Abstract
Living cells respond to various environmental cues and process them into a series of spatially and temporally regulated signaling events, which can be tracked in real time with an expanding repertoire of genetically encodable FRET-based biosensors. A series of these biosensors, designed to track dynamic activities of signaling enzymes such as protein kinases and small GTPases, have yielded invaluable information regarding the spatiotemporal regulation of these enzymes, shedding light on the orchestration of signaling pathways within the native cellular context. In this chapter, we first review the generalizable modular designs of FRET-based biosensors, followed by a detailed discussion about biosensors for reporting protein kinase activities and GTPase activation. Two general designs, uni- and bimolecular reporters, will be discussed with an analysis of their strengths and limitations. Finally, an example of using both uni- and bimolecular kinase activity reporters to visualize PKA activity in living cells will be presented to provide practical tips for using these biosensors to explore specific biological systems.
1. Introduction
In an ever changing environment, a living cell relies on exquisite spatial and temporal regulation of signal transduction machinery to make vital decisions, such as differentiation, migration, and apoptosis. For some of these signal transduction events, such as changes in pH, Ca2+ level, and membrane potential, specific fluorescent biosensors (Cohen et al., 1974; Grynkiewicz et al., 1985; Ross et al., 1974) have existed for a few decades to allow tracking of these events in real time. However, with the discovery of green fluorescent protein (GFP) and the advances in imaging technologies, the past decade has brought an escalation in both the development and the application of genetically encoded fluorescent biosensors for imaging signal transduction in complex biological systems such as living cells and organisms. These genetically encodable biosensors can be introduced in living cells by standard molecular and cellular biology techniques and targeted to specific cells or subcellular locations to monitor local dynamic signaling processes, such as changes in protein expression, localization, turnover, posttranslational modification or interactions with other proteins in the cellular milieu.
Many types of fluorescent biosensors have been developed for visualizing a variety of cellular and molecular events, such as those based on probe translocation, direct sensitization of a fluorescent protein (FP), or fragment complementation of FPs (Newman et al., 2011). In this chapter, we focus on Förster Resonance Energy Transfer (FRET)-based biosensors for tracking activities of signaling enzymes in living cells. FRET is a quantum mechanical phenomenon in which an excited donor fluorophore transfers energy in a nonradiative fashion to an acceptor fluorophore in its close proximity (i.e., <10 nm apart). For FRET (Forster, 1948), the efficiency of energy transfer is inversely proportional to the sixth power of the distance between the donor and acceptor and is also dependent on the relative orientation of the fluorophores. As FRET is particularly sensitive to variations in distance in the range of macromolecular dimension (from 10 to 100 nm), this technique has been applied to analyze the molecular dynamics of biologically relevant processes in a variety of different ways. In the context of FRET-based biosensors for the characterization of signaling enzymes such as protein kinases (PKs) and GTPases, which will be discussed here in detail, changes in the activation state or activity of the signaling enzymes are translated into changes in FRET. Importantly, FRET-based biosensors provide ratiometric readout, which is desirable to eliminate variations in probe concentration and cell thickness. Further, these biosensors have an established generalizable modular design, greatly facilitating the process of generating customized FRET-based biosensors for signaling enzymes in the same families. Consequently, this approach has the potential to be readily adopted to track a large number of dynamic signaling events in space and time.
Here, we first describe strategies to design genetically encodable FRET-based biosensors for signaling molecules, with a brief discussion about some differences between the two general classes, uni- and bimolecular reporters. We then discuss the development of FRET-based biosensors for two classes of signaling enzymes: PKs and small GTPases, and representative applications for each. We end with a specific example of FRET-based biosensors for tracking the activity of cAMP-dependent protein kinases, highlighting the experimental design and practical tips for using such sensors.
2. Generalizable Modular Designs
The generalizable modular design of genetically encodable FRET-based biosensors consists of two units: a signal sensing unit to recognize biologically relevant signals and a reporting unit, that is, FP pair, to convert the relevant signaling event into a change in FRET readout. There are two general classes of FRET-based biosensors, unimolecular and bimolecular which are based on intra- and intermolecular FRET, respectively. Importantly, both types of biosensors can be used to track signaling dynamics in real time, and each has its respective strengths and limitations. In this section, we discuss the general designs of FRET-based biosensors, with a focus on the makeup of both functional units.
2.1. Fluorescent protein pair
After being translated in cells, GFP and its derivatives form fluorophores via an autocatalytic mechanism (Chalfie et al., 1994; Cubitt et al., 1995; Tsien, 1998). Thus, as a general requirement for using FPs in living cells, FPs should express readily, mature rapidly, and should be stable and nontoxic when expressed in cells. Because of recent and extensive protein engineering efforts, not only have these above requirements for FPs been met, but there is also an abundance of FPs with distinct biophysical properties which can facilitate the design of an optimal biosensor to best suit the need of a specific study (Newman et al., 2011). Below we highlight some recommendations for selecting a FRET pair, which is typically used as the reporting unit in a FRET-based biosensor.
First of all, it is important to select a FP pair that has sufficient spectral overlap between donor emission and acceptor excitation to allow FRET to occur, as well as clearly separated excitation and emission peaks for both the donor and the acceptor to avoid cross-excitation and signal bleedthrough. Currently, the most widely used FRET pair in biosensor design is a cyan fluorescence protein (CFP)–yellow fluorescent protein (YFP) pair. Examples of cyan FPs include Cerulean, mTFP1, and CyPet, and yellow FPs include Venus, Citrine, and YPet. Cerulean exhibits superior brightness and efficient folding and maturation at 37 °C but has lower photostability compared to some of the other CFP variants (Rizzo et al., 2004). A more recent variant, mCerulean3, shows greatly reduced fluorescence photo-switching behavior and enhanced photostability (Markwardt et al., 2011). A different cyan variant, monomeric teal fluorescent protein (mTFP), also exhibits improved brightness and photostability and has a narrow emission peak (Henderson et al., 2007). Both Cerulean and mTFP have proven to be efficient donors for monomeric yellow FP variants such as Citrine and Venus. Moreover, in an effort to generate an optimal FRET pair, another CFP, cyan fluorescent protein for energy transfer (CyPet), was coevolved with yellow fluorescent protein for energy transfer (YPet), and this CyPet/YPet FRET pair has proven a useful FRET pair (Nguyen and Daugherty, 2005). While YPet is one of the brightest YFP variants and displays desirable pH- and photostability, CyPet suffers from poor folding efficiency, and it is thus often advantageous to use another suitable CFP variant, such as eCFP, as the donor for YPet. Further, circularly permutated FPs (cpFPs), which have identical secondary structure but rearranged N- and C-termini relative to the WT protein, can be introduced in a biosensor as a means to optimize the energy transfer efficiency between donor and acceptor by tweaking the relative orientation of the fluorophores in the basal and/or stimulated state (Allen et al., 2006; Allen and Zhang, 2006; Gao and Zhang, 2008; Nagai et al., 2004). In addition to the Cyan/Yellow FRET pair, other FRET pairs which have been utilized include Green/Red (EGFP/mCherry) (Albertazzi et al., 2009; Ni et al., 2011), Cyan/Orange (MiCy/mKO) (Karasawa et al., 2004), and Yellow/Red (mAmetrine/tdTomato) (Ai et al., 2008), and some of these spectrally distinct FRET pairs have been advantageous for simultaneous imaging of two or more signaling events.
In addition to the spectral considerations, an ideal FP should be bright, pH- and photostable, and monomeric. Some bright FPs have high extinction coefficients near 1000,000 M−1cm−1 and high quantum yields approaching the theoretic limit of 1. In the cellular context, factors like maturation speed, folding efficiency, and susceptibility to photobleaching affect the performance of a FP. In addition, in most cases, monomeric FPs are used in a biosensor, because dimerization or oligomerization may interfere with the function and localization of biosensors, and this can sometimes lead to the formation of protein aggregate. Therefore, FP selections are based on a comprehensive set of parameters. Many comprehensive reviews are available to aid in the optimal selection of an FP pair (Day and Schaufele, 2008; Newman et al., 2011; Shaner et al., 2005).
2.2. Sensing unit
The sensing unit of a FRET-based biosensor determines which biological event the biosensor monitors. The sensing unit senses a signaling activity within the intracellular environment, such as ligand binding or enzyme activation, and converts these signals into conformational changes or association/dissociation events that can alter FRET between the FP pair, the reporting unit.
When the signaling activity to be monitored generates a conformational change in a particular protein, such as a conformational change induced by ligand binding or covalent modification, this protein itself could be used as a sensing unit (Fig. 16.1A). For example, as cAMP binding induces an intrinsic conformational change in the Exchange Protein Activated by cAMP (Epac1), ICUE1 (Indicator of cAMP using Epac), a sensitive reporter of cAMP levels, was developed by sandwiching Epac1 between a FRET pair (DiPilato et al., 2004). Moreover, as the cAMP-induced conformational change is reversible, ICUE1 can be used to detect both the accumulation and degradation of cAMP. This strategy has been applied to many proteins that undergo a large conformational change upon activation (Calleja et al., 2007; Fujioka et al., 2006; Schleifenbaum et al., 2004). For instance, in the specific case of signaling enzymes, a kinase activation sensor can be designed by fusing the kinase between a FRET pair, as long as the kinase undergoes an inherent conformational change upon activation (Ananthanarayanan et al., 2007; Calleja et al., 2007).
Figure 16.1.
Modular designs of genetically encodable FRET-based unimolecular biosensors (A–C) and bimolecular biosensor (D). Two basic units include a reporting unit, consisting of CFP (blue cylinder) and YFP (yellow cylinder), and a sensing unit. The sensing unit can be a single conformationally responsive domain (orange U shape in a) or a “molecular switch” consisting of a sensing segment (purple oval in B–D) and a receiving segment (orange irregular shape in B–D) with different topology in (B) and (C); or in two different polypeptide chains in (D). The sensing unit recognizes the biologically relevant events, such as ligand binding and phosphorylation (depicted by adding the red triangles), and converts them into a change in FRET (orange arrows indicate the energy transfer from donor to acceptor).
In the case that a protein does not undergo a sufficient conformational rearrangement, a “molecular switch” can be engineered by coupling a “receiving” segment to a “sensing” segment to take advantage of signal-induced association/dissociation of the two segments (Fig. 16.1B–D). For example, the molecular switch of a kinase activity reporter (KAR) is designed by linking a peptide substrate which can be recognized and phosphorylated by a kinase-of-interest to a phosphoamino acid binding domain (PAABD). When phosphorylated, the substrate falls into the binding pocket of PAABD, and this engineered conformational change of the molecular switch can be read out as a change in FRET. Importantly, this design is also modular and can be applied to measure a variety of phosphor-ylation events by simply replacing the surrogate substrate in an existing biosensor with a substrate specific to the kinase under investigation (Ni et al., 2006). Moreover, this design strategy can be applied to study the activity of enzymes responsible for other posttranslational modifications, such as acetylation and O-GlcNAc glycosylation (Aye-Han et al., 2009). Similarly, this molecular switch concept can be applied to the engineering of FRET-based biosensors for small molecules. For example, a ligand-induced molecular switch can incorporate a pseudoligand, which binds to the sensing domain in the basal state and will be displaced when concentrations of the endogenous ligand are suitably increased, inducing the critical conformational change in the switch. This particular design is used in the biosensor, InPAkt (Indicator for phosphoinositides based on Akt), which probes the dynamics of membrane restricted lipid second messengers, phosphatidyl inositol (3,4,5) trisphosphate (PIP3) and phosphatidyl inositol (3,4) bisphosphate (PIP(3,4)2) (Ananthanarayanan et al., 2005). Specifically, a molecular switch was constructed by linking the pleckstrin homology (PH) domain from Akt, which specifically binds to PIP3 and PI(3,4)P2, to an acidic patch of amino acids from nucleolin 1 as the pseudoligand. When produced, 3′ phosphoinositides bind to the designed binding site within the PH domain, displacing the pseudoligand and inducing a conformational change in the molecular switch and a FRET change. As such, InPAkt serves as a FRET-based indicator for intracellular levels of 3′ phosphoinositides and, when combined with subcellular targeting sequences, can monitor 3′ phosphoinositde levels at different subcellular locations.
2.3. Unimolecular and bimolecular designs
In a unimolecular FRET-based biosensor, the fluorescent donor and acceptor are attached to the same protein to form a single-chain reporter. By design, this class of reporters relies on an intramolecular conformational change to alter the distance and/or orientation between the FRET pair (Fig. 16.1A–C). In contrast, in bimolecular reporters, the donor and acceptor FP are fused to either portion of the molecular switch (Fig. 16.1D) and expressed separately. With this design, the two FPs are physically separated in the basal state and are brought into close proximity only upon activation of the molecular switch, or vice versa. Each design has its strengths and limitations and special care should be taken to choose probes which are best suited to address the specific biological questions under study.
The design characteristics of each class of biosensor confer their unique features. First, the FRET response generated from a unimolecular reporter originates from the repositioning of two fluorophores with respect to one another, and this change in distance and orientation can be small due to structural constraints of the single peptide chain. However, the FRET change generated from a bimolecular system has the potential to be much larger since there is physical separation of the two FPs in the basal or unstimulated state. Consequently, bimolecular biosensors typically exhibit a larger dynamic range than their unimolecular counterparts, presumably due to lower FRET in the dissociated state.
Second, while bimolecular reporters can achieve superior sensitivity compared to unimolecular counterparts, they lack the fixed stoichiometry of the donor and the acceptor which is inherent to the unimolecular design. Thus special care needs to be taken to match the expression levels of the two components of bimolecular reporters to ensure proper analysis of signals. Unmatched and variable ratios between the two components will not only compromise the sensitivity of the biosensors but also complicate the quantification (Zhang et al., 2002).
Another consideration when using bimolecular probes is that, because the two components of the molecular switch are expressed independently, they are more likely to interact with endogenous molecules (Miyawaki, 2003), which may affect the performance of the biosensor or interfere with the signaling machinery. For example, in the case of bimolecular KARs, the designed PAABD may have to compete with endogenous phosphoamino acid binding proteins for the phosphopeptide, although this does not appear to be a concern in the case of bimolecular PKA and PKC activity reporters (Herbst et al., 2011). A final caveat of bimolecular sensors is that they are potentially less temporally sensitive than their unimolecular counterparts as they are more sensitive to diffusion constraints.
In many cases, both unimolecular (DiPilato et al., 2004; Miyawaki et al., 1997) and bimolecular FRET-based reporters (Herbst et al., 2011; Miyawaki et al., 1997; Zaccolo et al., 2000) have been effectively applied to monitor the same signaling process, for example, to track kinase activities, monitor the activation of small GTPases, and probe the intracellular dynamics of second messengers, such as Ca2+ and cAMP. Below we highlight the development of both uni- and bimolecular biosensors to probe the dynamics of signaling enzymes, with a focus on their designs and application.
3. FRET-Based Biosensors for Monitoring Signaling Enzymes
There are many biosensors available to track different enzymatic modifications, such as phosphorylation /dephosphorylation (Newman and Zhang, 2008; Ni et al., 2006), O-glycosylation (Carrillo et al., 2006), histone methylation (Lin et al., 2004) and acetylation (Sasaki et al., 2009), and protein ubiquitination (Perroy et al., 2004). However, kinase activation/activity biosensor and GTPase activation reporters represent two major classes where multiple examples exist (Aoki and Matsuda, 2009; Hodgson et al., 2008; Ni et al., 2006; Zhang and Allen, 2007). Here, we focus on these two families of biosensors, concentrating on their design and use in investigating specific signaling pathways.
3.1. Protein kinases
3.1.1. Introduction
Protein kinases are enzymes that catalyze the transfer of the γ-phosphate of ATP to the protein substrates, thus altering their functions. These signaling enzymes play a critical and complex role in regulating cellular signal transduction as they orchestrate multiple intracellular processes such as glycogen synthesis, hormone responses, and ion transport. Many signaling cascades involving protein kinases require dynamic control and spatial compartmentalization of kinase activity, which needs to be tracked continuously in different compartments and signaling microdomains in living cells; however, traditional methods to study protein kinases activity only provide static and limited snapshots of signaling events and fail to capture the dynamic changes of kinase activity. On the other hand, genetically encodable FRET-based biosensors offer a versatile and powerful approach to elucidate the spatiotemporal patterns of kinase signaling.
3.1.2. Various designs of kinase reporters
To fulfill their complex role in signal transduction, protein kinases are subjected to multiple levels of regulation, such as ligand binding, protein–protein interaction, and phosphorylation by other kinases or by themselves. To date, there are three families of biosensors, kinase activation biosensors, kinase activity reporters (KARs), and substrate-based reporters. A kinase activation biosensor usually consists of a kinase sandwiched between a FP pair, capturing the kinase “activation”; whereas a kinase activity reporter (KAR) monitors the activity of protein kinases by using the biosensor as a surrogate substrate for the kinase. Therefore, for the kinase activation biosensors, a linear relationship exists between the number of activated signaling molecules and the observed signals. However, one active kinase molecule can phosphorylate many KARs, and these biosensors thus benefit from enzymatic amplification and display a nonlinear, and typically logarithmic, relationship between the signal and activated kinase species. The third group, substrate-based biosensors employ similar design as KARs, however, report phosphorylation of a specific substrate by any of the multiple upstream kinases.
A kinase activation biosensor is generally constructed by flanking the kinase-of-interest with a fluorescent protein pair in such a way that the conformational change of the kinase induced by activation can be translated into a change in FRET signal. This strategy has been utilized to generate reporters for activation of kinases such as protein kinase C (PKC) (Schleifenbaum et al., 2004), Akt (Calleja et al., 2007), and extracellular regulated protein kinase (ERK) (Fujioka et al., 2006). As another example, the FRET-based reporter of Akt action (ReAktion) was constructed by flanking full-length Akt-1 with FP donor and acceptor, and the biosensor reports Akt-1 activation when it is phosphorylated on a threonine in the activation loop. This has led to a study of the regulatory role of this phosphorylation event in dissociation of Akt from the plasma membrane upon its activation (Ananthanarayanan et al., 2007). As the design of kinase activation biosensors usually requires incorporation of the full-length or a large portion of an active kinase, potential complication of this approach includes the perturbation of endogenous signal transduction due to presence of an active kinase. In this case, restricting the expression to a relatively low level becomes important. Alternatively, using a biologically inert mutant of the protein which retains its ability to undergo a conformational change is appropriate.
The second type of kinase reporters, KARs utilize an engineered molecular switch consisting of a phosphorylation-sensing domain (a substrate for the kinase-of-interest) with a suitable PAABD. There are three critical components to this design. First, it is critical to have a substrate domain that is specific for the kinase of interest. This is often designed based on the sequence of an endogenous substrate or the consensus substrate sequence for a kinase of interest, which is often derived from peptide library screening data. Second, a PAABD, such as phosphotyrosine binding Src homology 2 (SH2) domain, phosphoserine/threonine binding 14-3-3 and WW domain, or phosphothreonine binding forkhead-associated (FHA) domain, can be selected to be compatible with candidate substrate sequences. For instance, FHA1 domain specifically recognizes phosphothreonine with aspartate at the +3 position as respect to the phosphothreonine site, whereas WW domain recognizes phosphoserine/phosphothreonine with proline at the +1 position. Therefore, the substrate sequence may need to be further modified to fit the PAABD binding requirement by incorporating appropriate point mutations, but it is important to be sure that the kinase can still specifically recognize and phosphorylate the mutated substrate. For example, in Akt activity reporter (AktAR), the substrate sequence has been modified by mutating the residue at the +3 position with respect to the phosphothreonine site to aspartate for efficient binding to the FHA1 domain that is utilized in this reporter (Gao and Zhang, 2008). Because Akt recognizes a consensus sequence as RXRXXS/T, this change following the phosphoamino acid site unlikely affects the binding and phosphory-lation of AktAR by Akt. A third component of KAR design is the linker regions between individual segments (fluorescent proteins, substrate, PAABD). Such linkers can be optimized to appropriate length to improve the dynamic range and intracellular stability of KARs. If structural information regarding the binding domain and phosphor-substrate is available, optimization of linker lengths may be guided by examining the relative positioning of these components in the bound form. Alternatively, if structural information is unavailable, linker lengths can be optimized by empirical testing in a trial-and-error manner or through screening of libraries of biosensors containing randomized linker lengths; as a general starting point, flexible linkers such as GGSGG are used. In one example, a long 72-glycine linker is utilized in a reporter for ERK activity (Harvey et al., 2008).
Other design characteristics of KARs are optional but help to improve specificity and sensitivity in detecting certain signaling events. First, to study localized signaling events, a targeting sequence can be added to the sequence of the biosensor. For example, by targeting AktAR to different microdomains within the plasma membrane, a faster and larger AktAR response was observed within membrane rafts compared to nonraft regions of plasma membrane, indicating that Akt activity is differentially regulated in discrete membrane microdomains (Gao and Zhang, 2008). As untargeted KARs diffuse faster than membrane targeted probes (Lu et al., 2008), addition of a targeting sequence improves the signal-to-noise ratio in detecting the localized kinase activities (Lim et al., 2008). Second, in order to increase the specificity of kinase activity detection, a kinase docking domain can be added to the biosensor. This is especially important for kinases such as MAPKs which are similar in terms of substrate consensus sequences and often use docking domains to enhance the specificity of phosphorylating their endogenous substrates. For instance, such docking domains are essential in establishing the specificity of JNKAR and EKAR for the c-Jun N-terminal Kinase (JNK) and ERK, respectively (Fosbrink et al., 2010; Harvey et al., 2008).
As alluded to above, an effective approach to increase the dynamic range of KARs is to design bimolecular kinase activity reporters (BimKAR). This class of biosensors utilizes a kinase-inducible bimolecular switch (KIBS) in which a kinase-specific substrate and a PAABD are each fused to a FP and expressed as independent peptide chains. When phosphorylated by endogenous kinase, the phosphopeptide is bound by the PAABD, resulting in intermolecular FRET. With this design, FRET change largely depends on the ability of the KIBS to bring the two parts into close proximity, as the basal FRET is low when they are fully separated. Based on this concept, A-Kinase and C-Kinase inducible bimolecular switches have been generated (Herbst et al., 2011). While this design strategy is suitable for any KAR which contains a two-component molecular switch, in the case of biosensors such as EKAR and JNKAR which also contain kinase docking domains (Fosbrink et al., 2010; Harvey et al., 2008), it will be important to keep the docking domain on the portion of the KIBS that contains the substrate sequence to facilitate substrate recognition. Further, as aforementioned, targeted KARs can be used to probe compartmentalized kinase activity. Notably, in BimKAR, the two components of the KIBS need not be expressed in the same subcellular compartment; to monitor local signaling dynamics, it is only necessary to target the substrate portion of the switch. It is with this design that the most sensitive nonraft region targeted PKA activity reporter (BimAKAR-CAAX) was developed (Herbst et al., 2011). In the case that a unimolecular biosensor does not already exist, it is still possible to construct a KIBS for a kinase of interest. In fact, as the KIBS design relieves some of the structural constraints of a unimolecular biosensor, it may be a more suitable starting point for KAR design. As the KIBS design is modular, a BimKAR for a kinase of interest can be designed by replacing the PKA-specific substrate with a substrate sequence specific to the kinase of interest, and this was shown with the generation of BimCKAR, a bimolecular reporter of PKC activity. However, as is the case when generating any new biosensor, it is critical to test the specificity of the BimKAR for the kinase of interest by using suitable inhibitors and through generation of mutants in which the phosphoacceptor residues are mutated to Ala.
The third group of biosensors, substrate-based reporters, is designed to report the phosphorylation of a specific substrate, which can be mediated by multiple upstream kinases. As a proof-of-concept, the newly developed indicator of CREB activation due to phosphorylation (ICAP) was constructed to report the critical phosphorylation at serine 133 of Ca2+- and cAMP-responsive element-binding protein (CREB) that leads to its activation (Friedrich et al., 2010). Flanked by CFP/Citrine, the engineered molecular switch consists of a moiety from kinase-inducible domain (KID) of CREB, which contains the key residue serine 133 and a moiety from the KIX domain of CREB binding protein, which specifically recognizes phosphoserine 133. Upon given stimulations, the serine 133 in the sensor was phosphorylated by a number of upstream kinases, such as PKA and CaMK IV, leading to a conformational change in the molecular switch and a subsequent FRET change. Of note, this design has been used to generate functional sensors of ATF-1 or CREM activation by substitution of the KID domain of CREB in ICAP with the homologous KID domains of ATF-1 or CREM, respectively. Therefore, ICAP and analogous substrate-based reporters represent a new class of biosensors which allows the study of the combined control of phosphorylation of a specific protein, often a key node in signaling networks, by a group of protein kinases in living cells.
3.1.3. Application—Oscillations in kinase activity
Recently, KARs have revealed new modes of exquisite temporal controls of kinase activity in cells. It has been long acknowledged that the concentration of second messengers such as Ca2+ oscillates in a periodic fashion in response to certain stimuli (Berridge et al., 2003; Clapham, 2007), and fluorescent kinase biosensors have begun to reveal additional oscillatory signals that are involved in regulating various important cellular processes (Dunn et al., 2006; Ni et al., 2011; Violin et al., 2003).
In one such example, Dunn et al. investigated how waves of spontaneous electrical activity that spread across the retinal ganglion cell (RGC) layer are coupled to the cAMP/PKA pathway by using a cAMP indicator, ICUE2, and a PKA activity reporter, AKAR2.2. Previously, it had been demonstrated that such retinal waves produced by RGCs play an essential role in the early stages of retinal development. Although the cAMP/PKA pathway had previously been implicated in establishment and refinement of RGC axonal projections, the link between the retinal waves and cAMP/PKA signaling had not been characterized. Using the biosensor imaging approach, cAMP and PKA activities were shown to oscillate in response to depolarization-induced Ca2+ influx caused by spontaneous electrical activity in RGCs (Dunn et al., 2006).
Also using fluorescence imaging, a highly integrated oscillatory circuit consisting of Ca2+–cAMP–PKA has been discovered in MIN6 β cells. PKA is known to play important roles in regulating Ca2+-dependent exocytosis, which is critical for the pulsatile insulin secretion in pancreatic cells. In this study, fura-2, a fluorescent Ca2+ indicator, was simultaneously imaged with the genetically encoded FRET-based biosensor for PKA (AKAR-GR, green and red FP version) to illustrate the precise temporal correlation between Ca2+ dynamics and the oscillations in PKA activity, or with the cAMP probe (ICUE-YR, yellow and red FP version) to uncover the coordination between Ca2+ and cAMP dynamics (Ni et al., 2011). To probe the temporal relationship between PKA and cAMP activities, a single-chain, dual-specificity biosensor, ICUEPID, was also employed to simultaneously monitor cAMP levels and PKA activity (Ni et al., 2011). The observed synchronized oscillations of Ca2+, cAMP, and PKA, together with quantitative mathematic modeling data, demonstrate that these three signaling molecules form a highly integrated oscillatory circuit (Ni et al., 2011). In addition, PKA activity was found to be required for the oscillatory Ca2+–cAMP–PKA circuit, and it was not only capable of initiating the signaling oscillations but also able to modulate their frequency such that input signals can be integrated and output signals can be diversified to exquisitely control substrate phosphorylation in a context dependent manner.
3.2. Small GTPases
3.2.1. Introduction
Small GTPases are enzymes that catalyze the hydrolysis of guanosine tri-phosphate (GTP) to guanosine diphosphate (GDP). As the most well-known members, Ras GTPases play essential roles in regulating cell growth, cell differentiation, cell migration, and lipid vesicle trafficking. Ras GTPases cycle between the active GTP-bound state and the inactive GDP-bound state; they are inactivated by GTPase activating proteins (GAP) and activated by guanine nucleotide exchange factors (GEF). The former activate the intrinsic GTPase activity of Ras GTPases, which hydrolyze GTP to GDP, while the latter cause dissociation of GDP from Ras GTPases and association of GTP. Characterizing the spatial and temporal regulation of small GTPases using traditional biochemical methods have proven difficult (Walker and Lockyer, 2004), but these complications have been overcome by the development of genetically encodable biosensors.
3.2.2. Different GTPase activation reporters
GTPase biosensors, like KARs, are based off of a modular design composed of a molecular switch and a FRET pair. When GTP binds to Ras, it induces a conformational change in the effector region of Ras and consequently recruits and activates downstream molecules, including the serine/threo-nine kinase Raf. The first genetically encodable FRET-based biosensor for GTPase activation, Raichu-Ras (Ras and interacting protein chimeric unit for Ras), used this interaction between activated Ras and Raf as the basis for its molecular switch. Specifically, Raichu-Ras used H-Ras as the sensing segment and Ras Binding Domain (RBD) of Raf as the receiving segment to form an engineered molecular switch, which is sandwiched by CFP and YFP. With this design, activation of Ras can be monitored in living cells by changes in FRET. By simply replacing the H-Ras with Rap1, a biosensor for Rap1 activity, Raichu-Rap1, was generated. Using these biosensors in COS-7 cells, epidermal growth factor-induced activation of Ras was shown to occur at the plasma membrane, and activation of Rap1 at the perinuclear region, indicating spatially distinct regulation. Further, by using the fluorescence recovery after photobleaching (FRAP) technique, high Ras activity at the extending neurite in PC12 cells following nerve growth factor treatment was found to be due to high GTP/GDP exchange rate and/or low GTPase activity, rather than the retention of the active Ras (Mochizuki et al., 2001).
A different approach has been employed to design a biosensor for RhoA, another small GTPase involved in cytoskeletal regulation. In addition to GAP and GEF, Rho-family G proteins are regulated by guanine nucleotide dissociation inhibitor (GDI). The interaction of GDIs with Rho GTPases not only prevents the release of GDP from GTPases but also retains Rho G proteins in the cytosol. To maintain a free C-terminus of RhoA for interacting with Rho GDIs, a biosensor is constructed by fusing, from N-terminus to C-terminus, Rho-binding domain of the effector rhotekin fragment, which specifically binds to GTP-RhoA, to CFP, YFP, and RhoA (Pertz et al., 2006) (Fig. 16.1C). Thus this biosensor is sensitive to GDI and can be reversibly targeted from cytosol to membrane.
The bimolecular design has also been adopted to develop GTPase biosensors. For instance, a bimolecular molecular switch has been employed to generate activation reporter for Rac1, a GTPase that is involved in diverse cellular events including actin polymerization in cell adhesion. In this case, a fragment of p21-activated kinase (PAK) which binds to activated Rac1 was fused to YPet, and Rac1 was linked to CyPet, to generate a bimolecular FRET biosensor, Rac1 FLAIR.
3.2.3. Application—Coordination of small GTPases in migrating cells
Three GTPases, Rac, Rho, and cdc42, play essential roles in coordinating cytoskeleton dynamics during cell migration. Previously, all three GTPases were shown to activate at the front of migrating cells (Kraynov et al., 2000; Nalbant et al., 2004; Pertz et al., 2006; Ridley et al., 2003), but the spatiotemporal coordination between them is unknown. To unravel the activation dynamics of RhoA, Cdc42, and Rac1 during cell protrusion, first the activation of each GTPase was visualized in separate experiments using two single-chain biosensors for RhoA and cdc42, and a dual chain biosensor, Rac1 FLAIR for Rac. Then the activation profiles of the three GTPases were subsequently aligned using the timing of cell protrusion/retraction as a common reference and analyzed using a computational multiplexing methodology. This technique compares the activity maps of the three GTPases at different time points and different locations along the cell boundary and builds up correlations between distinct signaling activities and the morphological dynamics of the cell edge. Using this approach, it was demonstrated that RhoA activation at the cell edge is coincident with edge advancement, whereas cdc42 and Rac1 are activated 2 μm behind the edge with a 40 s time lag. The time shifts between GTPase activation suggested by separate imaging experiments with help of computational multiplexing was further confirmed by coimaging of a dye-labeled bimolecular Cdc42 biosensor and the unimolecular RhoA biosensor to directly reveal the temporal correlation between activation of these two signaling molecules (Machacek et al., 2009). These data have led to a model of distinct roles of the three GTPases in initiating, reinforcing, and stabilizing membrane protrusions with spatiotemporally coordinated antagonistic actions of Rac1 and RhoA involved. More recently, a similar approach has been utilized to elucidate the regulatory role of PKA as a pacemaker of protrusion–retraction cycle via phosphorylating RhoA and thus interfering with RhoA–RhoGDI interaction (Tkachenko et al., 2011), further demonstrating the great potential of the computationally aided biosensor imaging approach in dissecting complex signaling controls of cellular processes.
4. Example: A-kinase Activity Reporter (AKAR)
Here, we provide a specific example of A-kinase activity reporter (AKAR), with discussion of the chronicle of the design and development of AKARs, followed by a practical perspective of application of FRET-based enzyme biosensors.
4.1. Development of AKAR
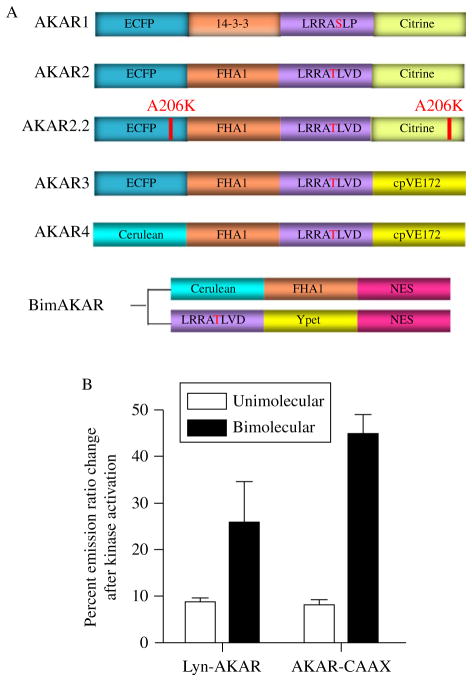
The initial AKAR1 reporter consists of ECFP, a phosphoserine/threonine binding domain (14-3-3) (Aitken et al., 1995; Fu et al., 2000), a PKA-specific peptide sequence (modified Kemptide, LRRA*SLP), and Citrine (Zhang et al., 2001) (Fig. 16.2A). One flexible linker, GGTGGS, replaced the C-terminal tail of 14-3-3 without disruption of the 14-3-3/substrate binding, and another linker, GTGGSEL was inserted between the substrate peptide and the YFP. With this design, AKAR1 has been successfully used to study compartmentalized PKA activity. Because of tight 14-3-3/substrate binding, however, the substrate of AKAR1 is inaccessible to phosphatases and thus irreversibly phosphorylated. Consequently, it does not report decreases in PKA activity.
Figure 16.2.
(A) Scheme representation of AKAR development; (B) Comparison of emission ratio change of plasma membrane targeted AKAR and BimAKAR following kinase activation.
Efforts to generate a reversible AKAR began with the hypothesis that reducing the affinity of the PAABD for the phosphosubstrate would render the biosensor more accessible to phosphatases and thus reversibility. To this end, the lower affinity PAABD, FHA1 was used. However, to facilitate the usage of FHA1 in AKAR1, the PKA-specific substrate also had to be changed (Zhang et al., 2005). As was the case with AktAR, to accommodate the preference of FHA1 binding for an Asp in the +3 position (Gao and Zhang, 2008), the PKA substrate was modified to LRRA*TLVD to give rise to AKAR2. AKAR2 was further optimized by introducing the mutation A206K into the FPs which reduced dimerization tendency of the FPs. The resulting reporter, AKAR2.2, displayed enhanced reversibility compared to it AKAR2 predecessor, however, it still suffered from a low dynamic range (Zacharias et al., 2002; Zhang et al., 2005).
To improve the dynamic range of AKAR2, different variants of CFP (ECFP and CyPet) and YFP (Citrine, YPet, Venus, circular permutated cpVN144, cpVK156, cpVE172, and cpVL194) were tested systematically. The highest dynamic change was observed in the construct harboring ECFP and cpVE172 as FRET pair, which was named AKAR3 (Allen and Zhang, 2006).
Engineering of new FP variants continues to provide opportunities for further improving biosensors. For instance, the most sensitive unimolecular reporter for PKA, AKAR4, was generated by replacing the ECFP of AKAR3 with Cerulean, a brighter variant of CFP. Compared to AKAR3, AKAR4 shows an improved dynamic range, with a response of 67% to the β-AR receptor agonist isoproterenol in HEK293T cells (Depry et al. 2011).
As has been emphasized throughout this review, switching from the unimolecular design to bimolecular design provides another strategy to improve the sensitivity of a KAR. In the case of PKA activity detection, generation of a bimolecular AKAR (BimAKAR) was achieved by fusing Cerulean to the N-terminus of FHA1 and the PKA-specific substrate to the N-terminus of YPet (Herbst et al., 2011) (Fig. 16.2A). In particular, two variants of BimAKAR, BimAKAR-CAAX and Lyn-BimAKAR, which report PKA activity in nonraft and lipid raft-like portions of the plasma membrane, respectively, show significantly enhanced dynamic range compared to their unimolecular counterparts. Specifically, in response to Fsk treatment, BimAKAR-CAAX shows a fivefold increase in dynamic range relative to AKAR4-CAAX while Lyn-BimAKAR shows a threefold increase in dynamic range compared to Lyn-AKAR4 (Herbst et al., 2011) (Fig. 16.2B).
4.2. Microscope setup
For wide-field fluorescence imaging, the instrumentation consists of an epifluorescence microscope with a shutter-controlled excitation light source, filter sets for the donor and acceptor, and a charge coupled device (CCD) camera. For a typical imaging setup, an Axiovert 200M microscopt (Carl Zeiss) with a cooled charged couple device (CCD) camera was controlled by Metafluor software (Universal imaging). For CFP and YFP, dual-emission ratio imaging uses two excitation filters (420DF20 and 495DF10), two dichroic mirrors (450DRLP and 515DRLP), and two emission filters (475DF40 for CFP and 535DF25 for YFP) switched by a Lambda 10-2 filter changer (Sutter Instruments).
4.3. Cellular expression of AKAR
Typically, HEK293T cells are maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum at 37 °C with 5% CO2. For imaging AKAR, cells are plated onto glass coverslip (No.1 thickness) in 35 mm dishes and transfected with a mammalian expression vector (pcDNA3) containing AKAR using Ca2+ phosphate-mediated transfection at 50–60% confluency, and then grown for approximately 24 h before imaging. In comparison, BimAKAR requires a double transfection. To increase the probability that both portions of the KIBS are expressed in cells, it is important to mix the DNA of both portions of the sensor prior to transfection. Typically, this is done at a 1:1 ratio.
To ensure consistency between imaging experiments, expression level of reporter should be examined before imaging. First, it is important to check if reporters are expressed in cells at sufficient level and also to ensure that cells have a healthy morphology. Second, proper subcellular localization of reporter should be verified. For example, uniform fluorescence throughout the cell should be expected for untargeted reporters, whereas targeted reporters should be expressed in their designed locale. Confirmation of localization can be verified by using colocalization markers. Finally, it is important to make sure that unimolecular reporters remain intact and that no severe proteolysis occurred. This also can be done by checking the intensity of both donor and acceptor fluorescence and by confirming that both are expressed in the same location.
4.4. Cell imaging
Before imaging, culture media is removed and cells are washed twice with Hank’s balanced salt solution buffer and maintained in the dark. Generally, healthy cells exhibiting normal morphology and expressing intermediate to high levels of fluorescence are selected. Cells with exceptionally high fluorescence intensity have too much reporter expressed, which may interfere with endogenous signaling pathways. Conversely, cells that are too dim do not have sufficient reporter expression and will be less sensitive.
In the case of imaging BimAKAR, special considerations are required for cell selection. As it is important that both halves of the KIBS be expressed at similar levels, both the donor and acceptor channels must be checked prior to the start of the experiment to ensure sufficient expression and proper localization of both components. In addition, one practical strategy to help select cells that have similar expression levels is to establish a narrow range of starting Y/C ratio. It is important to note that the starting ratio for each BimKAR will vary as this parameter is dependent on the imaging setup, the FPs used, and the basal activity of the kinase in the locale under study. Nonetheless, once an appropriate starting ratio range is determined, it will facilitate the cell selection process, reducing the variability in responses between cells.
For a typical time course imaging for CFP–YFP FRET pair, three image channels are acquired using an acquisition time of 10–1000 ms at time interval of 10–60 s: (1) CFP direct (excitation of donor CFP and acquisition of CFP emission), (2) YFP FRET (excitation of donor CFP and measurement of acceptor YFP emission), and (3) an optional YFP direct (excitation of acceptor YFP and acquisition of YFP emission) as a control to check for photobleaching or cellular morphological changes. It is critical to optimize the excitation exposure time since long exposure time increases the potential of photobleaching and phototoxicity whereas short exposure results in poor signal-to-noise ratio. In addition, time interval should be adjusted to be fast enough to capture the kinase activity dynamics while avoiding too frequent excitation exposure. For AKAR and BimAKAR, a 500-ms exposure for both the FRET and CFP channels and a 50-ms exposure for YFP channel, as well as 30 s time-lapse interval is used (Herbst et al., 2011).
4.5. Controls
An important control to verify that the emission ratio change originates from PKA phosphorylation of the designated threonine in the substrate region in AKAR is to generate and test an AKAR T/A mutant, in which the phosphorylation site is mutated to alanine. For example, all AKAR and BimAKAR T/A mutants do not show Fsk-induced FRET changes (Herbst et al., 2011; Zhang et al., 2005). Such a mutant should be used as a negative control to confirm that the FRET change is phosphorylation dependent when designing or testing any KAR.
It is also critical to test the specificity of KARs. In the case of AKAR and BimAKAR, this includes testing the cAMP-induced response of the sensors in the presence of PKA inhibitor. Specificity testing for a KAR should also be confirmed by testing the response of the biosensor to a compound which will activate cellular kinases other than the target kinase. For instance, BimAKAR does not show a FRET increase upon treatments which activate endogenous PKC, demonstrating the specificity of BimAKAR to detect PKA activity (Herbst et al., 2011). Such controls are critical to confirm the specificity of the biosensor for the event under study.
4.6. Data analysis and FRET quantification
After the imaging experiment, several cell regions, as well as control region without cells are selected within the images from each time point. For reporters which have fixed stoichiometry between the donor and acceptor fluorescent proteins, the change in emission ratio (YFP FRET/CFP) can be linked to FRET change, therefore representing the most experimental convenient readout as shown in Eq. (16.1), where FA, FD are the fluorescence intensity of acceptor emission upon donor excitation, and intensity of donor emission upon direct excitation, respectively. In the case of bimolecular reporters, the stoichiometry between the donor and acceptor fluorescent proteins is not fixed and will vary among different cells, thereby complicating the quantification and comparison between different cells. However, the emission ratio change (Eq. (16.1)) may still be used, for example, to get some kinetic information about acutely stimulated signaling activity. In addition, acceptor photobleaching can be used to quantify the FRET efficiency. By irreversibly photobleaching acceptor fluorophores, usually at the end of time-lapse experiments, the energy transfer from donor to acceptor is abolished and causes recovery of donor fluorescence intensity. Therefore, FRET efficiency can be represented by the relative fluorescence intensity of the donor before (FD) and after (FDA) acceptor photobleaching (Eq. (16.1)).
| (16.1) |
| (16.2) |
5. Summary and Perspectives
Genetically encoded FRET-based biosensors allow for the tracking of dynamic cellular signaling events in real time, thus representing versatile and powerful tools to study the regulation of signal transduction with unprecedented spatial and temporal resolution. The generalizable modular design has facilitated the development of an expanding repertoire of bio-sensors for visualizing the dynamics of several major classes of cellular signaling molecules, including protein kinases, GTPases, and second messengers, and have successfully uncovered specific molecular mechanisms regulating various signal transduction pathways in live cells.
Based on the highly modular and adaptable design strategies, it is anticipated that an increasing number of biosensors for signaling enzymes will be developed. By combining these versatile and powerful fluorescent biosensors with novel molecular tools that manipulate biological systems in a spatially and temporally restricted manner (Airan et al., 2009; Levskaya et al., 2009; Wu et al., 2009), biochemical events underlying signal transduction in the native context of cell or live organism can be studied. Moreover, quantitative fluorescence imaging data can be incorporated into mathematical models to obtain a systems understanding of the signaling network. Taken together, these technologies offer great potential to unravel the complex regulation of dynamic signaling processes in living biosystems.
Acknowledgments
Work in the lab was supported by NIH grant R01 DK073368 (JZ).
References
- Ai HW, et al. Fluorescent protein FRET pairs for ratiometric imaging of dual biosensors. Nat Methods. 2008;5:401–403. doi: 10.1038/nmeth.1207. [DOI] [PubMed] [Google Scholar]
- Airan RD, et al. Temporally precise in vivo control of intracellular signalling. Nature. 2009;458:1025–1029. doi: 10.1038/nature07926. [DOI] [PubMed] [Google Scholar]
- Aitken A, et al. 14-3-3 proteins: Biological function and domain structure. Biochem Soc Trans. 1995;23:605–611. doi: 10.1042/bst0230605. [DOI] [PubMed] [Google Scholar]
- Albertazzi L, et al. Quantitative FRET analysis with the EGFP-mCherry fluorescent protein pair. Photochem Photobiol. 2009;85:287–297. doi: 10.1111/j.1751-1097.2008.00435.x. [DOI] [PubMed] [Google Scholar]
- Allen MD, et al. Reading dynamic kinase activity in living cells for high-throughput screening. ACS Chem Biol. 2006;1:371–376. doi: 10.1021/cb600202f. [DOI] [PubMed] [Google Scholar]
- Allen MD, Zhang J. Subcellular dynamics of protein kinase A activity visualized by FRET-based reporters. Biochem Biophys Res Commun. 2006;348:716–721. doi: 10.1016/j.bbrc.2006.07.136. [DOI] [PubMed] [Google Scholar]
- Ananthanarayanan B, et al. Signal propagation from membrane messengers to nuclear effectors revealed by reporters of phosphoinositide dynamics and Akt activity. Proc Natl Acad Sci USA. 2005;102:15081–15086. doi: 10.1073/pnas.0502889102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananthanarayanan B, et al. Live-cell molecular analysis of Akt activation reveals roles for activation loop phosphorylation. J Biol Chem. 2007;282:36634–36641. doi: 10.1074/jbc.M706227200. [DOI] [PubMed] [Google Scholar]
- Aoki K, Matsuda M. Visualization of small GTPase activity with fluorescence resonance energy transfer-based biosensors. Nat Protoc. 2009;4:1623–1631. doi: 10.1038/nprot.2009.175. [DOI] [PubMed] [Google Scholar]
- Aye-Han NN, et al. Fluorescent biosensors for real-time tracking of post-translational modification dynamics. Curr Opin Chem Biol. 2009;13:392–397. doi: 10.1016/j.cbpa.2009.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berridge MJ, et al. Calcium signalling: Dynamics, homeostasis and remodelling. Nat Rev Mol Cell Biol. 2003;4:517–529. doi: 10.1038/nrm1155. [DOI] [PubMed] [Google Scholar]
- Calleja V, et al. Intramolecular and intermolecular interactions of protein kinase B define its activation in vivo. PLoS Biol. 2007;5:e95. doi: 10.1371/journal.pbio.0050095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo LD, et al. A cellular FRET-based sensor for beta-O-GlcNAc, a dynamic carbohydrate modification involved in signaling. J Am Chem Soc. 2006;128:14768–14769. doi: 10.1021/ja065835+. [DOI] [PubMed] [Google Scholar]
- Chalfie M, et al. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- Clapham DE. Calcium signaling. Cell. 2007;131:1047–1058. doi: 10.1016/j.cell.2007.11.028. [DOI] [PubMed] [Google Scholar]
- Cohen LB, et al. Changes in axon fluorescence during activity: Molecular probes of membrane potential. J Membr Biol. 1974;19:1–36. doi: 10.1007/BF01869968. [DOI] [PubMed] [Google Scholar]
- Cubitt AB, et al. Understanding, improving and using green fluorescent proteins. Trends Biochem Sci. 1995;20:448–455. doi: 10.1016/s0968-0004(00)89099-4. [DOI] [PubMed] [Google Scholar]
- Day RN, Schaufele F. Fluorescent protein tools for studying protein dynamics in living cells: A review. J Biomed Opt. 2008;13:031202. doi: 10.1117/1.2939093. [DOI] [PubMed] [Google Scholar]
- Depry C, et al. Visualization of PKA activity in plasma membrane microdomains. Mol Biosyst. 2011;7:52–58. doi: 10.1039/c0mb00079e. [DOI] [PubMed] [Google Scholar]
- DiPilato LM, et al. Fluorescent indicators of cAMP and Epac activation reveal differential dynamics of cAMP signaling within discrete subcellular compartments. Proc Natl Acad Sci USA. 2004;101:16513–16518. doi: 10.1073/pnas.0405973101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn TA, et al. Imaging of cAMP levels and protein kinase A activity reveals that retinal waves drive oscillations in second-messenger cascades. J Neurosci. 2006;26:12807–12815. doi: 10.1523/JNEUROSCI.3238-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster T. Intermolecular energy migration and fluorescence. Ann Phys. 1948;2:55–75. [Google Scholar]
- Fosbrink M, et al. Visualization of JNK activity dynamics with a genetically encoded fluorescent biosensor. Proc Natl Acad Sci USA. 2010;107:5459–5464. doi: 10.1073/pnas.0909671107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich MW, et al. Imaging CREB activation in living cells. J Biol Chem. 2010;285:23285–23295. doi: 10.1074/jbc.M110.124545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, et al. 14-3-3 proteins: Structure, function, and regulation. Annu Rev Pharmacol Toxicol. 2000;40:617–647. doi: 10.1146/annurev.pharmtox.40.1.617. [DOI] [PubMed] [Google Scholar]
- Fujioka A, et al. Dynamics of the Ras/ERK MAPK cascade as monitored by fluorescent probes. J Biol Chem. 2006;281:8917–8926. doi: 10.1074/jbc.M509344200. [DOI] [PubMed] [Google Scholar]
- Gao X, Zhang J. Spatiotemporal analysis of differential Akt regulation in plasma membrane microdomains. Mol Biol Cell. 2008;19:4366–4373. doi: 10.1091/mbc.E08-05-0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grynkiewicz G, et al. A new generation of Ca2+ indicators with greatly improved fluorescence properties. J Biol Chem. 1985;260:3440–3450. [PubMed] [Google Scholar]
- Harvey CD, et al. A genetically encoded fluorescent sensor of ERK activity. Proc Natl Acad Sci USA. 2008;105:19264–19269. doi: 10.1073/pnas.0804598105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson JN, et al. Structural basis for reversible photobleaching of a green fluorescent protein homologue. Proc Natl Acad Sci USA. 2007;104:6672–6677. doi: 10.1073/pnas.0700059104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbst KJ, et al. Luminescent kinase activity biosensors based on a versatile bimolecular switch. J Am Chem Soc. 2011;133:5676–5679. doi: 10.1021/ja1117396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson L, et al. Design and optimization of genetically encoded fluorescent biosensors: GTPase biosensors. Methods Cell Biol. 2008;85:63–81. doi: 10.1016/S0091-679X(08)85004-2. [DOI] [PubMed] [Google Scholar]
- Karasawa S, et al. Cyan-emitting and orange-emitting fluorescent proteins as a donor/acceptor pair for fluorescence resonance energy transfer. Biochem J. 2004;381:307–312. doi: 10.1042/BJ20040321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraynov VS, et al. Localized Rac activation dynamics visualized in living cells. Science. 2000;290:333–337. doi: 10.1126/science.290.5490.333. [DOI] [PubMed] [Google Scholar]
- Levskaya A, et al. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature. 2009;461:997–1001. doi: 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim CJ, et al. Integrin-mediated protein kinase A activation at the leading edge of migrating cells. Mol Biol Cell. 2008;19:4930–4941. doi: 10.1091/mbc.E08-06-0564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CW, et al. Genetically encoded fluorescent reporters of histone methylation in living cells. J Am Chem Soc. 2004;126:5982–5983. doi: 10.1021/ja038854h. [DOI] [PubMed] [Google Scholar]
- Lu S, et al. The spatiotemporal pattern of Src activation at lipid rafts revealed by diffusion-corrected FRET imaging. PLoS Comput Biol. 2008;4:e1000127. doi: 10.1371/journal.pcbi.1000127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machacek M, et al. Coordination of Rho GTPase activities during cell protrusion. Nature. 2009;461:99–103. doi: 10.1038/nature08242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markwardt ML, et al. An improved cerulean fluorescent protein with enhanced brightness and reduced reversible photoswitching. PLoS One. 2011;6:e17896. doi: 10.1371/journal.pone.0017896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyawaki A. Visualization of the spatial and temporal dynamics of intracellular signaling. Dev Cell. 2003;4:295–305. doi: 10.1016/s1534-5807(03)00060-1. [DOI] [PubMed] [Google Scholar]
- Miyawaki A, et al. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature. 1997;388:882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- Mochizuki N, et al. Spatio-temporal images of growth-factor-induced activation of Ras and Rap1. Nature. 2001;411:1065–1068. doi: 10.1038/35082594. [DOI] [PubMed] [Google Scholar]
- Nagai T, et al. Expanded dynamic range of fluorescent indicators for Ca(2+) by circularly permuted yellow fluorescent proteins. Proc Natl Acad Sci USA. 2004;101:10554–10559. doi: 10.1073/pnas.0400417101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalbant P, et al. Activation of endogenous Cdc42 visualized in living cells. Science. 2004;305:1615–1619. doi: 10.1126/science.1100367. [DOI] [PubMed] [Google Scholar]
- Newman RH, Zhang J. Visualization of phosphatase activity in living cells with a FRET-based calcineurin activity sensor. Mol Biosyst. 2008;4:496–501. doi: 10.1039/b720034j. [DOI] [PubMed] [Google Scholar]
- Newman RH, et al. Genetically encodable fluorescent biosensors for tracking signaling dynamics in living cells. Chem Rev. 2011;111:3614–3666. doi: 10.1021/cr100002u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen AW, Daugherty PS. Evolutionary optimization of fluorescent proteins for intracellular FRET. Nat Biotechnol. 2005;23:355–360. doi: 10.1038/nbt1066. [DOI] [PubMed] [Google Scholar]
- Ni Q, et al. Analyzing protein kinase dynamics in living cells with FRET reporters. Methods. 2006;40:279–286. doi: 10.1016/j.ymeth.2006.06.013. [DOI] [PubMed] [Google Scholar]
- Ni Q, et al. Signaling diversity of PKA achieved via a Ca2+-cAMP-PKA oscillatory circuit. Nat Chem Biol. 2011;7:34–40. doi: 10.1038/nchembio.478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perroy J, et al. Real-time monitoring of ubiquitination in living cells by BRET. Nat Methods. 2004;1:203–208. doi: 10.1038/nmeth722. [DOI] [PubMed] [Google Scholar]
- Pertz O, et al. Spatiotemporal dynamics of RhoA activity in migrating cells. Nature. 2006;440:1069–1072. doi: 10.1038/nature04665. [DOI] [PubMed] [Google Scholar]
- Ridley AJ, et al. Cell migration: Integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- Rizzo MA, et al. An improved cyan fluorescent protein variant useful for FRET. Nat Biotechnol. 2004;22:445–449. doi: 10.1038/nbt945. [DOI] [PubMed] [Google Scholar]
- Ross WN, et al. A large change in dye absorption during the action potential. Biophys J. 1974;14:983–986. doi: 10.1016/S0006-3495(74)85963-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K, et al. Real-time imaging of histone H4 hyperacetylation in living cells. Proc Natl Acad Sci USA. 2009;106:16257–16262. doi: 10.1073/pnas.0902150106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleifenbaum A, et al. Genetically encoded FRET probe for PKC activity based on pleckstrin. J Am Chem Soc. 2004;126:11786–11787. doi: 10.1021/ja0460155. [DOI] [PubMed] [Google Scholar]
- Shaner NC, et al. A guide to choosing fluorescent proteins. Nat Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- Tkachenko E, et al. Protein kinase A governs a RhoA-RhoGDI protrusion-retraction pacemaker in migrating cells. Nat Cell Biol. 2011;13:661–668. doi: 10.1038/ncb2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsien RY. The green fluorescent protein. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- Violin JD, et al. A genetically encoded fluorescent reporter reveals oscillatory phosphorylation by protein kinase C. J Cell Biol. 2003;161:899–909. doi: 10.1083/jcb.200302125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker SA, Lockyer PJ. Visualizing Ras signalling in real-time. J Cell Sci. 2004;117:2879–2886. doi: 10.1242/jcs.01285. [DOI] [PubMed] [Google Scholar]
- Wu YI, et al. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature. 2009;461:104–108. doi: 10.1038/nature08241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaccolo M, et al. A genetically encoded, fluorescent indicator for cyclic AMP in living cells. Nat Cell Biol. 2000;2:25–29. doi: 10.1038/71345. [DOI] [PubMed] [Google Scholar]
- Zacharias DA, et al. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- Zhang J, Allen MD. FRET-based biosensors for protein kinases: Illuminating the kinome. Mol Biosyst. 2007;3:759–765. doi: 10.1039/b706628g. [DOI] [PubMed] [Google Scholar]
- Zhang J, et al. Genetically encoded reporters of protein kinase A activity reveal impact of substrate tethering. Proc Natl Acad Sci USA. 2001;98:14997–15002. doi: 10.1073/pnas.211566798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, et al. Creating new fluorescent probes for cell biology. Nat Rev Mol Cell Biol. 2002;3:906–918. doi: 10.1038/nrm976. [DOI] [PubMed] [Google Scholar]
- Zhang J, et al. Insulin disrupts beta-adrenergic signalling to protein kinase A in adipocytes. Nature. 2005;437:569–573. doi: 10.1038/nature04140. [DOI] [PubMed] [Google Scholar]