Abstract
Dissecting the upstream regulatory architecture of rice genes and their cognate regulator proteins is at the core of network biology and its applications to comparative functional genomics. With the rapidly advancing comparative genomics resources in the genus Oryza, a reference genome annotation that defines the various cis-elements and trans-acting factors that interface each gene locus with various intrinsic and extrinsic signals for growth, development, reproduction and adaptation must be established to facilitate the understanding of phenotypic variation in the context of regulatory networks. Such information is also important to establish the foundation for mining non-coding sequence variation that defines novel alleles and epialleles across the enormous phenotypic diversity represented in rice germplasm. This review presents a synthesis of the state of knowledge and consensus trends regarding the various cis-acting and trans-acting components that define spatio-temporal regulation of rice genes based on representative examples from both foundational studies in other model and non-model plants, and more recent studies in rice. The goal is to summarize the baseline for systematic upstream sequence annotation of the rapidly advancing genome sequence resources in Oryza in preparation for genus-wide functional genomics. Perspectives on the potential applications of such information for gene discovery, network engineering and genomics-enabled rice breeding are also discussed.
Keywords: Cis-elements, Trans-acting factors, Spatio-temporal regulation, Regulatory network, Comparative genomics
Introduction
The ‘gene-centric’ and subsequently the ‘genome-enabled’ paradigms of functional biology have led to concerted efforts to understand the biochemical synergies in the plant cell, by examining how genes function in a spatio-temporal scale. Understanding the multi-layered complexity of gene regulation is central to this goal, from the process of chromatin remodeling to transcription initiation, post-transcriptional processing and modulation, and protein modification and interaction. The process of transcription initiation is a critical crossroad of such multi-layered complexity of gene regulation, because it is an early step in the process where coordination could be interfaced with various intrinsic and extrinsic signals for growth, development, reproduction and adaptation. Thus, the study of the architecture of promoters and the nature of the interacting general and specialized regulatory transcription factors has always been an important aspect of understanding the intricate wirings of gene function (Kaufmann et al., 2010; Mejia-Guerra et al., 2012).
A large number of rice genes of potential agronomic value are regulated largely at the transcriptional level. Looking back to as early as the mid-1980’s, rice geneticists and molecular biologists have always been interested in studying the architecture of promoters, initially for the primary objective of discovering regulatory sequences for spatio-temporal engineering of transgenes. Examples of seminal studies include the dissection of the regulatory elements for precise endosperm-specific expression of seed storage proteins, which also elucidated the mechanisms of how seed development and maturation are regulated, and led to applications in seed bioengineering (Takaiwa et al., 1987; Okita et al., 1989; Takaiwa et al., 1991).
Subsequently, the elucidation of the entire genome of a reference genotype of cultivated rice (O. sativa ssp. japonica, cv. Nipponbare) made the establishment of direct links between agronomically important physiological processes and gene regulation at the whole transcriptome level possible (Ohyanagi et al., 2006; Yun et al., 2010; Sato et al., 2011; Hamada et al., 2011). The enormous amount of transcriptome data generated to date has routinely spark the same old question that rice geneticists has long been trying to address even prior to the advent of the genome-enabled era of biology, that is the question of: What unifying DNA signals determine the spatio-temporal transcription patterns of rice genes and how do these elements work? In today’s context, we often ask a more elaborate question of: What cis-elements and trans-acting factors facilitate the coordinate expression of functionally related rice genes and what can we learn from such synergy to understand the network relationships among genes in a grander scale that can explain phenotypic variation? As we get more advanced in the era of integrative, predictive and comparative systems biology in rice research, such a question has become quite relevant in the context of subsequent translation of genomics to various applications in molecular breeding, allele mining and network engineering for the improvement of yield and nutritional value of rice.
This review addresses a fundamental question that is central to the understanding of how transcription of rice genes is programmed. Its primary motivation is the need to present a synthesis of the current state of knowledge and consensus trends regarding the cis-acting and trans-acting components of transcriptional regulatory modules using representative examples from both seminal and more recent studies. It is hoped that this review will catalyze concerted efforts to continuously compile and update information in the same dynamic pace as in the dicot model Arabidopsis. While the information contained in this review is by no means inclusive of the entire body of information available about this topic, it is to our knowledge the first attempt to integrate the results of seminal studies with more recent updates about the structure, organization and evolutionary conservation of cis-elements in rice. Integration of this body of knowledge should nucleate new concepts on the application of regulatory sequences to comparative genomics, allele mining, and genetic manipulation by network engineering.
Review
Core promoter architecture and basal transcription machinery
With the annotated reference genome sequence of O. sativa ssp. japonica (cv. Nipponbare) and a rapidly increasing volume of whole-genome resequencing data on representative germplasm diversity panel across the genus, one important feature that needs to be defined for a biologically meaningful interrogation of protein-coding genes is the proximal regulatory region or ‘core promoter’. The core promoter is at the very heart of the process of transcription, being the site for the assembly of various protein components of RNA polymerase II pre-initiation complex, and its cooperation with other cis-elements and trans-acting factors that facilitate precise spatio-temporal regulation.
So what constitutes a core promoter? The consensus core promoter of eukaryotic genes is a contiguous stretch of DNA comprised of various cis-acting elements (Figure 1). The most well characterized is the TATA-box, defined by the consensus sequence ‘TATAWAWAR’, where the upstream ‘T’ is often located around −30 relative to A +1 (or G +1) position of the initiator (Inr) sequence for the transcription start site or TSS (Smale and Baltimore, 1989). The TATA-box is recognized by a sub-unit of the general transcription factor TFIID called the TATA-binding protein (TBP) to facilitate the formation of pre-initiation complex (Butler and Kadonaga, 2001; Smale, 2001). Occasionally found upstream to the TATA-box is a less frequently occurring motif called BRE (TFIIB recognition element), which facilitates efficient binding of transcription factor TFIIB to the core promoter. About 30-nt downstream of Inr is the Downstream Promoter Element (DPE), which is recognized by the TBP-associated factor (TAF) to enhance TBP binding to the core promoter (Burke and Kadonaga, 1997). In some cases TATA-box, Inr and DPE all occur in the same promoter (Juven-Gershon and Kadonaga, 2010).
Figure 1.
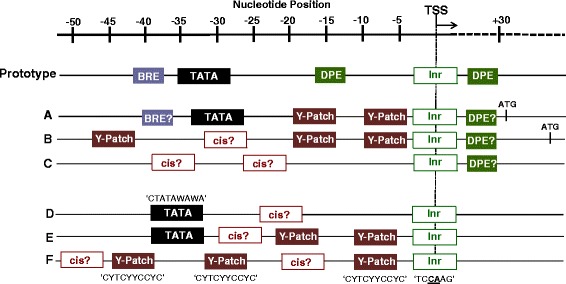
Comparison of the core promoter architectures of rice genes with the prototype core promoters of metazoan and Arabidopsis genes. In addition to the TATA-box, the composite prototype model of metazoan core promoter shows all previously identified cis-elements (BRE= TFIIB recognition element; DPE= Downstream promoter element; Inr= Initiator sequence) occurring in various combinations with the TATA-box. In Arabidopsis genes (A, B, C), TATA-box-containing core promoters (A) represent about 30% of all the protein-coding genes encoded by the genome. Non-TATA-box-containing promoters (B, C) represent a larger proportion of Arabidopsis genes. Small groups of genes contain novel motifs (cis?) that appear to be specific to higher plants. Evidence of the importance of DPE-like and/or BRE-like sequences in the functionality of Arabidopsis core promoters have not been established so far. In rice (D, E, F), TATA-box-containing core promoters (D) represent only about 18% of all protein-coding genes in the rice genome. Y-Patch is found in as much as 50% of the total protein-coding genes either in combination with or independent of TATA-box (E, F). DPE-like and BRE-like sequences are insignificantly represented in the core promoters of rice genes.
Structural conservation of TFIID in metazoans and plants suggests that the cis-acting elements involved in RNA polymerase II-dependent transcription are similar among eukaryotes (Vogel et al., 1993; Smale, 2001). Pioneering studies on the rice phenylalanine ammonia-lyase (PAL) promoter ZB8 showed that 5’-truncation up to −35 could maintain an accurate initiation of basal transcription (Zhu et al., 1995). The ‘TATTTAA’ motif within −35 to −28 that resembles the consensus sequence for eukaryotic TATA-box is critical for minimal promoter activity. Replacement of this sequence either with G-rich motif (‘GCGGGTT’) or with 2-nt substituted versions (TCGTTAA or TATGGAA) could lead to complete loss of promoter function. It has been shown that a recombinant TBP of rice (OsTBP2) interacts with OsTFIIB for efficient binding to the TATA-box in ZB8 promoter, causing highly efficient TATA-dependent basal transcription (Zhu et al., 2002). Moreover, the functionality of −35 to −28 region of ZB8 is in context with the consensus ‘TCCAAG’ Inr sequence. Substitution of the conserved core ‘CA’ motif at −1 and +1 positions of Inr leads to erroneous initiation at multiple locations.
Contrary to earlier notions that TATA-box is a universal feature of most eukaryotic core promoters, subsequent surveys of Arabidopsis, human and Drosophila genomes have shown that this was not the case (Molina and Grotewold, 2005; Lenhard et al., 2012). For instance, although the TATA-boxes among Arabidopsis genes were contextually positioned at the correct upstream location, their occurrence in such context was established only in less than 30% of the total validated genes. While the occurrence of TATA-related motifs was established reliably among highly expressed genes, other more prominent sequence motifs distinct from BRE and DPE have been identified in smaller subsets of TATA-less Arabidopsis genes (Figure 1).
Indeed, more recent surveys of TATA-box distribution in the rice genome established a trend similar to what has been established in Arabidopsis (Figure 1). Independent analysis with Gibbs Motif Sampler and Dragon Motif Builder showed that less than 20% of annotated rice genes contain TATA box-like motifs in the right context. Furthermore, DPE-like and/or BRE-like sequences are not significantly represented in rice core promoters (Clivan and Svec, 2009). The consensus sequence for rice TATA-box is defined as ‘CTATAWAWA’ located within a C-rich region around −49 to −20. However, much higher occurrence and broader distribution of a T/C-rich motif called Y-Patch (pyrimidine patch) was established in a global survey of rice core promoters. Although it has lesser degree of sequence conservation than TATA-box, Y-Patch elements occur in correct upstream context in more than 50% of annotated rice genes, either in combination with or independent of TATA-box. The consensus Y-Patch sequence has been defined as ‘CYTCYYCCYC’, occurring in one or more copies and with strict directional sensitivity around −49 to +1 region (Yamamoto et al., 2007; Clivan and Svec, 2009).
Key regulatory modules for constitutive expression of rice genes
A number of constitutive promoters have been particularly well investigated in rice including actin (OsAct), ubiquitin (OsUbi), cytochrome-c (OsCc1), ascorbate peroxidase (OsAPX), translation initiation factor-5 (OseIF5), phosphogluconate dehydrogenase (OsPGD), and R1G1-domain containing protein B (OsR1G1B) genes. The shared structural feature of these promoters is a short exon and large intron in the untranslated region (UTR) near the TSS, in addition to upstream regulatory modules (Figure 2, Table 1). The 5’-UTR introns are A/T-rich but share little sequence identity and with variable lengths (Jeong et al., 2006).
Figure 2.
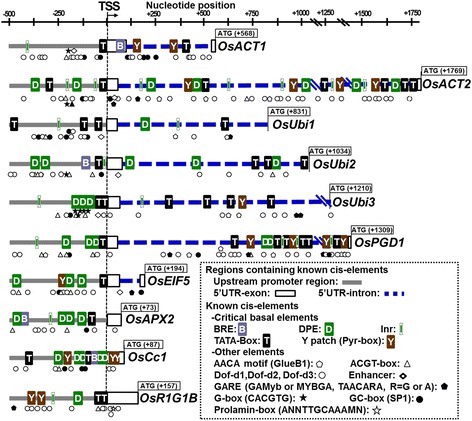
Models of the regulatory sequence architectures of well characterized promoters of constitutively expressed rice genes such as actin ( OsACT1, OsACT2 ), ubiquitin ( OsUbi1, OsUbi2, OsUbi3 ), phosphogluconate dehydrogenase ( OsPGD1 ), Initiation factor ( OsEIF5 ), ascorbic peroxidase ( OsAPX2 ), cytochrome-c ( OsCc1 ), and R1G1-domain-containing protein ( OsR1G1B ). Locations of critical cis-elements are indicated with colored symbols in both the upstream regions (gray lines) and downstream regions (5’UTR-exon, 5’UTR-intron; blue lines) relative to the location of the transcription start site (TSS).
Table 1.
List of experimentally or computationally defined upstream regulatory sequences, and their cognate transcriptional regulators that determine basal and constitutive expression of rice genes
| Cis-element | Core motif | Transcriptional regulator | Biological function |
|---|---|---|---|
| TATA-box | CTATAWAWA | TATA-binding protein (OsTBP2) | Core promoter; Pre-initiation complex |
| Y-Patch | CYTCYYCCYC | Unknown | Core promoter; Pre-initiation complex |
| (Pyrimidine patch) | |||
| Actin (Act) elements | CCCAA (tandem octa-repeats) | Unknown | Fine-tuner of constitutive OsAct genes |
| CAAT (CCAT-box) | Unknown | Positive regulator of constitutive OsAct genes | |
| GTGAC (intronic) | Unknown | Positive regulator of constitutive OsAct genes | |
| CCGCGTTGGC (ABA-responsive) | Unknown | Positive regulator of constitutive OsAct genes | |
| ATTAAT, CACGTA (light-responsive) | Unknown | Positive regulator of constitutive OsAct genes | |
| (A/T)GCC (dideca-repeats) | Unknown | Positive regulator of constitutive OsAct genes | |
| Ubiquitin (Ubi) elements | GTTGTGGTTTG | Unknown | Positive regulator of constitutive OsUbi genes |
List was based on seminal studies published in the literature and annotated in public databases.
The strong inducing effects of OsAct and OsUbi promoters are to a large degree due to the UTR introns. Intron-mediated enhancement (IME) of transcription requires the transcribed sequences near the TSS in the right orientation. In general, IME-type introns in rice are longer than non-enhancing introns and contain more regulatory elements (Bradnam and Korf, 2008; Parra et al., 2011). Dispersed within the IME-type intron are potential cis-elements including the predominant pentamer motif ‘CGATT’. It has been suggested that IME-type introns act through gene loops that facilitate steady reinitiation of transcription by RNA Pol II (Moabbi et al., 2011; Rose et al., 2011). The coding sequences of ubiquitin (OsUbi3) monomer often involving the first nine nucleotides enhance promoter activity over 4-fold (Clancy et al., 1994). In addition, the most commonly found elements in the 5’ upstream and UTR intron are Dof, GC-box, and ACGT-box (Figure 2, Table 1).
Typical cis-elements in canonical core promoters are also important elements required for strong constitutive expression of OsAct. For example, OsAct1 promoter contains a TATA-box at −35, a BRE-like motif at position +88 and two copies of Y-Patch element at positions +138 and +384. In addition, the region from position −50 are highly enriched with other potential regulatory elements including Sp1, light-responsive SORLIP, mesophyll-specific CACTFTPPCA1, Alfin1, a novel zinc-finger binding element found in the salt stress inducible genes, a signature element of nitrate reductase gene AMMORESIIUDCRNIA1 and a drought-responsive SBOXATRBCS (Figure 2, Table 1).
The critical regulatory elements of rice actin genes (OsAct1, OsAct2) include a 5’-upstream sequence, non-coding exon, 5’-UTR region intron, and a short non-coding region of the second exon (Figure 2). The first non-coding exon and intron are 79-bp and 58-bp, and 447-bp and 1,736bp for OsAct1 and OsAct2, respectively (McElroy et al., 1991; Zhang et al., 1991; He et al., 2009). Many cis-elements required for constitutive expression are located around the core promoter region, including a 38-bp poly(dA-dT) between positions −146 and −186 which act as a positive regulator, and eight tandemly repeated copies of imperfect pentamer motif with consensus ‘CCCAA’ at −294, acting as a negative regulator. In addition, the branch-point site (‘GTGAC’) for mRNA splicing located in the intron between the positions 359 and 363 is essential for promoter activity (McElroy et al., 1990).
A negative regulator between positions +96 and +274 within the intron is responsible for the repression of OsAct2 expression. On the other hand, the two CAAT-boxes between positions −448 and −445 and positions −635 and −632 act as possible positive regulators. Other cis-elements in the 5’-upstream region include ABA-responsive element (‘CCGCGTTGGC’), two different light responsive elements (‘ATTAAT’, ‘CACGTA’), and 20 repeated ‘A/TGCC’ triplets. Numerous cis-elements are also present between positions −50 and TSS, including CAATBOX1, SORLIP1AT, Alfin1, GATABOX, UPRMOTIFIIAT, C-rich Q, BOXCPSAS1, and ‘CGTGG’ motif.
Ubiquitin (OsUbi) promoter includes the 5’-upstream sequence and 5’-UTR containing a short first exon and intron (Wang and Oard, 2003; Bhattacharyya et al., 2012). Critical elements include a signature enhancer motif (‘GTTGTGGTTTG’) found in both OsUbi1 and OsUbi2 (Christensen and Quail, 1996; Wang and Oard, 2003). In addition to a TATA-box-like element, core promoters of OsUbi contain other signature cis-elements associated with inducible expression such as Alfin1, Dof, CARGCW8GAT, TAAAGSTKST1, PRECONSCRHSP70A, GT-box, POLLEN1LELAT52, WUSATAg, and ARR1AT (Figure 2, Table 1).
The promoter of cytochrome-c (OsCc1) includes the 5’-upstream region and 86-bp untranslated sequence without UTR intron (Figure 2). In addition to the signature core promoter elements such as TATA-box and CAAT box, it contains the cAMP response element (CRE), nuclear respiratory factor (NRF) binding site, and light responsive G-box (CACGTV) (Jang et al., 2002). Promoters of ascorbate peroxidase (OsAPX2), phosphoguconate dehydrogenase (OsPGD1), and R1G1-domain-containing protein (OsR1G1B) have similar constitutive strengths as the promoter of OsAct1, with various enhancers such as light responsive G-box and TGACG-element associated with high-level gene expression. Endosperm-specific regulatory elements such as ‘AACA’ and ‘ACGT’ are also important characteristics of OsPGD1 and OsR1G1B (Park et al., 2010a; Figure 2, Table 1). Based on the trends established from the analysis of housekeeping genes, it is apparent that constitutive expression requires many of the elements of the core promoter in various combinatorial complexities with various UTR structural features and a plethora of cis-elements involved in developmental and environmental signal perception.
Integration of hormonal, developmental and stress response signaling
Plant growth, development and adaptation are consequences of cooperative and antagonistic actions of different regulatory hormones, starting from germination to the development and maturation of seeds. Integration of hormonal and other intrinsic developmental and environmental signals leads to changes in gene expression.
Root development and auxin signaling
Auxin is recognized as the universal plant growth hormone because of its principal role in the regulation of cell expansion, division and differentiation (Kieffer et al., 2010). The principal receptor of auxin is a class of nuclear F-box protein that belongs to the TIR1/AFB family (Darmasiri et al., 2005). Auxin perception leads to rapid induction of three classes of auxin early responsive genes, which include the Aux/IAA, GH3 and the small auxin-up RNA or SAUR (Hagen and Guilfoyle, 2002). Aux/IAA regulates its own transcription, and functions as transcriptional regulator of other auxin responsive genes in cooperation with Auxin Response Factors (ARF) (Chapman and Estelle, 2009). ARF binds specifically to the promoters of GH3, SAUR and other auxin-responsive genes leading to rapid induction (Ulmasov et al., 1995; Ulmasov et al., 1997). In the absence of auxin, Aux/IAA inhibits ARF function and represses other auxin early responsive genes. Auxin binding activates TIR1/AFB, a component of ubiquitin E3 ligase SCF1, mediating ubiquitination and degradation of Aux/IAA. In effect ARF is relieved from repression, hence expression of auxin early responsive genes (Tiwari et al., 2003; Quint and Gray, 2006). In rice, 25 OsARF homologs have been identified, indicating a high degree of conservation across the dicot and monocot lineages (Wang et al., 2007).
Analysis of the japonica rice reference genome identified 31, 12 and 58 members of the OsAux/IAA, OsGH3 and OsSAUR gene families, respectively, many of which exhibit the characteristic rapid induction in response to auxin (Thakur et al., 2001; Jain et al., 2006a; Jain et al., 2006b, Jain et al., 2006c; Song et al., 2009). Moreover, it has been estimated that between 2% to 5% of the total genes in Arabidopsis and rice are either upregulated or downregulated by auxin (Nemhauser et al., 2004). The signature of the promoter of auxin early responsive genes is a 6-bp motif with the consensus sequence ‘TGTCTC’, often referred to as auxin-response element (AuxRE), occurring in either simple or composite configurations (Ulmasov et al., 1997). Simple AuxRE contains either direct or palindromic repeats of ‘TGTCTC’ motif separated by 3 to 7-nt spacer with no apparent consensus sequence. In contrast, composite AuxRE is functional only in modular form with CG-rich ‘coupling’ or ‘constitutive’ element (Table 2).
Table 2.
List of experimentally and computationally defined upstream regulatory sequences, and their cognate transcriptional regulators that determine spatio-temporal control of rice genes
| Cis-element | Consensus core motif | Transcriptional regulator | Biological function |
|---|---|---|---|
| Auxin responseelement (AuxRE1) | TGTCTC | Auxin responsefactor (ARF1, ARF2) | Primary regulator of auxin-regulated expression |
| Auxin responseelement (AuxRE2) | GACACA | Auxin responsefactor (ARF1, ARF2) | Primary regulator of auxin-regulated expression |
| Auxin responseelement (AuxRE3) | GACActGACA | Auxin responsefactor (ARF1) | Primary regulator of auxin-regulated expression |
| Gibberellin responseelement (GARE) | TAACCACC | GAMyb factors | Primary regulator of GA-regulated expression; primary regulator of sugar sensitivity of GA-responsive genes |
| Glut element | AACA/ACGT | Unknown | Regulator of endosperm-specific expression of rice seed storage protein glutelin (OsGlut) |
| Gt1/Gt2 box-I | ATATCATGAGTCACTTCA | GT1, GT2 | Positive regulator of endosperm-specific expression of rice glutelin (OsGlut) genes |
| Gt1/Gt2 box-II | CTTTCGTGTACCACA | GT1, GT2 | Positive regulator of endosperm-specific expression of rice glutelin (OsGlut) genes |
| Gt1/Gt2 box-III | ACAATGCTGCTCAATTA | GT1, GT2 | Positive regulator of endosperm-specific expression of rice glutelin (OsGlut) genes |
| Gt1/Gt2 box-IV | ATTATCCAATGTCATATTG | GT1/GT2 | Positive regulator of endosperm-specific expression of rice glutelin (OsGlut) genes |
| Gt1/Gt2 box-V | TAAGTCACGTTTGATGA | GT1, GT2 | Positive regulator of endosperm-specific expression of rice glutelin (OsGlut) genes |
| Gt3 box | ATATCATGAGTCACTTCA | GT1, GT2, GT3 | Positive regulator of endosperm-specific expression of rice glutelin (OsGlut) genes |
| PB-1 element | AAGCAACACACAAC | Dof-type P-boxBinding factor (PBF) | Positive regulator of endosperm-specific expression |
| Prolamin box | TGTAGAA, TGTTTTAATATGACGTGG | Dof-type P-boxbinding factor (PBF) | Positive regulator of endosperm-specific expression |
| SP8/SURE | TACTATT, TCACTATT, | Opaque-2 transcriptionfactor | Positive regulator of endosperm-specific expression |
| LPSE1 | ATTTGAGCTGCC | Unknown | Positive spatial regulator of expression in green leaves, stem and young panicle |
| LPSE2 | TTGATATATTTGT | Unknown | Positive regulator of expression on green leaves and negative regulator of expression in young panicle and roots |
| LPSRE1 | CGGCGCGCCAC | Unknown | Positive regulator of expression in green leaves and negative regulator of expression in panicle and stem |
| LPSRE2 | TTAGATAATGGA | Unknown | Positive spatial regulator of expression in green leaves, stem root, and young panicles |
| PSE1 | TTTATCTATTTCC | Unknown | Negative regulator of expression in young panicle and stem |
| PSE2 | TCTTTGGCAGAG | Unknown | Positive regulator of expression in green leaves and negative regulator of expression in stem and young panicles |
| Ethylene and jasmonic acid responsive element | GCC-CORE | Ethylene response factor | Regulator of senescence-induced gene expression (OsERF) |
| GS2 | TTCGAAATTGAACGTGCT-TAACCAAGAGAACAC | WRKY transcription factors | Senescence box |
| ABRE (ACGT-type) | CGMCACGTB | OsABI5, OREB1 | ABA-response element for ABA regulated gene expression during stress response and seed maturation |
| CE1 | TGCCACCGG | OsABI5, OREB1 | ABRE coupling factor |
| CE3 | ACGCGTGTCCTC | OsABI5, OREB1 | ABRE coupling factor |
| DRT/CRE | (a/g)CCGAC | OsDREB1, OsDREB2 | Dehydration response element for abiotic stress regulated gene expression independent of ABA |
| Abiotic/oxidative stress Element | CACG | OsNAC6, OsNAC5, OsSNAC2 | Regulator of abiotic stress-responsive gene expression independent of the DREB regulon |
| as1/ocs/TGA-like element | AATTTGAT, TAATTTGA | OsTGA10 | Regulator of chilling mediated oxidative stress signaling |
| Pyrimidine-box/Myb2-box element | AAAGAAAAA, TAGTTTTT | OsMyb4 | Regulator of chilling mediated oxidative stress signaling |
| JARE | CGTCA, TGACG, VCGCGB | Ethylene response factors (ERF) | Jasmonic acid response element; regulator of responses to pathogens and herbivores |
| GCC-box | TAAGAGCCGCC | Ethylene response factors (ERF) | Negative regulator of responses to Magnaporthe oryzae |
| P-box | CCGCCCTCCA | Ethylene response factors (ERF) | Disease responsive element |
| W-box | TTGAC element | OsWRKY45, OsWRKY71, OsWRKY03, OsWRKY53, OsWRKY31, OsWRKY30, OsWRKY76 | Disease responsive |
| E-box | CAGTCG, CACCTG | OsbHLH65 | Pathogen and herbivore response element; regulator of responses to Magnaporthe Oryzae and Nilaparvata lugens |
List was based on seminal studies published in the literature and annotated in public databases.
The analysis of the crown rootless (crl1) mutant, defective in auxin-mediated formation of crown roots facilitated the elucidation of cis-elements and trans-acting factors involved in auxin-regulated gene expression in rice (Inukai et al., 2005; Liu et al., 2005a). The crl1 mutation exhibits various auxin-related abnormalities including reduced lateral roots, insensitivity to auxin during lateral root initiation, and impaired root gravitropism. Wild-type Crl1 encodes an Assymetric leaves-2/Lateral Organ Boundaries (AS2/LOB) domain protein containing the N-terminus C-motif and GAS-motif at the C-terminus. The mutant protein contains a single amino acid substitution (A to T) within the AS2/LOB domain. The wild-type Crl1 is strongly induced by auxin, dependent on the degradation of Aux/IAA proteins, as shown by the effects of mutation in the degradation domain (domain II) of OsIAA3. The fact that Aux/IAA functions as a repressor of the transcriptional activator ARF has implied that Crl1 is a direct target of OsARF transcription factors. The promoter of wild-type Crl1 has two copies of AuxRE signature sequence. The first copy (AuxRE1) located upstream and distal from the TATA-box has the ‘TGTCTC’ sequence, while the second copy (AuxRE2) located upstream but more proximal to the TATA-box has the inverted sequence ‘GACACA’ (Table 2). The primary auxin-response transcriptional activator in rice (OsARF1) binds with high affinity with AuxRE2 but not with AuxRE1 (Waller et al., 2002).
Another crown rootless mutant in rice (crl5) has impaired initiation of crown root primordia (Kitomi et al., 2011). Mutants have abnormal root gravitropic response, delayed flowering, various types of floral anatomical abnormalities and small panicles. In wild-type plants, Crl5 is rapidly induced by exogenous auxin in the nodes or in regions of the stem where root growth is initiated. Like Crl1, rapid expression of Crl5 is also dependent upon the degradation of Aux/IAA proteins, thus Crl5 is downstream to OsAux/IAA and OsARF. The 642-aa residue CRL5 protein contains two copies of AP2 DNA-binding domain, related to AP2/ERF transcription factor AINTEGUMENTA (ANT). The promoter of Crl5 contains three copies of the ‘TGTC’ core motif of AuxRE. The first copy (RE1), which is upstream and most distal to the TATA-box consists of a tandem array of three inverted ‘TGTC’ motifs (‘GACAGACAGACA’). The second copy (RE2) is a single base substituted version (‘TGTCGC’) of the consensus for AuxRE. The third copy (RE3), which is upstream but most proximal to the TATA-box has two tandem ‘TGTC’ motifs with a dinucleotide spacer in between, i.e., ‘GACActGACA’ (Table 2). Direct binding of recombinant OsARF1 was demonstrated for the most proximal wild-type RE3 but not for its mutated version ‘GATActGATAct’, indicating that Crl5 is directly activated by OsARF1 like Crl1. The consensus from the analysis of Crl1 and Crl5 is that integration of auxin-responsive gene expression during root development requires a proximal AuxRE.
Seed germination and gibberellic acid signaling
First discovered in rice, the signature effect of gibberellic acid (GA) is the ability to promote stem elongation. Bioactive GAs affect virtually all aspects of plant growth and development from germination to hypocotyl elongation, stem growth, circadian rhythm, and reproductive organ and seed development (Lovegrove and Hooley, 2000; Hartweck and Olszewski, 2006).
Discovered first in rice, GID1/GID2 genes encode the nuclear localized primary receptor of GA (Gomi et al., 2004; Ueguchi-Tanaka et al., 2005; Griffiths et al., 2006; Ueguchi-Tanaka et al., 2007). Another important component of GA signaling are the DELLA proteins encoded by SLR1 in rice and GAI/RGA-like genes in Arabidopsis, which function as negative regulators of GA response (Ikeda et al., 2001; Fleet and Sun, 2005; Itoh et al., 2005). The rice DELLA protein is stable in the absence of GA ensuring that transcription of GA-early responsive genes are repressed by blocking the activity of a transcriptional activator bound to the promoters of target genes. GA binds to the nuclear localized GID1 receptor, triggering its interaction with the DELLA repressor. The GID1-DELLA complex recruits the GID2 F-box protein, a component of the SCF ubiquitin ligase complex, resulting in the degradation of the DELLA repressor and subsequent transcription of GA-early responsive genes (Hartweck, 2008). Analysis of the GA-response transcriptome estimated that about 250 and 300 genes are regulated by GA in Arabidopsis and rice, respectively (Ogawa et al., 2003; Yazaki et al., 2004).
In rice and other cereals, available sugars in the embryo are rapidly depleted at the onset of germination, triggering synthesis of GA which is then released to the aleurone layer. Subsequent expression of hydrolases ensures sufficient supply of sugar from the breakdown of starch to support seedling growth (Kaneko et al., 2002). Expression of α-amylases during rice germination is repressed by high sugar and abscisic acid (ABA) in the embryo, while they are induced by GA in the endosperm. Dissection of the promoter architecture of α-amylases established a clear picture of the interacting cis-elements that mediate differential regulation (Lu et al., 1998; Chen et al., 2002; Sutoh and Yamauchi, 2003; Chen et al., 2006). Among the members of the germination-associated α-amylase gene family, αAmy3 (RAmy3D) and αAmy8 (RAmy3E) provide an interesting contrast because both genes are synchronously induced by sugar depletion in the embryo, while in the endosperm αAmy3 is induced only by sugar depletion and αAmy8 only by GA. Induction of αAmy8 by GA is also correlated with the expression of GAMyb transcription factors.
The sugar response complex (SRC) was identified within the −186 to −82 region of αAmy3, comprised of a GC-box (~30-nt GC-rich motif) distal to the TATA-box, two tandemly repeated copies of TA-box (‘TATCCA’) upstream but more proximal to TATA-box, and G-box (‘CTACGTGG’) in between the two elements. The sugar response complex/GA-response complex (SRC/GARC) of αAmy8 is located within −318 to −89, comprised of a GC-box upstream and distal to TATA-box, single copy of proximal TA-box, and a gibberellic acid response element GARE (‘TAACCACC’) in between the two elements (Table 2). Mutation of GARE in the promoter of αAmy8 confirmed its critical role for GA-regulation as well as sugar-insensitivity (Table 2). Therefore, differential regulation of α-amylases by sugar depletion and GA in embryo and endosperm is facilitated by the interaction of GARE with GC-box and TA-box.
Expression of OsGAMyb transcription factors and αAmy8 are highly coordinated in the endosperm. In rice gamyb-2 mutant with impaired GA-dependent expression of α-amylases in the endosperm, αAmy8 becomes non-responsive to GA and repressible by glucose. Similarly, overexpression of OsGAMyb transcription factor has no effect on glucose repression of αAmy3, while an increase in expression of OsGAMyb renders αAmy8 insensitive to glucose repression. All these results established that OsGAMyb is a key regulator allowing differential responses of α-amylase genes to sugar depletion and GA in the endosperm and embryo by virtue of its interaction with GARE in GARC-promoters (Table 2).
Seed storage protein regulation during seed development
Making up as much as 80% of the total endosperm proteins of rice, glutelins serve as major nitrogen and carbon reservoir for the initial energy requirements during germination (Okita et al., 1989). Rice glutelins are classified into three groups: GluA, GluB, and GluC. The spatio-temporal regulation of three representative members of GluA sub-subfamily (Gt1, Gt2, Gt3) have been extensively investigated in an effort to understand their roles in the regulation of seed development. Gt1 and Gt2 are transcribed about 5 days after anthesis and continue virtually throughout the entire duration of seed development. In contrast, Gt3 transcripts accumulate maximally between 5 to 10 days post-anthesis followed by a steady decline (Okita et al., 1989; Zhao et al., 1994).
Promoter studies established that both 5’ distal and proximal cis-elements in Gt1 promoter are critical for precise spatio-temporal regulation (Zheng et al., 1993). The distal region (−5.1 to −1.8 kb) contains positive elements for temporal regulation and confer as much as 20-fold increase in expression. Leaky expression in leaves, stems, sheaths and roots was observed in promoter-GUS reporter driven by −507 or −154 truncated fragments of Gt1 promoter. This indicates that while the distal positive element within the −5.1 kb promoter contributes to temporal control of Gt1, proximal elements are critical for spatial specification in the endosperm.
DNase-I footprinting identified five protein-binding motifs (box-I to box-V) in Gt1 promoter ordered sequentially downstream from −434 position: ‘TAAGTCACGTTTGATGA’ (box-V), ‘ATTATCCAATGTCATATTG’ (box-IV), ‘ACAATGCTGCTCAATTA’ (box-III), ‘CTTTCGTGTACCACA’ (box-II), and ‘ATATCATGAGTCACTTCA’ (box-I) (Table 2). Promoter activity is completely abolished when boxes I to IV are mutated simultaneously. Box-II is the most critical, causing about 35-fold reduction in expression when mutated. Consistent with similar spatio-temporal patterns of Gt1 and Gt2, boxes-I to V are highly conserved between the two promoters, except that box-V has varied locations (Kim and Wu, 1990).
Similar to Gt1, the functional elements responsible for spatial, temporal and quantitative effects of Gt3 promoter are located at distal (−945 to −726) and proximal (−346 to −263) regions (Zhao et al., 1994; Croissant-Sych and Okita 1996). The critical elements responsible for precise temporal control post-anthesis are located within +7 to −181 and −278 to −320 regions. Removal of either of these proximal elements causes as much as 50% reduction in expression. The −106 to −88 and −125 to −111 regions of Gt3 promoter contain sequences similar to box-I and box-II in Gt1 and Gt2 promoters, with the same relative positions. Box-I contains the signature binding sequence (‘ATGAc/gTCAT’) for the yeast GCN4, AP-1, Jun and Fos transcription factors, while box-II contains another motif found in seed-specific α-amylase genes. Gt1, Gt2 and Gt3 promoters also share the signature motif ‘AACA’ around −75 to −64, which is essential for endosperm-specific expression (Table 2).
Gt3 promoter differed from Gt1 and Gt2 promoters by the absence of boxes-III, IV and V. However, its −482 to −278 region contains several functional elements that may be critical for its unique expression including the ‘AAGCAACACACAAC’ motif (−279 to −259) known as the binding site for nuclear factor PB-1, two prolamin-box-like (P-box-like) signals ‘TGTAGAA’ (−302 to −295) and ‘TGTTTTAA’ (−322 to −314) known to interact with Dof-type P-box binding factor (PBF), and ‘TATGACGTGG’ motif, which resembles the consensus binding sequence for the seed-development transcription factor Opaque-2 (Table 2). Gt3 promoter also contains additional sequences around −482 region that suppresses its expression in the leaves and other vegetative organs. Studies on other glutelin gene subfamilies also indicated the critical roles of GCN4, AACA-motif and P-box for endosperm-specific expression of GluB-3 and GluB-5 (Washida et al., 1999; Qu et al., 2008; Table 2).
Tissue/organ-specific gene expression
In rice, the nature of cis-elements and trans-acting factors that confer tissue or organ-specific programs is not yet well understood. However, results of the analysis of few candidate rice genes are starting to reveal interesting trends. One good example is Oshox1, encoding a homeo-domain leucine zipper (HD-Zip) transcription factor that plays an important role in vascular differentiation. Oshox1 expression initiates when procambial cells have already acquired distinctive anatomical properties but before achieving terminal differentiation (Scarpella et al., 2000).
Upstream region (−1,600) of Oshox1 directs vascular-specific expression through a process that involves auxin and sucrose (Scarpella et al., 2005). Oshox1 promoter drives expression in a tissue-specific fashion, showing particularly high expression in the apical region of primary and secondary procambial strands of cotyledon, but not in the axis of mature embryo. It also directs expression in the vascular cylinder of the basal mature region of the root, and in the apical hydathode of developing first leaves. Various proximal and distal sections of the −1,600 upstream sequence of Oshox1 confer different patterns of vascular cell, tissue and organ-specific expression and these sections contain modular combinations of ‘CCA/TTG’ repeats with stretches of ‘CCCC’ or ‘AACA’ motifs (Table 2). The auxin-responsive complex of Oshox1 promoter is located within −1,621 to −898 region defined by 11 copies of ‘AAGG’ Dof element and ‘GGTCCAT’ signature element typical of many known auxin-responsive genes. The same region (−1,621 to −898) is also critical for sucrose-regulated expression, consistent with the occurrence of several known sucrose responsive elements including SP8 element (‘TACTATT’), three copies of SURE (‘TACTATT’, ‘TCACTATT’, ‘TACTAT’) and one copy of B-box (‘CTAAAC’) (Grierson et al., 1994; Ishiguro and Nakamura, 1994; Baumann et al., 1999; Zourelidou et al., 2002).
Another example of organ/tissue-specific expression is illustrated by a photosystem-II 10-kDa polypeptide-encoding gene (Os08g10020), which is expressed strongly in leaves and sheaths but very weakly in non-green vegetative tissues of the root, stem, mature panicle and seed (Cai et al., 2007). The promoter of this gene spans the −2,134 to +41 region, punctuated by proximal core promoter signals such as ‘TATA-box’ and ‘CAAT-box’. The full-length promoter confers 14-fold, 90-fold, 14-fold and 129-fold higher expression in the leaves than in the stem, endosperm/embryo, panicle and root, respectively. As positive regulator, the ‘leaf and panicle/stem-specific element’ LPSE1 (‘ATTTGAGCTGCC’) within −106 to −96 defines the expression in leaves and young panicles. As negative regulators, the ‘leaf, panicle/stem and root element’ LPSRE2 (‘TTAGATAATGGA’) within −545 to −534 suppresses expression in the leaf, root, young panicle and stem, while the ‘panicle/stem-specific element’ PSE1 (‘TTTATCTATTTCC’) within −708 to −696 suppresses expression in young panicle and stem. Elements with dual roles as positive and negative regulators include the LPSRE1 (‘CGGCGCGCCAC’) within −222 to −214, LPSE2 (‘TTGATATATTTGT’) within −383 to −371, and PSE2 (‘TCTTTGGCAGAG’) within −670 to −659. All three elements function as enhancers in the leaf while LPSRE1, LPSE2 and PSE2 are silencers in the stem, young panicle/root, and young panicle/stem, respectively (Table 2).
Aging and senescence
Leaf senescence is a consequence of either developmentally programmed or stress-induced cell deterioration and declining rate of photosynthesis (Buchanan-Wollaston, 1997; Gan and Amasino, 1997; Chandlee, 2001). Cytokinin and ethylene are two major hormones that regulate the process of leaf senescence by virtue of their promoting and inhibiting effects, respectively. Other hormones and signaling molecules such as jasmonic acid, salicylic acid, abscisic acid (ABA) and reactive oxygen species also play either major or minor role singly or in combination (Nam, 1997). In rice, age-dependent leaf senescence is an important process for grain filling and seed maturation while developmentally uncoupled programmed cell death (premature senescence) is often induced by environmental stresses (Buchanan-Wolaston et al., 2003; Lim et al., 2007).
The ability to delay senescence has always been a major goal of plant biology for yield improvements by enhancing photosynthesis, i.e., ‘stay green’ phenotype (Thomas and Howart, 2000). Discovery of several classes of senescence-associated genes (SAG) provided a window to the mechanisms of senescence-regulated gene expression (Robatzek and Somssick, 2002). It was estimated that about 40 different types of transcription factors are involved in the regulation of SAGs including several Myb, zinc finger, MADS-box, WRKY and bZIP transcription factors (Chen et al., 2002). Knock-out mutation of senescence-associated WRKY6 has been shown to alter the spatio-temporal expression of certain SAGs delaying onset of senescence.
The cysteine protease-encoding AtSAG12 of Arabidopsis, which has a functional homolog in rice (OsSAG39) is highly expressed in senescing leaves, downregulated by auxin, cytokinin, and sucrose but not affected by desiccation, dark incubation, wounding, ABA or ethylene (Noh and Amasino, 1999). The senescence box or ‘GS2’ in the promoter of AtSAG2 is defined by the motif ‘TTCGAAATTGAACGTGCTTAACCAAGAGAACAC’ within −603 to −571 region that binds nuclear protein extracts from senescing leaves (Table 2). Recently, it was found that the upstream region of OsSAG39 contains positive regulatory elements within −2,021 to +24 for maximal expression when chlorophyll content is down to about 40% (Liu et al., 2010). Within its proximal and distal regions are arrays of known cis-elements such as ‘GCCCORE’ for ethylene and jasmonic acid regulated expression, ABA-boxes RYREPEATBNNAPA, MYCATRD22 and DPBFCOREDCDC3, drought elements MYBCORE and MYBST1, WRKY71OS site for OsWRKY71, and elicitor responsive element HBOXCONSENSUSPVCHS (Urao et al., 1993; Baranowskij et al., 1994; Abe et al., 1997). OsSAG39 promoter is inducible by the ethylene precursor ACC, ABA, GA, and low temperature. Protein extracts from senescing leaves bind to the HBOXCONSENSUSPVCHS and WRKY71OS elements (Table 2).
Abiotic stress and abscisic acid signaling
Drought, low temperature and salinity are essentially overlapping forms of stresses characterized by similar types of physiological perturbations and responses. At the cellular level, they trigger ‘physiological dehydration’, which affects almost every aspect of cellular physiology. As a major stress hormone in plants, the various components of abscisic acid (ABA) signal transduction pathway has been elucidated using Arabidopsis as a model (Razem et al., 2006). Seminal studies in Arabidopsis also led to the elucidation of major cis and trans components of stress response mechanisms that are evolutionarily conserved in flowering plants. Exposure to low temperature, dehydration and salinity stresses induces a common subset of genes referred to as cor, cold regulated or rd, responsive to desiccation genes through an ABA-dependent or ABA-independent pathway (Liu et al., 1998; Thomashow, 1999; Shinozaki and Yamaguchi-Shinozaki, 2000; Yamaguchi-Shinozaki and Shinozaki, 2005).
Subsequent transduction of ABA towards gene expression involves phospholipase-D-dependent signaling and expression of various MYB, MYC and bZIP transcription factors that control the expression of different ABA responsive genes through the various combinations of cis-elements including MybR, MycR and the most predominant ABRE (ABA Response Element) (Finkelstein, 2006). ABRE interacts with bZIP-type ABRE-binding factors or ABF that function as regulators of stress response and seed maturation (Giraudat et al., 1994; Guedes Correa et al. 2008). The ABF-target genes include many cor/rd genes with the ABRE core motif ‘ACGT’, also referred to as G-box or C-box (Guiltinan et al., 1990). Additionally, ABRE functionality is combinatorial with other types of ‘non-ACGT’ coupling elements such as CE1, CE3 and DRE (drought response element), comprising the minimal ABA-responsive complex or ABRC (Shen and Ho, 1995; Shen et al., 1996; Hobo et al., 1999; Narusaka et al., 2003).
In rice, the dominant ABRE-binding transcription factors have been identified as OsABI5 and OREB1 (Nakashima et al., 2009; Todaka et al., 2012). A recent genome-wide survey of co-expressed rice genes estimated that more than 10% (>400) of total genes are regulated through the ABA-signaling pathway, based on over-representation of the consensus sequence ‘CGMCACGTB’ within −1,000 upstream regions (Cheng et al., 2007; Lenka et al., 2009; Yun et al., 2010; Table 2). Included among the ABRE-enriched genes are various components of stress signaling and defense mechanisms, further reiterating the important role of ABA as a central signal that integrates various types of stress defenses in rice (Cheng et al., 2007; Yun et al., 2010). Furthermore, genome-wide comparative analysis of ABRC composition between in rice and Arabidopsis revealed striking differences between the two species (Gomez-Porras et al., 2007; Xu et al., 2012a). While ABRCs defined by ABRE-ABRE duplex occurred at similarly high frequencies among ABA-regulated genes in both species, the occurrence of CE3 coupling element with the consensus sequence ‘(c/a)CG(c/g)(c/g)g(c/a)gC(t/g)’ is nearly specific to the ABA-responsive genes of rice (Table 2). These trends indicate that ABA-regulated genes in rice and Arabidopsis have evolved distinct combinatorial cis-element modules.
The drought, low temperature and salt-inducible expression of cor/rd genes in Arabidopsis requires a class of element defined by the core motif ‘(a/g)CCGAC’ named as Dehydration Responsive Element/C-repeat (DRE/CRT) (Yamaguchi-Shinozaki and Shinozaki, 1994; Table 2). This element does not function as ABRE but could act as a coupling element for ABRE to form a functional ABRC (Hobo et al., 1999; Yazaki et al., 2004). The trans-activators DREB1/CBF and DREB2 that bind to the DRE/CRT (i.e., DREB for DRE-Binding factor or CBF for C-repeat Binding Factor) are members of the AP2/ERF family of transcription factors that are highly conserved between the dicot and monocot groups of plants (Stockinger et al., 1997; Liu et al., 1998; Stockinger et al., 2001). Constitutive expression of DREB1/CBF and DREB2 have led to the coordinated activation of target cor/rd genes, and subsequently increased stress tolerance (Jaglo-Ottosen et al., 1998; Kasuga et al., 1999; Gilmour et al., 2000). Overexpression studies estimated that more >300 genes encoded by the Arabidopsis genome are regulated through the DREB/CBF pathway with >100 genes confirmed as direct targets (Seki et al., 2001; Fowler and Thomashow, 2002).
The DREB/CBF regulon is highly conserved across the temperate and tropical groups of flowering plants as evidenced by the occurrence of functional DREB1/CBF and DREB2 orthologs in both monocots (including rice) and dicots (Jaglo et al., 2001; Choi et al., 2002; Dubouzet et al., 2003; Nakashima et al., 2009; Yun et al., 2010; Todaka et al., 2012). Genome-wide survey of the enrichment of ‘(a/g)CCGAC’ core motifs with correct spatial distribution revealed that DRE/CRT are as ubiquitous among the stress-regulated genes encoded by the rice genome as ABRE, i.e., significantly enriched in >10% of total rice genes, similar to what has been observed in Arabidopsis (Rabbani et al., 2003; Fowler and Thomashow, 2002; Yazaki et al., 2004; Vogel et al., 2005; Cheng et al., 2007; Lenka et al., 2009). For example, DRE/CRT is the dominant class of cis-elements that regulates a late embryogenesis abundant protein (OsLEA) network involved in dehydration, low temperature and salinity stress mechanisms in japonica rice (Meier et al., 2008).
The promoter of Arabidopsis CBF/DREB (e.g., CBF3) is characterized by a highly conserved 125-bp module as binding sites (ICE Boxes I, II, III, IV, V) for a Myc-like bHLH transcription factor ICE1 (Shinwari et al., 1998; Chinnusamy et al., 2003; Zarka et al., 2003). ICE1 is regulated by phosphorylation and its activated form binds to the c-Myc target element (‘CANNTG’) located in the highly conserved 125-bp region of CBF promoters. Box IV within the critical 125-bp region of CBF2 promoter contains the motif ‘CACATG’. Similar survey of upstream regions of all members of the orthologous DREB/CBF gene families in rice and sorghum revealed common signatures that include cis-elements like Ca/Calmodulin element (‘VCGCGB’), ABRE (‘CGTGG’, ‘MACGYGB’), jasmonic acid signaling (‘CGTCA’, ‘TGACG’) and MYC (‘CANNTG’) (Srivastav et al., 2010).
While the DREB/CBF network appears to play the dominant role low temperature (4°C and below) and drought response mechanisms in rice, several recent studies revealed other regulons with likely overlap with biotic stress response networks through oxidative signaling (Table 2). An example is the NAC regulon controlled by the NAC-type (NAM, ATAF, CUC) transcription factors OsNAC6/SNAC2 and OsNAC5, with target cis-elements having the core motif ‘CACG’ (Nakashima et al., 2007; Hu et al., 2008; Nakashima et al., 2009). Another example is the network of genes regulated by bZIP transcription factor OsTGA10 (Os06g41100) and R2R3-type Myb transcription factor OsMyb4 (Os04g43680), which is activated primarily in by oxidative signal stress during exposure to chilling and configures various components of oxidative defenses (Jakoby et al., 2002; Cheng et al., 2007; Park et al., 2010b; Yun et al., 2010; Xu et al., 2012). This network is comprised of a large number of target genes defined by several combinations of as1/ocs/TGA-like elements (‘AATTTGAT’, ‘TAATTTGA’) and GARE/Pyr-box/Myb2-box-like elements (‘AAAGAAAAA’, ‘TAGTTTTT’).
Biotic stress and ethylene, salicylic acid and jasmonic acid signaling
Ethylene, salicylic acid and jasmonic acid are key components of host plant signaling and defenses against pathogens and insect herbivores (Abeles et al. 1992; Pieterse et al., 1998; Johansson et al., 2006; Dempsey and Klessig, 2012). Ethylene signaling during host-pathogen or host-herbivore interactions lead to changes in gene expression through various types of ERF (Ethylene Response Factor) transcription factors, while the salicylic acid and jasmonic acid mediated gene expression involves the action of various types of WRKY and bHLH transcription factors, respectively (Helliwell and Yang 2013). For instance, the early responses of rice against the striped stemborer Chilo suppressalis is known to involve ethylene, salicylic acid, jasmonic acid and H2O2, mediating a MAPK-phosphorylation cascade that regulates the expression of several OsWRKY and OsERF3 transcription factors (Lu et al., 2011). Similarly, the early responses of rice to both virulent and avirulent pathovars of Magnaporthe oryzae (blast) is mediated by ethylene leading to the upregulation of OsERF922 transcription factor and its suite of target defense-related genes (Singh et al., 2004; Liu et al., 2012).
Examples of known targets of defense-related ERF transcription factors are the pathogenesis-related proteins PR3, PR4 and PDF1.2. A common feature shared by many of these PR-protein genes and other types of ethylene-regulated defense genes is the canonical GCC-box cis-element defined by the core motif ‘AGCCGCC’ (Hart et al., 1993; Ohme-Takagi and Shinshi, 1995; Manners et al., 1998; Suzuki et al., 1998; Table 2). A more recent example of ERF-type transcription factors that directly target defense-related genes in rice is OsERF922, which acts on GCC-box containing genes to negatively regulate resistance to Magnaporthe oryzae (Liu et al., 2012). In Arabidopsis, other types of ERF transcription factors such as Pti4/5/6 have also been shown to bind to the canonical GCC-box as well as other GCC-box related core-motifs such as ‘TAAGAGCCGCC’ (Table 2). These ERF transcription factors and their cognate cis-elements are highly conserved in both dicot and monocot plants including rice (Hao et al., 1998; Gu et al., 2000). Additionally, the conserved ‘GCC-box’ has also been shown to function as JA-responsive cis-element involved in both pathogen and herbivore defense mechanisms in Arabidopsis and rice (Brown et al., 2003). Many such ERF-type transcription factors that act on ‘GCC-box’ cis-element have been shown to modulate the expression of their target ethylene-responsive and jasmonic acid-responsive genes either positively or negatively, by virtue of their dual activation and repression domains (Rojo et al.,1999; Fujimoto et al., 2000; Ohta et al., 2001; Ohme-Takagi et al., 2000; Bodenhausen and Reymond, 2007).
Recent studies revealed other defense-related ERF transcription factors that regulate ethylene-inducible gene expression through the P-box element, defined by the core motif ‘CCGCCCTCCA’ (Table 2). Initially identified in the promoter of ethylene and pathogen-inducible putrescine-N-methyltransferase-2 (PMT2) gene of tobacco, P-box element binds various ERF- transcription factors that are not involved in GCC-box-mediated gene expression (e.g., ERF189, ERF115, ERF179, ERF163, ORCA3, AtERF13). P-box-like motifs have also been shown to be highly enriched among putative ERF-target defense-related genes in rice (Shoji et al., 2010; Shoji and Hashimoto 2012).
Salicylic acid plays a major role in defense mechanisms particularly against biotrophic pathogens that parasitize a living host (Alvarez, 2000; Desveaux et al., 2004; Garcion and Métraux, 2006; Ciolkowski et al., 2008; Vlot et al., 2009). In rice, salicylic acid-mediated gene expression has been shown to involve similar types of cis-elements and transcriptional regulators that have been identified in other dicot plants such as Arabidopsis and tobacco (Silverman et al., 1995; Yang et al., 2004). Primarily, salicylic acid-mediated defense involves WRKY transcription factors acting on the SA-responsive element defined by the core motif ‘TGACG’ and W-box element defined by the core motif ‘TTGAC(c/t)’ (Chen et al., 2002; Li et al., 2004). In rice, several WRKY transcription factors (OsWRKY45, OsWRKY71, OsWRKY03) have been shown to bind to W-box of several PR-protein genes such as OsPR10a (Liu et al. 2005b; Shimono et al. 2007; Hwang et al. 2008; Table 2).
Many members of WRKY family of transcription factors in rice are transcriptionally activated in response to M. oryzae, the causal agent of blast and Xanthomonas oryzae pv. oryzae, the causal agent of bacterial leaf blight (Ryu et al., 2006; Bagnaresi et al., 2012).
For example, OsWRKY45 has been shown to play a dual role in mechanisms of resistance to both blast and bacterial leaf blight, while many other WRKY transcription factors such as OsWRKY53, OsWRKY31, and OsWRKY30 have been shown to function specifically as positive regulators of defenses against blast by virtue of their ability to activate a suite of target genes with common features of having either W-box and/or SA-response elements (Zhang et al., 2008; Peng et al., 2012; Shimono et al.,2012). Some WRKY transcription factors such as OsWRKY76 function as negative regulators of disease resistance also by virtue of their interaction with the W-box. This indicates that the W-box functions both as activator and repressor of SA-mediated gene expression in rice (Yokotani et al. 2013).
Jasmonic acid is an important regulator of responses to pathogens, insect herbivores and other related factors such as wounding and mechanical injury (Creelman and Mullet, 1995; Farmer et al., 2003; Wasternack, 2007; Koo and Howe, 2009). The primary activators of jasmonic acid-mediated signaling are bHLH-type transcription factors that are conserved between Arabidopsis and rice. The precise identity of the associated activator and repressor cis-elements for jasmonic acid-mediated gene expression are just starting to be revealed, but a number of candidates have recently been suggested. For example, three copies of an E-box element defined by the core motif ‘CANNTG’ has been suggested to function as JA-responsive enhancer within −515 to −265 region of rice chitinase gene OsChia4a (Table 2). The E-box element has been suggested as the binding site for the bHLH-type RERJ1 transcription factor (Nishizawa et al., 1999; Miyamoto et al., 2012; Wager and Browse 2012; Santino et al., 2013). Recently, another bHLH-type transcription factor OsbHLH65 that responds to Magnaporthe grisea, Nilaparvata lugens, salicylic acid and jasmonic acid directly binds to several potential E-boxes with the signature motifs ‘CAGCTG’ and ‘CACCTG’ (Santino et al., 2013).
Cis-element and transcription factor databases
Systematic curation of cis-elements and their cognate trans-acting factors that define regulated transcription in eukaryotes is central to computational modelling of transcriptional networks (Buchler et al., 2003; Xie et al., 2005; Mohanty et al., 2012). Our recent survey of regulatory sequences of rice genes across the available public databases indicate that rice information has been steadily increasing given the level of global activity in rice functional genomics (Tables 2 and 3). During the last two decades, several data repositories have been developed to provide an annotated reference of cis-elements and their cognate trans-acting factors established from the results of wet-lab experiments. The first generation information repositories which include TRANSFAC, PLACE, PlantCARE and AGRIS served as initial foundation for large-scale computational prediction of the unifying regulatory features among co-expressed genes revealed by genome-wide transcriptome studies (Wingender et al., 1996; Higo et al., 1999; Lescot et al., 2002; Davuluri et al., 2003; Matys et al., 2003). However, while TRANSFAC has been utilized for the analysis of rice data, its predictive power for novel plant-specific elements is quite limited. On the other hand, PLACE, PlantCARE and AGRIS have offered relatively higher resolutions for the analysis of rice data, given their stronger emphasis on monocot and dicot plants.
Table 3.
Partial list of recently annotated regulatory sequences from rice based on recent updates in public cis-element and transcription factor databases
| Cis-element | Consensus core motif | Transcriptional regulator | Biological function |
|---|---|---|---|
| AACA element | AACAAAC | OsMYB5 | Regulator of endosperm-specific expression |
| ABRE | GTACGTGGCGC | Unknown | Positive regulator of GA response; Negative regulator of ABA response |
| ACGTGKC | Unknown | ABA-response element for ABA regulated gene expression | |
| GCCGCGTGGC | Unknown | ABA-response element for ABA regulated gene expression | |
| AGTACGTGGC | Unknown | ABA-response element for ABA regulated gene expression | |
| ABRE/hex-3 | GGACGCGTGGC | OsABF1 | ABA-response element for ABA regulated gene expression |
| ABRE-like | TACGTGTC | OsTRAB1 | ABA-response element for ABA regulated gene expression |
| G-box | RTACGTGGCR | OsTAF-1 | Positive regulator for response to ABA and desiccation in vegetative tissues |
| CTTCCACGTGGCA | OsBZ8 | ABA-response element for ABA regulated gene expression | |
| RY repeat | CATGCATG | Unknown | ABA-response element for ABA regulated gene expression |
| AH1 element | CAATTATTG | OsHox1 | Enhancer of provascular-specific expression |
| ARR1 element | AGATT | OsARR1 | Non-symbiotic hemoglobin-2 promoter |
| as1-like | GCATCTTTACTTTAGCATC | as1-like box binding factor | Phloem-specific expression |
| IIb element | TGTGGGACCATG | OsPCF1, OsPCF2 | Enhancer of meristem-specific expression |
| BIHD1OS element | TGTCA | OsBIHD1 | Pathogen-regulated expression |
| BP-73 element | AACGT | OsBP-73 | Regulation of cell proliferation |
| E2F element | GCGGGAAA | OsE2F | Enhancer in actively dividing cells |
| GATA box | GATA | OsASF-2 | Light regulated expression |
| GCC-box | AGCCGCC | OsEREBP1 | Pathogen-regulated expression |
| CATAAGAGCCGCCACT | OsBIERF3 | Response to biotic and abiotic stress | |
| IDE1 element | ATCAAGCATGCTTCTTGC | OsIDE1, OsIDE2 | Iron deficiency regulated expression |
| OsIRO2 element | CACGTGG | OsIRO2 | Iron deficiency regulated expression |
| PR2 element | ACGCTGCCG | OsWOX9 | Root meristem-specific expression |
| TGTCA element | TGTCA | OsBIHD1 | Pathogen-regulated expression |
| W-box | TTGAC | OsWRKY13, OsWRKY51, OsWRKY71 | Pathogen-regulated expression |
More recent generations of cis-element databases like the Database of Arabidopsis Transcription Factors (DATF), Database of Rice Transcription Factor (DRTF), Plant Transcription Factor Database (Plant TFDB), and Arabidopsis Stress Responsive Transcription Factor Database (STIFDB) have significantly improved the resolution of regulatory sequence prediction in rice genomic sequences given the broader emphasis on plant physiological processes (Guo et al., 2005; Gao et al., 2006; Riaño-Pachón et al., 2007; Shameer et al., 2009; Perez-Rodriguez et al., 2010; Wang et al., 2010; Zhang et al., 2011). DRTF in particular holds information on more than 2,000 putative transcription factors and their computationally predicted target cis-elements anchored to the reference genome sequence of rice. Other more recent additions to available databases include the Plant Transcription Factor database (PlantTFDB),GRASSIUS and Osiris (Guo et al., 2008; Morris et al., 2008; Yilmaz et al., 2009; Zhang et al., 2011).
In summary, the number of public databases that curate information about regulatory sequences and transcription factors in plants has increased substantially over the last decade. The information contained in these resources are evolving constantly and their maximum utility for high-resolution computational prediction of transcriptional networks in rice relies on systematic annotation based on experimentally validated data. What is currently necessary for more biologically meaningful analysis of regulatory sequence variation in the genus Oryza is a systematic means to integrate and synthesize the data from all resources in order to resolve inconsistencies in annotation, redundancy, and lack of uniform nomenclature.
Conclusions and perspectives
Applications of upstream sequence variation to genus-wide gene discovery, allele mining and network engineering
The fundamental question about the architecture of rice promoters and the nature of their cognate regulator proteins remains very relevant in the era of comparative functional genomics. Our recent survey of the major public eukaryotic cis-element and transcription factor databases have indicated that sequences and annotation of cis-elements and their cognate transcriptional regulators in the reference rice genome are still far from being comprehensive to guide a systematic scrutiny of regulatory networks at a genome-wide and comparative scales. The ‘finished’ reference genome sequence of rice has been available to the community now for a decade, serving as nucleator for hypothesis-driven interrogation of gene function and evolution. As we enter the next decade of reference genome-enabled experimentation in rice genetics, which has directed its focus on genotypic comparisons, allele and novel gene mining, the importance of a comprehensive annotation of cis-acting elements and trans-acting factors that define regulatory networks cannot be over-emphasized.
Creating novel combinations of superior alleles by integrative use of conventional and biotechnological methods is the overarching goal of 21st century rice breeding. The genome-enabled research paradigm in rice biology has contributed to this goal through the understanding of gene function and regulation. Moreover, at the very core of the reference genome-guided research paradigm is the quest to pinpoint useful allelic variation in the germplasm that explains simple or quantitative traits to guide targeted introgression and selection. Broadly, gene function can be described in the context of a gene’s specific molecular, biochemical and/or biological roles in the cell and in context of its role in a broader molecular or biochemical network. The upstream regulatory sequence signature of a gene is a window to the combinatorial complexity of its regulation and reflects how it is interfaced with the various intrinsic and extrinsic signals for growth, development, reproduction and adaptation (Figure 3A). In these contexts, a truly biologically meaningful annotation of the reference genomes of rice that could facilitate a holistic understanding of the nature of allelic and epiallelic variation for phenotypes of interest must include information on the critical cis-elements and trans-acting factors that define the spatio-temporal regulatory properties of each gene. Establishing a modular map of regulatory elements within the upstream regions of each gene locus as part of reference genome annotations in a similar manner that we understand protein domain architectures of coding sequences will be an important tool for various applications in comparative functional genomics both at the intraspecific and interspecific levels.
Figure 3.
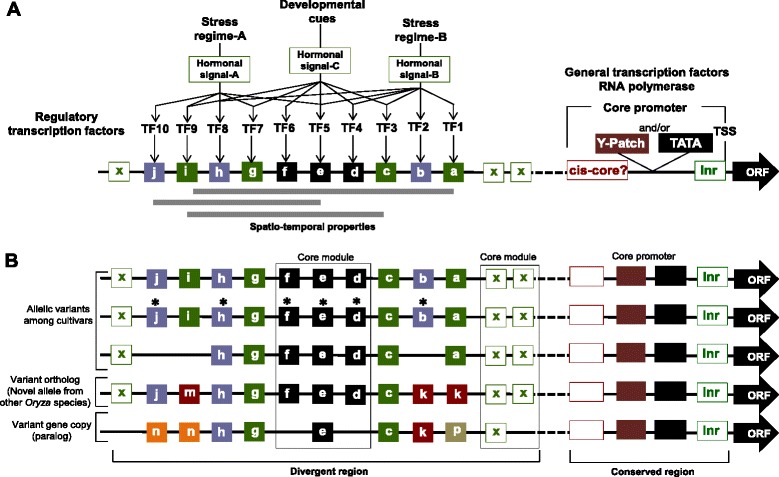
Hypothetical and simplistic models of upstream regulatory information content of spatio-temporally regulated genes of rice. (A) Prototype inducible promoter is comprised of combinations of cis-elements (i.e., colored boxes upstream from the core promoter identified by lowercase letters) that directly interface gene induction/represssion with various environmental and developmental signals. This occurs by virtue of their respective cognate regulatory transcription factors (i.e., TF1, TF2, etc.) that respond to hormonal or other physico-chemical changes in the cell. The spatio-temporal properties of genes are therefore defined by the integration of external and developmental cues through the synergistic interactions of various cis-elements and their cognate regulatory transcription factors. (B) Example of a pattern/trend that could be revealed by phylogenetic footprinting of upstream regulatory sequences of rice genes with potential significance to comparative functional genomics and allele mining. Phylogenetic footprinting has suggested that functional differences between the promoters of genes that vary in spatio-temporal regulation may be due to combinations of divergent regulatory fine-tuner elements (colored squares) and highly conserved core module elements (black squares). Variation in cis-regulatory information content may therefore reveal the genomic basis for variant alleles with unique spatio-temporal properties. This concept may be extrapolated for interspecific comparisons of orthologs and paralogs towards the discovery of potential novel alleles in the genus Oryza. Differences in expression in the absence of apparent upstream sequence variation may be used as basis for targeted analysis of differential methylation (shown with asterisks*) of homologous promoters. This concept may have potential applications for the discovery of epialleles that determine phenotypic variation.
The specific applications of upstream sequence annotation are numerous and some of the more important concepts are discussed in this review. First, in-depth understanding of the modular architectures of upstream regulatory regions for each annotated gene locus (i.e., open-reading frame) in the reference genome could nucleate direct comparison of orthologs and paralogs by phylogenetic footprinting across a meaningful germplasm diversity panel. Phylogenetic footprinting is based on the assumption that conserved sequence motifs or modular combinations of motifs within the upstream sequences of homologous genes represent functional cis-elements that define the spatio-temporal regulation of that gene (Hardison, 2000; Hong et al., 2003; Zhang and Gerstein, 2003; De Bodt et al., 2006; Freeling and Subramaniam, 2009; Xu et al., 2012a). In other words, patterns of cis-element conservation or divergence revealed by phylogenetic footprints define the integrated effects of various developmental and external signals on the expression potential of a given gene (Figure 3A). An offshoot of that concept is that upstream sequences containing critical regulatory elements can be the subject of targeted scrutiny to reveal meaningful sequence variation both at the intraspecific and interspecific levels for the purpose of uncovering conserved or divergent patterns associated with expression variability and elite phenotypes (Figure 3B). This is an area in rice comparative functional genomics that has not been actively pursued given the rapidly growing resources for comparative functional genomics for mining novel alleles. As more germplasm resequencing data become available, meaningful comparative analysis of upstream regulatory sequence variation by phylogenetic footprinting must be pursued towards the discovery of novel alleles and associated networks with potential applications in rice breeding.
An example of a seminal study that highlights the potential of phylogenetic footprinting for novel gene discovery and allele mining was a recent comparison of upstream regulatory sequence signatures across orthologous and paralogous groups of stress-associated bZIP transcription factors of rice, Arabidopsis and sorghum, three species representing more than 140 million years of evolutionary history and unique geo-climatic distribution (Xu et al., 2012a). It was revealed that while orthologs shared very similar basal developmental programming by virtue of highly conserved ‘core modules’ of cis-elements, they also exhibit unique cis-element footprints hypothesized to act as ‘regulatory fine tuners’ in conjunction with the ‘core modules’. It was inferred that differences in upstream sequence footprints contributed by the ‘regulatory fine tuners’ may serve to highlight the unique spatio-temporal characteristics of orthologs and paralogs, underscoring the potential uniqueness of each ortholog and paralog in a regulatory context hence expression variability. Indeed, the trends established from such comparison has been supported by similar patterns established based on phylogenetic footprints of upstream non-coding sequences between rice and other syntenic cereal genomes including maize, sorghum and barley, suggesting that phylogenetic footprints could be a powerful tool for revealing some of the major determinants of novel allelic variation (Guo and Moose, 2003; Wang et al., 2009).
The genus Oryza represents about 15 million years of evolutionary divergence that has been established as a ‘one-of-its-kind’ tractable comparative genomics system. Capitalizing on the ‘finished’ reference sequence of O. sativa ssp. japonica, an international consortium is currently developing a genus-wide platform for the discovery of novel genes and for mining novel alleles with potential use in genomics-enabled introgression breeding (Wing et al., 2005; Jacquemin et al., 2012). To this date, a comprehensive set of sequence scaffolds representing all diploid genomes (i.e., AA, BB, CC, EE, FF, and GG) has been developed and in various stage of sequence assembly and completion, with few already released including the African cultivated species O. glaberrima (AA) and the wild species O. barthii (AA), O. longistaminata (AA), O. nivara (AA), O. punctata (AA), O. rufipogon (AA), and O. brachyantha (FF) (Rounsley et al., 2009; Xu et al., 2012b; Chen et al., 2013; Jacquemin et al., 2014). Given this rapidly unfolding genomics resources, it would soon be possible to conduct a meaningful global and cross-genomic analysis of orthologous and paralogous upstream sequence and expression variation in Oryza. The resulting knowledge may provide an important window to the nature of allelic variation for rice genes that are regulated at the transcriptional level, and could set the stage for targeted gene introgression and allele replacement in the future.
Second, it is important to articulate that the true essence of functional genomics in the context of rice breeding must be deeply rooted to the ability to identify meaningful sequence variation across an allelic series, understanding how such sequence variation contributes to differences in gene function, and whether differences in gene function are due simply to sequence variation (alleles) or differential DNA methylation in the absence of drastic sequence variation (epialleles). More and more examples of homologous genes that vary in expression in the absence of meaningful sequence variation are being reported in the literature (Peng and Zhang, 2009; Hai et al., 2013; Zheng et al., 2013). Understanding the organization and distribution of critical regulatory element modules within the upstream sequences of intraspecific or interspecific homologs has the potential to provide another layer of information for understanding the contribution of ‘epiallelic variation’ to phenotypic variation. In this context, homologous genes that differ in expression characteristics despite the absence of any meaningful differences in upstream regulatory sequence architecture should justify examination of the contribution of differential upstream DNA methylation in the expression of the desired phenotype (Figure 3B). While the concept of epiallelic introgression to phenotypic gain is still vague and yet to be explored in rice genetics and breeding, the establishment of reference genome sequences with well annotated upstream regulatory sequences should set the stage for future large-scale association between DNA methylation patterns and upstream sequence variation. Certainly, the rapidly unfolding reference genome sequence resources across the genus Oryza will serve as foundation for the discovery of potential epiallelic series that are due to differential upstream sequence methylation.
Third, it is well established that the major transcriptional regulatory networks involved in stress response mechanisms are highly conserved or universal in flowering plants. For instance, the DREB/CBF regulon has functioning equivalents in plant species with contrasting sensitivities to low temperature such as rice (intolerant) and Arabidopsis (tolerant), or to drought such as rice (intolerant) and sorghum (tolerant). DREB/CBF regulons are also equally functional even between any two rice genotypes that differ significantly in regard to relative stress tolerance. What remains unclear is how variation in the activity of the DREB/CBF regulatory network contributes to phenotypic gradient for stress tolerance at the intraspecific and interspecific levels. One theory is that regulatory networks like the DREB/CBF regulon may contribute to phenotypic gradient by virtue of variation in size and compositional complexity of downstream genes that are controlled by the master regulator. It has been proposed that this may be dependent on the relative distribution of the target cis-elements across individual genomes (Yun et al., 2010; Zhang et al., 2004). Testing this hypothesis in rice will require a systematic survey of genome-wide distribution of bona-fide DREB/CBF target cis-element modules across a meaningful diversity panel that represents the gradient of stress sensitivity in the germplasm.
The outcome of such type of analysis could also contribute to the understanding of how regulatory networks can be reconfigured by altering the distribution of critical cis-regulatory modules by homologous recombination and genome shuffling during the process of parental intercrossing. This concept could also lead to future exploration of the potential of transcription factor and target gene cluster complementation during introgression. For instance, a novel transcription factor homolog from a donor may be combined with a much more elaborate array of downstream target genes that contain the critical cis-element in the recipient genome. The possible effect could be the further elaboration of the regulon hence novel configuration of the transcriptional network due to such novel combination of master switch and downstream target cis-elements. This may lead to physiological reconfiguration and may be explored as foundation for the creation of novel and/or transgressive phenotypes by breeding.
Finally, one major application of understanding cis-element function that has been explored since the early days of rice biotechnology is the concept of ‘designer promoters’ (Mehrotra et al., 2011). Given the growing comparative genomics resources in rice, this concept may be revisited with a slightly different set of questions. The classical concept asked the question regarding the feasibility of finely optimizing the spatio-temporal expression of a regulon that defines the expression of a complex phenotype such as drought tolerance by driving its major regulatory gene (i.e., regulatory hub) under the control of a novel and ideal chimeric promoter. Conversely in today’s context, given the emerging technology for gene editing and replacement, it may be relevant to ask the question on the feasibility of using our knowledge of cis-element architecture to alter the distribution of critical cis-elements in the genome to create a more elaborate transcriptional regulatory network and novel phenotypes. These are some of the forward-looking questions in rice regulon engineering that would require in-depth understanding of the nature and distribution cis-elements and their cognate regulators in a genome-wide scale. The rapid progress in rice genetics facilitated by reference sequence-enabled comparative genomics in Oryza should nucleate future studies addressing some of these concepts.
Acknowledgement
This study was supported by a grant to BGDR from the USDA-National Research Initiative (2006-35604-1669) and base funding from the Maine Agricultural and Forest Experiment Station (Publication 3392). BM and DYL were supported by the National Research Foundation of Singapore (NRF-CRP5-2009-03) and Next-Generation BioGreen 21 Program (PJ009520), Rural Development Administration, Republic of Korea. MRP was supported by Korea Research Foundation Postdoctoral Fellowship (KRF-2006-352-F00002). SJY was supported by a grant from the Regional Subgenebank Support Program of Rural Development Administration, Republic of Korea.
Abbreviations
- TSS
Transcription start site
- TBP
TATA-binding protein
- BRE
TFIIB recognition element
- DPE
Downstream promoter element
- IME
Intron-mediated enhancement
- IAA
Indole 3-acetic acid or auxin
- ABA
Abscisic acid
- GA
Gibberellic acid
- SRC
Sugar response complex
- JA
Jasmonic acid
- SA
Salicylic acid
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
BGDR was responsible for the overall concept, data integration and synthesis of data, and wrote the manuscript. BM and DYL contributed information and to the writing of the sections on biotic stress and databases. SJY and MRP contributed information and to the writing of the sections on constitutive promoters. All authors read and approved the final manuscript.
Contributor Information
Benildo G de los Reyes, Email: benildo.de@maine.edu.
Bijayalaxmi Mohanty, Email: chebijay@nus.edu.sg.
Song Joong Yun, Email: sjyun@jbnu.ac.kr.
Myoung-Ryoul Park, Email: mrp3711@louisiana.edu.
Dong-Yup Lee, Email: cheld@nus.edu.sg.
References
- Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D, Shinozaki K. Role of Arabidopsis MYC and MYB homologs in drought and abscisic acid regulated gene expression. Plant Cell. 1997;9:1859–1868. doi: 10.1105/tpc.9.10.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abeles FB, Morgan PW, Saltvet ME. Ethylene in Plant Biology. 2. San Diego: CA, Academic Press Inc.; 1992. [Google Scholar]
- Alvarez ME. Salicylic acid in the machinery of hypersensitive cell death and disease resistance. Plant Mol Biol. 2000;44:429–442. doi: 10.1023/A:1026561029533. [DOI] [PubMed] [Google Scholar]
- Bagnaresi P, Biselli C, Orrù L, Urso S, Crispino L, Abbruscato P, Piffanelli P, Lupotto E, Cattivelli L, Valè G. Comparative transcriptome profiling of the early response to Magnaporthe oryzae in durable resistant vs susceptible rice (Oryza sativa L.) genotypes. PLoS ONE. 2012;7(12):e51609. doi: 10.1371/journal.pone.0051609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann K, De Paolis A, Constantino P, Gualberti G. The DNA binding site of the Dof protein NtBBF1 is essential for tissue-specific and auxin-regulated expression of the rolB oncogene in plants. Plant Cell. 1999;11:323–334. doi: 10.1105/tpc.11.3.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranowskij N, Frohberg C, Prat S, Willmitzer L. A novel DNA binding protein with homology to Myb oncoproteins containing only one repeat can function as a transcriptional activator. EMBO J. 1994;13:5383–5392. doi: 10.1002/j.1460-2075.1994.tb06873.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya J, Chowdhury AH, Ray S, Jha JK, Das S, Gayen S, Chakraborty A, Mitra J, Maiti MK, Basu A, Sen SK. Native polyubiquitin promoter of rice provides increased constitutive expression in stable transgenic rice plants. Plant Cell Rep. 2012;31:271–279. doi: 10.1007/s00299-011-1161-4. [DOI] [PubMed] [Google Scholar]
- Bodenhausen N, Reymond P. Signaling pathways controlling induced resistance to insect herbivores in Arabidopsis. Mol Plant-Micr Interact. 2007;20:1406–1420. doi: 10.1094/MPMI-20-11-1406. [DOI] [PubMed] [Google Scholar]
- De Bodt S, Thiessen G, Van de Peer Y. Promoter analysis of MADS-box genes in eudicots through phylogenetic footprinting. Mol Biol Evol. 2006;23:1293–1303. doi: 10.1093/molbev/msk016. [DOI] [PubMed] [Google Scholar]
- Bradnam KR, Korf I. Longer first introns are a general property of eukaryotic gene structure. PLoSOne. 2008;3(8):e3093. doi: 10.1371/journal.pone.0003093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RL, Kazan K, McGrath KC, Maclean DJ, Manners JM. A role for the GCC-box in jasmonate-mediated activation of the PDF1.2 gene of Arabidopsis. Plant Physiol. 2003;132:1020–1032. doi: 10.1104/pp.102.017814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan-Wolaston V, Earl S, Harrison E, Mathas E, Navabpour S, Page T, Pink D. The molecular analysis of leaf senescence- A genomics approach. Plant Biotech J. 2003;1:3–22. doi: 10.1046/j.1467-7652.2003.00004.x. [DOI] [PubMed] [Google Scholar]
- Buchanan-Wollaston V. The molecular biology of leaf senescence. J Exptl Bot. 1997;48:181–199. doi: 10.1093/jxb/48.2.181. [DOI] [Google Scholar]
- Buchler NE, Gerland U, Hwa T. On schemes of combinatorial transcription logic. Proc Natl Acad Sci U S A. 2003;100:5136–5141. doi: 10.1073/pnas.0930314100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke TW, Kadonaga JT. The downstream core promoter element DPE is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes Dev. 1997;11:3020–3031. doi: 10.1101/gad.11.22.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler JEF, Kadonaga JT. Enhancer-promoter specificity mediated by DPE or TATA core promoter motifs. Genes Dev. 2001;15:2515–2519. doi: 10.1101/gad.924301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai M, Wei J, Li X, Caiguo X, Wang S. A rice promoter containing both novel positive and negative cis-elements for regulation of green tissue-specific gene expression in transgenic plants. Plant Biotech J. 2007;5:664–674. doi: 10.1111/j.1467-7652.2007.00271.x. [DOI] [PubMed] [Google Scholar]
- Chandlee JM. Current molecular understanding of the genetically programmed process of leaf senescence. Physiol Plant. 2001;113:1–8. doi: 10.1034/j.1399-3054.2001.1130101.x. [DOI] [Google Scholar]
- Chapman EJ, Estelle M. Mechanism of auxin-regulated gene expression in plants. Annu Rev Genet. 2009;43:265–285. doi: 10.1146/annurev-genet-102108-134148. [DOI] [PubMed] [Google Scholar]
- Chen J, Huang Q, Gao D, Wang J, Lang Y, Liu T, Li B, Bai Z, Goicoechea JL, Liang C, Chen C, Zhang W, Sun S, Liao Y, Zhang X, Yang L, Song C, Wang M, Shi J, Liu G, Liu J, Zhou H, Zhou W, Yu Q, An N, Chen Y, Cai Q, Wang B, Liu B, Min J, Huang Y, Wu H, Li Z, Zhang Y, Yin Y, Song W, Jiang J, Jackson S, Wing R, Wang J, Chen M, et al. Whole-genome sequencing of Oryza brachyantha reveals mechanisms underlying Oryza genome evolution. Nature Comm. 2013;4:1595. doi: 10.1038/ncomms2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PW, Lu CA, Yu TS, Tseng TH, Wang CS, Yu SM. Rice α-amylase transcriptipnal enhancers direct multiple mode regulation of promoters in transgenic rice. J Biol Chem. 2002;277:13641–13649. doi: 10.1074/jbc.M109722200. [DOI] [PubMed] [Google Scholar]
- Chen PW, Chiang CM, Tseng TH, Yu SM. Interaction between rice MYBGA and the gibberellin response element controls tissue-specific sugar sensitivity of a-amylase genes. Plant Cell. 2006;18:2326–2340. doi: 10.1105/tpc.105.038844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, Yun KY, Ressom H, Mohanty B, Bajic VB, Jia Y, Yun SJ, De los Reyes BG. An early response regulatory cluster induced by low temperature and hydrogen peroxide in seedlings of chilling-tolerant japonica rice. BMC Genomics. 2007;8:e175. doi: 10.1186/1471-2164-8-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Ohta M, Kanrar S, Lee B, Hong X, Agarwal M, Zhu JK. ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003;17:1043–1054. doi: 10.1101/gad.1077503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi DW, Rodriguez EM, Close TM. Barley Cbf3 gene identification, expression pattern and map location. Plant Physiol. 2002;129:1781–1787. doi: 10.1104/pp.003046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen AH, Quail PH. Ubiquitin promoter-based vectors for high levels of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Res. 1996;5:213–218. doi: 10.1007/BF01969712. [DOI] [PubMed] [Google Scholar]
- Ciolkowski I, Wanke D, Birkenbihl RP, Somssich IE. Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function. Plant Mol Biol. 2008;68:81–92. doi: 10.1007/s11103-008-9353-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy M, Vasil V, Hannah LC, Vasil IK. Maize shrunken-1 intron and exon regions increase gene expression in maize protoplasts. Plant Sci. 1994;98:151–161. doi: 10.1016/0168-9452(94)90005-1. [DOI] [Google Scholar]
- Clivan P, Svec M. Genome-wide analysis of rice (O. sativa ssp. japonica) TATA box and Y-patch promoter elements. Genome. 2009;52:294–297. doi: 10.1139/G09-001. [DOI] [PubMed] [Google Scholar]
- Creelman RA, Mullet JE. Jasmonic acid distribution and action in plants: Regulation during development and response to biotic and abiotic stress. Proc Natl Acad Sci U S A. 1995;92:4114–4119. doi: 10.1073/pnas.92.10.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croissant-Sych Y, Okita T. Identification of positive and negative regulatory cis-elements of the rice glutelin Gt3 promoter. Plant Sci. 1996;116:27–35. doi: 10.1016/0168-9452(96)04360-9. [DOI] [Google Scholar]
- Darmasiri N, Darmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435:441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- Davuluri RV, Sun H, Palaniswamy SK, Matthews N, Molina C, Kurtz M, Grotewold E. AGRIS: Arabidopsis Gene Regulatory Information Server, an information resource of Arabidopsis cis-regulatory elements and transcription factors. BMC Bioinfor. 2003;4:e25. doi: 10.1186/1471-2105-4-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey DA, Klessig DF. SOS - too many signals for systemic acquired resistance? Trends Plant Sci. 2012;17:538–545. doi: 10.1016/j.tplants.2012.05.011. [DOI] [PubMed] [Google Scholar]
- Desveaux D, Subramaniam R, Despres C. A ‘Whirly’ transcription factor is required for salicylic acid-dependent disease resistance in Arabidopsis. Dev Cell. 2004;6:229–240. doi: 10.1016/S1534-5807(04)00028-0. [DOI] [PubMed] [Google Scholar]
- Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, Miura S, Seki M, Shinozaki K, Yamaguchi-Shinozaki Y. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high salt-, and cold-responsive gene expression. Plant J. 2003;33:751–763. doi: 10.1046/j.1365-313X.2003.01661.x. [DOI] [PubMed] [Google Scholar]
- Farmer EE, Alméras E, Krishnamurthy V. Jasmonates and related oxylipins in plant responses to pathogenesis and herbivory. Curr Opin Plant Biol. 2003;6:372–378. doi: 10.1016/S1369-5266(03)00045-1. [DOI] [PubMed] [Google Scholar]
- Finkelstein R. Studies of abscisic acid perception finally flower. Plant Cell. 2006;18:786–791. doi: 10.1105/tpc.106.041129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleet CM, Sun T. A DELLAcate balance: The role of gibberellins in plant morphogenesis. Curr Opin Plant Biol. 2005;8:77–85. doi: 10.1016/j.pbi.2004.11.015. [DOI] [PubMed] [Google Scholar]
- Fowler S, Thomashow MF. Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell. 2002;14:1675–1690. doi: 10.1105/tpc.003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M, Subramaniam S. Conserved non-coding sequences (CNSs) in higher plants. Cur Opin Plant Biol. 2009;12:126–132. doi: 10.1016/j.pbi.2009.01.005. [DOI] [PubMed] [Google Scholar]
- Fujimoto SY, Ohta M, Usui A, Shinshi H, Ohme-Takagi M. Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell. 2000;12:393–404. doi: 10.1105/tpc.12.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan S, Amasino RM. Making sense of senescence. Plant Physiol. 1997;133:313–319. doi: 10.1104/pp.113.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G, Zhong Y, Guo A, Zhu Q, Tang W, Zheng W, Gu X, Wei L, Luo J. DRTF: A database of rice transcription factors. Bioinformatics. 2006;22(10):1286–1287. doi: 10.1093/bioinformatics/btl107. [DOI] [PubMed] [Google Scholar]
- Garcion C, Métraux JP. Salicylic acid. Plant Hormone Signal. 2006;24:229–255. [Google Scholar]
- Gilmour SJ, Sebolt AM, Salazar MP, Everard JD, Thomashow MF. Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol. 2000;124:1854–1865. doi: 10.1104/pp.124.4.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudat J, Parcy F, Bertandre N, Gosti F, Leung J, Morris PC, Bouvier-Durand M, Vartanian N. Current advances in abscisic acid action and signaling. Plant Mol Biol. 1994;26:1557–1577. doi: 10.1007/BF00016490. [DOI] [PubMed] [Google Scholar]
- Gomez-Porras JL, Riano-Pachon DM, Dreyer I, Mayer JE, Mueller-Roeber B. Genome-wide analysis of ABA-responsive elements ABRE and CE3 reveals divergent patterns in Arabidopsis and rice. BMC Genomics. 2007;8:260. doi: 10.1186/1471-2164-8-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomi K, Sasaki A, Itoh H, Ueguchi-Tanaka M, Ashikari M, Kitano H, Matsuoka M. GID2, an F-box subunit of the SCF E3 complex, specifically interacts with phosphorylated SLR1 protein and regulates the gibberellin-dependent degradation of SLR1 in rice. Plant J. 2004;37:626–634. doi: 10.1111/j.1365-313X.2003.01990.x. [DOI] [PubMed] [Google Scholar]
- Grierson C, Du JS, De Torres ZM, Beggs K, Smith C, Holdsworth M, Bevan M. Separate cis sequences and trans factors direct metabolic and developmental regulation of a potato tuber storage protein gene. Plant J. 1994;5:815–826. doi: 10.1046/j.1365-313X.1994.5060815.x. [DOI] [PubMed] [Google Scholar]
- Griffiths J, Murase K, Rieu I, Zentella R, Zhang ZL, Powers SJ, Gong F, Philips AL, Hedden P, Sun TP, Thomas SG. Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell. 2006;18:3399–3414. doi: 10.1105/tpc.106.047415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Yang C, Thara YK, Zhou J, Martin GB. Pti4 is induced by ethylene and salicylic acid, and its product is phosphorylated by the Pto kinase. Plant Cell. 2000;12:771–786. doi: 10.1105/tpc.12.5.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guedes Correa LG, Riano-Pachon DM, Schrago CG, Vincentini dos Santos R, Mueller-Roeber B, Vincents M. The role of bZIP transcription factors in green plants evolution: Adaptive features emerging from four founder genes. PLoSOne. 2008;8:e2994. doi: 10.1371/journal.pone.0002944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiltinan MJ, Marcotte WR, Jr, Quatrano RS. A plant leucine zipper protein that recognizes and abscisic acid response element. Science. 1990;250:267–271. doi: 10.1126/science.2145628. [DOI] [PubMed] [Google Scholar]
- Guo AY, Chen X, Gao G, Zhang H, Zhu QH, Liu XC, Zhong YF, Gu X, He K, Luo J. PlantTFDB: a comprehensive plant transcription factor database. Nucleic Acids Res. 2008;36:D966–D969. doi: 10.1093/nar/gkm841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo A, He K, Liu D, Bai S, Gu X, Wei L, Luo J. DATF: A Database of Arabidopsis Transcription Factors. Bioinformatics. 2005;21:2568–2569. doi: 10.1093/bioinformatics/bti334. [DOI] [PubMed] [Google Scholar]
- Guo H, Moose SP. Conserved noncoding sequences among cultivated cereal genomes indentify candidate regulatory sequence elements and patterns of promoter evolution. Plant Cell. 2003;15:1143–1158. doi: 10.1105/tpc.010181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen G, Guilfoyle T. Auxin-responsive gene expression: Genes, promoters and regulatory factors. Plant Mol Biol. 2002;49:373–385. doi: 10.1023/A:1015207114117. [DOI] [PubMed] [Google Scholar]
- Hai P, Guanghai J, Jing Z, Weixiong Z, WenXue Z. DNA methylation polymorphism and stability in Chinese indica hybrid rice. Sci China Life Sci. 2013;56:1097–1106. doi: 10.1007/s11427-013-4576-z. [DOI] [PubMed] [Google Scholar]
- Hamada K, Hongo K, Suwabe K, Shimizu A, Nagayama T, Abe R, Kikuchi S, Yamamoto N, Fujii T, Yokoyama K, Tsuchida H, Sano K, Mochizuki T, Oki N, Horiuchi Y, Fujita M, Watanabe M, Matsuokoa M, Kurata N, Yano K. OryzaExpress: An integrated database of gene expression networks and omics annotations in rice. Plant Cell Physiol. 2011;52:220–229. doi: 10.1093/pcp/pcq195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao D, Ohme-Takagi M, Sarai A. Unique mode of GCC-box recognition by the DNA-binding domain of ethylene-responsive element-binding factor (ERF domain) in plant. J Biol Chem. 1998;273:26857–26861. doi: 10.1074/jbc.273.41.26857. [DOI] [PubMed] [Google Scholar]
- Hardison RC. Conserved noncoding sequences are reliable guides to regulatory elements. Trends Genet. 2000;16:369–372. doi: 10.1016/S0168-9525(00)02081-3. [DOI] [PubMed] [Google Scholar]
- Hart CM, Nagy F, Meins F., Jr A 61-bp enhancer element of the tobacco beta-1,3-glucanase B gene interacts with one or more regulated nuclear proteins. Plant Mol Biol. 1993;21:121–131. doi: 10.1007/BF00039623. [DOI] [PubMed] [Google Scholar]
- Hartweck LM. Gibberellin signaling. Planta. 2008;229:1–13. doi: 10.1007/s00425-008-0830-1. [DOI] [PubMed] [Google Scholar]
- Hartweck LM, Olszewski NE. Rice gibberellin insensitive dwarf1 is a gibberellin receptor that illuminates and raises questions about GA signaling. Plant Cell. 2006;18:278–282. doi: 10.1105/tpc.105.039958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Lin Z, McElroy D, Wu R. Identification of a rice Actin2 gene regulatory region for high-level expression of transgenes in monocots. Plant Biotech J. 2009;7:227–239. doi: 10.1111/j.1467-7652.2008.00393.x. [DOI] [PubMed] [Google Scholar]
- Helliwell EE, Yang Y. Molecular strategies to improve rice disease resistance. Methods Molec Biol. 2013;956:285–309. doi: 10.1007/978-1-62703-194-3_21. [DOI] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database. Nucl Acids Res. 1999;27:297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobo T, Kowyama Y, Hatori Y. A bZIP factor TRAB1 interacts with VP1 and mediates abscisic acid induced transcription. Proc Natl Acad Sci USA. 1999;96:15348–15353. doi: 10.1073/pnas.96.26.15348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong RL, Hamaguchi L, Busch MA, Weigel D. Regulatory elements of the floral homeotic gene AGAMOUS identified by phylogenetic footprinting and shadowing. Plant Cell. 2003;15:1296–1309. doi: 10.1105/tpc.009548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, You J, Fang Y, Zhu X, Xiong L. Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Mol Biol. 2008;67:169–181. doi: 10.1007/s11103-008-9309-5. [DOI] [PubMed] [Google Scholar]
- Hwang SH, Lee IA, Yie SW, Hwang DJ. Identification of an OsPR10a promoter region responsive to salicylic acid. Planta. 2008;227:1141–1150. doi: 10.1007/s00425-007-0687-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inukai Y, Sakamoto T, Ueguchi-Tanaka M, Shibata Y, Gomi K, Umemura I, Hasegawa Y, Ashikari M, Kitano H, Matsuoka M. Crown rootless1, which is essential for crown root formation in rice, is a target of an auxin response factor in auxin signaling. Plant Cell. 2005;17:1387–1396. doi: 10.1105/tpc.105.030981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda A, Ueguchi-Tanaka M, Sonoda Y, Kitano H, Koshioka M, Futshuhara Y, Matsuoka M, Yamaguchi J. Slender rice, a constitutive gibberellins response mutant is caused by a null mutation in the SLR1 gene, an orthologue of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell. 2001;13:999–1010. doi: 10.1105/tpc.13.5.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro S, Nakamura K. Characterization of a cDNA encoding a novel DNA-binding protein SPF1 that recognizes SP8 sequences in the 5’ upstream regions of genes coding for sporamin and beta-amylase from sweet potato. Mol Gen Genet. 1994;244:563–571. doi: 10.1007/BF00282746. [DOI] [PubMed] [Google Scholar]
- Itoh H, Sasaki A, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Hasegwa Y, Minami E, Ashikari M, Matsuoka M. Dissection of the phosphorylation of rice DELLA protein SLENDER RICE1. Plant Cell Physiol. 2005;46:1392–1399. doi: 10.1093/pcp/pci152. [DOI] [PubMed] [Google Scholar]
- Jacquemin J, Bhatia D, Singh K, Wing RA. The International Oryza Map Alignment Project: Development of a genus-wide comparative genomics platform to help solve the 9 billion-people question. Curr Opin Plant Biol. 2012;16:147–156. doi: 10.1016/j.pbi.2013.02.014. [DOI] [PubMed] [Google Scholar]
- Jacquemin J, Ammiraju SS, Haberer G, Billheimer DD, Yu Y, Liu LC, Rivera LF, Mayer K, Chen M, Wing RA. Fifteen million years of evolution in the Oryza genus shows extensive gene family expansion. Molec Plant. 2014;7(4):642–656. doi: 10.1093/mp/sst149. [DOI] [PubMed] [Google Scholar]
- Jaglo KR, Kleff S, Amundsen KL, Zhang X, Haake V, Zhang JZ, Deits T, Thomashow MF. Components of the Arabidopsis C-repeat/dehydration-responsive element binding factor cold-response pathway are conserved in Brassica napus and other plant species. Plant Physiol. 2001;127:910–917. doi: 10.1104/pp.010548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaglo-Ottosen KR, Gilmour SJ, Zarka DG, Schabenberger O, Thomashow MF. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science. 1998;280:104–106. doi: 10.1126/science.280.5360.104. [DOI] [PubMed] [Google Scholar]
- Johansson A, Staal J, Dixelius C. Early responses in the Arabidopsis-Verticillium longisporum pathosystem are dependent on NDR1, JA- and ET-associated signaling via cytosolic NPR1 and RFO1 Mol. Plant Microbe Interact. 2006;19:958–969. doi: 10.1094/MPMI-19-0958. [DOI] [PubMed] [Google Scholar]
- Jain M, Kaur N, Tyagi AK, Khurana JP. The auxin-responsive GH3 gene family in rice (Oryza sativa) Funct Integr Genomics. 2006;6:36–46. doi: 10.1007/s10142-005-0142-5. [DOI] [PubMed] [Google Scholar]
- Jain M, Tyagi AK, Khurana JP. Genome-wide analysis, evolutionary expansion, and expression of early auxin-responsive SAUR gene family in rice (Oryza sativa) Genomics. 2006;88:360–371. doi: 10.1016/j.ygeno.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Jain M, Kaur N, Garg R, Thakur JK, Tyagi AK, Khurana JP. Strcuture and expression analysis of early auxin-responsive Aux/IAA gene family in rice (Oryza sativa) Funct Integr Genomics. 2006;6:47–59. doi: 10.1007/s10142-005-0005-0. [DOI] [PubMed] [Google Scholar]
- Jakoby M, Weisshaar B, Droge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002;7:106–111. doi: 10.1016/S1360-1385(01)02223-3. [DOI] [PubMed] [Google Scholar]
- Jang IC, Choi WB, Lee KH, Song SI, Nahm BH, Kim JK. High-level and ubiquitous expression of the rice cytochrome c gene OsCc1 and its promoter activity in transgenic plants provides a useful promoter for transgenesis of monocots. Plant Physiol. 2002;129:1473–1481. doi: 10.1104/pp.002261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong YM, Mun JH, Lee I, Woo JC, Hong CB, Kim SG. Distinct roles of the first introns on the expression of Arabidopsis profilin gene family members. Plant Physiol. 2006;140:196–209. doi: 10.1104/pp.105.071316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juven-Gershon T, Kadonaga JT. Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev Biol. 2010;339:225–229. doi: 10.1016/j.ydbio.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko M, Itoh H, Euguchi-Tanaka M, Ashikari M, Matsuoka M. The α-amylase induction in endosperm during rice seed germination is caused by gibberellin synthesized in epithelium. Plant Physiol. 2002;128:1264–1270. doi: 10.1104/pp.010785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga M, Kiu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Improving plant drought, salt and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nature Biotech. 1999;17:287–291. doi: 10.1038/7036. [DOI] [PubMed] [Google Scholar]
- Kaufmann K, Pajoro A, Angenent GC. Regulation of transcription in plants: Mechanisms controlling developmental switches. Nature Rev Genet. 2010;11:830–842. doi: 10.1038/nrg2885. [DOI] [PubMed] [Google Scholar]
- Kieffer M, Neve J, Kepinski S. Defining auxin response contexts in plant development. Cur Opin Plant Biol. 2010;13:12020. doi: 10.1016/j.pbi.2009.10.006. [DOI] [PubMed] [Google Scholar]
- Kim SY, Wu R. Multiple protein factors bind to a rice glutelin promoter region. Nucl Acids Res. 1990;18:6845–6852. doi: 10.1093/nar/18.23.6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitomi Y, Ito H, Hobo T, Aya K, Kitano H, Inikai Y. The auxin responsive AP2/ERF transcription actor CROWN ROOTLESS5 is involved in crown root initiation in rice through the induction of OsRR1, a type-A response regulator of cytokinin signaling. Plant J. 2011;67:472–484. doi: 10.1111/j.1365-313X.2011.04610.x. [DOI] [PubMed] [Google Scholar]
- Koo AJK, Howe GA. The wound hormone jasmonate. Phytochem. 2009;70:1571–1580. doi: 10.1016/j.phytochem.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenhard B, Sandelin A, Carninci P. Metazoan promoters: Emerging characteristics and insights into transcriptional regulation. Nature Rev Genet. 2012;13:233–245. doi: 10.1038/nrg3163. [DOI] [PubMed] [Google Scholar]
- Lenka SK, Lohia B, Kumar A, Chinnusamy V, Bansal KC. Genome-wide targeted prediction of ABA responsive genes in rice based on over-represented cis-motif in co-expressed genes. Plant Molec Biol. 2009;69:261–271. doi: 10.1007/s11103-008-9423-4. [DOI] [PubMed] [Google Scholar]
- Lescot M, Déhais P, Thijs G, Marchal M, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim PO, Kim HJ, Nam HG. Leaf senescence. Ann Rev Plant Biol. 2007;58:115–136. doi: 10.1146/annurev.arplant.57.032905.105316. [DOI] [PubMed] [Google Scholar]
- Li J, Brader G, Palva ET. The WRKY70 transcription factor: a node of convergence for jasmonate mediated and salicylate-mediated signals in plant defense. Plant Cell. 2004;16:319–331. doi: 10.1105/tpc.016980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Wang S, Yu X, Yu J, He X, Zhang S, Wu P. ARL1, a LOB domain protein required for adventitious root formation in rice. Plant J. 2005;43:47–56. doi: 10.1111/j.1365-313X.2005.02434.x. [DOI] [PubMed] [Google Scholar]
- Liu XQ, Bai XQ, Qian Q, Wang XJ, Chen MS, Chu CC. OsWRKY03, a rice transcriptional activator that functions in defense signaling pathway upstream of OsNPR1. Cell Res. 2005;15:593–603. doi: 10.1038/sj.cr.7290329. [DOI] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low temperature-responsive gene expression, respectively in Arabidopsis. Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Zhou Y, Szczerba MW, Li S, Lin Y. Identification and application of a rice senescence-associated promoter. Plant Physiol. 2010;153:1239–1249. doi: 10.1104/pp.110.157123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Chen X, Liu J, Ye J, Guo Z. The rice ERF transcription factor OsERF922 negatively regulates resistance to Magnaporthe oryzae and salt tolerance. J Exp Bot. 2012;63:3899–3911. doi: 10.1093/jxb/ers079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovegrove A, Hooley R. Gibberellin and abscisic acid signaling in aleurone. Trends Plant Sci. 2000;5:102–110. doi: 10.1016/S1360-1385(00)01571-5. [DOI] [PubMed] [Google Scholar]
- Lu J, Ju HP, Zhou GX, Zhu CS, Erb M, Wang XP, Wang P, Lou YG. An EAR-motif-containing ERF transcription factor affects herbivore-induced signaling, defense and resistance in rice. Plant J. 2011;68:583–596. doi: 10.1111/j.1365-313X.2011.04709.x. [DOI] [PubMed] [Google Scholar]
- Lu CA, Lim EK, Yu SM. Sugar response sequence in the promoter of a rice a-amylase gene serves as a transcriptional enhancer. J Biol Chem. 1998;273:10120–10131. doi: 10.1074/jbc.273.17.10120. [DOI] [PubMed] [Google Scholar]
- Manners JM, Penninckx IAMA, Vermaere K, Kazan K, Brown RL, Morgan A, Maclean DJ, Curtis MD, Cammue BPA, Broekaert WF. The promoter of the plant defensin gene PDF1.2 from Arabidopsis is systemically activated by fungal pathogens and responds to methyl jasmonate but not to salicylic acid. Plant Mol Biol. 1998;38:1071–1080. doi: 10.1023/A:1006070413843. [DOI] [PubMed] [Google Scholar]
- Matys V, Fricke E, Geffers R, GoBling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, Kloos DU, Land S, Lewicki-Potapov B, Michael H, Munch R, Reuter I, Rotert S, Saxel H, Scheer M, Thiele S, Wingender E. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucl Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElroy D, Blowers AD, Jenes B, Wu R. Construction of expression vectors based on the rice actin1 (Act1) 5’ region for use in monocot transformation. Mol Gen Genet. 1991;231:150–160. doi: 10.1007/BF00293832. [DOI] [PubMed] [Google Scholar]
- McElroy D, Zhang W, Wu R. Isolation of an efficient actin promoter for in rice transformation. Plant Cell. 1990;2:163–171. doi: 10.1105/tpc.2.2.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrotra R, Gupta G, Sethi R, Bhalothia P, Kumar N, Mehrotra S. Designer promoter: An artwork of cis engineering. Plant Mol Biol. 2011;75:527–536. doi: 10.1007/s11103-011-9755-3. [DOI] [PubMed] [Google Scholar]
- Meier S, Gehring C, MacPherson CR, Kaur M, Maqungo M, Reuben S, Mayunga S, Shih MD, Wei FJ, Wanchana S, Mauleon R, Radovanovic A, Bruskiewich R, Tanaka T, Mohanty B, Itoh T, Wing R, Gojobori T, Sasaki T, Swarup S, Hsing Y, Bajic VB. The promoter signatures in rice LEA genes can be used to build a co-expressing LEA gene network. Rice. 2008;1:177–187. doi: 10.1007/s12284-008-9017-4. [DOI] [Google Scholar]
- Mejia-Guerra MK, Pomeranz M, Morohashi K, Grotewold E. From plant gene regulatory grids to network dynamics. Biochim Biophys Acta. 2012;1819:454–465. doi: 10.1016/j.bbagrm.2012.02.016. [DOI] [PubMed] [Google Scholar]
- Miyamoto K, Shimizu T, Lin F, Sainsbury F, Thuenemann E, Lomonossoff G, Nojiri H, Yamane H, Okada K. Identification of an E-box motif responsible for the expression of jasmonic acid-induced chitinase gene OsChia4a in rice. J Plant Physiol. 2012;169:621–627. doi: 10.1016/j.jplph.2011.12.008. [DOI] [PubMed] [Google Scholar]
- Moabbi AM, Agarwal N, Kaderi BE, Ansari A. Role for gene looping in intron-mediated enhancement of transcription. Proc Natl Acad Sci U S A. 2011;109:8505–8510. doi: 10.1073/pnas.1112400109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty B, Hearth V, Wijaya E, Yeo HC, De los Reyes BG, Lee DY. Patterns of cis-element enrichment revealed potential regulatory modules involved in the transcriptional regulation of anoxia response in japonica rice. Gene. 2012;511:235–242. doi: 10.1016/j.gene.2012.09.048. [DOI] [PubMed] [Google Scholar]
- Molina C, Grotewold E. Genome-wide analysis of Arabidopsis core promoters. BMC Genomics. 2005;6:25e. doi: 10.1186/1471-2164-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris RT, O’Connor TR, Wyrick JJ. Osiris: an integrated promoter database for Oryza sativa L. Bioinformatics. 2008;24:2915–2917. doi: 10.1093/bioinformatics/btn537. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Tran LSP, Van Nguyen D, Fujita M, Maruyama K, Todaka D, Ito Y, Hayashi N, Shinozaki K, Yamaguchi-Shinozaki K. Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 2007;51:617–630. doi: 10.1111/j.1365-313X.2007.03168.x. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Ito Y, Yamaguchi-Shinozaki K. Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol. 2009;149:88–95. doi: 10.1104/pp.108.129791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam HG. The molecular genetic analysis of leaf senescence. Curr Opin Biotech. 1997;8:200–207. doi: 10.1016/S0958-1669(97)80103-6. [DOI] [PubMed] [Google Scholar]
- Narusaka Y, Nakashima K, Shinwari ZK, Sakuma Y, Furihata T, Abe H, Narusaka M, Shinozaki K, Yamaguchi-Shinozaki K. Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J. 2003;34:137–144. doi: 10.1046/j.1365-313X.2003.01708.x. [DOI] [PubMed] [Google Scholar]
- Nemhauser JL, Mockler TC, Chory J. Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol. 2004;2:e258. doi: 10.1371/journal.pbio.0020258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa Y, Kawakami A, Hibi T, He DY, Shibuya N, Minami E. Regulation of the chitinase gene expression in suspension cultured rice cells by N-acetylchitooligosaccharides: differences in the signal transduction pathways leading to the activation of elicitor-responsive genes. Plant Mol Biol. 1999;39:907–914. doi: 10.1023/A:1006161802334. [DOI] [PubMed] [Google Scholar]
- Noh YS, Amasino RM. Identification of a promoter region responsible for the senescence-specific expression of SAG12. Plant Molec Biol. 1999;41:181–194. doi: 10.1023/A:1006342412688. [DOI] [PubMed] [Google Scholar]
- Ogawa M, Hanada A, Yamauchi Y, Kuwahara A, Kamiya Y, Yamaguchi S. Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell. 2003;15:1591–1604. doi: 10.1105/tpc.011650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohme-Takagi M, Shinshi H. Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell. 1995;7:173–182. doi: 10.1105/tpc.7.2.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohme-Takagi M, Suzuki K, Shinshi H. Regulation of ethylene-induced transcription of defence genes. Plant and Cell Physiol. 2000;41:1187–1192. doi: 10.1093/pcp/pcd057. [DOI] [PubMed] [Google Scholar]
- Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M. Repression domains of class II ERF transcriptional repressor share an essential motif for active repression. Plant Cell. 2001;13:1959–1968. doi: 10.1105/tpc.13.8.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyanagi H, Tanaka T, Sakai H, Shigemoto Y, Yamaguchi K, Habara T, Fujii Y, Antonio BA, Nagamura Y, Imanishi T, Ikeo K, Itoh T, Gajobori T, Sasaki T. The rice annotation project (RAP-DB) Hub for Oryza sativa ssp. japonica genome information. Nucl Acids Res. 2006;34:D741–D744. doi: 10.1093/nar/gkj094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita TW, Whang YS, Huilo J, Kim WT, Aryan AP, Larsen R, Krishnan HB. Structure and expression of the rice glutelin multigene family. J Biol Chem. 1989;264:12573–12581. [PubMed] [Google Scholar]
- Parra G, Bradnam K, Rose AB, Korf I. Comparative and functional analysis of intron-mediated enhancement signals reveals conserved features among plants. Nucl Acids Res. 2011;39:5328–5337. doi: 10.1093/nar/gkr043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, Yi N, Kim YS, Jeong MH, Bang SW, Choi YD, Kim JK. Analysis of five novel putative constitutive gene promoters in transgenic rice plants. J Exptl Bot. 2010;61:2459–2467. doi: 10.1093/jxb/erq076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MR, Yun KY, Herath V, Mohanty B, Xu F, Bajic VB, Yun SJ, De los Reyes BG. Supra-optimal expression of the cold-regulated OsMyb4 transcription factor in transgenic rice changes the complexity of transcriptional network with major effects on stress tolerance and panicle development. Plant Cell Environ. 2010;33:2209–2230. doi: 10.1111/j.1365-3040.2010.02221.x. [DOI] [PubMed] [Google Scholar]
- Peng H, Zhang J. Plant genomic DNA methylation in response to stresses: Potential applications and challenges in plant breeding. Prog Nat Sci. 2009;19:1037–1045. doi: 10.1016/j.pnsc.2008.10.014. [DOI] [Google Scholar]
- Peng X, Hu Y, Tang X, Zhou P, Deng X, Wang H, Guo Z. Constitutive expression of rice WRKY30 gene increases the endogenous jasmonic acid accumulation, PR gene expression and resistance to fungal pathogens in rice. Planta. 2012;236:1485–1498. doi: 10.1007/s00425-012-1698-7. [DOI] [PubMed] [Google Scholar]
- Perez-Rodriguez P, Riano-Pachon DM, Correa LG, Rensing SA, Kersten B, Mueller-Roeber B. PlnTFDB: Updated content and new features of the plant transcription factor database. Nucl Acids Res. 2010;38:D822–D827. doi: 10.1093/nar/gkp805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse CMJ, Van Wees SCM, Van Pelt JA, Knoester M, Laan R, Gerrits H, Weisbeek PJ, Van Loon LC. A novel signalling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell. 1998;10:1571–1580. doi: 10.1105/tpc.10.9.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu LQ, Xing YP, Liu W, Xu XP, Song YR. Expression pattern and activity of six glutelin gene promoters in transgenic rice. J Exptl Bot. 2008;59:2417–2424. doi: 10.1093/jxb/ern110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quint M, Gray WM. Auxin signaling. Cur Opin Plant Biol. 2006;9:448–453. doi: 10.1016/j.pbi.2006.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabbani MA, Maruyama K, Abe H, Khan MA, Katsura K, Ito Y, Yoshiwara K, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Monitoring expression profiles of rice genes under cold, drought, and high salinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analyses. Plant Physiol. 2003;133:1755–1767. doi: 10.1104/pp.103.025742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razem FA, El Kereamy A, Abrams SR, Hill RD. The RNA-binding protein FCA is an abscisic acid receptor. Nature. 2006;439:290–294. doi: 10.1038/nature04373. [DOI] [PubMed] [Google Scholar]
- Riaño-Pachón DM, Ruzicic S, Dreyer I, Mueller-Roeber B. PlnTFDB: An integrative plant transcription factor database. BMC Bioinformatics. 2007;8:e42. doi: 10.1186/1471-2105-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robatzek S, Somssick IE. Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev. 2002;16:1139–1149. doi: 10.1101/gad.222702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo E, Leon J, Sanchez-Serrano JJ. Cross-talk between wound signaling pathways determines local versus systemic gene expression in Arabidopsis thaliana. Plant J. 1999;20:135–142. doi: 10.1046/j.1365-313x.1999.00570.x. [DOI] [PubMed] [Google Scholar]
- Rose AB, Emami S, Bradnam K, Korf I. Evidence for a DNA-based mechanism of intron-mediated enhancement. Front Plant Sci. 2011;2:1–9. doi: 10.3389/fpls.2011.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rounsley S, Reddy Marri P, Yu Y, He R, Sisneros N, Goicoechea JL, Lee SJ, Angelova A, Kudrna D, Luo M, Affourtit J, Desany B, Knight J, Niazi F, Egholm M, Wing RA. De novo next generation sequencing of plant genomes. Rice. 2009;2:35–43. doi: 10.1007/s12284-009-9025-z. [DOI] [Google Scholar]
- Ryu HS, Han M, Lee SK, Cho JI, Ryoo N, Heu S, Lee YH, Bhoo SH, Wang GL, Hahn TR, Jeon JS. A comprehensive expression analysis of the WRKY gene superfamily in rice plants during defense response. Plant Cell Rep. 2006;25:836–847. doi: 10.1007/s00299-006-0138-1. [DOI] [PubMed] [Google Scholar]
- Santino A, Taurino M, DeDomenico S, Bonsegna S, Poltronieri P, Pastor V, Flors V. Jasmonate signaling in plant development and defense response to multiple (a)biotic stresses. Plant Cell Rep. 2013;32:1085–1098. doi: 10.1007/s00299-013-1441-2. [DOI] [PubMed] [Google Scholar]
- Sato K, Shimizu T, Kondoh H, Hirguri A, Sasaya T, Choi IR, Omura T, Kikuchi S. Relationship between symptoms and gene expression induced by the infection of three strains of rice dwarf virus. PLoSOne. 2011;6:3e18094. doi: 10.1371/journal.pone.0018094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpella E, Rueb S, Boot KJM, Hoge JHC, Meijer AH. A role of the rice homeobox gene OsHox1 in provascular fate commitment. Development. 2000;127:3655–3669. doi: 10.1242/dev.127.17.3655. [DOI] [PubMed] [Google Scholar]
- Scarpella E, Simons EJ, Meijer AH. Multiple regulatory elements contribute to the vascular-specific expression of the rice HD-ZIP gene OsHox1 in Arabidopsis. Plant Cell Physiol. 2005;46:1400–1410. doi: 10.1093/pcp/pci153. [DOI] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Abe H, Kasuga M, Yamaguchi-Shinozaki K, Caminci P, Hayashizaki Y, Shinozaki K. Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell. 2001;13:61–72. doi: 10.1105/tpc.13.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shameer K, Ambika S, Varghese SM, Karaba N, Udayakumar M, Sowdhamini R. (2009) STIFDB–Arabidopsis Stress-responsive Transcription Factor DataBase. Int J Plant Genomics ID583429: doi:10.1155/2009/583429. [DOI] [PMC free article] [PubMed]
- Shen QJ, Zhang P, Ho TH. Modular nature of abscisic acid (ABA) response complexes: Composite promoter units that are necessary and sufficient for ABA induction of gene expression in barley. Plant Cell. 1996;8:1107–1119. doi: 10.1105/tpc.8.7.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen QJ, Ho TH. Functional dissection of an abscisic acid (ABA)-inducible gene reveals two independent ABA-responsive complexes each containing G-box and a novel cis-acting element. Plant Cell. 1995;7:295–307. doi: 10.1105/tpc.7.3.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimono M, Sugano S, Nakayama A, Jiang CJ, Ono K, Toki S, Takatsuji H. Rice WRKY45 plays a crucial role in benzothiadiazole-inducible blast resistance. Plant Cell. 2007;19:2064–2076. doi: 10.1105/tpc.106.046250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimono M, Koga H, Akagi A, Hayashi N, Goto S, Sawada M, Kurihara T, Matsushita A, Sugano S, Jiang CJ, Kaku H, Inoue H, Takatsuji H. Rice WRKY45 plays important roles in fungal and bacterial disease resistance. Mol Plant Pathol. 2012;13:83–94. doi: 10.1111/j.1364-3703.2011.00732.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. Molecular responses to dehydration and low temperature: Differences and cross-talk between two stress signaling pathways. Curr Opin Plant Biol. 2000;3:217–223. doi: 10.1016/S1369-5266(00)00067-4. [DOI] [PubMed] [Google Scholar]
- Shinwari ZK, Nakashima K, Miura S, Masuga M, Seki M, Yamaguchi-Shinozaki K, Shinozaki K. An Arabidopsis gene family encoding DRE/CRT binding proteins involved in low temperature responsive gene expression. Biochem Biophys Res Comm. 1998;250:161–170. doi: 10.1006/bbrc.1998.9267. [DOI] [PubMed] [Google Scholar]
- Shoji T, Kajikawa M, Hashimoto T. Clustered transcription factor genes regulate nicotine biosynthesis in tobacco. Plant Cell. 2010;22:3390–3409. doi: 10.1105/tpc.110.078543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji T, Hashimoto T. DNA-binding and transcriptional activation properties of tobacco NIC2-locus ERF189 and related transcription factors. Plant Biotechnol. 2012;29:35–42. doi: 10.5511/plantbiotechnology.11.1216a. [DOI] [Google Scholar]
- Singh MP, Lee FN, Counce PA, Gibbons JH. Mediation of partial resistance to rice blast through anaerobic induction of ethylene. Phytopathology. 2004;94:819–825. doi: 10.1094/PHYTO.2004.94.8.819. [DOI] [PubMed] [Google Scholar]
- Silverman P, Seskar M, Kanter D, Schweizer P, Metraux JP, Raskin I. Salicylic acid in rice. Biosynthesis, conjugation, and possible role. Plant Physiol. 1995;108:633–639. doi: 10.1104/pp.108.2.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smale ST. Core promoters: Active contributors to combinatorial gene regulation. Genes Dev. 2001;15:2503–2508. doi: 10.1101/gad.937701. [DOI] [PubMed] [Google Scholar]
- Smale ST, Baltimore D. The ‘initiator’ as a transcription control element. Cell. 1989;57:103–113. doi: 10.1016/0092-8674(89)90176-1. [DOI] [PubMed] [Google Scholar]
- Song Y, You J, Xiong L. Characterization of OsIAA gene, a member of rice Aux/IAA family involved in auxin and brassinosteroid hormone responses and plant morphogenesis. Plant Mol Biol. 2009;70:297–309. doi: 10.1007/s11103-009-9474-1. [DOI] [PubMed] [Google Scholar]
- Srivastav AQ, Mehta S, Lindlof A, Bhargava S. Over-represented promoter motifs in abiotic stress-induced DREB genes of rice and sorghum and their probable role in regulation of gene expression. Plant Signal Behav. 2010;5:775–784. doi: 10.4161/psb.5.7.11769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger EJ, Yaopan M, Regier MK, Triezenberg SJ, Thomashow MF. Transcriptional adaptor and histone acetyltransferase proteins in Arabidopsis and their interactions with CBF1, a transcriptional activator involved in cold-regulated gene expression. Nucl Acids Res. 2001;29:1524–1533. doi: 10.1093/nar/29.7.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger EJ, Gilmour SJ, Thomashow MF. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcription activator element that binds to the C-repeat/DRE, a cis-acting regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci U S A. 1997;94:1035–1040. doi: 10.1073/pnas.94.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutoh K, Yamauchi D. Two cis-acting elements necessary and sufficient for gibberellin-upregulated proteinase expression in rice seeds. Plant J. 2003;34:635–645. doi: 10.1046/j.1365-313X.2003.01753.x. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Suzuki N, Ohme-Takagi M, Shinshi H. Immediate early induction of mRNAs for ethylene-responsive transcription factors in tobacco leaf strips after cutting. Plant J. 1998;15:657–665. doi: 10.1046/j.1365-313x.1998.00243.x. [DOI] [PubMed] [Google Scholar]
- Takaiwa F, Kikuchi S, Oono K. Nucleotide sequence of a rice glutelin gene. FEBS Lett. 1987;206:33–35. doi: 10.1016/0014-5793(86)81335-7. [DOI] [Google Scholar]
- Takaiwa F, Oono K, Wing D, Kato A. Sequence of three members and expression of a new major subfamily of glutelin genes from rice. Plant Molec Biol. 1991;17:875–885. doi: 10.1007/BF00037068. [DOI] [PubMed] [Google Scholar]
- Thakur JK, Tyagi AK, Khurana JP. OsIAA1, an Aux/IAA cDNA from rice and changes in its expression as influenced by auxin and light. DNA res. 2001;8:193–203. doi: 10.1093/dnares/8.5.193. [DOI] [PubMed] [Google Scholar]
- Thomas H, Howart CJ. Five ways to stay green. J Expt Bot. 2000;5:329–337. doi: 10.1093/jexbot/51.suppl_1.329. [DOI] [PubMed] [Google Scholar]
- Thomashow MF. Plant cold acclimation: Freezing tolerance genes and regulatory mechanisms. Ann Rev Plant Physiol Plant Molec Biol. 1999;50:571–599. doi: 10.1146/annurev.arplant.50.1.571. [DOI] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle T. The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell. 2003;43:265–285. doi: 10.1105/tpc.008417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todaka D, Nakashima K, Shinozaki K, Yamaguchi-Shinozaki K. Toward understanding transcriptional regulatory networks in abiotic stress responses and tolerance in rice. Rice. 2012;5:e6. doi: 10.1186/1939-8433-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Nakajima M, Katoh E, Ohmiya H, Asano K, Saji S, Hongyu X, Ashikari M, Kitano H, Yamaguchi I, Matsuoka M. Molecular interactions of a soluble gibberellins receptor, GID1 with a rice DELLA protein SLR1 and gibberellins. Plant Cell. 2007;19:2140–2155. doi: 10.1105/tpc.106.043729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Ashikari M, Nakajima M, Itoh H, Katoh E, Kobayashi M, Chow TY, Hsing YI, Kitano H, Yamaguchi I, Matsuokoa M. GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature. 2005;437:693–698. doi: 10.1038/nature04028. [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. ARF1, a transcription factor that binds to auxin response elements. Science. 1997;276:1865–1868. doi: 10.1126/science.276.5320.1865. [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Liu ZB, Hagen G, Guilfoyle TJ. Composite structure of auxin response elements. Plant Cell. 1995;7:1611–1623. doi: 10.1105/tpc.7.10.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urao T, Yamaguchi-Shinozaki K, Urao S, Shinozaki K. An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell. 1993;5:1529–1539. doi: 10.1105/tpc.5.11.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlot AC, Dempsey DMA, Klessig DF. Salicylic acid, a multifaceted hormone to combat disease. Annu Rev Phytopathol. 2009;47:177–206. doi: 10.1146/annurev.phyto.050908.135202. [DOI] [PubMed] [Google Scholar]
- Vogel JT, Zarka DG, Van Buskirk HA, Fowler SG, Thomashow MF. Roles of CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J. 2005;41:195–211. doi: 10.1111/j.1365-313X.2004.02288.x. [DOI] [PubMed] [Google Scholar]
- Vogel JM, Roth B, Cigan M, Freeling M. Expression of the two maize TATA binding protein genes and functions of the encoded TBP proteins by complementation in yeast. Plant Cell. 1993;5:1627–1638. doi: 10.1105/tpc.5.11.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wager A, Browse J. Social network: JAZ protein interactions expand our knowledge of jasmonate signaling. Front Plant Sci. 2012;3:1–11. doi: 10.3389/fpls.2012.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waller F, Furuya M, Nick P. OsARF1, an auxin response factor from rice is an auxin-regulated and classifies as a primary auxin responsive gene. Plant Mol Biol. 2002;50:415–425. doi: 10.1023/A:1019818110761. [DOI] [PubMed] [Google Scholar]
- Wang Z, Libault M, Joshi T, Valliyodan B, Nguyen HT, Xu D, Stacey G, Cheng J. SoyDB: A knowledge database of soybean transcription factors. BMC Plant Biol. 2010;10:e14. doi: 10.1186/1471-2229-10-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Pei K, Fu Y, Sun Z, Li S, Liu H, Tang K, Han B, Tao Y. Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa) Gene. 2007;394:13–24. doi: 10.1016/j.gene.2007.01.006. [DOI] [PubMed] [Google Scholar]
- Wang X, Haberer G, Mayer KFX. Discovery of cis-elements between sorghum and rice using co-expression and evolutionary conservation. BMC Genom. 2009;10:284e. doi: 10.1186/1471-2164-10-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Oard JH. Rice ubiquitin promoters: Deletion analysis and potential usefulness in plant transformation systems. Plant Cell Rep. 2003;22:129–134. doi: 10.1007/s00299-003-0657-y. [DOI] [PubMed] [Google Scholar]
- Washida H, Wu CY, Suzuki A, Yamanouchi U, Akihama T, Harada K, Takaiwa F. Identification of cis-regualtory elements required for endosperm expression of the rice storage protein glutelin gene GluB-1. Plant Molec Biol. 1999;40:1–12. doi: 10.1023/A:1026459229671. [DOI] [PubMed] [Google Scholar]
- Wasternack C. Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann Bot. 2007;100:681–697. doi: 10.1093/aob/mcm079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wing RA, Ammiraju SS, Luo M, Kim HR, Yu Y, Kudrna D, Goicoechea JL, Wang W, Nelson W, Rao K, Brar DS, Mackill DJ, Han B, Soderlund C, Stein L, SanMiguel P, Jackson S. The Oryza Map Alignment Project: the golden path to unlocking the genetic potential of wild rice species. Plant Molec Biol. 2005;59:53–62. doi: 10.1007/s11103-004-6237-x. [DOI] [PubMed] [Google Scholar]
- Wingender E, Dietze P, Karas H, Knüppel R. TRANSFAC: a database on transcription factors and their DNA binding sites. Nucl Acids Res. 1996;24:238–241. doi: 10.1093/nar/24.1.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Lu J, Kullbokas EJ, Golub T, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematics discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Park MR, Mohanty B, Herath V, Kitazumi A, Yun SJ, De los Reyes BG. Cis-regulatory signatures among orthologous groups of stress-associated bZIP transcription factors from rice, sorghum and Arabidopsis based on phylogenetic footprints. BMC Genomics. 2012;13:e497. doi: 10.1186/1471-2164-13-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Liu X, Ge S, Jensen JD, Hu F, Li X, Dong Y, Gutenkunst RN, Fang L, Huang L, Li J, He W, Zhang G, Zheng X, Zhang F, Li Y, Yu C, Kristiansen K, Zhang X, Wang J, Wright M, McCouch S, Nielsen R, Wang J, Wang W. Resequencing 50 accessions of cultivated and wild rice yields markers for identifying agronomically important genes. Nature Biotech. 2012;30:105–111. doi: 10.1038/nbt.2050. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. Organization of cis-acting regulatory elements in osmotic and cold stress responsive promoters. Trends Plant Sci. 2005;10:88–94. doi: 10.1016/j.tplants.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. A novel cis-acting element in an Arabidopis gene is involved in responsiveness to drought, low temperature or high salt stress. Plant Cell. 1994;6:251–264. doi: 10.1105/tpc.6.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto YY, Ichida H, Matusi M, Obokata J, Sakurai T, Satou M, Seki M, Shinozaki K, Abe T. Identification of plant promoter constituents by analysis of local distribution of short sequences. BMC Genomics. 2007;8:e67. doi: 10.1186/1471-2164-8-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Qi M, Mei C. Endogenous salicylic acid protects rice plants from oxidative damage caused by aging as well as biotic and abiotic stress. Plant J. 2004;40:909–919. doi: 10.1111/j.1365-313X.2004.02267.x. [DOI] [PubMed] [Google Scholar]
- Yazaki J, Shimatani Z, Hashimoto A, Nagata Y, Fujii F, Kojima K, Suzuki K, Taya T, Tonouchi M, Nelson C, Nakagawa A, Otomo Y, Murakami K, Matsubara K, Kawai J, Carninci P, Hayashizaki Y, Kikuchi S. Transcriptional profiling of genes responsive to abscisic acid and gibberellin in rice: phenotyping and comparative analysis between rice and Arabidopsis. Physiol Genomics. 2004;17:87–100. doi: 10.1152/physiolgenomics.00201.2003. [DOI] [PubMed] [Google Scholar]
- Yilmaz A, Nishiyama MY, Jr, Fuentes BG, Souza GM, Janies D, Gray J, Grotewold E. GRASSIUS: a platform for comparative regulatory genomics across the grasses. Plant Physiol. 2009;149:171–180. doi: 10.1104/pp.108.128579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokotani N, Sato Y, Tanabe S, Chujo T, Shimizu T, Okada K, Yamane H, Shimono M, Sugano S, Takatsuji H, Kaku H, Minami E, Nishizawa Y. WRKY76 is a rice transcriptional repressor playing opposite roles in blast disease resistance and cold stress tolerance. J Expt Bot. 2013;64:5085–5097. doi: 10.1093/jxb/ert298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun KY, Park MR, Mohanty B, Herath V, Xu F, Mauleon R, Wijaya E, Bajic VB, Bruskiewich R, De los Reyes BG. Transcriptional regulatory network triggered by oxidative signals configures the early response mechanisms of japonica rice to chilling stress. BMC Plant Biol. 2010;10:e16. doi: 10.1186/1471-2229-10-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarka DG, Vogel JT, Cook D, Thomashow MF. Cold induction of Arabidopsis CBF genes involves multiple ICE (Inducer of CBF Expression) promoter elements and a cold-regulatory circuit that is desensitized by low temperature. Plant Physiol. 2003;133:910–918. doi: 10.1104/pp.103.027169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Gerstein M. Of mice and men: Phylogenetic footprinting aids the discovery of regulatory elements. BMC J Biol. 2003;2:e11. doi: 10.1186/1475-4924-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Fowler SG, Cheng H, Lou Y, Rhee SY, Stockinger EJ, Thomashow MF. Freezing-sensitive tomato has a functional CBF cold response pathway, but a CBF regulon that differs from that of freezing-tolerant Arabidopsis. Plant J. 2004;39:905–919. doi: 10.1111/j.1365-313X.2004.02176.x. [DOI] [PubMed] [Google Scholar]
- Zhang H, Jin JP, Tang L, Zhao Y, Gu XC, Gao G, Luo JC. PlantTFDB 2.0: update and improvement of the comprehensive plant transcription factor database. Nucleic Acids Res. 2011;39:D1114–D1117. doi: 10.1093/nar/gkq1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Peng Y, Guo Z. Constitutive expression of pathogen-inducible OsWRKY31 enhances disease resistance and affects root growth and auxin response in transgenic rice plants. Cell Res. 2008;18:508–521. doi: 10.1038/cr.2007.104. [DOI] [PubMed] [Google Scholar]
- Zhang W, McElroy D, Wu R. Analysis of rice Act1 5’ region activity in transgenic rice plants. Plant Cell. 1991;3:1155–1165. doi: 10.1105/tpc.3.11.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Leisy DJ, Okita TW. Tissue-specific expression and temporal regulation of the rice glutelin Gt3 gene are conferred by at least two spatially separated cis-regulatory elements. Plant Molec Biol. 1994;25:429–436. doi: 10.1007/BF00043871. [DOI] [PubMed] [Google Scholar]
- Zheng X, Chen L, Li M, Lou Q, Xia H, Wang P, Li T, Liu H, Luo L. Transgenerational variations in DNA methylation induced by drought stress in two rice varieties with distinguished difference to drought resistance. PLoSOne. 2013;8:e80253. doi: 10.1371/journal.pone.0080253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Kawagoe Y, Xiao S, Li Z, Okita T, Hau TL, Lin A, Murai N. 5’ distal and proximal cis-acting regulator elements are required for developmental control of a rice seed storage protein glutelin gene. Plant J. 1993;4:357–366. doi: 10.1046/j.1365-313X.1993.04020357.x. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Dabi T, Lamb C. TATA-box and initiator functions in the accurate transcription of a plant minimal promoter in vitro. Plant Cell. 1995;7:1681–1689. doi: 10.1105/tpc.7.10.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Ordiz MI, Dabi T, Beachy RN, Lamb C. Rice TATA binding protein interacts functionally with transcription factor IID and the RF2a bZIP transcriptional activator in an enhanced plant in vitro transcription system. Plant Cell. 2002;14:795–803. doi: 10.1105/tpc.010364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zourelidou M, De-Torres Zabala M, Smith C, Bevan MW. Store-keeper defines a new class of plant-specific DNA binding proteins and is a putative regulator of patatin expression. Plant J. 2002;30:489–497. doi: 10.1046/j.1365-313X.2002.01302.x. [DOI] [PubMed] [Google Scholar]


