Piperacillin/tazobactam resistance | Piperacillin/tazobactam |
| Imipenem resistance | Imipenem |
| Ceftazidime resistance | Ceftazidime |
| Fastidious organism detection | AST-S |
| Acinetobacter spp. detection | Sulbactam |
| Stenotrophomonas maltophilia/Pseudomonas aeruginosa differentiation | Trimethoprim/sulfamethoxazole |
| Growth control | Mueller-Hinton agar |
Definition of abbreviations: AST-S = Phoenix AST-S Broth/Indicator (BD Diagnostics, Sparks Glencoe, MD); MRSA = methicillin-resistant Staphylococcus aureus.
Method Concordance and Timing Comparison
Technicians were blinded to patient characteristics and clinical laboratory results. Results were compared between microscopy and clinical laboratory cultures for concordance (quantitation, species, resistance phenotype) and time to diagnosis from the time of specimen arrival in each laboratory (adjusted for projected turnaround time for microscopy analysis). Discordant results were resolved by use of a suitable reference method.
Results
Thirty-three patients were enrolled in the study. Patient demographics are summarized in Table 2. Patients were critically ill (median APACHE II score was 21 (IQR, 16–24) and the majority had radiological pulmonary infiltrates. Of the 33 patients, one patient (patient 5) was diagnosed with VAP by clinical criteria during study enrollment.
Table 2.
Patient Demographics and Baseline Characteristics
| Demographic/Characteristic | Value |
|---|---|
| Patients, n | 33 |
| Age (yr), median (IQR) | 55 (41–60) |
| Female sex | 12 (36.4%) |
| Race | |
| White | 14 (42.4%) |
| Hispanic | 14 (42.4%) |
| African American | 4 (12.1%) |
| Native American | 1 (3.0%) |
| APACHE II score, median (IQR) | 21 (16–24) |
| Smoker | |
| Ever | 27 (81.8%) |
| Current | 17 (51.5%) |
| Alcohol use (AUDIT score), median (IQR) | 7 (0–18) |
| Chest radiograph findings | |
| Infiltrate | 11 (33.3%) |
| Diffuse infiltrate | 20 (60.6%) |
| No infiltrate | 3 (9.1%) |
| Days mechanically ventilated, median (IQR) | 4 (6–10) |
| Days in ICU, median (IQR) | 10.5 (6.5–18.2) |
| ICU discharge status | |
| Sent home | 18 (54.5%) |
| Deceased | 11 (33.3%) |
| Transferred to skilled nursing facility | 3 (9.1%) |
| Transferred to acute care hospital | 1 (3.0%) |
Definition of abbreviations: APACHE = Acute Physiology and Chronic Health Evaluation; AUDIT = Alcohol Use Disorders Identification Test; ICU = intensive care unit; IQR = interquartile range.
Mini-BAL Surveillance and Safety
Table 3 summarizes results concerning mini-BAL safety. The median number of surveillance mini-BAL procedures performed per patient was 2 (IQR, 1–4; range, 1–7), with a total of 77 procedures performed (66 with a Combicath catheter [Plastimed, Le Plessis Bouchard, France] and 11 with an AirLife catheter [CareFusion, Vernon Hills, IL]). The mean ± SEM volume of sample returned by mini-BAL was 5.2 ± 0.5 ml (P = not significant for comparison between catheters).
Table 3.
Mini–Bronchoalveolar Lavage Surveillance and Safety in 33 Enrolled Patients
| Number | |
|---|---|
| Surveillance mini-BAL performed, n | 77 |
| Combicath (Plastimed) | 66 |
| AirLife catheter (Carefusion) | 11 |
| BAL per patient, median (IQR, range) | 2 (1–4, 1–7) |
| BAL return (ml), average ± SEM | 5.2 ± 0.5 |
| Surveillance mini-BAL adverse events, n (%) | |
| Desaturation requiring increased FiO2 | 2 (3) |
| Tachycardia | 1 (1) |
| Agitation after mini-BAL (60 min) | 2 (3) |
| Bloody return | 4 (5) |
| Total | 9 (12) |
Definition of abbreviations: BAL = bronchoalveolar lavage; FiO2 = fraction of inspired oxygen; IQR = interquartile range.
Nine adverse events were associated with the surveillance mini-BAL procedure (12%), of which none were considered serious. Bloody lavagate occurred in four cases (5%), desaturation requiring an increase in fraction of inspired oxygen occurred in two cases (3%), agitation within the 60 minutes after the procedure occurred in two cases (3%), and there was one instance of transient sinus tachycardia (1%).
Of 77 specimens obtained for analysis, 3 were rejected because of quality issues, and 1 additional specimen could not be analyzed because of a technical failure with the microscopy system. A total of 73 specimens were tested by both microscopy and conventional clinical microbiological cultures.
Clinical Culture Results and Impact on Prescribing
Semiquantitative clinical culture identified 15 samples acquired from 11 patients as microbiologically positive with at least 1 bacterial type at or exceeding 104 cfu/ml (21% of 73 specimens, 33% of 33 patients). Eleven (15%) of these samples contained mixed respiratory bacteria and 7 (10%) samples from 5 patients contained respiratory bacterial pathogens: 4 Staphylococcus aureus, 2 Stenotrophomonas maltophilia, and 1 Klebsiella pneumoniae.
Routine culture results were used by clinical staff to guide changes to antimicrobial prescribing for two of five (40%) microbiologically positive patients (Table 4). For patient 6, identification of S. maltophilia in the mini-BAL sample taken on Day 1 (6-D1), resulted in initiation of trimethoprim-sulfamethoxazole therapy on Day 3, and subsequent discontinuation of caspofungin, piperacillin-tazobactam, and vancomycin. Levofloxacin therapy initiated on Day 2 was continued on the basis of AST results indicating susceptibility to trimethoprim-sulfamethoxazole and levofloxacin.
Table 4.
Impact of Clinical Culture Surveillance Results on Antimicrobial Prescribing
| Patient No. | Sample | Discontinued | Started | Summary |
|---|---|---|---|---|
| 3 | Day 1 | No change | ||
| 6 | Day 1 | Caspofungin, piperacillin-tazobactam, vancomycin | Trimethoprim-sulfamethoxazole | |
| Day 3 | No change | |||
| 8 | Day 7 | No change | ||
| Day 10 | No change | |||
| 17 | Day 1 | Ceftriaxone | Cefazolin | |
| 22 | Day 3 | No change* |
Confirmed methicillin-resistant Staphylococcus aureus obtained from mini–bronchoalveolar lavage obtained 1 day before start of study.
For patient 17, identification of K. pneumoniae by conventional culture of Day 1 mini-BAL sample (17-D1) resulted in the initiation of treatment with ceftriaxone and cefazolin on Day 2. AST results indicating susceptibility to both drugs, but available only more than 48 hours after sample acquisition, resulted in discontinuation of ceftriaxone treatment and continuation of cefazolin treatment by Day 3.
Comparison between Clinical Culture and Automated Microscopy Results and Timing
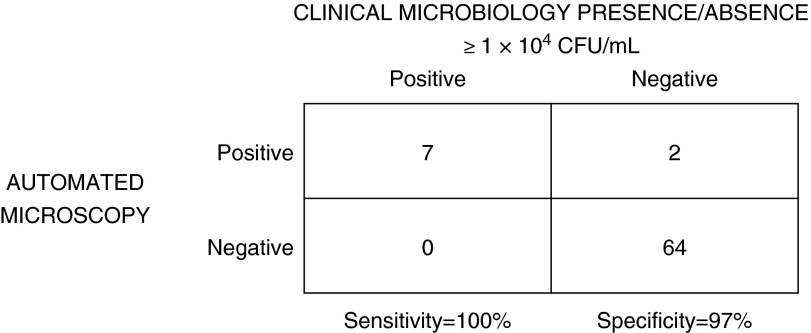
Clinical culture and automated microscopy identification and quantitation results for microbiologically positive samples are summarized in Table 5. Five of nine samples were polymicrobial, but no sample contained more than one VAP-associated bacterial pathogen. Two patients had positive pathogen isolates by conventional culture and microscopy in more than one specimen. Microscopy-based microbiological quantitation was positive in six of the seven samples that were microbiologically positive by culture (sensitivity, 86%). The single false negative (3-D1) contained S. aureus that grew too slowly in Mueller-Hinton broth but had a high growth rate in fastidious medium. Primary (blinded) analysis did not include images from the fastidious medium. Reanalysis after unblinding including images from the fastidious medium identified S. aureus, resulting in 7 of 7 microbiologically positive samples (sensitivity, 100%) and 64 of 66 microbiologically negative samples (specificity, 97%) (Figure 1). There were two false-positive results including one sample determined to contain an enteric species (5-D7, 1.28 × 105 cfu/ml) by microscopy, but that was consistently negative by clinical culture. That patient had an initial CPIS of 3 at the time of specimen collection, but was diagnosed 2 days later with VAP on the basis of clinical criteria. A second false positive occurred in a cultured sample (33-D7) that included Candida and mixed respiratory flora. The clinical laboratory culture report noted “rare” gram-positive cocci on Gram stain. Further analysis by a coagulase assay determined that a coagulase-negative Staphylococcus organism was present. The microscopy assay detected cocci, but incorrectly identified this organism as S. aureus at a microbiologically positive density (6.64 × 104 cfu/ml). These false-positive results would not have significantly increased the annual ICU VAP rate of 2.8–2.9 cases per 1,000 ventilator days.
Table 5.
Microbiologically Positive Microorganism Identifications Made by Clinical Culture and Automated Microscopy
| Specimen (Patient No.-Day) | CPIS | Discharge Status | Clinical Culture |
Automated Microscopy |
||
|---|---|---|---|---|---|---|
| Identification | Concentration (cfu/ml) | Identification | Concentration (cfu/ml) | |||
| 3-D1 | 4 | SNF | Staphylococcus aureus*,† | 104–105 | Fastidious organism‡ | 1.07 × 104 |
| 5-D7 | 3 | Died | No isolate§ | — | Enteric|| | 1.28 × 105 |
| 6-D1 | 6 | Home | Stenotrophomonas maltophilia | >105 | S. maltophilia | 7.68 × 105 |
| 6-D3 | 9 | Home | S. maltophilia | 104–105 | S. maltophilia | 1.60 × 104 |
| 8-D7 | 9 | Died | S. aureus*,** | >105 | S. aureus | 1.11 × 106 |
| 8-D10 | 9 | Died | S. aureus | 104–105 | S. aureus | 1.42 × 105 |
| 17-D1 | 7 | SNF | Klebsiella pneumoniae* | 104–105 | Unknown/enteric | 1.87 × 104 |
| 22-D3 | 8 | Home | S. aureus* | 104–105 | S. aureus | 4.00 × 104 |
| 33-D7 | 6 | Home | Candida**,†† | 104–105 | S. aureus‡‡ | 6.64 × 104 |
Definition of abbreviations: cfu = colony-forming units; CPIS = Clinical Pulmonary Infection Score; SNF = skilled nursing facility.
Polymicrobial sample containing mixed respiratory bacteria.
Polymicrobial sample containing lactose-fermenting gram-negative bacillus (103–104 cfu/ml).
False-negative microscopy result; organism grew only in flowcell containing fastidious growth media, but S. aureus identification test was not activated in that flowcell. S. aureus was correctly identified when the specimen was analyzed further after unblinding.
Negative result by clinical culture; no growth to date.
False-positive microscopy result; ventilator-associated pneumonia was diagnosed in this patient on the basis of clinical criteria.
Polymicrobial sample containing yeast (104–105 cfu/ml) (not Candida albicans or Cryptococcus neoformans).
Negative result by clinical culture; colonization by Candida species in a patient who was not immunosuppressed.
False-positive microscopy result; however, the patient presented with a CPIS equal to or greater than 6 and diffuse infiltrates on chest radiograph. Gram-positive clustered cocci were identified. Further analysis determined that a coagulase-negative Staphylococcus species was present.
Figure 1.
Performance characteristics of automated microscopy compared with clinical culture for qualitative presence or absence of ventilator-associated pneumonia (VAP)-associated bacteria above the VAP diagnostic threshold of 1 × 104 colony-forming units (cfu)/ml.
AST results were available for comparison with four patient specimens. Microscopy results characterized two specimens containing S. aureus as methicillin-resistant S. aureus (MRSA) phenotypes (8-D7, 22-D3) and two specimens from one patient as amikacin-resistant S. maltophilia (6-D1, 6-D3), concordant with culture results. One specimen (8-D7) contained S. aureus classified as clindamycin resistant by microscopy but clindamycin susceptible by culture. D-test confirmed that the specimen did not exhibit inducible clindamycin resistance.
The time to obtain results from hospital clinical microbiology laboratory cultures averaged 31 ± 3 hours (SEM) for identification and quantitation and 50 ± 7 hours (SEM) for AST (total time elapsed after starting cultures). Time to obtain identification, quantitation, and AST results from microscopy analysis included 1.5 hours of sample preparation plus a 3-hour fixed duration for image acquisition (P < 0.0001 for comparison of total elapsed time). Microscopy times were determined as the interval between the start of specimen processing and final image acquisition (approximately 5 h). Image analysis was performed offline after data collection and was not included in the timing calculation for this study.
Simulated Impact of Automated Microscopy Results on Prescribing
Simulated application of automated microscopy results would have resulted in a change in antimicrobials for three of seven patients (43%) found microbiologically positive by automated microscopy (Table 6). Microscopy results for specimen 5-D7 (patient 5, Day 7) identified an enteric species (1.28 × 105 cfu/ml) that was susceptible to amikacin and imipenem, and resistant to piperacillin-tazobactam. This specimen, however, showed no growth by clinical culture. The patient was not receiving any antibiotics at the time the specimen was collected (Day 7). Two days later (Day 9), the patient was clinically diagnosed with VAP. Treatment with vancomycin and piperacillin-tazobactam was initiated and continued for 3 days. The patient died 2 months later. Simulated application of automated microscopy results would have initiated treatment with amikacin and/or imipenem on Day 7, and eliminated 3 days of inadequate treatment with vancomycin and piperacillin-tazobactam.
Table 6.
Simulated Impact of Automated Microscopy Surveillance Results on Antimicrobial Prescribing
| Patient No. | Sample | Discontinued | Started | Summary |
|---|---|---|---|---|
| 3 | Day 1 | No change | ||
| 5 | Day 7 | Piperacillin-tazobactam, vancomycin | Amikacin, imipenem | |
| 6 | Day 1 | Imipenem, levofloxacin, vancomycin | Trimethoprim-sulfamethoxazole | |
| Day 3 | No change | |||
| 8 | Day 7 | Metronidazole, piperacillin-tazobactam | Vancomycin | |
| Day 10 | No change | |||
| 17 | Day 1 | No change | ||
| 22 | Day 3 | No change | ||
| 33 | Day 7 | No change |
S. maltophilia was identified by automated microscopy in two specimens from patient 6 (6-D1 and 6-D3). In simulation, trimethoprim-sulfamethoxazole treatment would have been initiated on Day 1 instead of on Day 3, eliminating 1 day of unnecessary treatment with imipenem, 3 days of inadequate treatment with caspofungin and piperacillin-tazobactam, and 5 days of inadequate treatment with vancomycin and levofloxacin.
Vancomycin was initiated for patient 8 on Day 11 of clinical care. In simulation, vancomycin would have been initiated on Day 7 based on microscopy identification of MRSA in specimen 8-D7 (patient 8, Day 7) instead, eliminating 1 day of inadequate treatment with piperacillin-tazobactam and 4 days of inadequate treatment with metronidazole.
Discussion
This prospective single-center cohort study applied two diagnostic methods to analyze lower respiratory tract specimens obtained before VAP clinical diagnosis: standard laboratory culturing with 2- to 3-day turnaround (50 ± 7 h), and a novel automated microscopy method that has potential for same-day phenotypic antibiotic susceptibility analysis. The study determined, in mechanically ventilated adults with bilateral pulmonary infiltrates on portable chest X-ray, the frequency of microbiologically positive mini-BAL specimens before a clinical VAP diagnosis. We assessed the attending physician’s use of clinically available surveillance culture results in timing and selection of antibiotics. The study also compared time to diagnosis and accuracy for automated microscopy results with standard clinical culture results performed with paired aliquots from a single surveillance mini-BAL specimen. To our knowledge, this is the first clinical assessment of a polymicrobial diagnostic technology potentially capable of same-day quantitative identification and major antibiotic resistance phenotyping directly from mini-BAL specimens.
The clinical microbiological diagnosis of VAP is controversial and risk prediction scoring is of variable value in determining pretest probability of VAP. However, CPIS has previously been evaluated in this type of study, providing a useful benchmark.
Of the 33 patients, 1 patient (patient 5) was diagnosed with VAP by clinical criteria. Microbiologically positive (>104 cfu/ml) quantities of VAP-associated bacteria were detected by culture in 5 of the 32 patients (16%) not diagnosed with VAP. Four of these five patients (80%) had a CPIS equal to or greater than 6 but fell short of meeting additional criteria for VAP clinical diagnosis. The fifth patient had a CPIS of 4 despite having a positive pathogen burden. These results indicate that microbiological surveillance can detect potentially lethal asymptomatic infections in settings of low clinical suspicion as proposed by Zhuo and colleagues (20). A majority of critically ill, mechanically ventilated patients are treated with broad-spectrum antibiotics, even in the absence of proven bacterial infection (11). In the current study, clinical culture results resulted in changes to empiric antimicrobial prescribing for two of five patients (40%) found to be microbiologically positive for a VAP-associated pathogen but negative for VAP clinical diagnosis. Appropriate therapy was initiated sooner, and ineffective therapy was discontinued in each case. Microbiological surveillance differentiates infection from colonization on the basis of a quantitative criterion, that is, organism specimen density. This information is important not only for initiating antimicrobial treatment in infected patients, but also for optimal antimicrobial stewardship by avoiding inappropriate antimicrobial treatment in patients without pneumonia.
Adverse events associated with the surveillance mini-BAL procedure and sample collection were infrequent (9 of 77 mini-BALs, 12%) and were transient in nature, requiring specific therapy (transient increase in fraction of inspired oxygen post-procedure) in few cases. These data support the relative safety of this procedure as previously reported (23). Bronchial aspirates were not used in this study, as their use may be associated with overdiagnosis of VAP.
In comparison with clinical culture results, pathogen identification by automated microscopy was both sensitive (100%) and specific (97%), according to reanalysis after unblinding. One S. aureus sample (3-D1) grew only on fastidious medium. Although the blinded study phase did not include data from the channel that contained the fastidious medium, reanalysis after unblinding resolved this discordant result. Including data for fastidious media in future studies would detect such organisms. Of the two false positives, one was a coagulase-negative S. aureus that automated microscopy misidentified as S. aureus. Improved identification algorithms could prevent this error in the future by applying appropriate criteria for differential growth rate.
The second false positive was an enteric organism that may have been undercounted by conventional culturing. This patient (patient 5) was diagnosed with VAP 2 days after the specimen was obtained, suggesting that conventional culture was insufficiently sensitive to identify the true etiological pathogen that microscopy detected at microbiologically positive density. Microscopy AST confirmed that the organism was susceptible to amikacin and imipenem.
Post hoc simulations using microscopy results at the end of the 5-hour specimen preparation and data collection period to inform clinical decisions indicated that treatment changes could have been made for three of seven patients found microbiologically positive by automated microscopy. Adequate therapy could have been initiated 2–3 days sooner in each case, and 12 total antimicrobial days of inadequate therapy could have been eliminated for the three culture-positive patients. In addition, in simulation, one could have received targeted antibiotic treatment 2 days sooner than the empiric therapy initiated on clinical diagnosis of VAP. Although these data do not prove a broad necessity for surveillance screening, the usefulness of this approach for early detection of MDRO pathogens in very high-risk patients is of significant potential interest. The current findings are consistent with those of a previous simulation study (24) that used automated microscopy of banked isolates and retrospective chart review for S. aureus infections. That study categorized actual drug choices as inactive, optimal, or suboptimal (excessively broad spectrum or unnecessary), using the patient-care clinical laboratory culture result as the reference. The previous study found a potential reduction of inactive therapy from 27 to 0%; reduction of suboptimal therapy from 26 to 3%; and potential increase of optimal therapy from 46 to 97% after applying the observed analytical accuracy of the automated microscopy system.
The possible clinical benefits of this new rapid diagnostic technique include reducing inappropriate antimicrobial use by decreasing the rate of inadequate antimicrobials shortly after initiation of empirical antibiotics and guiding early and aggressive deescalation. These in turn could potentially result in improved patient outcomes including decreased mortality rates and lengths of stay as well as reduced hospital costs.
One limitation of our study in estimating the potential impact of bacteriological surveillance was the absence of a control arm for the current usual care approach in which cultures are not obtained unless and until a patient meets clinical criteria for VAP diagnosis. However, our study design is consistent with those previously published. Brusselaers and coworkers concluded from a systematic review that sequential lower respiratory tract surveillance cultures to predict bacterial VAP were useful (sensitivity, ∼0.75; specificity, ∼0.92 in culture-positive VAP) (25). However, 13 of the 14 included studies used only endotracheal aspirates for surveillance. In only one study was nonbronchoscopic, mini-BAL used as one of the strategies for sequential surveillance (26). Microbiological sensitivity in that study of mini-BAL was only 74% with a specificity of 70%, significantly lower performance characteristics than we found. That study also lacked a usual care arm for comparison.
A second limitation was the use of a custom automated microscopy system and offline analyses performed with expert interpretation assisted by automated growth analysis. These conditions enabled feasibility assessment by means of simulated clinical application. In future studies, the performance of a fully automated system with real-time image analysis will be determined. A fully automated system could be used to prospectively assess the impact of surveillance with same-day microbiological diagnosis on choice of therapy and on surrogate outcome variables. A third limitation was that the simulation method looked only at microbiological data alone to modify treatment decisions rather than taking additional factors into account as would occur in a clinical setting. A fourth limitation was that all specimens were obtained from a single institution. This restriction significantly limited the cumulative number of cases available within the study period. The use of prospective informed consent significantly limited the number of enrolled cases. As a fifth limitation, the small number of positive diagnoses limited the statistical usefulness in assessing the potential value of microbiological surveillance in ICU patients at risk of acquiring VAP. The number of specimens acquired did, however, support the relative safety of using mini-BAL for surveillance.
We conclude that despite these limitations, the usefulness of mini-BAL surveillance and clinical cultures to detect lung infection and inform treatment options is feasible and safe. A significant proportion of surveillance patients did exhibit microbiologically positive pathogen burden before or in the absence of a positive clinical diagnosis. We also conclude that automated microscopy has the potential to further accelerate rapid microbiological diagnosis in patients at risk for VAP and before clinical diagnosis. Earlier availability of identification and AST results obtained through targeted surveillance has the potential to improve clinical outcomes for patients. Early detection and specific antibiotic guidance is also critical for improving antimicrobial stewardship by decreasing the inadequate or suboptimal use of broad-spectrum antibiotic therapy. Future expanded trials using fully automated analysis of automated microscopy results are needed to determine the clinical impact of same-day microbiological diagnostics on treatment optimization and outcomes for emergent VAP.
Acknowledgments
Acknowledgment
The authors thank Alena Shamsheyeva of Accelerate Diagnostics for developing computer algorithms that assisted with the image analysis, and Christina Chantell for manuscript preparation.
Footnotes
Supported by NIH/NCRR Colorado CTSI grant UL1 RR025780 (Accelerate Diagnostics, Inc.).
Author Contributions: I.S.D. conceived the study, analyzed and interpreted the data, performed the statistical analysis, drafted the manuscript, and reviewed the manuscript for important intellectual content. C.S.P. conceived the study, analyzed and interpreted the data, drafted the manuscript, and reviewed the manuscript for important intellectual content. K.H.O. conceived the study, acquired the clinical data, and reviewed the manuscript for important intellectual content. R.F.W. conceived the study, acquired the clinical data, and reviewed the manuscript for important intellectual content. S.W.M. developed and validated the automated microscopy device, acquired the data, analyzed and interpreted the data, drafted the manuscript, and reviewed the manuscript for important intellectual content. K.R.H. derived the data analysis rules, performed visual image analysis, interpreted the data, drafted the manuscript, and reviewed the manuscript for important intellectual content. D.C.H. conceived the study, developed and validated the automated microscopy device, analyzed and interpreted the data, drafted the manuscript, and reviewed the manuscript for important intellectual content.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.201408-1468OC on January 13, 2015
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Dudeck MA, Weiner LM, Allen-Bridson K, Malpiedi PJ, Peterson KD, Pollock DA, Sievert DM, Edwards JR. National Healthcare Safety Network (NHSN) report, data summary for 2012, device-associated module. Am J Infect Control. 2013;41:1148–1166. doi: 10.1016/j.ajic.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Skrupky LP, McConnell K, Dallas J, Kollef MH. A comparison of ventilator-associated pneumonia rates as identified according to the National Healthcare Safety Network and American College of Chest Physicians criteria. Crit Care Med. 2012;40:281–284. doi: 10.1097/CCM.0b013e31822d7913. [DOI] [PubMed] [Google Scholar]
- 3.Kollef MH. What is ventilator-associated pneumonia and why is it important? Respir Care. 2005;50:714–721, discussion 721–724. [PubMed] [Google Scholar]
- 4.Kollef MH, Shorr A, Tabak YP, Gupta V, Liu LZ, Johannes RS. Epidemiology and outcomes of health-care-associated pneumonia: results from a large US database of culture-positive pneumonia. Chest. 2005;128:3854–3862. doi: 10.1378/chest.128.6.3854. [DOI] [PubMed] [Google Scholar]
- 5.Cosgrove SE. The relationship between antimicrobial resistance and patient outcomes: mortality, length of hospital stay, and health care costs. Clin Infect Dis. 2006;42:S82–S89. doi: 10.1086/499406. [DOI] [PubMed] [Google Scholar]
- 6.Iregui M, Ward S, Sherman G, Fraser VJ, Kollef MH. Clinical importance of delays in the initiation of appropriate antibiotic treatment for ventilator-associated pneumonia. Chest. 2002;122:262–268. doi: 10.1378/chest.122.1.262. [DOI] [PubMed] [Google Scholar]
- 7.Kollef MH, Hamilton CW, Ernst FR. Economic impact of ventilator-associated pneumonia in a large matched cohort. Infect Control Hosp Epidemiol. 2012;33:250–256. doi: 10.1086/664049. [DOI] [PubMed] [Google Scholar]
- 8.Restrepo MI, Anzueto A, Arroliga AC, Afessa B, Atkinson MJ, Ho NJ, Schinner R, Bracken RL, Kollef MH. Economic burden of ventilator-associated pneumonia based on total resource utilization. Infect Control Hosp Epidemiol. 2010;31:509–515. doi: 10.1086/651669. [DOI] [PubMed] [Google Scholar]
- 9.Luna CM, Aruj P, Niederman MS, Garzón J, Violi D, Prignoni A, Ríos F, Baquero S, Gando S Grupo Argentino de Estudio de la Neumonía Asociada al Respirador Group. Appropriateness and delay to initiate therapy in ventilator-associated pneumonia. Eur Respir J. 2006;27:158–164. doi: 10.1183/09031936.06.00049105. [DOI] [PubMed] [Google Scholar]
- 10.Chastre J, Trouillet J-L, Combes A, Luyt C-E. Diagnostic techniques and procedures for establishing the microbial etiology of ventilator-associated pneumonia for clinical trials: the pros for quantitative cultures. Clin Infect Dis. 2010;51:S88–S92. doi: 10.1086/653054. [DOI] [PubMed] [Google Scholar]
- 11.American Thoracic Society; Infectious Diseases Society of America. Guidelines for the management of adults with hospital-acquired, ventilator-associated, and healthcare-associated pneumonia. Am J Respir Crit Care Med. 2005;171:388–416. doi: 10.1164/rccm.200405-644ST. [DOI] [PubMed] [Google Scholar]
- 12.Mentec H, May-Michelangeli L, Rabbat A, Varon E, Le Turdu F, Bleichner G. Blind and bronchoscopic sampling methods in suspected ventilator-associated pneumonia: a multicentre prospective study. Intensive Care Med. 2004;30:1319–1326. doi: 10.1007/s00134-004-2284-7. [DOI] [PubMed] [Google Scholar]
- 13.Muscedere J, Dodek P, Keenan S, Fowler R, Cook D, Heyland D VAP Guidelines Committee and the Canadian Critical Care Trials Group. Comprehensive evidence-based clinical practice guidelines for ventilator-associated pneumonia: diagnosis and treatment. J Crit Care. 2008;23:138–147. doi: 10.1016/j.jcrc.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 14.Shorr AF, Sherner JH, Jackson WL, Kollef MH. Invasive approaches to the diagnosis of ventilator-associated pneumonia: a meta-analysis. Crit Care Med. 2005;33:46–53. doi: 10.1097/01.ccm.0000149852.32599.31. [DOI] [PubMed] [Google Scholar]
- 15.Luna CM, Vujacich P, Niederman MS, Vay C, Gherardi C, Matera J, Jolly EC. Impact of BAL data on the therapy and outcome of ventilator-associated pneumonia. Chest. 1997;111:676–685. doi: 10.1378/chest.111.3.676. [DOI] [PubMed] [Google Scholar]
- 16.Micek ST, Heuring TJ, Hollands JM, Shah RA, Kollef MH. Optimizing antibiotic treatment for ventilator-associated pneumonia. Pharmacotherapy. 2006;26:204–213. doi: 10.1592/phco.26.2.204. [DOI] [PubMed] [Google Scholar]
- 17.Chastre J, Wolff M, Fagon J-Y, Chevret S, Thomas F, Wermert D, Clementi E, Gonzalez J, Jusserand D, Asfar P, et al. PneumA Trial Group. Comparison of 8 vs 15 days of antibiotic therapy for ventilator-associated pneumonia in adults: a randomized trial. JAMA. 2003;290:2588–2598. doi: 10.1001/jama.290.19.2588. [DOI] [PubMed] [Google Scholar]
- 18.Boyce JM, Pop O-F, Abreu-Lanfranco O, Hung WY, Fisher A, Karjoo A, Thompson B, Protopapas Z. A trial of discontinuation of empiric vancomycin therapy in patients with suspected methicillin-resistant Staphylococcus aureus health care–associated pneumonia. Antimicrob Agents Chemother. 2013;57:1163–1168. doi: 10.1128/AAC.01965-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niederman MS. Use of broad-spectrum antimicrobials for the treatment of pneumonia in seriously ill patients: maximizing clinical outcomes and minimizing selection of resistant organisms. Clin Infect Dis. 2006;42:S72–S81. doi: 10.1086/499405. [DOI] [PubMed] [Google Scholar]
- 20.Zhuo H, Yang K, Lynch SV, Dotson RH, Glidden DV, Singh G, Webb WR, Elicker BM, Garcia O, Brown R, et al. Increased mortality of ventilated patients with endotracheal Pseudomonas aeruginosa without clinical signs of infection. Crit Care Med. 2008;36:2495–2503. doi: 10.1097/CCM.0b013e318183f3f8. [DOI] [PubMed] [Google Scholar]
- 21.Scholte JBJ, van Mook WNKA, Linssen CFM. Surveillance cultures in healthcare-associated pneumonia: sense or nonsense? Curr Opin Pulm Med. 2014;20:259–271. doi: 10.1097/MCP.0000000000000044. [DOI] [PubMed] [Google Scholar]
- 22.Douglas IS, Price CS, Overdier K, Thompson K, Wolken B, Metzger S, Howson D. Rapid microbiological identification and major drug resistance phenotyping with novel multiplexed automated digital microscopy (MADM) for ventilator-associated pneumonia (VAP) surveillance [abstract] Am J Respir Crit Care Med. 2011;183:A3928. [Google Scholar]
- 23.Kollef MH, Bock KR, Richards RD, Hearns ML. The safety and diagnostic accuracy of minibronchoalveolar lavage in patients with suspected ventilator-associated pneumonia. Ann Intern Med. 1995;122:743–748. doi: 10.7326/0003-4819-122-10-199505150-00002. [DOI] [PubMed] [Google Scholar]
- 24.Cooper S, Metzger S, Saveli C, Howson D, Price CS.Potential impact of rapid phenotype identification on antimicrobial prescribing [abstract]48th ICAAC, October 25–28, 2008Washington, DC. Poster D-4013. [Google Scholar]
- 25.Brusselaers N, Labeau S, Vogelaers D, Blot S. Value of lower respiratory tract surveillance cultures to predict bacterial pathogens in ventilator-associated pneumonia: systematic review and diagnostic test accuracy meta-analysis. Intensive Care Med. 2013;39:365–375. doi: 10.1007/s00134-012-2759-x. [DOI] [PubMed] [Google Scholar]
- 26.Flanagan PG, Findlay GP, Magee JT, Ionescu A, Barnes RA, Smithies M. The diagnosis of ventilator-associated pneumonia using non-bronchoscopic, non-directed lung lavages. Intensive Care Med. 2000;26:20–30. doi: 10.1007/s001340050007. [DOI] [PubMed] [Google Scholar]