Abstract
The mechanistic target of rapamycin (mTOR) complex 1 (mTORC1) integrates environmental and intracellular signals to regulate cell growth. Amino acids stimulate mTORC1 activation at the lysosome in a manner thought to be dependent on the Rag small guanosine triphosphatases (GTPases), the Ragulator complex, and the vacuolar H+-adenosine triphosphatase (v-ATPase). We report that leucine and glutamine stimulate mTORC1 by Rag GTPase-dependent and -independent mechanisms, respectively. Glutamine promoted mTORC1 translocation to the lysosome in RagA and RagB knockout cells and required the v-ATPase but not the Ragulator. Furthermore, we identified the adenosine diphosphate ribosylation factor-1 GTPase to be required for mTORC1 activation and lysosomal localization by glutamine. Our results uncover a signaling cascade to mTORC1 activation independent of the Rag GTPases and suggest that mTORC1 is differentially regulated by specific amino acids.
Cells sense environmental nutrient flux and respond by tightly controlling anabolic and catabolic processes to best coordinate cell growth with nutritional status. The mechanistic target of rapamycin (mTOR), a conserved serine-threonine kinase, is part of the mTOR complex 1 (mTORC1), which helps coordinate cell growth with nutritional status. Dysregulation of mTORC1 is common in human diseases, including cancer and diabetes (1). Amino acids are essential for mTORC1 activation (2, 3); however, it remains unclear how specific amino acids are sensed. Leucine (Leu) (2, 4, 5), glutamine (Gln) (5–7), and arginine (Arg) (2) have been implicated in mTORC1 activation. In one model, mTORC1 indirectly senses amino acids within the lysosomal lumen that requires the Rag guanosine triphosphatases (GTPases), which are regulated by the pentameric Ragulator complex, the vacuolar H+–adenosine triphosphatase (v-ATPase), and the Gator complex (8, 9). When activated, the Rag GTPases bind to and recruit mTORC1 to the lysosome, where the Rheb GTPase activates mTORC1 (4). In mammals, there are four Rag proteins: RagA and RagB, which are functionally redundant; and RagC and RagD, which are also functionally equivalent. The formation of a heterodimer between RagA or RagB with RagC or RagD, and the guanine nucleotide state of the Rag proteins determines mTORC1 recruitment to the lysosome and subsequent activation (4, 10, 11). Under amino acid sufficiency, RagA and RagB complexes are guanosine triphosphate (GTP)–loaded and capable of binding Raptor. Somehow the v-ATPase detects the buildup of lysosomal amino acids (12), stimulates Ragulator guanine nucleotide exchange factor (GEF) activity, and inhibits Gator GTPase-activating protein (GAP) activity (9, 13). This loads RagA-RagB complexes with GTP and recruits mTORC1 to the lysosome, where it encounters Rheb, a potent mTORC1 activator that mediates growth factor signals. The tuberous sclerosis complex (TSC) tumor suppressor is also localized at the lysosome, and it negatively regulates mTORC1 by acting as a GAP for Rheb (14).
We generated mouse embryonic fibroblasts that lack both RagA and RagB [RagA/B knockout (KO) MEFs] (Fig. 1A and fig. S1). RagA-RagB complexes bind directly to mTORC1 (15), and overexpression of a constitutively active version of one of the two proteins renders mTORC1 insensitive to amino acid starvation (fig. S2) (4, 10). Deletion of RagA/B diminished the abundance of RagC, consistent with RagA and RagB stabilizing RagC and RagD by forming heterodimers (Fig. 1A) (4, 16). Unexpectedly, deletion of RagA and RagB reduced (∼30%), but did not abolish, mTORC1 activity, as judged by the phosphorylation state of its substrates ribosomal S6 kinase 1 (S6K1) and eukaryotic translation initiation factor 4E–binding protein 1 (4EBP1). Phosphorylation of S6K1 and 4EBP1 was abolished when the RagA/B KO cells were treated with the mTOR inhibitors Torin1 and Rapamycin or were depleted of the mTORC1 subunit Raptor with short hairpin RNA (shRNA) (fig. S3). Thus, mTORC1 is active in the absence of RagA and RagB.
Fig. 1. Gln, but not Leu, activates mTORC1 independently of RagA and RagB.
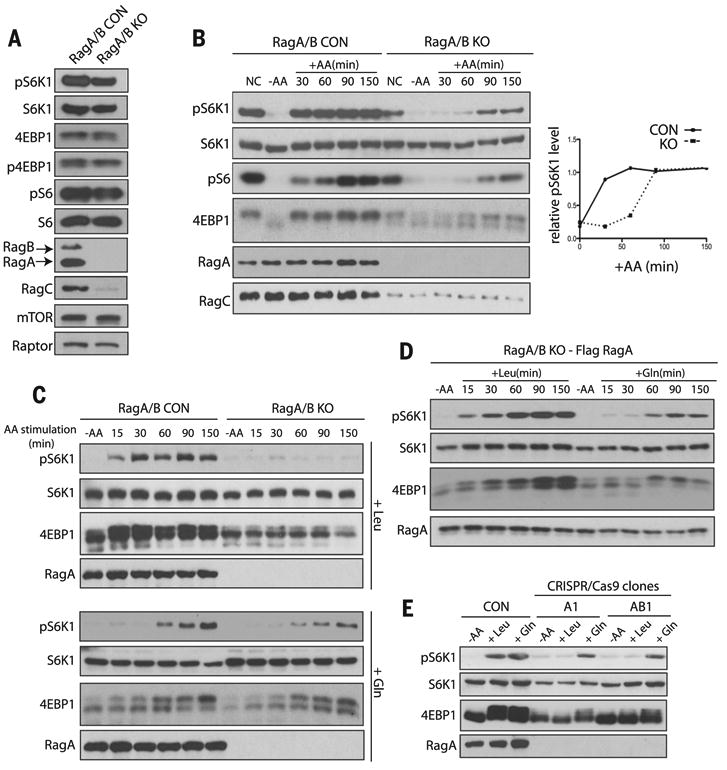
mTORC1 activity was analyzed by the phosphorylation of S6K1 (pS6K1), S6 (pS6), and 4EBP1 (p4EBP1) and the mobility shift of 4EBP1. AA, amino acids. (A) mTORC1 activity was analyzed in control (CON) and RagA/B KO MEFs under normal conditions (NC). mTOR, Raptor, RagA, RagB, and RagC protein were also analyzed. (B) mTORC1 activity was analyzed in CON and RagA/B KO MEFs under NC, in the absence of amino acids (–AA) and at the indicated times after the addition of amino acids (+AA) (left). Relative abundance of pS6K1 is plotted (right). (C) mTORC1 activity after stimulation with Leu (top) or Gln (bottom) in CON and RagA/B KO MEFs. (D) mTORC1 activity was analyzed after stimulation with Leu or Gln in RagA/B KO MEFs stably expressing Flag-tagged RagA at the indicated times. (E) mTORC1 activity was analyzed in CON, RagA KO (A1) and RagA/B KO (AB1) HEK293A cells that were starved of amino acids or stimulated with Leu or Gln for 150 min.
To investigate the amino acid response of the RagA/B KO MEFs, we stimulated cells with amino acids and analyzed the kinetics of mTORC1 activation. Both the magnitude and rate at which mTORC1 was activated by amino acids were reduced in cells lacking RagA and RagB (Fig. 1B and fig. S4). Likewise, mTORC1 activity was reduced in RagA/B KO MEFs upon amino acid withdrawal (fig. S5). To exclude the possibility that some cells lacking RagA and RagB spontaneously mutated to compensate for decreased mTORC1 activity, we analyzed individual clones derived from the RagA/B KO MEF population. Single clones displayed an increase in mTORC1 activity in response to amino acids (fig. S6). To determine which amino acids activate mTORC1 in the absence of RagA and RagB, we individually stimulated RagA/B KO MEFs with each of the 20 standard amino acids (fig. S7). Leu and Arg stimulated mTORC1 activation in control, but not RagA/B KO cells (Fig. 1C and figs. S7 and S8). Gln-stimulated activation of mTORC1 in RagA/B KO cells displayed kinetics similar to that of control cells and when RagA/B KO cells were stimulated with the 20 standard amino acids (Fig. 1, B and C, and fig. S4). Stable reexpression of Flag-tagged RagA in the RagA/B KO MEFs restored mTORC1 activation in response to Leu (Fig. 1D), which confirmed that RagA and RagB are required for mTORC1 activation by Leu but not Gln.
We performed genome editing by means of clustered regularly interspaced short palindromic repeats (CRISPR/Cas9) to inactivate the RagA and RagB genes in human embryonic kidney 293A (HEK293A) cells (17, 18) (fig. S9). In HEK293A cells and in MEFs, RagA is more abundant than RagB (Fig. 1A and fig. S9E). Loss of RagA alone or both RagA and RagB in these cells prevented Leu-, but not Gln-induced, activation of mTORC1 (Fig. 1E and fig S9F). Thus, Gln can stimulate mTORC1 activation independently of RagA and RagB or cell type.
The lysosome is essential in the amino acidsensing pathway to mTORC1 and is thought to be a platform for optimal mTORC1 activation that integrates effects of growth factors, such as insulin, through Rheb and those of amino acids through the Rags (19). Because Gln can activate mTORC1 in the absence of RagA and RagB (Fig. 1, C and E, fig. S7, and fig. S9F), we investigated whether lysosomal localization of mTORC1 was required for Gln-induced activation of mTORC1 in cells lacking RagA and RagB. In control cells, mTOR translocated to lysosomal membranes identified by the presence of the marker protein lysosome-associated membrane protein 2 (LAMP2) as early as 50 min and remained at the lysosome 150 min after amino acid stimulation (fig. S10A) (4, 11). In contrast, mTOR did not localize to lysosomal membranes in RagA/B KO cells after 50 min of amino acid stimulation (fig. S10B). However, by 150 min, we observed lysosomal localization of mTOR in a subset of cells that also showed activation of mTORC1 (Fig. 2A and fig. S10B). Gln, but not Leu, induced lysosomal localization of mTOR in RagA/B KO MEFs (Fig. 2, B and C). Furthermore, synergistic activation of mTOR by amino acids and insulin was observed in RagA/B KO cells (fig. S11). Thus, Gln appears to induce mTORC1 activation through translocation to the lysosome in a manner independent of RagA and RagB.
Fig. 2. Gln-induced mTORC1 lysosomal localization in the absence of RagA and RagB.
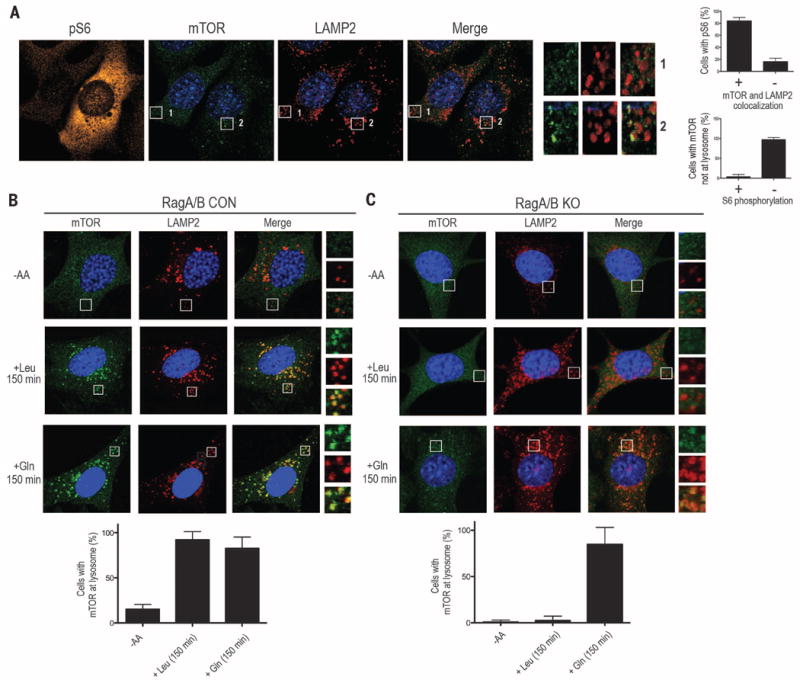
(A) Immunofluorescence (IF) analysis depicting mTORC1 activation by phosphorylation of S6 (pS6; orange) in RagA/B KO MEFs. mTOR (green) and LAMP2 (red) are also shown. Quantification of the percentage of pS6 cells with mTOR and LAMP2 colocalization (top right) and the percentage of cells with mTOR not at lysosome that also contain S6 phosphorylation (bottom right). (B and C) IF analysis depicting mTOR and LAMP2 in CON (B) or RagA/B KO MEFs (C), without amino acids or stimulated with Leu or Gln for 150 min (top). Quantification of the percent of cells with mTOR at the lysosome without amino acids or stimulated with Leu or Gln (bottom). Higher-magnification images (A) to (C) of the area depicted by the inset and their overlays are shown on the right.
Amino acid transporters (5, 12) and the Ragulator complex (11, 13) have been implicated in mTORC1 activation. We analyzed mTORC1 activity in cells depleted of several amino acid transporters and in MEFs lacking p14 (p14 KO MEFs), an essential subunit of the Ragulator complex. Gln activated mTORC1 in cells depleted of some amino acid transporters and in p14 KO MEFs, which indicated that these transporters and the Ragulator are not required for Gln-induced activation of mTORC1 (fig. S12 and Fig. 3A).
Fig. 3. Gln-induced mTORC1 activation requires the v-ATPase but not the Ragulator.
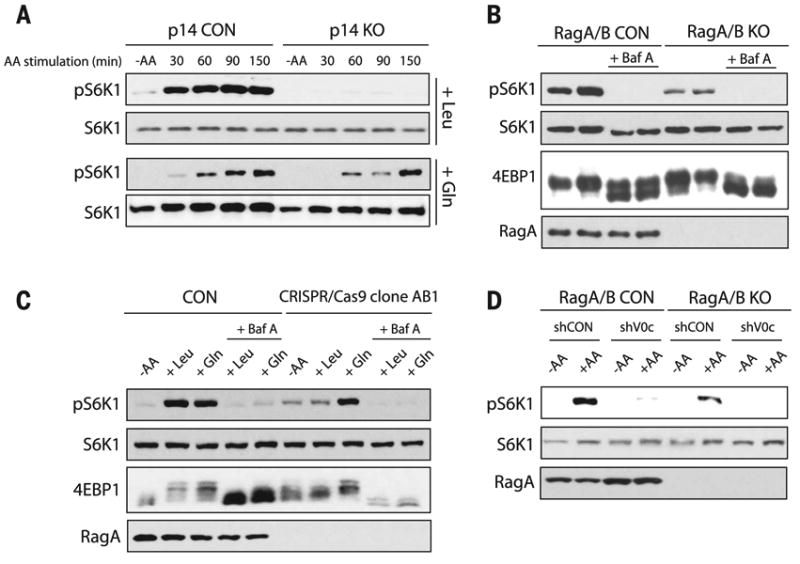
mTORC1 activity was analyzed by the phosphorylation of S6K1 (pS6K) and the mobility shift of 4EBP1. (A) mTORC1 activity was analyzed in CON and p14 KO MEFs that were starved of amino acids, then stimulated with Leu (top) or Gln (bottom) at the indicated times. (B and C) Analysis of mTORC1 activity in CON and RagA/B KO cells that were starved of amino acids; pretreated with or without 1 μM Baf A; followed by amino acid, Leu, or Gln stimulation for 150 min. (D) CON and RagA/B KO MEFs were treated with shRNA CON (shCON) or shRNA targeting the v-ATPase V0c subunit (shV0c). CON and RagA/B KO MEFs were starved of amino acids, followed by amino acid stimulation, and mTORC1 activity was assessed.
The v-ATPase is essential for the acidification of the lysosome and interacts with the Rags and Ragulator to stimulate mTORC1 activation in response to amino acids (12). We treated control and RagA/B KO cells with the v-ATPase inhibitor bafilomycin A (Baf A) (20). Baf A inhibited mTORC1 activation in both control and RagA/B KO cells when the cells were stimulated either with all amino acids (Fig. 3B and fig. S13A) or with Leu or Gln individually (Fig. 3C). Baf A also inhibited lysosomal localization of mTORC1 in RagA/B KO cells (fig. S13, B and C). Furthermore, inhibition of the v-ATPase by concanamycin A or inhibition of the lysosomal pH gradient by chloroquine also blocked Gln-induced lysosomal localization and activation of mTORC1 in RagA/B KO cells (fig. S13, D to H). Moreover, depletion of the v-ATPase V0c subunit, which interacts with the Ragulator and controls mTORC1 activity (12), largely prevented amino acid–induced activation of mTORC1 in control and RagA/B KO MEFs (Fig. 3D and fig. S13I). Furthermore, depletion of several lysosomal proteins had no effect on Gln-induced activation of mTORC1 and localization in the absence of RagA and RagB, which indicated that modification of the v-ATPase was not secondary to a general disruption in lysosomal structure and function (fig. S13, K and L). Taken together, Gln-induced activation of mTORC1 appears to require the v-ATPase and lysosomal function.
In Drosophila S2 cells, TORC1 activity is inhibited in cells depleted of the Drosophila ADP ribosylation factor-Arf1 (dArf1) (21), and we observed a further decrease in amino acid–induced TORC1 activation when both dRagA and dArf1 were depleted (Fig. 4A). We used small interfering RNA (siRNA) to deplete Arf1 from HEK293A RagA/B KO cells. Gln stimulated mTORC1 activation in RagA/B KO cells treated with a control siRNA; however, it failed to induce mTORC1 activation in RagA/B KO cells depleted of Arf1 (Fig. 4B). Depletion of other Arf family members failed to inhibit Gln-induced activation of mTORC1 in RagA/B KO cells (fig. S14A). Treatment of RagA/B KO cells with brefeldin A (BFA), an Arf1 GEF inhibitor (22), at high doses blocked amino acid signaling to mTORC1, whereas BFA caused only a small decrease in mTORC1 activation in response to amino acids in control cells (Fig. 4C and fig. S14, B and C). Consistently, BFA blocked Gln-induced activation of mTORC1 in RagA/B KO cells (Fig. 4D and fig. S14D). In addition, depletion of Arf1 or BFA treatment did not inhibit Leu-induced activation of mTORC1 in control cells, nor did they affect lysosomal pH (fig. S14, E to G).
Fig. 4. Arf1 is required for Gln signaling to mTORC1 in the absence of RagA and RagB.
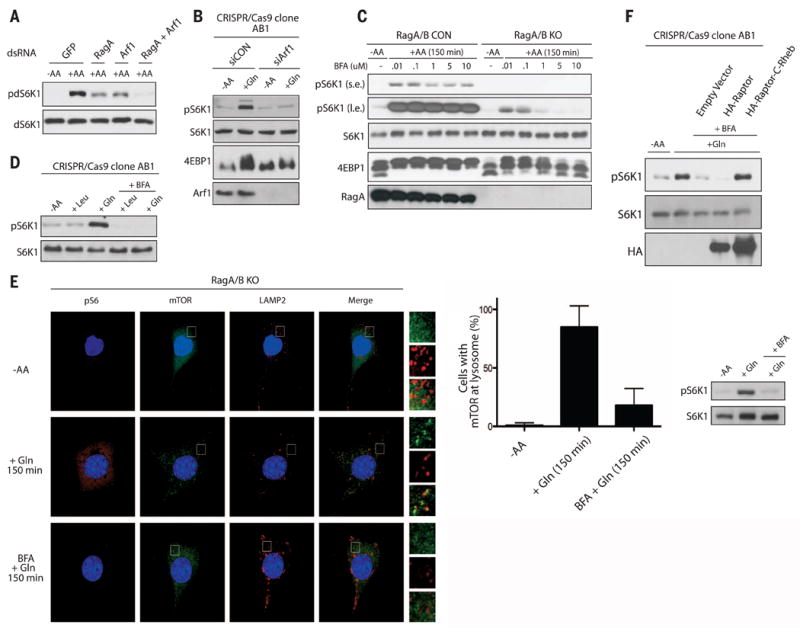
(A) TORC1 activity was analyzed for the phosphorylation of Drosophila S6K1 (pdS6K1) in Drosophila S2 cells treated with double-stranded RNA (dsRNA) targeting GFP, RagA, or Arf1. Cells were starved of amino acids for 1 hour, then stimulated with or without amino acids for 30 min. (B) mTORC1 activity was analyzed by the phosphorylation of S6K1 (pS6K1) or mobility shift of 4EBP1 in RagA/B KO HEK293A cells treated with control (siCON) or Arf1 siRNA (siArf1). Cells were starved of amino acids then stimulated with Gln for 150 min. (C) mTORC1 activity was analyzed as in (B) in CON and RagA/B KO MEFs starved of amino acids, then pretreated with the indicated concentrations of BFA, and stimulated with amino acids for 150 min. Labels s.e. and l.e. denote shorter exposure and longer exposure, respectively. (D) mTORC1 activity was analyzed as in (B) in RagA/B KO HEK293A cells starved of amino acids, then pretreated with or without 1 μM BFA, and stimulated with Leu or Gln for 150 min. (E) IFanalysis depicting mTORC1 activation (pS6; orange) and lysosomal localization (LAMP2; red, mTOR; green) in RagA/B KO MEFs. Cells were starved of amino acids, pretreated with or without 1 μM BFA, followed by stimulation with Gln for 150 min. Higher magnification images of the area depicted by the inset and their overlays are shown on the right of the images. Quantification of the percentage of cells with mTOR at the lysosome under different conditions and corresponding Western blot (right). (F) mTORC1 activity was analyzed as in (B) in RagA/B KO HEK293A cells transfected with HA-Raptor or HA-Raptor containing the C-terminal CAAX motif of Rheb (HA-Raptor-C-Rheb). Cells were starved of amino acids, pretreated with or without 1 μM BFA, and stimulated with Gln for 150 min.
Leu or Gln stimulation did not appear to affect the guanine nucleotide state of Arf1 (fig. S14H). Overexpression of a constitutively active Arf1-GTP failed to restore mTORC1 activation under amino acid deficiency (fig. S14I). Further, green fluorescent protein-tagged Arf1 (Arf1-GFP) localization was unaffected by amino acid starvation or stimulation (fig. S15). These results indicate that GTP hydrolysis or nucleotide cycling of Arf1, or both, is required for mTORC1 activation.
Arf1 regulates vesicular trafficking, so we tested whether bidirectional inhibition of trafficking between the endoplasmic reticulum (ER) and Golgi would affect Gln-induced activation of mTORC1 (23). We depleted proteins involved in anterograde trafficking and treated RagA/B KO cells with Golgicide A (24), yet did not observe an effect on Gln-induced activation of mTORC1 (fig. S16). These results support that Arf1 signaling to mTORC1 is specific and independent of ER-Golgi vesicular transport.
RagA/B KO MEFs treated with BFA were analyzed for mTOR localization in response to Gln stimulation. Gln-induced mTOR localization to the lysosome (Fig. 4E and Fig. 2C); however, pretreatment of cells with BFA inhibited the effect of Gln (Fig. 4E). Artificially targeting mTORC1 to the lysosomal surface by adding the C-terminal lysosomal targeting motif of Rheb to Raptor (11) activated mTORC1 in RagA/B KO cells, even in the presence of BFA (Fig. 4F). Thus, BFA inhibits mTORC1 by interfering with its lysosomal localization, which implicates Arf1 in the signaling pathway that links Gln to mTORC1 localization and activation at the lysosome.
In conclusion, we show that mTORC1 is differentially regulated by Gln and Leu (fig. S17). Our results demonstrate that RagA and RagB are essential for mTORC1 activation by Leu, but not by Gln, and this appears to be evolutionarily conserved in Saccharomyces cerevisiae (25). We identified the Arf1 GTPase to be involved in a signaling pathway that connects Gln to mTORC1 activation at the lysosome in the absence of the Rag GTPases. Many cancer cell lines have increased mTORC1 activity and show a high dependence on Gln for growth. Therefore, Gln-induced mTORC1 activation may be important for the growth of both normal and tumor cells.
Supplementary Material
Materials and Methods
Figs. S1 to S17
References (26-35)
Acknowledgments
We thank members of the Guan laboratory for critical comments; L. Huber for p14 null cells; and F. Flores, J. Zhou, and C. Worby for technical help. Supported by grants from NIH (R01GM051586 and R01CA108941 to K.-L.G.; T32CA121938 to J.L.J.; T32GM007752 to S.W.P.; and K99DK099254 to V.S.T.), Department of Defense (W81XWH-13-1-055) to K.-L.G., The Hartwell Foundation to J.L.J., and a Canadian Institutes of Health Research grant to R.C.R.
Footnotes
SUPPLEMENTARY MATERIALS
References And Notes
- 1.Laplante M, Sabatini DM. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hara K, et al. J Biol Chem. 1998;273:14484–14494. doi: 10.1074/jbc.273.23.14484. [DOI] [PubMed] [Google Scholar]
- 3.Wang X, Campbell LE, Miller CM, Proud CG. Biochem J. 1998;334:261–267. doi: 10.1042/bj3340261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sancak Y, et al. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nicklin P, et al. Cell. 2009;136:521–534. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim SG, et al. Mol Cell. 2013;49:172–185. doi: 10.1016/j.molcel.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Durán RV, et al. Mol Cell. 2012;47:349–358. doi: 10.1016/j.molcel.2012.05.043. [DOI] [PubMed] [Google Scholar]
- 8.Jewell JL, Russell RC, Guan KL. Nat Rev Mol Cell Biol. 2013;14:133–139. doi: 10.1038/nrm3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bar-Peled L, et al. Science. 2013;340:1100–1106. doi: 10.1126/science.1232044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Nat Cell Biol. 2008;10:935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sancak Y, et al. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zoncu R, et al. Science. 2011;334:678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bar-Peled L, Schweitzer LD, Zoncu R, Sabatini DM. Cell. 2012;150:1196–1208. doi: 10.1016/j.cell.2012.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Menon S, et al. Cell. 2014;156:771–785. doi: 10.1016/j.cell.2013.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gong R, et al. Genes Dev. 2011;25:1668–1673. doi: 10.1101/gad.16968011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sekiguchi T, Hirose E, Nakashima N, Ii M, Nishimoto T. J Biol Chem. 2001;276:7246–7257. doi: 10.1074/jbc.M004389200. [DOI] [PubMed] [Google Scholar]
- 17.Cong L, et al. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mali P, et al. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jewell JL, Guan KL. Trends Biochem Sci. 2013;38:233–242. doi: 10.1016/j.tibs.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forgac M. Nat Rev Mol Cell Biol. 2007;8:917–929. doi: 10.1038/nrm2272. [DOI] [PubMed] [Google Scholar]
- 21.Li L, et al. J Biol Chem. 2010;285:19705–19709. doi: 10.1074/jbc.C110.102483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Helms JB, Rothman JE. Nature. 1992;360:352–354. doi: 10.1038/360352a0. [DOI] [PubMed] [Google Scholar]
- 23.D’Souza-Schorey C, Chavrier P. Nat Rev Mol Cell Biol. 2006;7:347–358. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- 24.Sáenz JB, et al. Nat Chem Biol. 2009;5:157–165. doi: 10.1038/nchembio.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stracka D, Jozefczuk S, Rudroff F, Sauer U, Hall MN. J Biol Chem. 2014;289:25010–25020. doi: 10.1074/jbc.M114.574335. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Materials and Methods
Figs. S1 to S17
References (26-35)


