Abstract
We designed Lactococcus lactis to detect Enterococcus faecalis. Upon detection, L. lactis produce and secrete antienterococcal peptides. The peptides inhibit enterococcal growth and reduce viability of enterococci in the vicinity of L. lactis. The enterococcal sex pheromone cCF10 serves as the signal for detection. Expression vectors derived from pCF10, a cCF10-responsive E. faecalis sex-pheromone conjugative plasmid, were engineered in L. lactis for the detection system. Recombinant host strains were engineered to express genes for three bacteriocins, enterocin A, hiracin JM79 and enterocin P, each with potent antimicrobial activity against E. faecalis. Sensitive detection and specific inhibition occur both in agar and liquid media. The engineered L. lactis also inhibited growth of multidrug-resistant E. faecium strains, when induced by cCF10. The presented vectors and strains can be components of a toolbox for the development of alternative antibiotic technologies targeting enterococci at the site of infection.
Keywords: lactic acid bacteria, Lactococcus lactis, Enterococcus faecalis, drug delivery vector, recombinant plasmids
Enterococci have undergone a remarkable metamorphosis in the last quarter century, from low grade-pathogens to a major burden to health care systems.1 Before the first reports of vancomycin-resistant enterococcus (VRE) strains in the late 1980s, these microbes were considered harmless, if not helpful, commensals of the human gastrointestinal (GI) tract. Now enterococci are also viewed as a major cause of hospital-acquired infections.2 According to these recent reports, VRE colonization in health-care institutions has reached endemic proportions. VRE are easily spread through direct contact and are resilient and versatile organisms thriving on nearly every type of environmental surface tested. As a consequence, patients with VRE infection have increased mortality, morbidity, length of hospital stay, and hospital costs, in comparison to uninfected groups.3 The enterococcal strains that dominate pathogenesis in the human GI tract are E. faecalis and E. faecium. Most enterococcal infections are caused by E. faecalis; yet, E. faecium is currently the most difficult to treat. As of 2007, 80% of E. faecium isolates were vancomycin resistant compared to just over 5% of E. faecalis.4
Since the source for many enterococcal infections is the intestinal tract, antibiotic technologies that can effectively prevent or treat enterococcal intestinal colonization may limit the public health harm caused by these microorganisms.1
Lactic acid bacteria (LAB) have emerged as potential vehicles for drug delivery in the GI tract.5 They are bile-resistant organisms and have the ability to survive passage to the GI tract. Importantly, some of these LAB are generally recognized as safe (GRAS) for human and animal consumption.6L. lactis is a model LAB, being one of the most amenable expression cell factories for heterologous protein secretion.7 Indeed, L. lactis is being explored as the first genetically modified organism used alive for the treatment of human Crohn’s disease by the production of interleukin-10 in the GI tract.8
Several expression systems have been developed in LAB. One of the most successful and widely used inducible systems is the NIsin Controlled gene Expression (NICE) system. The NICE system is based on the nisA operon, which is involved in the biosynthesis of nisin A, a post-translationally modified antimicrobial peptide produced by several strains of L. lactis.9 In previous experiments, induction is initiated with nisin A in culture, before LAB are supplied to animals. Consequently, the recombinant bacteria entering the host are activated, producing the protein of interest, not unlike in the case of a constitutive promoter.10 The difference, of course, is that the NICE system will be turned off once the intracellular nisin A is degraded. It may then be difficult to control the duration of protein production or the amount produced.
Other LAB expression systems are inducible under various conditions such as phage attack, temperature or pH shift, or the presence of specific sugars.10 Regulation of gene expression with these promoters is challenging, often because of leaky expression. To say that there are limits in these expression systems is not to say that they have little or no value. Nonetheless, an ideal system would be one that remains inactive when administered to the host, survives passage to the GI tract and then produces the proteins of interest only when needed, for example, only in the presence of pathogens. Here, we discuss such a LAB-based system that acts as a sensitive enterococcal detection device and then, upon detection, produces and secretes a molecular arsenal of antimicrobial peptides to specifically target the sensed enterococci.
We designed a set of vectors based on the pheromone-mediated intercellular signaling system pCF10 that regulates expression of conjugative plasmid transfer genes in Enterococcus faecalis. This plasmid contains genes for resistance to tetracycline, virulence factors, along with genes for conjugative plasmid transfer.11 Conjugation and plasmid transfer between donor cells and receiver cells is stimulated by pheromone cCF10. This heptapeptide (LVTLVFV) is produced from a conserved chromosomal gene present in E. faecalis.12 cCF10 induces high gene expression in donor cells carrying pCF10.13 When donor cells import cCF10 into the cytoplasm, the pheromone binds to the pCF10-encoded master protein regulator PrgX. This binding abolishes repression of transcription of the prgQ operon, encoding conjugation genes.14,15 The donor cells produce reduced but still active levels of cCF10.16 This residual pheromone activity is neutralized by the production of peptide iCF10 from the prgQ locus of pCF10.17 Peptide iCF10 (AITLIFI) competes with cCF10 for binding to PrgX, opposes cCF10 effects, and causes PrgX to repress gene expression.18 While the direct effects of the regulatory peptides on transcription initiation from the prgQ promoter in vivo are modest, the multiple layers of post-transcriptional regulation via antisense RNAs and interference between colliding RNA polymerase elongation complexes (illustrated in the Supporting Information Figures S1 and S2) endow the system with extremely tight control and a high dynamic range of expression.18−20
We engineered cCF10-inducible vectors in L. lactis to express bacteriocins. Bacteriocins are defined as ribosomally synthesized peptides produced by bacteria that inhibit the growth of other closely related bacteria. We focus on class IIa enterocins, particularly, enterocin A (EntA), enterocin P (EntP), and hiracin JM79 (HirJM79). These are antimicrobial peptides (AMPs) produced by enterococci, with strong antimicrobial activity against other enterococci, including VRE.21−23 The two organisms share common protein expression machinery allowing the transfer of components from one to the other. In Methods, we discuss previous examples of successful cloning of enterococcal components into L. lactis.
Experiments reveal that the recombinant LAB are able to detect the cCF10 naturally produced by E. faecalis and to respond by producing the bacteriocins with high activity against enterococcal strains.
Results and Discussion
E. faecalis and E. faecium are threats to human health partly because of their remarkable ability to acquire and spread determinants of antibiotic resistance via horizontal gene transfer.24E. faecalis employ peptide-pheromones as signals to communicate with each other and trigger the expression of genes involved in conjugation and plasmid transfer.13 We turned their capacity to detect their own cell population density against enterococci by hijacking components of plasmid pCF10 to engineer recombinant LAB capable of sensing and responding to enterococcal pheromones. In the following, we detail how (1) we designed a set of inducible and constitutive expression vectors for controlled expression of proteins by L. lactis, (2) the engineered L. lactis-based systems are induced in the presence of E. faecalis, (3) the recombinant L. lactis produce and secrete bacteriocins with activity against E. faecalis and E. faecium, and (4) we combined sensing and inhibition components so recombinant LAB can be used to detect E. faecalis and then produce a molecular arsenal of antimicrobial peptides to target the sensed enterococci. To our knowledge, this is the first LAB-based system engineered for both detection and inhibition of specific pathogens.
Function and Optimization of the Detection System in LAB
We designed and tested a set of expression plasmids in L. lactis, originally characterized in E. faecalis,18,25 for their ability to confer expression of a transcriptional lacZ reporter gene in response to exogenously supplied cCF10 peptide (Supporting Information Figure S1). Plasmid pBK2 encodes the key components of the pCF10 genetic switch. Plasmid pBK1 contains a deletion of the prgX, and thus shows no repression of the prgQ promoter. In plasmid pBK2idT, the deletion of three nucleotides from prgQ leads to the production of an inactive iCF10-derivative, which can no longer bind to PrgX, reducing PrgX repression (Supporting Information Figures S1 and S2).
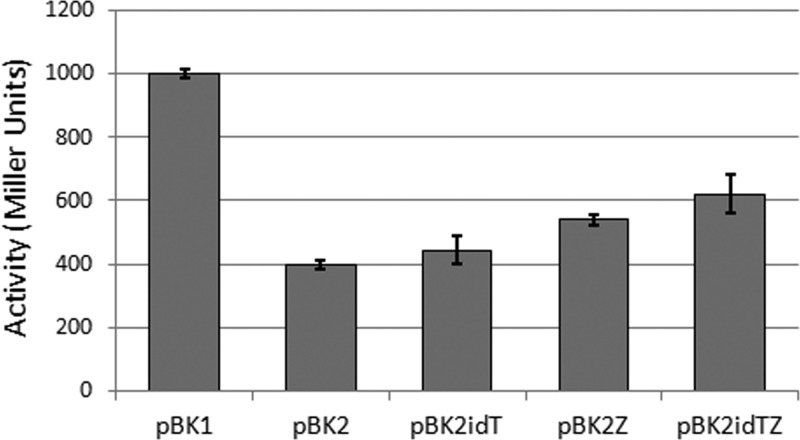
As expected, highest levels of β-gal production were observed for L. lactis-pBK1, where the absence of prgX prevents the repression of PQ by PrgX, and therefore PQ works as a constitutive promoter, with the expression of lacZ always on (Figure 1). Strains carrying either pBK2 or pBK2idT both produced β-gal upon induction with the pheromone cCF10, with the latter strain showing higher levels of induced expression (Figure 1). Although we presented induction results using 50 ng/mL of cCF10, we found that lower levels of pheromone (25 ng/mL and 10 ng/mL) resulted in comparable expression.
Figure 1.
β-gal activity (Miller Units) from recombinant L. lactis strains carrying engineered plasmids (pBK1, pBK2, pBK2iDT, pBK2Z, pBK2idTZ). Exponential cultures of each strain were induced with 100 ng/mL cCF10 90 min prior to harvesting cells for β-gal assay. The results are shown as averaged results from two independent experiments, each done in triplicate. The error bars represent the standard deviation.
To increase sensitivity to exogenous cCF10, we cloned and expressed prgZ (coding for PrgZ, a surface cCF10-binding protein known to mediate pheromone import26) under the control of the strong constitutive promoter P32, in the plasmids pBK2 and pBK2idT. The β-gal activity from recombinant strains originated, L. lactis-pBK2Z and L. lactis-pBK2idTZ, was approximately 15% higher than that from strains lacking prgZ (L. lactis-pBK2 and L. lactis-pBK2idT, respectively). Overall, pheromone-detecting L. lactis produced the highest β-gal activity when carrying vector pBK2idTZ. This vector presents two major improvements over the original pBK2. The first is the elimination of the repressor molecule iCF10. There is then no iCF10-cCF10 competition for PrgX. Consequently, the PQ promoter is on, until cCF10 is sufficiently degraded. The second difference is the presence of prgZ regulated by the strong constitutive promoter P23. PrgZ is an extracellular peptide-binding protein, acting as a strong receptor of cCF10.26 PrgZ delivers cCF10 to the oligopeptide permease, Opp, which subsequently imports it inside the cell for induction. It is worth noting that deletion of prgZ in E. faecalis decreases the sensitivity of the donor cells to cCF10.26 We have also observed that the presence of PrgZ is not necessary for pheromone import, presumably because OppA can fulfill this role, albeit with lower affinity.26 Nevertheless, the overexpression of prgZ markedly augments cCF10 transport inside L. lactis, in effect enhancing the activity of PQ.
Current results confirm previous studies, demonstrating that the PrgX-PrgQ system is tightly regulated.18,19,25,26 In the absence of cCF10, we observed no measurable β-gal production. As discussed previously, out of the opposing and overlapping orientation of the PQ and PX promoters emerges a synergy of transcriptional interference with antisense regulation.19 This synergy results in a tightly regulated bistable switch behavior. Other LAB inducible systems, such as the NICE or the SakP-based system (pSIP), have been shown to be efficient and well regulated when using L. lactis or Lactobacillus as hosts, respectively. However, they often exhibit significantly leaky basal activity in other hosts.27,28 Furthermore, these other systems typically rely on the autoregulation of the inducer via a two-component system composed of a membrane histidine protein kinase (HPK) and a response regulator (RR).29 In some cases, these systems can only be used by specific strains containing the regulators integrated in the chromosome or by employing two different plasmids. In our system, the inducer cCF10 is transported to the cytoplasm where it directly interacts with the repressor PrgX, to modulate gene expression by PQ. All the components involved in the regulation are located within a single plasmid. In addition, the cCF10 inducer peptide is nontoxic, allowing the use of very high inducer levels, without impacting growth of the producer strain. These results demonstrate the function of the pCF10 pheromone sensing system in L. lactis and how the natural system can be manipulated to vary the sensitivity to an inducing signal.
Detection of E. faecalis in Coculture
To determine whether the natural production of pheromone cCF10 by E. faecalis is sufficient to induce the expression of lacZ by recombinant L. lactis, we carried two different sets of experiments, in semisolid and liquid medium.
When E. faecalis and recombinant L. lactis were grown together on agar plates, we observed three different levels of β-gal production: high, medium, and low. As observed in Figure 2A, L. lactis-pBK1 exhibited high levels of β-gal production on the plate. The interaction between recombinant L. lactis-pBK2Z or L. lactis-pBK2idTZ with E. faecalis OG1RF triggered the expression of lacZ by these two strains. β-gal production was higher when using L. lactis-pBK2idTZ. No β-gal production was detected in those colonies located outside the area of interaction with E. faecalis OG1RF.
Figure 2.
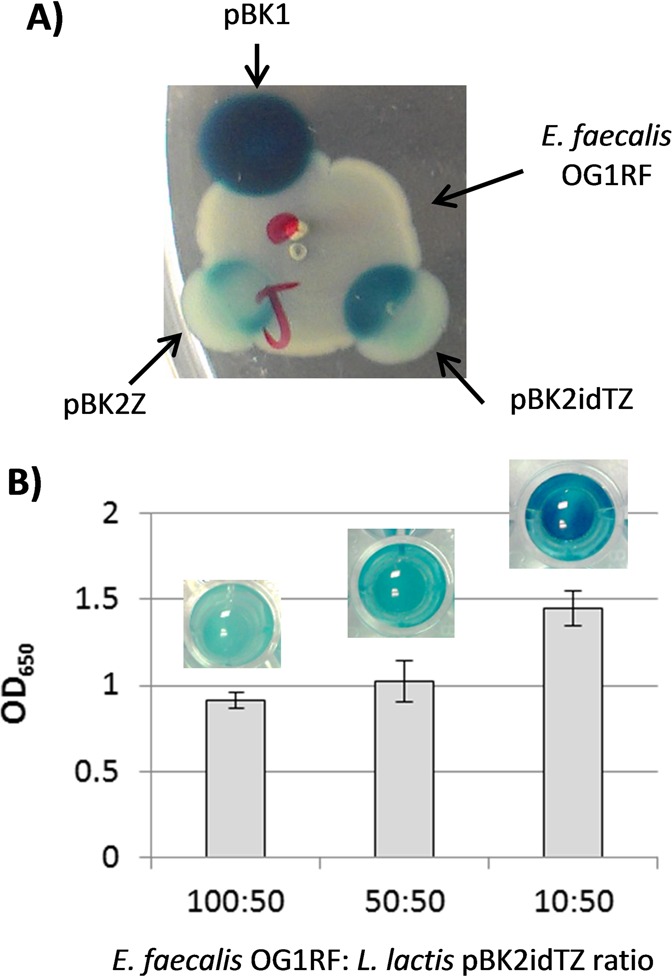
lacZ expression of recombinant L. lactis in coculture with E. faecalis. (A) Plate experiments: 10 μL dot of E. faecalis OG1RF and 5 μL dot of recombinant L. lactis (pBK1, pBK2Z and pBK2idTZ) grown in BHI plates supplemented with X-gal. The blue staining corresponds to the expression of lacZ by recombinant L. lactis strains. These are representative images of an experiment that was repeated several times. (B) Absorbance at OD650 and image of E. faecalis OG1RF and L. lactis-pBK2idTZ grown in cocultures at different ratios (v/v) in M9X media. The results shown are averaged from two independent experiments, each done in triplicate. The error bars represent the standard deviation.
The observable response to cCF10 was markedly improved when the experiments were repeated in liquid medium. We mixed either E. faecalis OG1RF or E. faecalis JRC101 (a cCF10 deficient strain) with L. lactis-pBK2idTZ in cocultures at different ratios. Substantial induction of lacZ reporter expression was observed at different E. faecalis OG1RF/L. lactis-pBK2idTZ ratios (Figure 2B). Furthermore, no β-gal production was detected when growing L. lactis-pBK2idTZ together with E. faecalis JRC101, or when any of the strains was grown alone, confirming that the promoter controlling lacZ is tightly regulated by cCF10, secreted by E. faecalis OG1RF.
To probe the specificity of the system toward E. faecalis, we repeated the liquid experiments using instead different E. faecium spp., that are naturally non producers of cCF10. No significant β-gal production was detected in the media, which confirms the high specificity of the pheromone cCF10 against PrgX.
cCF10-Inducible Bacteriocin Production by L. lactis
We constructed three separate constructs, replacing lacZ in pBK2idTZ with the genes SPusp45:entA,30hirJM79 or entP,31 coding for the bacteriocins enterocin A, hiracin JM79, and enterocin P, respectively. In each case, along with one of the enterocins, we cloned the corresponding immunity gene (entiA, hiriJM79, and entiP, respectively30,31). Furthermore, we constructed a polycistronic sequence with the 6 genes (three AMPs and three corresponding immunity genes), and cloned it into pBK2idTZ (dubbed pBK2idtZ:Bac).
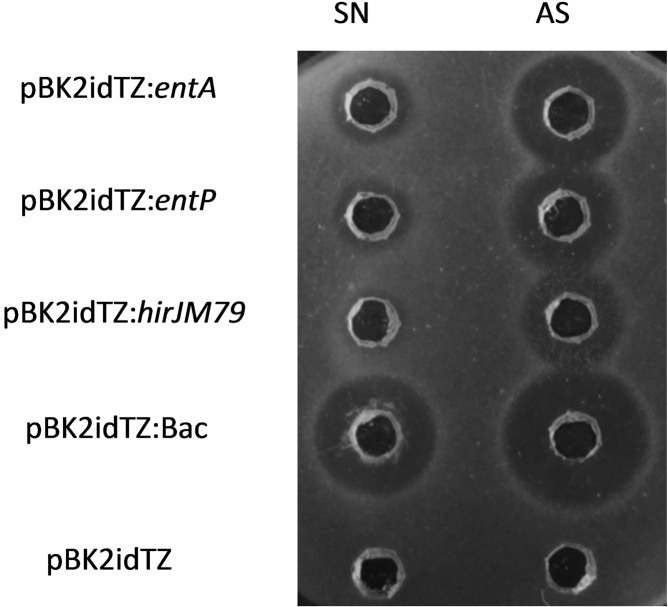
In order to quantify bacteriocin inhibition with our expression vectors, we first induced the recombinant L. lactis cultures with cCF10, collected the supernatants and then tested them against various E. faecalis and E. faecium indicator strains. We had previously determined that the maximum or near-maximum expression levels were obtained 3 h after inducing L. lactis with cCF10 at an OD600 of 0.4. In order to increase the bacteriocin concentration in the medium and avoid false negatives, we also performed protein precipitation of the supernatants with ammonium sulfate. Both the concentrated and nonconcentrates supernatants from recombinant L. lactis showed antimicrobial activity against all the strains tested in an agar diffusion test (ADT) (see Figure 3 for results using E. faecium 8 as indicator).
Figure 3.
Antimicrobial activity of supernatants (SN) and ammonium sulfate precipitated peptides (AS) from recombinant L. lactis strains. Exponential cultures of each strain were induced with 100 ng/mL cCF10 120 min prior collecting the supernatants. The antimicrobial activity was determined by an agar well diffusion test (ADT) with E. faecium 8 as the indicator microorganism.
The activity against enterococci was most accentuated when all three bacteriocins were expressed from the same strain, although modest antimicrobial activity was exhibited by the supernatants containing individual bacteriocins. No activity was observed in the supernatant of the control strain L. lactis NZ9000-pBK2idTZ.
Subsequently, we tested the activity of supernatants from L. lactis-pBK2idTZ:Bac by a 96 well plate assay. We observed inhibition against all E. faecalis and E. faecium indicator strains used in the study (Table 1). Notably, the antimicrobial activity from the supernatants was higher against E. faecium compared to E. faecalis. Three of the E. faecium strains tested had been identified by the University of Minnesota Clinical Microbiology Laboratory as exhibiting high-level Vancomycin resistance with minimum inhibitory concentrations >256 mg/mL whereas one strain (7a) was sensitive to Vancomycin (Dr. Patricia Ferrieri, personal communication). All four isolates were equally susceptible to killing by bacteriocin-producing LAB, suggesting the potential utililty of this system in control of multiresistant enterococci in clinical settings.
Table 1. Antimicrobial Activity (Bacteriocin Units/mL) of Supernatants from Recombinant Lactococcus lactis NZ9000-pBK2idTZ:Bac, against Different E. faecalis and E. faecium Indicator Strains.
| indicator | BU/mLa |
|---|---|
| E. faecalis | |
| V583 | 41 (±4) |
| CH116 | 50 (±18) |
| OG1RF | 68 (±21) |
| JH2-2 | 54 (±11) |
| E. faecium | |
| 6 | 224 (±88) |
| 7a | 106 (±34) |
| 8 | 269 (±50) |
| 9b | 208 (±51) |
Exponential cultures of each strain were induced with 100 ng/mL cCF10 120 min prior collecting the supernatants. The results are shown as averaged results from two independent experiments that were each done in triplicate. ± represents the standard deviation.
In this work, we used the bacteriocins enterocin A, hiracin JM79, and enterocin P, all of them naturally produced by enterococcus spp.21−23 All three peptides have been previously cloned and expressed in L. lactis individually, showing antimicrobial activity against E. faecium and Listeria monocytogenes spp.30,31 We observed a substantial increase of antimicrobial activity when the three were produced and secreted together. The synergistic effect of combining different bacteriocins has been previously studied and exploited to reduce the count of food borne and pathogenic bacteria in various applications.32
This type of synergism is presumably the result of the lower chance for an individual enterococcus cell to resist all three molecules. Genes for bacteriocin production as well as for immunity against these three bacteriocins are common among enterococci, but apparently only in a few cases do the genes for the three bacteriocins appear within the same enterococcal strain.33 Nevertheless, and perhaps not surprisingly, we observed the appearance of mutants resistant to all three bacteriocins in our plate experiments. The appearance of resistant mutants to class IIa bacteriocins has been previously documented.34 A potential future exploration can combine the use of class IIa bacteriocins with other bacteriocins or peptides having different modes of action.35
Coculture Detection and Inhibition
The ultimate goal of this work was to engineer L. lactis strains to detect E. faecalis in the environment and then produce a set of antimicrobial peptides. Once we established that both detection of E. faecalis and production of active bacteriocins by recombinant L. lactis were separately functional, we tested whether L. lactis could perform both tasks concurrently. The lactococcal strain selected for this set of experiments was L. lactis pBK2idTZ:Bac, the one with the highest sensitivity to cCF10 as well as the highest antimicrobial activity against E. faecalis. We grew this strain in coculture with E. faecalis OG1RF, under two different conditions, with or without an external addition of cCF10, and measured the enterococcal viability over time. We also cocultured L. lactis pBK1:Bac with E. faecalis OG1RF, in order to determine the inhibition caused by the constitutive system. To further verify that the inhibition of E. faecalis is caused by the expression and production of the three bacteriocins and discard any antimicrobial activity from the native lactococcal strain, we cultured the control strain L. lactis-pBK2idTZ together with E. faecalis OG1RF. To show that the induction of the AMPs is specifically triggered by cCF10, we cultured L. lactis pBK2idTZ:Bac together with E. faecalis JRC101, an E. faecalis OG1RF-derivative with a knockout in the gene coding for cCF10. As observed in Figure 4, the highest reduction of E. faecalis OG1RF counts was achieved by L. lactis-pBK1:Bac and L. lactis-pBK2idTZ:Bac induced with cCF10. Under these conditions we observed reductions of the enterococcal strain of up to 6 and 4 logarithmic units, respectively, in the first 2 h postinoculation. However, when using uninduced L. lactis-pBK2idtZ:Bac we observed a reduction of up to 2-fold in the total number of enterococcal CFU in the coculture in comparison with the control (Figure 4). These differences were not significant during the first 3 h, and reached the highest levels after 5–9 h of coculture.
Figure 4.
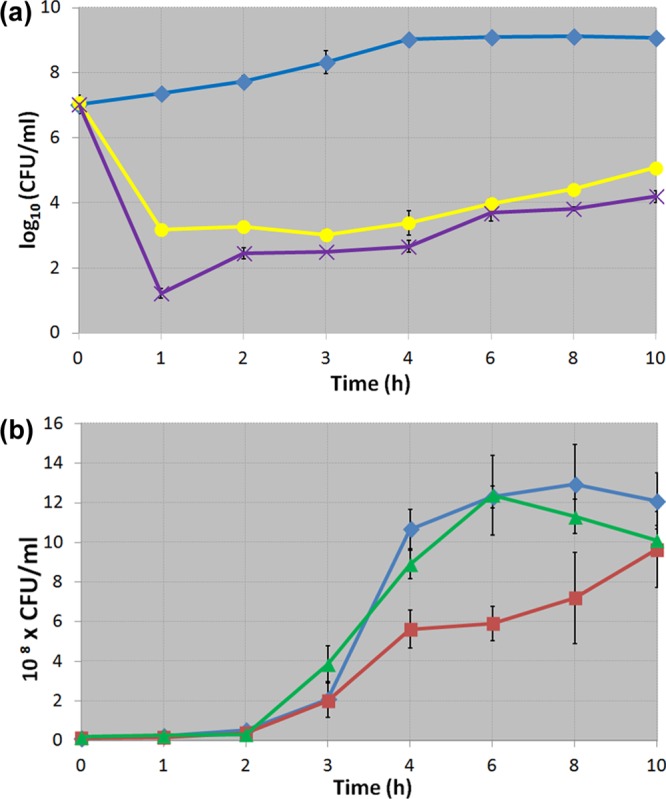
E. faecalis viability when grown in coculture with recombinant L. lactis strains. (A) E. faecalis OG1RF viability at different time points when cocultured with L. lactis-pBK2idTZ (blue diamonds), L. lactis-pBK1:Bac (purple x’s) or L. lactis-pBK2idTZ:Bac induced with 100 ng/mL cCF10 at time 0 (yellow circles). The viability is expressed as the log10 of the total E. faecalis OG1RF colony forming units per ml (CFU/mL). (B) E. faecalis OG1RF viability at different time points when cocultured with L. lactis-pBK2idTZ:Bac (red squares) and, shown again for cross-reference to Figure 4A, L. lactis-pBK2idTZ (blue diamonds). Also shown is E. faecalis JRC101 (a cCF10 deficient strain) viability when cocultured with L. lactis-pBK2idTZ:Bac (green triangles). The viability is expressed as the total number of E. faecalis CFU/mL. Cell ratios are as described in the text. All results are shown as averaged results from two independent experiments, each done in triplicate. The error bars represent the standard deviation.
A useful comparison is one between the AMP-containing supernatants and commonly used antibiotics (Ampicillin, Vancomycin, and Streptomycin). In Figure 5, E. faecium 8 growth curves are presented when the bacteria are treated with supernatant from L. lactis-pBK2idtZ:Bac and with each of the three antibiotics. The results are indicative of the inhibitory activity of secreted bacteriocins on E. faecium. We note that we observe similar profiles for other enterococcal strains studied here.
Figure 5.
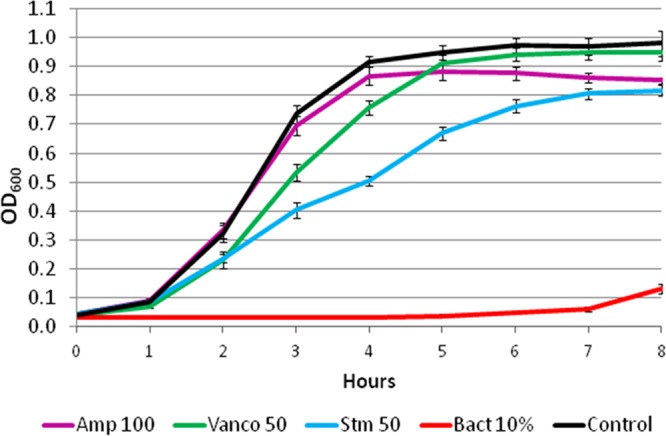
E. faecium inhibitory activity comparison between secreted bacteriocins and commonly used antibiotics. Growth curves of E. faecium 8 treated with different antibiotics and the supernatant of L. lactis-pBK2idTZ:Bac. An overnight culture of E. faecium 8 was diluted to an optical density at 600 nm (OD 600) of 0.1 in BHI broth supplemented with different antibiotics: Ampicillin 100 μg/mL (Amp 100), Vancomycin 50 μg/mL (Vanco 50) and Streptomycin 50 μg/mL (Stm 50); and with a 10% (v/v) supernatant of L. lactis pBK2idTZ:Bac (Bact 10%). E. faecium 8 grown in the absence of any treatment was used as control. The cultures were incubated at 37 °C and the OD600 was measured at various time periods. The experiments were repeated three times. The error bars represent the standard deviation.
The inhibition of E. faecalis OG1RF by uninduced L. lactis-pBK2idtZ:Bac is significant compared to the inhibition by the control lactococcal strain. Because the pheromone-levels produced naturally by E. faecalis are low (around 2 × 10–11 M), we can conclude that the systems described herein are notably sensitive to cCF10. Certainly, there is room for improvement, as exogenous cCF10 in higher quantities induces much stronger expression of genes in L. lactis. Future strategies may involve transcriptional autofeedback loops to amplify initial signals, for example, modifying L. lactis to express additional cCF10 in the presence of enterococcus-produced cCF10. Another direction may involve the design of other bacteriocins. In Figure 4A, it can be observed that E. faecalis OG1RF is starting to grow again, albeit slowly, after approximately 5–6 h. As a first explanation this may be attributed to the stability of the pheromone and antimicrobial peptides. As E. faecalis counts decrease, there is likely less cCF10, which may result in less bacteriocins expressed from the prgX/prgQ system. The finite stability of these peptides may then result in a dynamic interplay between the two species in coculture. More stable peptides may then further tip the balance in favor of L. lactis.
To conclude, we note that there is an increasing volume of scientific evidence for the beneficial role some LAB play in human health. The pBK systems described here exhibit several advantages in comparison to expression systems previously described for LAB. Arguably the most significant advantage is pathogen detection. These detection-based promoter systems allow for spatiotemporal control, ideally releasing proteins at the site and time of infection. In our experiment, we used other closely related bacteria, such as E. faecium, and found no detection responses. This reinforces the idea that the production of recombinant peptides by LAB would specifically be triggered upon detecting E. faecalis spp. This concept can potentially be explored with other pheromone systems, endogenous to different enterococcal, or other bacterial, strains.
We note that there are significant regulatory, ethical, environmental, public health, and public policy aspects for the development and use of a technology that is based on genetically modified organisms. Yet, we believe that it would be out of the scope of the current manuscript to discuss in length these important aspects of technology use. Nonetheless, we note that technology applications that face surmountable obstacles may be related to animal health. Certainly, there is a need to explore alternative technologies to antibiotics used in agriculture. We can envision important benefits to farm animals and humans that may be realized with successful development of the proposed technology. This intervention may result in healthy animals, decrease the emergence of antibiotic-resistance in foodborne pathogens, decrease the supply of antibiotics to animals, and reduce the rates of transmission of infectious diseases from animal farms to humans.
Methods
Bacterial Strains, Growth Conditions, and Plasmids
Bacteria used in this study are listed in Table 2. L. lactis NZ9000 was cultured at 30 °C in M17 broth (Oxoid Ltd., Basingstoke, U.K.) supplemented with 0.5% (w/v) glucose (GM17). The enterococcal strains were grown in BHI broth (Oxoid) at 37 °C. E. coli strains were grown in LB broth (Fisher Scientific, Fair Lawn, NJ, U.S.A.) at 37 °C, with shaking. Agar plates were made by the addition of 1.5% (wt/vol) agar (Oxoid) to the liquid media. When necessary, Rifampicin (Sigma Chemical Co., St. Louis, MO, U.S.A.) was added to the media at 50 μg/mL, and chloramphenichol at 5 μg/mL or 20 μg/mL, for L. lactis or E. coli, respectively.
Table 2. Bacterial Strains Used in This Study.
| strains | description | source |
|---|---|---|
| Lactococcus lactis NZ9000 | plasmid-free strain, derivative of MG1363; pepN::nisRK, nonbacteriocin producer | Mobitec |
| Enterococcus faecalis | ||
| V583 | ATCC 700802; St. Louis, MO, U.S.; first isolated vancomycin-resistant and first sequenced E. faecalis genome | UMN |
| CH116 | Boston, MA, U.S.; Gent/Kan/Strep/Tet/Erm/Pen resistant, β-lactamase-producing isolate | UMN |
| OG1RF | ATCC 47077; plasmid-free, Rif/Fus resistant mutant of OG1; common laboratory strain | UMN |
| JRC101 | OG1RF, ccfA- (not producer of cCF10) | UMN |
| JH2-2 | U.K.; Rif/Fus resistant mutant; common laboratory strain | UMN |
| Enterococcus faecium | ||
| 6 | UMNa isolate, Amp/Vanc/Linezolid resistant | UMN |
| 7a | UMN isolate, Amp/Linezolid resistant | UMN |
| 8 | UMN isolate, Amp/Vanc/Linezolid resistant | UMN |
| 9b | UMN isolate, Amp/Vanc/Linezolid resistant | UMN |
UMN, University of Minnesota.
Construction of Plasmids and L. lactis Transformation
Plasmids were constructed using standard molecular cloning techniques.36 All restriction enzymes were purchased from New England Biolabs (Beverly, MA, U.S.A.). Primers used in PCR reactions are listed in Supporting Information Table S1. Fragments obtained and plasmids used are listed in Table 3. Detailed information on the plasmids constructed and the structure of engineered promoters can be found in the suporting information (Supporting Information Materials and Methods, Figures S1 and S2).
Table 3. Plasmids and Fragments Used in This Study.
| plasmids | description | source |
|---|---|---|
| pBK2 | Cmr; inducible expression vector carrying the prgX/prgQ system and lacZ | (25) |
| pBK1 | Cmr; constitutive expression vector; pBK2 derivative with a deletion of the entire prgX | (25) |
| pBK2idT | Cmr; inducible expression vector; pBK2 derivative with 1 amino acid deletion (19Tdel) in the prgQ region coding for iCF10 | (18) |
| pBK2P23 | pBK2 derivative with P23 | this work |
| pBK2idTP23 | pBK2idT derivative with P23 | this work |
| pBK2Z | pBK2 derivative with P23::PrgZ | this work |
| pBK2idTZ | pBK2idT derivative with P23::PrgZ | this work |
| pBac | Spcr; Source of Bac | GeneArt |
| pBK2idtZ:Bac | pBK2idTZ derivative with Bac | this work |
| pBK2idTZ:EntA | pBK2idTZ derivative with EntA | this work |
| pBK2idTZ:HirJM79 | pBK2idTZ derivative with HirJM79 | this work |
| pBK2idTZ:EntP | pBK2idTZ derivative with EntP | this work |
| pBK1:Bac | pBK1 derivative with Bac | this work |
| fragments | description |
|---|---|
| P23 | 202-bp PstI fragment containing P23 constitutive promoter and a restriction site for KpnI |
| PrgZ | 1,805-bp KpnI fragment containing prgZ |
| Bac | 1,628-bp BamHI-EcoRI fragment containing the enterocin A structural gene (entA) with its immunity gene (entiA), the enterocin P structural gene (entP) with its immunity gene (entiP) and the hiracin JM79 structural gene (hirJM79) with its immunity gene (hiriJM79) |
| EntA | 538-bp BamHI-EcoRI fragment containing entA + entiA |
| HirJM79 | 546-bp BamHI-EcoRI fragment containing hirJM79 + hiriJM79 |
| EntP | 504-bp BamHI-EcoRI fragment containing entP + entiP |
Electrocompetent L. lactis cells were transformed with a Gene Pulser XCell (Bio-Rad Laboratories, Hercules, CA, U.S.A.) as described previously.37,38
β-Galactosidase Assays
To determine the sensitivity of the expression vectors to cCF10, we used a β-galactosidase (β-gal) assay to measure the levels of lacZ expression from recombinant L. lactis upon induction with pure cCF10 added externally.
Strains were grown overnight in GM17 medium and then diluted 1:5 into fresh GM17 medium and grown for an additional hour (OD = 0.4–0.5) before addition of cCF10 (50 ng/mL). After induction the cultures were incubated for 90 min. β-gal assays were performed as described previously.39 Activity was expressed as Miller Unit equivalents (MU).
Expression of lacZ by L. lactis in Coculture with E. faecalis
β-gal activity of L. lactis-pBK2idTZ was also measured when grown in coculture with cCF10 producing E. faecalis OG1RF.
For liquid experiments the strains E. faecalis OG1RF (OG), E. faecalis JRC101 (JR) and L. lactis NZ9000-pBK2idTZ (NZ) were grown overnight in their respective liquid media. The cultures were reinoculated and grown to an OD600 of 1. Then OG and NZ or JR and NZ were inoculated into 1 mL M9 media and M9 media supplemented with X-gal (250 μg/mL) (M9X) at different ratios (v/v) (OG/NZ and JR/NZ at 10:50; 50:50; and 100:50). As positive control NZ was grown in M9 and M9X media with cCF10 (50 ng/mL). The cocultures were incubated overnight at 32 °C for 16 h and then β-gal activity was measured by subtracting the OD650 values from the cocultures grown in M9X and M9.
For plate experiments OG, NZ, L. lactis NZ9000-pBK2iZ and L. lactis NZ9000-pBK1 were grown overnight in their respective liquid-media. The cultures were reinoculated and grown to an OD600 of 1, then 10 μL of OG were spotted in BHI-agar plates supplemented with X-gal (250 μg/mL). Once dried, 5 μL of the L. lactis cultures were spotted into the edges of the OG drop and let dry. The plates were incubated overnight at 32 °C for 16h and then lacZ expression was observed by the growth of blue staining colonies.
Antimicrobial Activity Assays
The antimicrobial activity of individual colonies was examined by the stab-on-agar test (SOAT), as previously described.30 Recombinant LAB cultures, transformed with either pBK2idTZ:Bac, pBK2idTZ:EntA, pBK2idTZ:HirJM79, and pBK2idTZ:EntP were induced for bacteriocin production at an OD600 of 0.5, using 50 ng/mL cCF10. The induced cultures were grown at 32 °C during 2 h. Cell-free culture supernatants (SN) were obtained by centrifugation of cultures at 12 000g, 4 °C for 10 min, filtered through 0.2 μm pore-size filters (Whatman Int. Ltd., Maidstone, U.K.), and stored at −20 °C until use. Ammonium sulfate precipitation (AS) was performed to increase peptide concentration. Ammonium sulfate (4 g; Sigma) was added to 10 mL SN, mixed and let stand on ice for 1 h. Then the tubes were centrifuged at 12 000g, 4 °C for 10 min, the SN discarded and the precipitated pellet eluted in 1 mL fresh GM17. The antimicrobial activity of the supernatants was examined by an agar well diffusion test (ADT) and a microtiter plate assay (MPA), as previously described30 using E. faecium 8 as indicator microbe. With the MPA, growth inhibition of the sensitive culture was measured spectrophotometrically at 620 nm with a microplate reader (SpectraMax Plus384; Molecular Devices, Sunnyvale, CA). One bacteriocin unit (BU) was defined as the reciprocal of the highest dilution of the bacteriocin causing 50% growth inhibition (50% of the turbidity of the control culture without bacteriocin). The antimicrobial activity of recombinant L. lactis-pBK2idTZ:Bac was also tested against different E. faecalium and E. faecalis strains using the MPA.
E. faecalis Growth-Inhibition in Coculture with Recombinant L. lactis
Overnight cultures of recombinant L. lactis-pBK2idTZ, L. lactis-pBK2iDTZ:Bac, and L. lactis-pBK1:Bac were inoculated (20%) in 20 mL fresh media until an OD600 of 0.5. The cultures were then inoculated with 2 × 108 CFU (Colony Forming Units) (200 μL) of E. faecalis OG1RF and let grow at 32 °C. One of the L. lactis-pBK2idTZ:Bac/E. faecalis OG1RF cocultures was also induced with 50 ng/mL cCF10 at time 0. In parallel, and as negative control, L. lactis-pBK2idTZ:Bac was inoculated with the non-cCF10 producer E. faecalis JRC101. Samples were taken every hour for 10 h, and the number of CFU per mL (CFU/mL) was determined by plating 10 μL of serial dilutions of the cocultures on GM17 agar plates supplemented with Rifampicin. The plates were incubated at 37 °C for 16 h, and the number of viable E. faecalis OG1RF cells was assessed by counting CFUs.
Acknowledgments
We thank Dr. Patricia Ferrieri of the University of Minnesota Medical School for supplying clinical isolates of E. faecium with varied spectra of antibiotic resistance for these studies. This work was supported by grants from the National Institutes of Health (American Recovery and Reinvestment Act grant GM086865 to Y.N.K. and GM049530 to G.M.D.) and by a grant from the National Science Foundation (CBET-0644792 to Y.N.K.). Support from the University of Minnesota Biotechnology Institute is gratefully acknowledged.
Supporting Information Available
This material available free of charge via the Internet at http://pubs.acs.org
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Gilmore M. S.; Lebreton F.; van Schaik W. (2013) Genomic transition of enterococci from gut commensals to leading causes of multidrug-resistant hospital infection in the antibiotic era. Curr. Opin. Microbiol. 16, 10–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias C. A.; Murray B. E. (2012) The rise of the Enterococcus: Beyond vancomycin resistance. Nat. Rev. Microbiol. 10, 266–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubeda C.; Bucci V.; Caballero S.; Djukovic A.; Toussaint N. C.; Equinda M.; Lipuma L.; Ling L.; Gobourne A.; No D.; Taur Y.; Jenq R. R.; van den Brink M. R.; Xavier J. B.; Pamer E. G. (2013) Intestinal microbiota containing Barnesiella species cures vancomycin-resistant Enterococcus faecium colonization. Infect. Immun. 81, 965–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidron A. I.; Edwards J. R.; Patel J.; Horan T. C.; Sievert D. M.; Pollock D. A.; Fridkin S. K. (2008) National Healthcare Safety Network Team; and Participating National Healthcare Safety Network Facilities. Infect. Control Hosp. Epidemiol. 29, 996–1011NHSN annual update: Antimicrobial-resistant pathogens associated with healthcare-associated infections. Annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention. [DOI] [PubMed] [Google Scholar]
- LeBlanc J. G.; Aubry C.; Cortes-Perez N. G.; de Moreno de LeBlanc A.; Vergnolle N.; Langella P.; Azevedo V.; Chatel J. M.; Miyoshi A.; Bermúdez-Humarán L. G. (2013) Mucosal targeting of therapeutic molecules using genetically modified lactic acid bacteria: An update. FEMS. Microbiol. Lett. 344, 1–9. [DOI] [PubMed] [Google Scholar]
- Wagner R. D.; Warner T.; Roberts L.; Farmer J.; Balish E. (1997) Colonization of congenitally immunodeficient mice with probiotic bacteria. Infect. Immun. 65, 3345–3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermúdez-Humarán L. G.; Kharrat P.; Chatel J. M.; Langella P. (2011) Lactococci and lactobacilli as mucosal delivery vectors for therapeutic proteins and DNA vaccines. Microb. Cell. Fact. 10(Suppl 1), S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steidler L.; Hans W.; Schotte L.; Neirynck S.; Obermeier F.; Falk W.; Fiers W.; Remaut E. (2000) Treatment of murine colitis by Lactococcus lactis secreting interleukin-10. Science 289, 1352–1355. [DOI] [PubMed] [Google Scholar]
- Mierau I.; Kleerebezem M. (2005) 10 years of the nisin-controlled gene expression system (NICE) in Lactococcus lactis. Appl. Microbiol. Biotechnol. 68, 705–717. [DOI] [PubMed] [Google Scholar]
- Benbouziane B.; Ribelles P.; Aubry C.; Martin R.; Kharrat P.; Riazi A.; Langella P.; Bermúdez-Humarán L. G. (2013) Development of a stress-inducible controlled expression (SICE) system in Lactococcus lactis for the production and delivery of therapeutic molecules at mucosal surfaces. J. Biotechnol. 168, 120–129. [DOI] [PubMed] [Google Scholar]
- Chuang O. N.; Schlievert P. M.; Wells C. L.; Manias D. A.; Tripp T. J.; Dunny G. M. (2009) Multiple functional domains of Enterococcus faecalis aggregation substance Asc10 contribute to endocarditis virulence. Infect. Immun. 77, 539–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M.; Sakagami Y.; Ishii Y.; Isogai A.; Kitada C.; Fujino M.; Adsit J. C.; Dunny G. M.; Suzuki A. (1998) Structure of cCF10, a peptide sex pheromone which induces conjugative transfer of the Streptococcus faecalis tetracycline resistance plasmid, pCF10. J. Biol. Chem. 263, 14574–14578. [PubMed] [Google Scholar]
- Dunny G. M. (2013) Enterococcal sex pheromones: Signaling, social behavior, and evolution. Annu. Rev. Genet. 47, 457–482. [DOI] [PubMed] [Google Scholar]
- Shi K.; Brown C. K.; Gu Z. Y.; Kozlowicz B. K.; Dunny G. M.; Ohlendorf D. H.; Earhart C. A. (2005) Structure of peptide sex pheromone receptor PrgX and PrgX/pheromone complexes and regulation of conjugation in Enterococcus faecalis. Proc. Natl. Acad. Sci. U.S.A. 102, 18596–18601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlowicz B. K.; Shi K.; Gu Z. Y.; Ohlendorf D. H.; Earhart C. A.; Dunny G. M. (2006) Molecular basis for control of conjugation by bacterial pheromone and inhibitor peptides. Mol. Microbiol. 62, 958–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antiporta M. H.; Dunny G. M. (2002) ccfA, the genetic determinant for the cCF10 peptide pheromone in Enterococcus faecalis OG1RF. J. Bacteriol. 184, 1155–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J.; Ruhfel R. E.; Dunny G. M.; Isogai A.; Suzuki A. (1994) The prgQ gene of the Enterococcus faecalis tetracycline resistance plasmid pCF10 encodes a peptide inhibitor, iCF10. J. Bacteriol. 176, 7405–7408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A.; Kaznessis Y. N.; Hu W. S. (2008) Tweaking biological switches through a better understanding of bistability behavior. Curr. Opin. Biotechnol. 19, 475–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A.; Johnson C. M.; Shu C. C.; Kaznessis Y. N.; Ramkrishna D.; Dunny G. M.; Hu W. S. (2011) Convergent transcription confers a bistable switch in Enterococcus faecalis conjugation. Proc. Natl. Acad. Sci. U.S.A. 108, 9721–9726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C. M.; Haemig H. H.; Chatterjee A.; Wei-Shou H.; Weaver K. E.; Dunny G. M. (2011) RNA-mediated reciprocal regulation between two bacterial operons is RNase III dependent. MBio. 2, e00189-11. 10.1128/mBio.00189-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aymerich T.; Holo H.; Håvarstein L. S.; Hugas M.; Garriga M.; Nes I. F. (1996) Biochemical and genetic characterization of enterocin A from Enterococcus faecium, a new antilisterial bacteriocin in the pediocin family of bacteriocins. Appl. Environ. Microbiol. 62, 1676–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cintas L. M.; Casaus P.; Håvarstein L. S.; Hernández P. E.; Nes I. F. (1997) Biochemical and genetic characterization of enterocin P, a novel sec-dependent bacteriocin from Enterococcus faecium P13 with a broad antimicrobial spectrum. Appl. Environ. Microbiol. 63, 4321–4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez J.; Diep D. B.; Herranz C.; Nes I. F.; Cintas L. M.; Hernández P. E. (2007) Amino acid and nucleotide sequence, adjacent genes, and heterologous expression of hiracin JM79, a sec-dependent bacteriocin produced by Enterococcus hirae DCH5, isolated from Mallard ducks (Anas platyrhynchos). FEMS. Microbiol. Lett. 270, 227–236. [DOI] [PubMed] [Google Scholar]
- Huycke M. M.; Sahm D. F.; Gilmore M. S. (1998) Multiple-drug resistant enterococci: The nature of the problem and an agenda for the future. Emerg. Infect. Dis. 4, 239–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlowicz B. K. (2005) The molecular mechanism and peptide signaling response of PrgX used to control pheromone-induced conjugative transfer of pCF10. Ph.D. dissertation. University of Minnesota, Minneapolis, MN. [Google Scholar]
- Leonard B. A. B.; Podbielski A.; Hedberg P. J.; Dunny G. M. (1996) Enterococcus faecalis pheromone binding protein, PrgZ, recruits a chromosomal oligopeptide permease system to import sex pheromone cCF10 for induction of conjugation. Proc. Natl. Acad. Sci. U.S.A. 93, 260–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørvig E.; Mathiesen G.; Naterstad K.; Eijsink V. G.; Axelsson L. (2005) High-level, inducible gene expression in Lactobacillus sakei and Lactobacillus plantarum using versatile expression vectors. Microbiology 151, 2439–2449. [DOI] [PubMed] [Google Scholar]
- Kaswurm V.; Nguyen T. T.; Maischberger T.; Kulbe K. D.; Michlmayr H. (2013) Evaluation of the food grade expression systems NICE and pSIP for the production of 2,5-diketo-d-gluconic acid reductase from Corynebacterium glutamicum. AMB. Express. 3, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diep D. B.; Mathiesen G.; Eijsink V. G.; Nes I. F. (2009) Use of lactobacilli and their pheromone-based regulatory mechanism in gene expression and drug delivery. Curr. Pharm. Biotechnol. 10, 62–73. [DOI] [PubMed] [Google Scholar]
- Borrero J.; Jiménez J. J.; Gútiez L.; Herranz C.; Cintas L. M.; Hernández P. E. (2011) Protein expression vector and secretion signal peptide optimization to drive the production, secretion, and functional expression of the bacteriocin enterocin A in lactic acid bacteria. J. Biotechnol. 156, 76–86. [DOI] [PubMed] [Google Scholar]
- Borrero J.; Jiménez J. J.; Gútiez L.; Herranz C.; Cintas L. M.; Hernández P. E. (2011) Use of the usp45 lactococcal secretion signal sequence to drive the secretion and functional expression of enterococcal bacteriocins in Lactococcus lactis. Appl. Microbiol. Biotechnol. 89, 131–143. [DOI] [PubMed] [Google Scholar]
- Gálvez A.; Abriouel H.; López R. L.; Ben Omar N. (2007) Bacteriocin-based strategies for food biopreservation. Int. J. Food Microbiol. 120, 51–70. [DOI] [PubMed] [Google Scholar]
- Almeida T.; Brandão A.; Muñoz-Atienza E.; Gonçalves A.; Torres C.; Igrejas G.; Hernández P. E.; Herranz C.; Cintas L. M.; Poeta P. (2011) Identification of bacteriocin genes in enterococci isolated from game animals and saltwater fish. J. Food Prot. 74, 1252–1260. [DOI] [PubMed] [Google Scholar]
- Kjos M.; Nes I. F.; Diep D. B. (2011) Mechanisms of resistance to bacteriocins targeting the mannose phosphotransferase system. Appl. Environ. Microbiol. 77, 3335–3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter P. D.; Ross R. P.; Hill C. (2013) Bacteriocins—A viable alternative to antibiotics?. Nat. Rev. Microbiol. 11, 95–105. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E. F., and Maniatis T. (1989) In Molecular Cloning, Cold Spring Harbor Laboratory Press, New York. [Google Scholar]
- Holo H.; Nes I. F. (1995) Transformation of Lactococcus by electroporation. Methods Mol. Biol. 47, 195–199. [DOI] [PubMed] [Google Scholar]
- Volzing K.; Borrero J.; Sadowsky M.; Kaznessis Y. (2013) Antimicrobial peptides targeting Gram-negative pathogens, produced and delivered by lactic acid bacteria. ACS. Synth. Biol. 2, 643–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlowicz B. K.; Bae T.; Dunny G. M. (2004) Enterococcus faecalis pheromone-responsive protein PrgX: Genetic separation of positive autoregulatory functions from those involved in negative regulation of conjugative plasmid transfer. Mol. Microbiol. 54, 520–532. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.