Abstract
Rationale: Up to 20% of cases of idiopathic interstitial pneumonia cluster in families, comprising the syndrome of familial interstitial pneumonia (FIP); however, the genetic basis of FIP remains uncertain in most families.
Objectives: To determine if new disease-causing rare genetic variants could be identified using whole-exome sequencing of affected members from FIP families, providing additional insights into disease pathogenesis.
Methods: Affected subjects from 25 kindreds were selected from an ongoing FIP registry for whole-exome sequencing from genomic DNA. Candidate rare variants were confirmed by Sanger sequencing, and cosegregation analysis was performed in families, followed by additional sequencing of affected individuals from another 163 kindreds.
Measurements and Main Results: We identified a potentially damaging rare variant in the gene encoding for regulator of telomere elongation helicase 1 (RTEL1) that segregated with disease and was associated with very short telomeres in peripheral blood mononuclear cells in 1 of 25 families in our original whole-exome sequencing cohort. Evaluation of affected individuals in 163 additional kindreds revealed another eight families (4.7%) with heterozygous rare variants in RTEL1 that segregated with clinical FIP. Probands and unaffected carriers of these rare variants had short telomeres (<10% for age) in peripheral blood mononuclear cells and increased T-circle formation, suggesting impaired RTEL1 function.
Conclusions: Rare loss-of-function variants in RTEL1 represent a newly defined genetic predisposition for FIP, supporting the importance of telomere-related pathways in pulmonary fibrosis.
Keywords: genetics, idiopathic pulmonary fibrosis, telomere
At a Glance Commentary
Scientific Knowledge on the Subject
Approximately one in five cases of pulmonary fibrosis run in families; however, the genetic cause of disease in most families is unknown. Short telomeres are commonly identified in peripheral blood mononuclear cells in patients with sporadic and familial idiopathic interstitial pneumonia in the absence of known mutations in telomere-related genes.
What This Study Adds to the Field
Using next-generation sequencing technology in kindreds with familial interstitial pneumonia, we identified heterozygous rare variants in the gene encoding for regulator of telomere elongation helicase 1 (RTEL1) that segregate with disease in nine unrelated families. The presence of RTEL1 rare variants was associated with severe telomere shortening and increased T-circle formation in peripheral blood mononuclear cells. These findings underscore the importance of genetic variants in telomere-related pathways in the pathogenesis of pulmonary fibrosis.
Idiopathic pulmonary fibrosis (IPF), the most common and severe form of idiopathic interstitial pneumonia (IIP), is a progressive lung disease that affects approximately 1 million patients worldwide (1). Up to one in five patients with IPF have a family history of interstitial lung disease, comprising the syndrome of familial interstitial pneumonia (FIP) (2). Analysis of FIP pedigrees indicates that the typical age of disease onset is 50–70 years old and suggests an autosomal-dominant mode of inheritance in most kindreds (3). Previous studies using linkage and candidate gene approaches have identified rare heterozygous mutations in surfactant protein A2 (SFTPA2) (4), surfactant protein C (SFTPC) (5, 6), telomerase reverse transcriptase (TERT), telomerase RNA component (TERC) (7, 8), and dyskerin (DKC1) (9, 10) that account for at most 10–15% of FIP cases.
Identification of rare loss-of-function genetic variants in telomerase pathway components TERT and TERC first implicated telomere maintenance as an important component of the disease process in FIP (7, 8). Even in the absence of identified telomerase mutations, short telomeres are frequently observed in peripheral blood mononuclear cells (PBMCs) and lung tissue of patients with FIP and sporadic IPF with at least one-third of these patients having PBMC telomere length less than 10th percentile for age (11, 12). The common finding of short telomeres in individuals affected with FIP suggests that genetic variants in other telomere pathway genes could be responsible for disease.
Whole-exome sequencing (WES) has become a frequently used approach for investigation of rare Mendelian diseases and has been successful in identifying the genetic basis of over 100 disorders (13, 14). In this study, we combined WES with a candidate gene/pathway approach to identify novel FIP genes. We identified nine kindreds carrying heterozygous rare variants (RVs) in the gene encoding for regulator of telomere elongation helicase 1 (RTEL1), which is a DNA helicase involved in telomere replication and stability, which has recently been implicated in severe cases of dyskeratosis congenita (DC) (15–19). These RTEL1 RVs segregate with disease, appear functionally deleterious, and are associated with severe telomere shortening in PBMCs. Together, our findings identify heterozygous RTEL1 mutations as a genetic predisposition for FIP and further support the conclusion that telomere-related pathways play critical roles in pulmonary fibrosis.
Methods
For additional information, see the online supplement.
Subjects and Specimens
Fifty-four affected subjects from 25 FIP kindreds were initially selected for WES from the registry of the Familial Interstitial Pneumonia Consortium, which includes families enrolled at Vanderbilt University, the University of Colorado, Duke University, and National Jewish Health. This study was approved by the institutional review boards at each participating institution. FIP was defined by the presence of IIP in two or more family members with at least one affected individual diagnosed with IPF by radiographic and/or histologic criteria. Within families, more distantly related affected individuals were prioritized for WES to maximize power of segregation analysis. Pedigrees were constructed using Progeny (Progeny Software, Delray Beach, FL). DNA was isolated from blood and/or paraffin-embedded lung tissue using a PureGene Kit (Gentra Systems, Minneapolis, MN). Subsequently, probands from an additional 163 FIP kindreds were screened by WES for variants in candidate genes identified in the initial WES analysis. Ascertainment of extrapulmonary clinical phenotypes within families was performed by review of all available medical records, including imaging and pathology. Control DNA was obtained from family members (spouses) who were not in the bloodline of disease in FIP kindreds.
Whole-Exome Sequencing
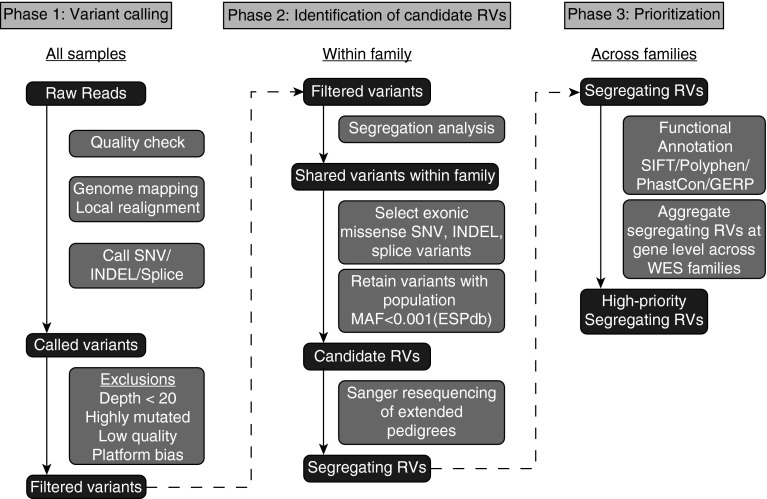
WES of genomic DNA was performed on an Illumina HiSeq2000 (Illumina, San Diego, CA) per the manufacturer’s protocol (see the online supplement). The analysis pipeline is outlined in Figure 1. Following initial quality checking, genome mapping, and local realignment, single-nucleotide variants, inserts and deletions, and splice variants were identified. After variant calling, quality control filtering was done to eliminate low-quality sequence, platform bias, and highly mutated regions. The resulting filtered variants underwent cosegregation analysis to exclude variants that were not present in all family members for whom WES data were available. Within families, variants with a high likelihood of affecting the amino acid coding sequence were retained, including exonic inserts and deletions, splicing, nonsense, and missense single-nucleotide variants. From the remaining shared variants, we further restricted candidate variants to those with a minor allele frequency (MAF) of less than 0.001 among 4,300 Caucasian subjects (8,600 alleles) in the NHLBI GO Exome Sequencing Project (evs.gs.washington.edu) (20). This procedure was performed for all kindreds in the study. The resulting candidate RVs were confirmed by Sanger resequencing and additional segregation analysis was performed in extended pedigrees by Sanger sequencing to generate a list of segregating RVs within families.
Figure 1.
Whole-exome sequencing (WES) analysis pipeline. WES analysis occurred through three phases. In phase 1, raw reads from all samples underwent quality checking, genome mapping, local realignment, and variant calling using Varscan 2.0 to call variants. Following exclusionary quality-control filter, in phase 2 only variants that were found in all affected WES subjects within a family were retained. From among these segregating variants, only those highly likely to affect protein quantity or function (nonsynonymous exonic single-nucleotide variants [i.e., missense or nonsense], insertions and deletions, splice variants) were selected for further analysis. From among these variants, those with minor allele frequency (MAF) greater than 0.001 in the Exome Sequencing Project database (ESPdb) were excluded to yield candidate rare variants (RVs). Sanger requencing was then performed in extended pedigrees encompassing all affected subjects for whom DNA was available to exclude variants that did not segregate fully with disease. These segregating RVs from each family were functionally annotated and aggregated at the gene level across families for further study. GERP = genomic evolutionary rate profiling; INDEL = insertion and deletion; SIFT = Sorting Intolerant from Tolerant; SNV = single-nucleotide variants.
Next, we queried probands from an additional 163 kindreds to ascertain additional families with candidate RVs in genes of interest and performed functional annotation (genomic evolutionary rate profiling [GERP], PhastCon, Sorting Intolerant from Tolerant [SIFT], Polyphen) of variants. Variants were then prioritized for further study based on the population frequency of the variant, number of segregating RVs in the gene, and degree of conservation at the site across species.
Telomere Analysis
PBMC telomere length was determined by terminal restriction fragment length analysis using the TeloTAGGG Telomere Length Assay kit (Roche Applied Science, Indianapolis, IN). The mean terminal restriction fragment lengths were estimated as the weighted average of the optical density according to manufacturer’s instructions (see online supplement).
T-Circle Analysis
T-circle analysis was performed as previously described (21) with minor modifications (see online supplement). T-circle percentage was calculated as the optical density of T-circles relative to total telomeric DNA.
Statistical Analysis
Differences between groups were assessed by analysis of variance or two-tailed nonparametric tests. Results are presented as mean ± standard deviation. The programming language R version 2.15.2 (22) and GraphPad Prism version 5.0 (GraphPad Software, San Diego, CA) were used to make plots and to perform the statistical analyses.
Results
Fifty-four affected subjects with FIP (mean age, 66.5 yr; 59% male) in 25 kindreds were selected for initial WES (Table 1). After calling of variants, filtering, and initial cosegregation analysis within families, our findings indicated that RVs in a single gene (or small group of genes) was unlikely to account for disease in most of these FIP kindreds. Therefore, we analyzed families carrying RVs in genes related to telomere biology (23) because this pathway has previously been implicated in FIP.
Table 1.
Demographics of FIP WES Cohort
| Subjects | 54 |
| Families | 25 |
| Affected subjects sequenced per family | |
| 3 | 4 (16) |
| 2 | 21 (84) |
| Age | 66.5 (8.8) |
| Sex (male) | 32 (59.2) |
| Ethnicity | |
| European | 52 (96.3) |
| African | 0 (0) |
| Hispanic | 2 (3.7) |
| Asian | 0 (0) |
| Native American | 0 (0) |
Definition of abbreviations: FIP = familial interstitial pneumonia; WES = whole-exome sequencing.
Data are expressed as mean (SD) or number (percent).
In one large multiplex family (Family A), both individuals undergoing WES were found to have profound telomere shortening in PBMCs (Figure 2). In this family, there were eight affected individuals, ranging in age from 45 to 72 years of age at the time of diagnosis; four of the eight subjects had a history of tobacco use (Table 2). Across four affected individuals in this family for whom DNA was available for sequencing, a total of 11 heterozygous nonsynonymous RVs were found to segregate with disease including C9orf72, CNTNAP4, DMTF1, EFCAB6, GFRA1, HPS4, MYH11, RBM45, SLC7A14, ZNF326, and RTEL1. Of the RVs shared by affected individuals in this family, only RTEL1 had known interactions with telomere pathways. Offspring of two additional affected subjects also carried this RTEL1 variant (A.III.8 and A. III.12), indicating their affected parent (A.II.5 and A.II.6, respectively) was an obligate carrier of this RV. RTEL1 was recently implicated as a locus associated with telomere length by genome-wide association studies (24), and homozygous and compound heterozygous mutations in RTEL1 have been reported to cause Hoyeraal-Hreidarsson syndrome (HHS), a severe form of DC (15–19). The RTEL1 variant identified in this kindred was a novel three amino acid in-frame deletion (not present in the NHLBI GO Exome Sequencing Project database) that occurs in the helicase domain, in the same region as mutations linked to HHS (15–19).
Figure 2.
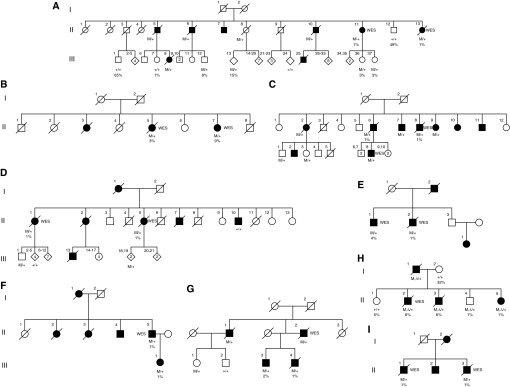
Pedigrees of familial interstitial pneumonia (FIP) families with RTEL1 rare variants. (A–I) Pedigrees of kindreds with heterozygous RTEL1 rare variants that segregate with FIP are depicted. Roman numerals indicate generation and Arabic numerals indicate subject number within a generation. Diamonds are used to indicate multiple offspring including males and females or at the request of subjects and families to maintain confidentiality. Heterozygous RTEL1 rare variant carriers are denoted as M/+; wild-type RTEL1 subjects are denoted as +/+. In family H, the missense variant is denoted as M, and the 35-base-pair deletion is denoted as Δ. Mutation status and telomere percentile (adjusted for age) are depicted below subjects. Whole-exome sequencing (WES) indicates subjects included in the WES cohort; genotypes for all other subjects were obtained by Sanger sequencing.
Table 2.
Characteristics of Affected Subjects
| Kindred |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | H | I | Total | |
| Number of affected | 8 | 3 | 9 | 7 | 4 | 6 | 4 | 4 | 4 | 49 |
| Age at diagnosis | 45–72 | 76–87 | 57–75 | 68–77 | 66–75 | 45–78 | 65–73 | 49–63 | 58–82 | 64.8 (10.6) |
| Tobacco use | 4 | 1 | 3 | 3 | 1 | 1 | 2 | 3 | 1 | 19 (38.8%) |
| Diagnosis | ||||||||||
| Definite | 6 | 4 | 1 | 2 | 3 | 1 | 17 (34.7%) | |||
| Probable | 2 | 2 | 2 | 3 | 2 | 1 | 2 | 1 | 1 | 16 (32.6%) |
| Possible | 2 | 4 | 2 | 4 | 2 | 14 (28.6%) | ||||
| Other ILD | 1 | 1 | 2 (4.1%) | |||||||
| FVC, % | 58–75 | 55–86 | 58–84 | 77–79 | 60 | 90–102 | 31–79 | 42–63 | 70.3 (16.9) | |
| DlCO, % | 45–82 | 33–58 | 19–85 | 34–89 | 33–85 | 53 | 34–94 | 73–93 | 33–55 | 54.1 (21.9) |
Definition of abbreviations: DlCO = diffusing capacity of the lung for carbon monoxide; ILD = interstitial lung disease.
Data are presented as mean (SD) or number (percent). Pulmonary function tests were not available for all subjects.
After identification of RTEL1 as a candidate gene for FIP, we screened an additional 163 FIP kindreds for RTEL1 RVs by WES. Using the approach described in Figure 1, we identified eight additional kindreds (Families B–I) with nonsynonymous heterozygous RTEL1 variants that segregated with disease (Figure 2). In addition, we identified five protein-altering RVs that did not segregate with disease in families and eight benign (synonymous or intronic) RVs in RTEL1 (see Table E1 in the online supplement). Of the eight benign variants, six did not segregate with disease; the other two were found in subjects who were the only sequenced individuals from their family. Intermediate (0.001 < MAF < 0.05) and common (MAF > 0.05) variants in RTEL1 among WES subjects are shown Table E2. Cumulatively, nine families with segregating, protein-altering RTEL1 RVs were identified from among 188 screened families (4.7%).
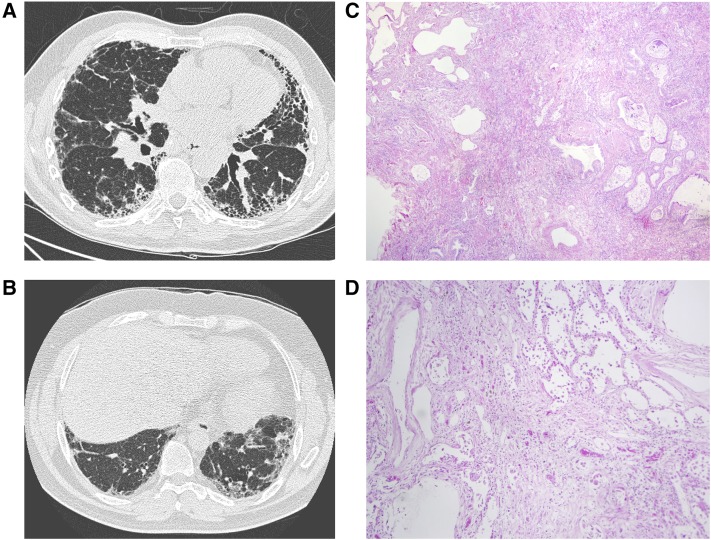
Representative high-resolution computed tomography images and biopsy images from affected individuals carrying disease-associated RTEL1 RVs are shown in Figure 3. Characteristics of affected subjects from the nine RTEL1 RV families (shown in Table 2) were similar to others in the WES cohort, and previously described FIP cohorts (3). Notably, we found no evidence for a family history or clinical features of DC or HHS, including hematopoietic malignancy or aplastic anemia (25).
Figure 3.
Radiographic and histopathologic changes associated with RTEL1 rare variants. (A, B) Representative high-resolution computed tomography images from affected subjects carrying RTEL1 rare variants show typical usual interstitial pneumonia features. (C, D) Hematoxylin and eosin–stained lung biopsies from RTEL1 rare variant carriers show dense interstitial fibrosis, temporal heterogeneity, and microscopic honeycomb cysts consistent with usual interstitial pneumonia. Original magnification ×40 (C) and ×100 (D).
Disease-associated RVs in RTEL1 encompass several classes of mutations (Figure 4A, Table 3), including a 9-bp deletion that leads to loss of three amino acids in the helicase domain, four missense mutations, one nonsense mutation (Arg974X) that has previously been reported to cause DC (16, 19) and HHS (17) in homozygous or compound heterozygous state with other loss-of-function variants, and three splice mutations. Splice variants were confirmed to lead to alternative messenger RNA splicing of RTEL1 by complementary DNA sequencing. Six RVs lie within or near the predicted helicase domain (Figure 4B). R974X is a nonsense mutation whose transcript is subject to nonsense-mediated decay. R1264H is similar to an alternative splice variant of RTEL1 (NM_001283009) previously reported to cause HHS in homozygous or complex heterozygous form (15, 19) and lies near a reported proliferating cell nuclear antigen interacting domain that seems to be crucial for global genome replication (26). During confirmation of the WES missense variant V516L by Sanger sequencing, we identified a 35-bp deletion 3 bp downstream that was determined to be in cis with V516L. This deletion was too large to be detected by WES and is predicted to cause a frameshift in the messenger RNA sequence.
Figure 4.
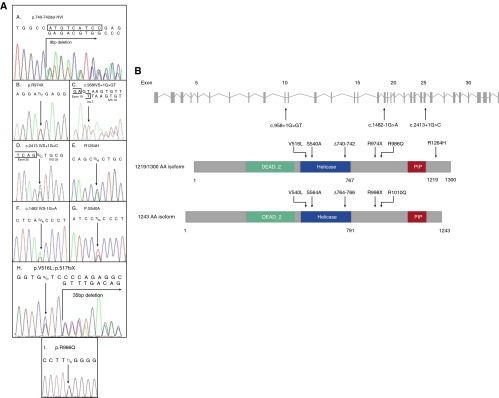
RTEL1 rare variants detected by whole-exome sequencing. (A) Modified Sanger sequencing confirmed the RTEL1 rare variants detected by whole-exome sequencing in families A–I. (B) Schematic depiction of the location of RTEL1 rare variants in the 1,219-amino-acid isoform and an alternatively spliced 1,300-amino-acid isoform. PIP = PCNA (proliferating cell nuclear antigen)-interacting protein.
Table 3.
RTEL1 Rare Variants That Segregate with FIP
| Position | Type | Ref > alt | rsID | Function GVS | Amino Acids | Protein Position (1,219/1,243 AA) | Complementary DNA Position (1,219/1,243 AA) | GERP | Poly Phen | ESP Allele Counts | References |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 20:62309537 | Indel | G > GT | 0 | Splice-5 | None | NA | c.958 IVS + 1G > GT c.1030 IVS + 1G > GT | 4.32 | NA | None | |
| 20:62319289 | SNP | G > A | 0 | Splice-3 | None | NA | c.1482 IVS − 1G > A c.1554 IVS − 1G > A | 5.48 | NA | G = 12,986 | |
| 20:62319354 | SNP | G > C & 35 bp del | 0 | Missense/DEL | VAL/LEU | 516/1,219, 540/1,243 | NA | NA | None | ||
| 20:62319514 | SNP | T > G | 0 | Missense | SER/ALA | 540/1,219, 564/1,243 | c.1618 c.1690 | 4.27 | 0.931 | T = 12,994 | |
| 20:62321517 | Indel | ATGTCATCC > del | 0 | 9 bp DEL | HVI/Del | 740_742del/1219 764_766del/1243 | c.2219_2227del c.2291_2299del | NA | NA | None | |
| 20:62321795 | SNP | G > C | 0 | Splice-5 | None | NA | c.2413 IVS + 1G > C c.2485 IVS + 1G > C | 4.16 | NA | G = 12,938 | |
| 20:62324564 | SNP | C > T | 0 | Stop-gained | ARG/stop | 974/1,219, 998/1,243 | c.2920 c.2992 | 4.82 | NA | C = 12,990 | 20, 21, 23 |
| 20:62324601 | SNP | G > A | 146221660 | Missense | ARG/GLN | 986/1,219, 1,010/1,243 | c.2957 c.3029 | −9.64 | 0.037 | A = 1/G = 12,971 | |
| 20:62326972 | SNP | G > A | 201540674 | Missense (1,300 AA) | ARG/HIS | 1,264/1,300 | c.3791G > A (1,300 AA) | 3.73 | NA | G = 12,936 | 19, 23 |
Definition of abbreviations: AA = amino acid; alt = base in sample; DEL = deletion; ESP = NHLBI GO Exome Sequencing Project; FIP = familial interstitial pneumonia; Function GVS = function predicted by genome variation server; GERP = genomic evolutionary rate profiling; Indel = insertion and deletion; IVS = intervening sequence; NA = not applicable; Poly Phen = polymorphism phenotyping; Ref = reference base; rsID = reference SNP cluster ID; SNP = single-nucleotide polymorphism.
Telomere length in PBMCs from probands in each RTEL1 mutation kindred was extremely short, similar to PBMC telomere length in probands from other kindreds with known telomerase pathway mutations (TERT, TERC, and DKC1) (Figure 4A). Consistent with loss-of-function RTEL1 mutations, all affected and unaffected RTEL1 mutation carriers had PBMC telomere length less than 10th percentile for age (Figures 2 and 5A). In addition, several offspring of RTEL1 mutation carriers who did not inherit their parent’s RTEL1 mutation had short telomeres (A.III.7 and H.II.1), as has been previously reported in families with TERT mutations associated with short telomeres (27).
Figure 5.
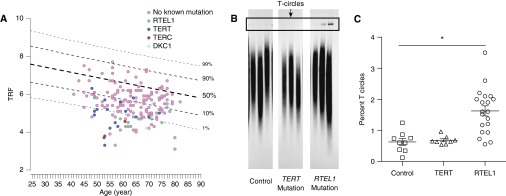
Carriers of RTEL1 rare variants have short telomeres and increased T-circle formation. (A) Telomere restriction fragment (TRF) is plotted versus age for probands from families in the familial interstitial pneumonia (FIP) registry. All probands in RTEL1-mutation families were less than or equal to 10th percentile for age, similar to what is seen for TERT (n = 18), TERC (n = 2), and DKC1 (n = 1). (B) A 1-D electrophoresis gel from T-circle assay comparing age-matched unaffected married-in control subjects, TERT mutation carriers, and RTEL1 mutation carriers. (C) Quantification of T-circle formation in peripheral blood mononuclear cells, defined as the percentage of T-circles compared with total telomeric DNA in eight unaffected control subjects, nine TERT mutation carriers, and 20 RTEL1 mutation carriers. *P < 0.001 across groups and pairwise RTEL1 carrier versus control and RTEL1 carriers versus TERT carriers.
Previous reports of RTEL1 mutations in DC and HHS indicate that loss of RTEL1 function can lead to increased T-circles in telomeric DNA fractions. To determine whether the heterozygous RTEL1 mutations we identified resulted in increased T-circle formation, we performed T-circle assays using DNA isolated from PBMCs obtained from RTEL1 mutation carriers, TERT mutation carriers, and healthy age-matched control subjects. We found that among RTEL1 mutation carriers, T-circle formation was significantly increased compared with both TERT mutation carriers and healthy control subjects (Figures 5B and 5C), suggesting that these heterozygous RTEL1 mutations are functionally deleterious. In addition, it seems that increased T-circle formation is not a generalizable finding in all patients with FIP with telomere-pathway defects.
Discussion
Although a substantial proportion of IIP cases cluster in families, the genetic basis of disease in most FIP kindreds remains uncertain. Using next-generation sequencing technologies to facilitate genetic discovery, we identified RTEL1 as a novel gene associated with FIP. By focusing our search on genes known to interact with telomere pathways, we found nine unrelated FIP kindreds in which all affected individuals had very short telomeres in PBMCs and were heterozygous for RVs in RTEL1. Functional testing revealed increased T-circle formation by PBMCs, further suggesting the functional importance of these RTEL1 variants. Together, our findings indicate that rare loss-of-function variants in RTEL1 represent an important genetic risk for development of FIP, particularly in the subgroup with short telomeres in PBMCs.
The mechanisms by which telomerase dysfunction and short telomeres lead to lung fibrosis has been studied extensively but remains incompletely understood. Telomeres are protective DNA repeat (TTAGGG) structures, which are required for the proper replication of chromosome ends, and play important roles in regulating the replicative capacity of human somatic cells and in maintaining genomic stability (28). In somatic cells, successive rounds of cell replication lead to progressive shortening of telomere length because DNA polymerase cannot fully replicate the 3′ overhanging end of the DNA strand. Ultimately, telomeres can become critically short and trigger the cell to enter replicative senescence (28). In contrast, following replication in germ cells and other stem cells that require renewal, telomere length is maintained by telomerase. It is possible that telomerase pathway mutations lead to premature senescence of progenitor cells of the distal lung, resulting in a proliferative defect and failure of repair mechanisms following injury to the alveolar epithelium.
RTEL1 is reported to act as a DNA helicase crucial for unwinding the T-loop structure at the telomeric ends of chromosomes (29). Loss of functional RTEL1 leads to cleavage of the telomeric end of the chromosome proximal to the T-loop by endonuclease SLX4 (30), leading to release of circular fragments of telomeric DNA (T-circles) and progressive telomere shortening through successive rounds of DNA replication. In addition to telomere shortening, loss of RTEL1 function has been shown to lead to global genome replication defects and genomic instability (26, 30–32), suggesting that cell-cycle progression is disrupted in the presence of altered RTEL1 expression or function. Further studies are necessary to clarify the mechanistic role of RTEL1 in the pathogenesis of lung fibrosis.
Several recent reports have described heterozygous, compound heterozygous, and homozygous RTEL1 mutations as the cause of a rare, severe form of DC known as HHS (15–19). However, we ascertained no history or symptoms suggestive of DC or HHS in any of our FIP families with heterozygous RTEL1 RVs. Although it was not possible to perform exhaustive clinical phenotyping of all affected subjects, the age of presentation of the youngest affected subject with an RV in RTEL1 was 45 years, suggesting that heterozygous RTEL1 mutations present primarily with pulmonary manifestations and much later in life than individuals with homozygous/compound heterozygous mutations. However, because extrapulmonary clinical phenotyping was limited in this study, it is possible subtle features of DC or HHS may have been present in some subjects. Similar to reports in DC and HHS, we found that RTEL1 mutation carriers had telomere shortening (16, 19). Many RTEL1 mutation carriers also had increased T-circle formation, suggesting this finding is common but may not be uniform or entirely specific for RTEL1 mutations.
Mutations in TERT, TERC, DKC1, and RTEL1 seem to explain a substantial proportion of the one-third of patients with FIP with short telomeres in PBMCs (<10th percentile for age); however, the explanation for genetic susceptibility in the remaining FIP families with short telomeres requires further study. Segregating, protein-altering RVs in other candidate genes in the telomerase pathway were not identified in this study cohort. These candidates included genes whose products are associated with the telomere complex and loci associated with differential telomere length in the general population (33), including ACYP2, NAF1, OBFC1, or ZNF208. Variants with higher allele frequency (34, 35) or those found in intronic or regulatory regions excluded by our present analysis could contribute to disease in some families (see Table E2). In addition to RVs, common variants in TERT, TERC, and OBFC1 have been associated with IIPs in a recent genome-wide association study (34), suggesting that common genetic variants alone or in combination with environmental factors, such as tobacco smoke exposure (36), could be responsible for telomere shortening in some FIP families.
There are several limitations to the strategy we used in this study. The power of WES to determine the contribution of individual RVs to disease is limited in many FIP kindreds because of small numbers of affected individuals available for study. The late onset of FIP, well into adulthood, limits the ability to ascertain disease across multiple generations and reduces the genetic informativeness of FIP families, thus making it challenging to identify and prove causative variants without functional studies. To overcome this inherent limitation, we combined WES with a targeted pathway approach using a recognized phenotypic distinction (telomere shortening) among a subset of FIP families. Technical constraints of WES limit the ability to detect large insertions, deletions (see family H, Figures 2 and 4), or inversions. In addition, WES does not capture intronic variants that may impact splicing, and may not identify promoter or other regulatory variants. RTEL1 variants with allele frequencies above our MAF threshold of 0.001 could also be functional and confer disease risk (see Table E1), thus it is possible that the global contribution of genetic variants in RTEL1 to risk of FIP may be underestimated by our current WES analysis.
Like other telomere-pathway RVs, the penetrance of RTEL1 RVs is not known but is likely to be incomplete and dependent on age. It is likely that not all RTEL1 RVs are pathogenic because we identified RTEL1 missense RVs that did not segregate with disease in several subjects. However, these variants were generally in regions of low conservation and/or were not predicted to adversely affect RTEL1 function. Future studies are critical to more fully characterize the functional importance of individual RTEL1 RVs.
Rather than sequencing an independent group of control subjects, we used publically available large databases (which sequenced subjects on the same platform) to define allele frequency. The depth of these public databases is incomplete and in selected populations, variants we define as rare may have allele frequencies higher than our prespecified threshold (<0.001). For example, R1264H has been reported to have an allele frequency of nearly 0.005 in Ashkenazi Jewish subjects (37). Because this variant has been previously implicated in HHS in a compound heterozygous state, we propose this variant is nonetheless likely to deleteriously affect RTEL1 function. Although it was not possible to exclude all other candidate variants in each individual family, these RTEL1 RVs segregated with disease across all nine families. This segregation, combined with short telomeres and impaired RTEL1 function, provides compelling evidence that RTEL1 mutations are a genetic basis of FIP in these kindreds. Additional study is required to identify the total contribution of all RTEL1 variants to FIP and determine if RTEL1 plays a role in sporadic IPF.
Conclusions
Rare loss-of-function RTEL1 variants are associated with the inherited form of IIP. These RTEL1 variants segregate with disease in affected kindreds and are associated with telomere shortening in PBMCs and increased T-circle formation. Further studies are needed to better understand the total contribution of RTEL1 variants to FIP and define the mechanisms through which RTEL1 and telomerase pathway mutations lead to lung fibrosis.
Acknowledgments
Acknowledgment
The authors thank the patients and families who contributed data and samples to make this work possible.
Footnotes
Supported by NIH NHLBI (HL92870 and HL085317 [T.S.B.]; HL105479 [W.E.L.]; HL094296 [J.A.K.]; HL0097163 [D.A.S.]), Vanderbilt University Clinical and Translational Science Award (UL1 RR024975 [J.D.C.]), Department of Veterans Affairs (T.S.B., W.E.L., and D.A.S.), and American Thoracic Society Foundation/Boehringer Ingelheim Pharmaceuticals Career Award in Idiopathic Pulmonary Fibrosis (J.A.K.). Sequencing was provided by the University of Washington Center for Mendelian Genomics and was funded by the National Human Genome Research Institute and the NHLBI grant 1U54HG006493 to Drs. Debbie Nickerson, Jay Shendure, and Michael Bamshad.
Author Contributions: J.D.C. designed the study, performed experiments, analyzed and interpreted data, and wrote and revised the manuscript. J.A.K. designed the study, collected, analyzed, and interpreted data, and wrote and revised the manuscript. M.Z. analyzed and interpreted data and revised the manuscript. D.B.M. and L.R. performed experiments and analyzed and interpreted data. C.M., E.T.G., K.H.M., W.R.M., D.F.M., J.P., and E.M. collected and analyzed data. L.M.O. and L.C. analyzed and interpreted data. D.-S.C., U.W.C.M.G., and E.M.B. performed experiments and analyzed data. L.R.Y. designed the study and analyzed and interpreted data. L.H.L. designed the study and collected, analyzed, and interpreted data. M.P.S., K.K.B., M.I.S., T.E.F., D.A.S., W.E.L., J.E.L., and Z.Z. designed the study, collected, analyzed, and interpreted data, and revised the manuscript. J.A.P. and T.S.B. designed the study, collected, analyzed, and interpreted data, and wrote and revised the manuscript.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.201408-1510OC on January 21, 2015
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.King TE, Jr, Pardo A, Selman M. Idiopathic pulmonary fibrosis. Lancet. 2011;378:1949–1961. doi: 10.1016/S0140-6736(11)60052-4. [DOI] [PubMed] [Google Scholar]
- 2.Garcia-Sancho C, Buendia-Roldan I, Fernandez-Plata MR, Navarro C, Perez-Padilla R, Vargas MH, Loyd JE, Selman M. Familial pulmonary fibrosis is the strongest risk factor for idiopathic pulmonary fibrosis. Respir Med. 2011;105:1902–1907. doi: 10.1016/j.rmed.2011.08.022. [DOI] [PubMed] [Google Scholar]
- 3.Steele MP, Speer MC, Loyd JE, Brown KK, Herron A, Slifer SH, Burch LH, Wahidi MM, Phillips JA, III, Sporn TA, et al. Clinical and pathologic features of familial interstitial pneumonia. Am J Respir Crit Care Med. 2005;172:1146–1152. doi: 10.1164/rccm.200408-1104OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y, Kuan PJ, Xing C, Cronkhite JT, Torres F, Rosenblatt RL, DiMaio JM, Kinch LN, Grishin NV, Garcia CK. Genetic defects in surfactant protein A2 are associated with pulmonary fibrosis and lung cancer. Am J Hum Genet. 2009;84:52–59. doi: 10.1016/j.ajhg.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nogee LM, Dunbar AE, III, Wert SE, Askin F, Hamvas A, Whitsett JA. A mutation in the surfactant protein C gene associated with familial interstitial lung disease. N Engl J Med. 2001;344:573–579. doi: 10.1056/NEJM200102223440805. [DOI] [PubMed] [Google Scholar]
- 6.Thomas AQ, Lane K, Phillips J, III, Prince M, Markin C, Speer M, Schwartz DA, Gaddipati R, Marney A, Johnson J, et al. Heterozygosity for a surfactant protein C gene mutation associated with usual interstitial pneumonitis and cellular nonspecific interstitial pneumonitis in one kindred. Am J Respir Crit Care Med. 2002;165:1322–1328. doi: 10.1164/rccm.200112-123OC. [DOI] [PubMed] [Google Scholar]
- 7.Armanios MY, Chen JJ, Cogan JD, Alder JK, Ingersoll RG, Markin C, Lawson WE, Xie M, Vulto I, Phillips JA, III, et al. Telomerase mutations in families with idiopathic pulmonary fibrosis. N Engl J Med. 2007;356:1317–1326. doi: 10.1056/NEJMoa066157. [DOI] [PubMed] [Google Scholar]
- 8.Tsakiri KD, Cronkhite JT, Kuan PJ, Xing C, Raghu G, Weissler JC, Rosenblatt RL, Shay JW, Garcia CK. Adult-onset pulmonary fibrosis caused by mutations in telomerase. Proc Natl Acad Sci USA. 2007;104:7552–7557. doi: 10.1073/pnas.0701009104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kropski JA, Mitchell DB, Markin C, Polosukhin VV, Choi LA, Johnson JE, Lawson WE, Phillips JA, III, Cogan JD, Blackwell TS, et al. A novel dyskerin (DKC1) mutation is associated with familial interstitial pneumonia. Chest. 2014;146:e1–e7. doi: 10.1378/chest.13-2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alder JK, Parry EM, Yegnasubramanian S, Wagner CL, Lieblich LM, Auerbach R, Auerbach AD, Wheelan SJ, Armanios M. Penetrant telomere phenotypes in females with heterozygous mutations in the dyskeratosis congenita 1 (DKC1) gene. Hum Mutat. 2013;34:1481–1485. doi: 10.1002/humu.22397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alder JK, Chen JJ, Lancaster L, Danoff S, Su SC, Cogan JD, Vulto I, Xie M, Qi X, Tuder RM, et al. Short telomeres are a risk factor for idiopathic pulmonary fibrosis. Proc Natl Acad Sci USA. 2008;105:13051–13056. doi: 10.1073/pnas.0804280105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cronkhite JT, Xing C, Raghu G, Chin KM, Torres F, Rosenblatt RL, Garcia CK. Telomere shortening in familial and sporadic pulmonary fibrosis. Am J Respir Crit Care Med. 2008;178:729–737. doi: 10.1164/rccm.200804-550OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Y, Muzny DM, Reid JG, Bainbridge MN, Willis A, Ward PA, Braxton A, Beuten J, Xia F, Niu Z, et al. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N Engl J Med. 2013;369:1502–1511. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rabbani B, Mahdieh N, Hosomichi K, Nakaoka H, Inoue I. Next-generation sequencing: impact of exome sequencing in characterizing Mendelian disorders. J Hum Genet. 2012;57:621–632. doi: 10.1038/jhg.2012.91. [DOI] [PubMed] [Google Scholar]
- 15.Ballew BJ, Joseph V, De S, Sarek G, Vannier JB, Stracker T, Schrader KA, Small TN, O'Reilly R, Manschreck C, et al. A recessive founder mutation in regulator of telomere elongation helicase 1, RTEL1, underlies severe immunodeficiency and features of Hoyeraal Hreidarsson syndrome. PLoS Genet. 2013;9:e1003695. doi: 10.1371/journal.pgen.1003695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ballew BJ, Yeager M, Jacobs K, Giri N, Boland J, Burdett L, Alter BP, Savage SA. Germline mutations of regulator of telomere elongation helicase 1, RTEL1, in dyskeratosis congenita. Hum Genet. 2013;132:473–480. doi: 10.1007/s00439-013-1265-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deng Z, Glousker G, Molczan A, Fox AJ, Lamm N, Dheekollu J, Weizman OE, Schertzer M, Wang Z, Vladimirova O, et al. Inherited mutations in the helicase RTEL1 cause telomere dysfunction and Hoyeraal-Hreidarsson syndrome. Proc Natl Acad Sci USA. 2013;110:E3408–E3416. doi: 10.1073/pnas.1300600110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le Guen T, Jullien L, Touzot F, Schertzer M, Gaillard L, Perderiset M, Carpentier W, Nitschke P, Picard C, Couillault G, et al. Human RTEL1 deficiency causes Hoyeraal-Hreidarsson syndrome with short telomeres and genome instability. Hum Mol Genet. 2013;22:3239–3249. doi: 10.1093/hmg/ddt178. [DOI] [PubMed] [Google Scholar]
- 19.Walne AJ, Vulliamy T, Kirwan M, Plagnol V, Dokal I. Constitutional mutations in RTEL1 cause severe dyskeratosis congenita. Am J Hum Genet. 2013;92:448–453. doi: 10.1016/j.ajhg.2013.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tennessen JA, Bigham AW, O'Connor TD, Fu W, Kenny EE, Gravel S, McGee S, Do R, Liu X, Jun G, et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zellinger B, Akimcheva S, Puizina J, Schirato M, Riha K. Ku suppresses formation of telomeric circles and alternative telomere lengthening in arabidopsis. Mol Cell. 2007;27:163–169. doi: 10.1016/j.molcel.2007.05.025. [DOI] [PubMed] [Google Scholar]
- 22.Dean CB, Nielsen JD. Generalized linear mixed models: a review and some extensions. Lifetime Data Anal. 2007;13:497–512. doi: 10.1007/s10985-007-9065-x. [DOI] [PubMed] [Google Scholar]
- 23.Mason PJ, Perdigones N. Telomere biology and translational research. Transl Res. 2013;162:333–342. doi: 10.1016/j.trsl.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Codd V, Nelson CP, Albrecht E, Mangino M, Deelen J, Buxton JL, Hottenga JJ, Fischer K, Esko T, Surakka I, et al. Identification of seven loci affecting mean telomere length and their association with disease. Nat Genet. 2013;45:422–427, 427e421–422. doi: 10.1038/ng.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jonassaint NL, Guo N, Califano JA, Montgomery EA, Armanios M. The gastrointestinal manifestations of telomere-mediated disease. Aging Cell. 2013;12:319–323. doi: 10.1111/acel.12041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vannier JB, Sandhu S, Petalcorin MI, Wu X, Nabi Z, Ding H, Boulton SJ. RTEL1 is a replisome-associated helicase that promotes telomere and genome-wide replication. Science. 2013;342:239–242. doi: 10.1126/science.1241779. [DOI] [PubMed] [Google Scholar]
- 27.Diaz de Leon A, Cronkhite JT, Katzenstein AL, Godwin JD, Raghu G, Glazer CS, Rosenblatt RL, Girod CE, Garrity ER, Xing C, et al. Telomere lengths, pulmonary fibrosis and telomerase (TERT) mutations. PLoS ONE. 2010;5:e10680. doi: 10.1371/journal.pone.0010680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blackburn EH, Greider CW, Szostak JW. Telomeres and telomerase: the path from maize, Tetrahymena and yeast to human cancer and aging. Nat Med. 2006;12:1133–1138. doi: 10.1038/nm1006-1133. [DOI] [PubMed] [Google Scholar]
- 29.Vannier JB, Sarek G, Boulton SJ. RTEL1: functions of a disease-associated helicase. Trends Cell Biol. 2014;24:416–425. doi: 10.1016/j.tcb.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 30.Vannier JB, Pavicic-Kaltenbrunner V, Petalcorin MI, Ding H, Boulton SJ. RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell. 2012;149:795–806. doi: 10.1016/j.cell.2012.03.030. [DOI] [PubMed] [Google Scholar]
- 31.Barber LJ, Youds JL, Ward JD, McIlwraith MJ, O'Neil NJ, Petalcorin MI, Martin JS, Collis SJ, Cantor SB, Auclair M, et al. RTEL1 maintains genomic stability by suppressing homologous recombination. Cell. 2008;135:261–271. doi: 10.1016/j.cell.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ding H, Schertzer M, Wu X, Gertsenstein M, Selig S, Kammori M, Pourvali R, Poon S, Vulto I, Chavez E, et al. Regulation of murine telomere length by Rtel: an essential gene encoding a helicase-like protein. Cell. 2004;117:873–886. doi: 10.1016/j.cell.2004.05.026. [DOI] [PubMed] [Google Scholar]
- 33.Fuentes Fajardo KV, Adams D, Mason CE, Sincan M, Tifft C, Toro C, Boerkoel CF, Gahl W, Markello T. Detecting false-positive signals in exome sequencing. Hum Mutat. 2012;33:609–613. doi: 10.1002/humu.22033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fingerlin TE, Murphy E, Zhang W, Peljto AL, Brown KK, Steele MP, Loyd JE, Cosgrove GP, Lynch D, Groshong S, et al. Genome-wide association study identifies multiple susceptibility loci for pulmonary fibrosis. Nat Genet. 2013;45:613–620. doi: 10.1038/ng.2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Noth I, Zhang Y, Ma SF, Flores C, Barber M, Huang Y, Broderick SM, Wade MS, Hysi P, Scuirba J, et al. Genetic variants associated with idiopathic pulmonary fibrosis susceptibility and mortality: a genome-wide association study. Lancet Respir Med. 2013;1:309–317. doi: 10.1016/S2213-2600(13)70045-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Theall KP, McKasson S, Mabile E, Dunaway LF, Drury SS. Early hits and long-term consequences: tracking the lasting impact of prenatal smoke exposure on telomere length in children. Am J Public Health. 2013;103:S133–S135. doi: 10.2105/AJPH.2012.301208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fedick AM, Shi L, Jalas C, Treff NR, Ekstein J, Kornreich R, Edelmann L, Mehta L, Savage SA. Carrier screening of RTEL1 mutations in the Ashkenazi Jewish population. Clin Genet. (In press) [DOI] [PubMed]