Abstract
The objective of this study was to genetically characterize isolates of Giardia duodenalis and to determine if zoonotic potential of G. duodenalis could be found in stray cats from urban and suburban environments in Guangzhou, China. Among 102 fresh fecal samples of stray cats, 30 samples were collected in Baiyun district (urban) and 72 in Conghua district (suburban). G. duodenalis specimens were examined using light microscopy, then the positive specimens were subjected to PCR amplification and subsequent sequencing at 4 loci such as glutamate dehydrogenase (gdh), triose phosphate isomerase (tpi), β-giardin (bg), and small subunit ribosomal RNA (18S rRNA) genes. The phylogenetic trees were constructed using obtained sequences by MEGA5.2 software. Results show that 9.8% (10/102) feline fecal samples were found to be positive by microscopy, 10% (3/30) in Baiyun district and 9.7% (7/72) in Conghua district. Among the 10 positive samples, 9 were single infection (8 isolates, assemblage A; 1 isolate, assemblage F) and 1 sample was mixed infection with assemblages A and C. Based on tpi, gdh, and bg genes, all sequences of assemblage A showed complete homology with AI except for 1 isolate (CHC83). These findings not only confirmed the occurrence of G. duodenalis in stray cats, but also showed that zoonotic assemblage A was found for the first time in stray cats living in urban and suburban environments in China.
Keywords: Giardia duodenalis, multilocus genotyping, zoonotic assemblage, cat, China
Giardia duodenalis commonly causes enteric infections in humans as well as a wide variety of domestic animals and wildlife species worldwide. In Asia, Africa, and Latin America, approximately 200 million people present symptomatic giardiasis and 500,000 new cases are diagnosed per year [1]. G. duodenalis is a species complex comprising at least 8 assemblages or genotypes (A-H) [2]. The assemblages A and B are important in human infections; these assemblages have been detected not only in livestock but also in companion animals such as dogs and cats [1]. Assemblages C and D are found in dogs, while assemblage E have been identified in many different ruminants; assemblage F seems to be specific for cats, G for rats, and H for seals, respectively [3].
In China, the zoonotic assemblages (A and B) of G. duodenalis have been reported in human fecal samples in Henan and Anhui province [4,5] and nonhuman primates (Macaca fascicularis) in Guangxi province [6]. The G. duodenalis zoonotic assemblage A in canine fecal samples had been reported in Guangdong province [7,8]. However, there have been no reports on the zoonotic potential of G. duodenalis from cats in China.
Previous studies have shown that multilocus sequence typing may detect mixed assemblages attributed to mixed infections, while single locus typing is difficult to detect them, especially those with zoonotic potential where assemblage A was the predominant [9,10]. The objective of this study was to determine if zoonotic potential of G. duodenalis could be found in stray cats from Guangzhou, South China by multilocus genotyping at the tpi, gdh, bg, and 18S rRNA loci, in order to better understand the transmission mechanism of zoonotic genotypes of G. duodenalis.
A total of 102 feline fecal samples were obtained from 2 shelters during March to July 2013. These 2 shelters were located in Baiyun (urban, n=30) and Conghua (suburban, n=72) districts of Guangzhou City, China. Only 1 fecal submission was obtained from each cat. All fecal samples submitted to the laboratory, were preserved in 2.5% potassium dichromate and stored at 4˚C. Direct microscopic examination was done after centrifugation of 1.5 g feces and concentration by the flotation technique with 1.18 SG ZnSO4 [11].
DNAs were extracted directly from fecal samples using a commercial DNA extraction kit (QIAamp DNA Stool Mini Kit, QIAgen, Hilden, Germany) according to the manufacturer’s protocols. However, all samples were pretreated with 5 cycles of heating at 100˚C for 5 min and immediate freezing at -80˚C for 5 min. A negative control (water) was used in each extraction group. Extracted DNAs were stored at -20˚C.
The tpi, gdh, bg, and 18S rRNA genes from cat-associated G. duodenalis were analyzed by PCR assays. Portions of the 18S rRNA (291 bp), bg (515 bp), gdh (530 bp), and tpi (334) genes were individually amplified according to previously described protocols [12-15]. The PCR products were analyzed after electrophoresis in 1.5% agarose gels and stained with 0.2 g/ml of ethidium bromide and visualized on a UV transilluminator. Positive amplicons were purified and sequenced at an ABI 377 automated DNA sequencer (BigDye Terminator Chemistry). Obtained sequences were aligned and analyzed with MEGA5.2 software. Obtained nucleotide sequences have been deposited in the GenBank™ database under following accession nos.: KJ027439 to KJ027447 for tpi, KJ027434 to KJ027437 for gdh, KJ027408 to KJ027410, KJ027412, KJ027416, and KJ027424 for bg, and KJ027404 to KJ027406 for 18S rRNA, respectively.
The amplification fragments of the tpi (334 bp), gdh (530 bp), bg (515 bp), and 18S rRNA (291 bp) genes were obtained from 9, 4, 6, and 3 feline samples collected in Guangzhou, respectively (Fig. 1). The assemblages or sub-assemblages of G. duodenalis were identified by PCR assay of each gene in these fecal samples (Table 1). A 9.8% (10/102) of feline fecal samples were found to be positive by microscopy, 10% (3/30) in Baiyun district and 9.7% (7/72) in Conghua district. The tpi PCR was able to genotype 90.0% (9/10) of the Giardia-positive samples. Among them, 100% (9/9) of the samples were found to contain potentially zoonotic assemblage AI. The gdh PCR was able to genotype 40.0% (4/10) of the Giardia-positive samples. Among them, all the 4 samples were zoonotic assemblage AI. The bg PCR was able to genotype 60.0% (6/10) of the Giardia-positive samples. Among them, assemblage AI was the most prevalent, comprising of 66.7% (4/6) samples, followed by 1 assemblage C and 1 assemblage F. Only 30.0% (3/10) of the Giardia-positive samples were typed by the 18S rRNA PCR, sequence analysis revealed assemblage A in 2 isolates, and assemblage F in 1 isolate.
Fig. 1.
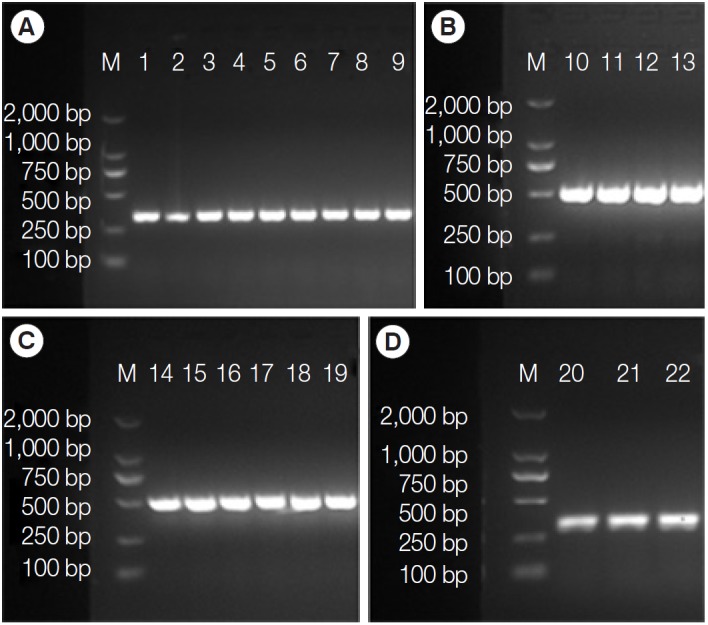
PCR amplification of tpi (A), gdh (B), bg (C), and 18S rRNA (D) genes from the cat-derived Giardia. M, DL2000 DNA marker; 1,15-CBY19; 2,16-CBY20; 3,17-CHC79; 4-CHC83; 5,11,21- CHC66; 6-CHC77; 7,13-CHC62; 8-CHC80; 9,19,22-CHC68; 10-CHC83; 12,18-CHC77; 14,20-CBY01.
Table 1.
Genotyping data of Giardia duodenalis isolates from cats in Guangzhou, China at the 4 loci; tpi, gdh, bg, and 18S rRNA
| Sample ID |
G. duodenalis assemblages identified by different gene loci |
|||
|---|---|---|---|---|
| tpi | gdh | bg | 18S | |
| CBY01 | Neg | Neg | F | F |
| CBY19 | AI | Neg | C | Neg |
| CBY20 | AI | Neg | AI | Neg |
| CHC79 | AI | Neg | AI | Neg |
| CHC83 | AI | AI | Neg | Neg |
| CHC66 | AI | AI | Neg | A |
| CHC77 | AI | AI | AI | Neg |
| CHC62 | AI | AI | Neg | Neg |
| CHC80 | AI | Neg | Neg | Neg |
| CHC68 | AI | Neg | AI | A |
Neg, negative.
Single infections with assemblage AI or assemblage A were found in 8 Giardia-positive samples. Among them, 3 samples (CHC66, CHC68, and CHC77) were typed at 3 loci (Table 1); 4 samples (CBY20, CHC62, CHC79, and CHC83) were typed at 2 loci; 1 sample (CHC80) was typed at 1 locus. Whereas, single infection with assemblage F was found in 1 sample (CBY01) which was typed at 2 loci. A mixed infection with 2 assemblages (assemblage A and C) was detected in 1 sample.
The sequences of the 4 genes were included in the phylogenetic tree and homologous analysis, which were performed by using neighbor joining method and nucleotide base alignment, respectively. All sequences of assemblage A were placed in 1 cluster with AI reference sequence (GU564275) separating from AII and AIII at tpi gene by the neighbor joining tree with high bootstrap support (Fig. 2A). At gdh gene, the neighbor joining tree placed the 4 sequences (CHC62, CHC66, CHC77, and CHC83) in 1 cluster with AI reference sequence (EF685696) with high bootstrap support (Fig. 2B). Assemblage A, C, and F were significantly separated from each other at bg gene (Fig. 2C). All sequences of assemblage A (CBY20, CHC68, CHC77, and CHC79) were placed in 1 cluster with AI reference sequence (X85958) separating from AII and AIII. Assemblage A and F were also significantly separated from each other at 18S rRNA gene (Fig. 2D). Based on tpi, gdh, and bg genes, all sequences of assemblage A showed complete homology with AI (GU564275, EF685696, and X85958, respectively) except for 1 isolate (CHC83) where substitution at 1 position was observed (T-G at position 25) (Table 2).
Fig. 2.
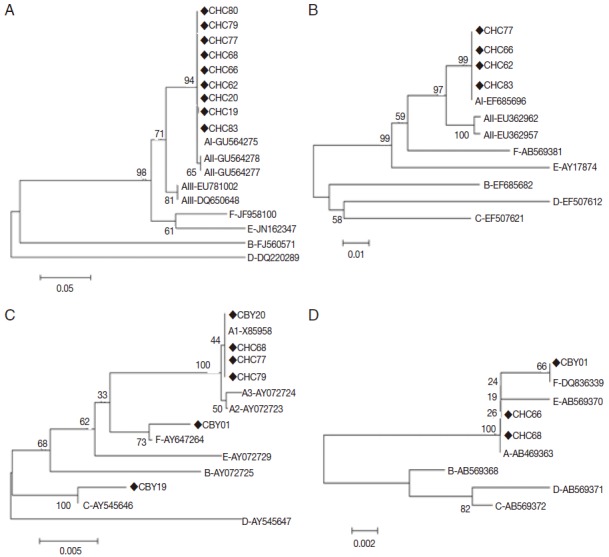
Phylogenetic relationships of Giardia duodenalis assemblages from cats inferred by the neighbor-joining analysis of 4 gene loci. (A) Triose phosphate isomerase (tpi) gene. (B) Glutamate dehydrogenase (gdh) gene. (C) β-giardin (bg) gene. (D) Small subunit ribosomal RNA (18S rRNA) gene. Bootstrap values obtained from 1,000 replicates are indicated on branches in percentage.
Table 2.
Summary on nucleotide variations of Giardia duodenalis A genotype sub-assemblages from cat isolates at the bg, tpi, and gdh genes
| Alignment position | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| bg | 482 | 490 | 628 | |||||||||||
| Reference sequences | AI-X85958 | C | T | C | ||||||||||
| A2-AY072723 | . | . | T | |||||||||||
| A3-AY072724 | T | C | T | |||||||||||
| Cat | CBY20/CHC68/CHC77/CHC79 | . | . | . | . | . | ||||||||
| gdh | 527 | 582 | 626 | 644 | 722 | 776 | 830 | 854 | 884 | 890 | 893 | 917 | 925 | |
| Reference sequences | AI-EF685696 | C | T | T | C | T | C | C | C | T | T | T | C | C |
| AII-EU362962 | A | . | C | T | C | T | T | T | C | C | C | T | T | |
| Cat | CHC62/CHC66/CHC77/CHC83 | . | . | . | . | . | . | . | . | . | . | . | . | . |
| tpi | 318 | 351 | 366 | 369 | 375 | 378 | 387 | 391 | 402 | 420 | 432 | 447 | 489 | |
| Reference sequences | AI-GU564275 | T | T | C | G | C | C | T | G | G | A | A | A | C |
| AII-GU564277 | . | . | . | . | C | |||||||||
| AIII-EU781002 | C | T | A | T | T | T | A | G | G | G | T | |||
| Cat | CHC83 | G | . | . | . | . | . | . | . | . | . | . | . | . |
| CHC80/CHC79/CHC77/CHC68/ | ||||||||||||||
| CHC66/CHC62/CBY20/CBY19 | . | . | . | . | . | . | . | . | . | . | . | . | . | |
Nucleotide substitutions are numbered from the start of the gene sequence which was amplified. Dots indicate identity to the A1 reference sequence.
The present study found that assemblage A was predominant in cat-associated G. duodenalis in Guangzhou, China, which is not consistent with Scorza et al. [16], who reported that 162 of 270 (60.0%) G. duodenalis isolates of cats worldwide were the cat-adapted assemblage F. This phenomenon of the dominant assemblage A may be related to the living environments of the stray cats in China. These stray cats were living in groups, which increased the possibility for transmission among the animals. Previous studies showed that cats with crowded breeding had higher levels of infection compared with household cats [17]. A similar variability has been noted among dogs in tea-growing communities in Assam, India, all isolates of G. duodenalis from dogs were identified as assemblage A (predominantly AII) and B [18]. There was a highly significant relationship between humans, dog ownership, and presence of a Giardia-positive dog in the same household.
In our study, the tpi primers had the highest amplification rate in cat isolates, followed by the bg, gdh, and 18S rRNA primers. They showed a greater success (n=9) in detecting assemblage AI which have zoonotic potential, than bg (n=4) and gdh (n=4) primers. Furthermore, assemblage AI was the dominant assemblage from cats in China, which is similar with the finding that the subassemblage AI is dominant among the limited numbers of assemblage A isolates from cats in Europe, United States, Brazil, Australia, and Japan [9,10,19-22]; while AII is found mainly in humans [3]. Also, 25 G. duodenalis-positive specimens belonging to AII were amplified in China [23]. Moreover, the assemblage F from 1 isolate was also typed by bg gene. However, the dog-specific assemblage C was identified by bg gene locus in 1 feline sample (CBY19), which was not expected. This may be due to contamination of the sample during collection, or actual infection of the cat by the assemblage C, which remains to be determined. However, similar results have been previously reported in cats in Australia and Europe [9,10] and humans in China [24]. Furthermore, CBY19 was mixed infection with assemblage C and assemblage AI, which provide further evidence that multiple primer sets should be used in G. duodenalis genotyping studies, as suggested previously [25,26]. Our results showed poorest amplification at the 18S rRNA gene locus, but with longer fragment (291 bp) than most of the previous studies (130 bp). Also the fragment of 130 bp is just too short to submit to GenBank. What’s more, we have been doing G. duodenalis studies for several years in our laboratory, no assemblage B was found in canine and feline isolates in Guangdong, southern China. It might be that the assemblages B of isolates do not exist in the region. Of course, more researches should be done to confirm it.
In conclusion, most of the stray cats we examined in urban and suburban environments of Guangzhou, China were infected with assemblage A or subassemblage AI of G. duodenalis with potentially zoonotic genotype. If these cats were previously in a home and not stray, they could have obtained the genotype from an infected family member and vice versa [25]. Our study not only confirmed the occurrence of G. duodenalis in stray cats, but also showed for the first time that zoonotic assemblages/sub-assemblages appeared frequently in cats living in urban and suburban environments in China. Humans may be involved in a zoonotic cycle of transmission. Cats should therefore be considered a potential reservoir for infection in humans.
Acknowledgments
This work was supported by grant from National Natural Science Foundation of China (grant no. 31272551 and 30972179). We thank the humane shelter' personnel for helping us in collecting all of the samples.
Footnotes
The authors declare that there is no conflict of interests regarding the publication of this paper.
REFERENCES
- 1.Thompson RC, Hopkins RM, Homan WL. Nomenclature and genetic groupings of Giardia infecting mammals. Parasitol Today. 2000;16:210–213. doi: 10.1016/s0169-4758(99)01624-5. [DOI] [PubMed] [Google Scholar]
- 2.Monis PT, Caccio SM, Thompson RC. Variation in Giardia : towards a taxonomic revision of the genus. Trends Parasitol. 2009;25:93–100. doi: 10.1016/j.pt.2008.11.006. [DOI] [PubMed] [Google Scholar]
- 3.Feng Y, Xiao L. Zoonotic potential and molecular epidemiology of Giardia species and giardiasis. Clin Microbiol Rev. 2011;24:110–140. doi: 10.1128/CMR.00033-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yong TS, Park SJ, Hwang UW, Yang HW, Lee KW, Min DY, Rim HJ, Wang Y, Zheng F. Genotyping of Giardia lamblia isolates from humans in China and Korea using ribosomal DNA sequences. J Parasitol. 2000;86:887–891. doi: 10.1645/0022-3395(2000)086[0887:GOGLIF]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 5.Wang R, Zhang X, Zhu H, Zhang L, Feng Y, Jian F, Ning C, Qi M, Zhou Y, Fu K, Wang Y, Sun Y, Wang Q, Xiao L. Genetic characterizations of Cryptosporidium spp. and Giardia duodenalis in humans in Henan, China. Exp Parasitol. 2011;127:42–45. doi: 10.1016/j.exppara.2010.06.034. [DOI] [PubMed] [Google Scholar]
- 6.Ye J, Xiao L, Li J, Huang W, Amer SE, Guo Y, Roellig D, Feng Y. Occurrence of human-pathogenic Enterocytozoon bieneusi, Giardia duodenalis and Cryptosporidium genotypes in laboratory macaques in Guangxi, China. Parasitol Int. 2014;63:132–137. doi: 10.1016/j.parint.2013.10.007. [DOI] [PubMed] [Google Scholar]
- 7.Li J, Zhang P, Wang P, Alsarakibi M, Zhu H, Liu Y, Meng X, Li J, Guo J, Li GQ. Genotype identification and prevalence of Giardia duodenalis in pet dogs of Guangzhou, Southern China. Vet Parasitol. 2012;188:368–371. doi: 10.1016/j.vetpar.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 8.Zheng GC, Alsarakibi M, Liu YJ, Hu W, Luo Q, Tan LP, Li GQ. Genotyping of Giardia duodenalis isolates from dogs in Guangdong, China based on multi-locus sequence. Korean J Parasitol. 2014;52:299–304. doi: 10.3347/kjp.2014.52.3.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Read CM, Monis PT, Thompson RC. Discrimination of all genotypes of Giardia duodenalis at the glutamate dehydrogenase locus using PCR-RFLP. Infect Genet Evol. 2004;4:125–130. doi: 10.1016/j.meegid.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 10.Sprong H, Caccio SM, van der Giessen JWB, Network Z. Partners identification of zoonotic genotypes of Giardia duodenalis. PLoS Negl Trop Dis. 2009;3:e558. doi: 10.1371/journal.pntd.0000558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dryden MW, Payne PA, Smith V. Accurate diagnosis of Giardia spp. and proper fecal examination procedures. Vet Ther. 2006;7:4–14. [PubMed] [Google Scholar]
- 12.Lalle M, Pozio E, Capelli G, Bruschi F, Crotti D, Cacciò SM. Genetic heterogeneity at the beta-giardin locus among human and animal isolates of Giardia duodenalis and identification of potentially zoonotic subgenotypes. Int J Parasitol. 2004;35:207–213. doi: 10.1016/j.ijpara.2004.10.022. [DOI] [PubMed] [Google Scholar]
- 13.Hopkins RM, Meloni BP, Groth DM, Wetherall JD, Reynoldson JA, Thompson RC. Ribosomal RNA sequencing reveals differences between the genotypes of Giardia isolates recovered from humans and dogs living in the same locality. J Parasitol. 1997;83:44–51. [PubMed] [Google Scholar]
- 14.Sulaiman IM, Jiang JL, Singh A, Xiao LH. Distribution of Giardia duodenalis genotypes and subgenotypes in raw urban wastewater in Milwaukee, Wisconsin. Appl Environ Microb. 2004;70:3776–3780. doi: 10.1128/AEM.70.6.3776-3780.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang P, Zhu HB, Li J, Liu YJ, Guo JC, Meng XL, Li GQ. Genotyping of two strains of Giardia from dogs by tpi gene. China Anim Husb Vet Med. 2011;38:59–63. (in Chinese) [Google Scholar]
- 16.Scorza AV, Ballweber LR, Tangtrongsup S, Panuska C, Lappin MR. Comparisons of mammalian Giardia duodenalis assemblages based on the beta-giardin, glutamate dehydrogenase and triose phosphate isomerase genes. Vet Parasitol. 2012;189:182–188. doi: 10.1016/j.vetpar.2012.04.032. [DOI] [PubMed] [Google Scholar]
- 17.Tangtrongsup S, Scorza V. Update on the diagnosis and management of Giardia spp. infections in dogs and cats. Top Companion Anim Med. 2010;25:155–162. doi: 10.1053/j.tcam.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 18.Traub RJ, Monis PT, Robertson I, Irwin P, Mencke N, Thompson RC. Epidemiological and molecular evidence supports the zoonotic transmission of Giardia among humans and dogs living in the same community. Parasitology. 2004;128:253–262. doi: 10.1017/s0031182003004505. [DOI] [PubMed] [Google Scholar]
- 19.Vasilopulos RJ, Rickard LG, Mackin AJ, Pharr GT, Huston CL. Genotypic analysis of Giardia duodenalis in domestic cats. J Vet Intern Med. 2007;21:352–355. doi: 10.1892/0891-6640(2007)21[352:gaogdi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 20.Volotão AC, Costa-Macedo LM, Haddad FS, Brandão A, Peralta JM, Fernandes O. Genotyping of Giardia duodenalis from human and animal samples from Brazil using beta-giardin gene: a phylogenetic analysis. Acta Trop. 2007;102:10–19. doi: 10.1016/j.actatropica.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 21.Souza SL, Gennari SM, Richtzenhain LJ, Pena HF, Funada MR, Cortez A, Gregori F, Soares RM. Molecular identification of Giardia duodenalis isolates from humans, dogs, cats and cattle from the state of São Paulo, Brazil, by sequence analysis of fragments of glutamate dehydrogenase (gdh) coding gene. Vet Parasitol. 2007;149:258–264. doi: 10.1016/j.vetpar.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 22.Suzuki J, Murata R, Kobayashi S, Sadamasu K, Kai A, Takeuchi T. Risk of human infection with Giardia duodenalis from cats in Japan and genotyping of the isolates to assess the route of infection in cats. Parasitology. 2011;138:493–500. doi: 10.1017/S0031182010001459. [DOI] [PubMed] [Google Scholar]
- 23.Wang L, Xiao L, Duan L, Ye J, Guo Y, Guo M, Liu L, Feng Y. Concurrent infections of Giardia duodenalis, Enterocytozoon bieneusi, and Clostridium difficile in children during a cryptosporidiosis outbreak in a pediatric hospital in China. PLoS Negl Trop Dis. 2013;7:e2437. doi: 10.1371/journal.pntd.0002437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu H, Shen Y, Yin J, Yuan Z, Jiang Y, Xu Y, Pan W, Hu Y, Cao J. Prevalence and genetic characterization of Cryptosporidium, Enterocytozoon, Giardia and Cyclospora in diarrheal outpatients in China. BMC Infect Dis. 2014;14:25. doi: 10.1186/1471-2334-14-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Covacin C, Aucoin DP, Elliot A, Thompson RC. Genotypic characterisation of Giardia from domestic dogs in the USA. Vet Parasitol. 2011;177:28–32. doi: 10.1016/j.vetpar.2010.11.029. [DOI] [PubMed] [Google Scholar]
- 26.Beck R, Sprong H, Pozio E, Caccio SM. Genotyping Giardia duodenalis isolates from dogs: lessons from a multilocus sequence typing study. Vector Borne Zoonotic Dis. 2012;12:206–213. doi: 10.1089/vbz.2011.0751. [DOI] [PubMed] [Google Scholar]


