Abstract
The capability of the fungi Nigrospora sp. CBMAI 1328 and Arthopyrenia sp. CBMAI 1330 isolated from marine sponge to synthesise laccases (Lcc) in the presence of the inducer copper (1–10 μM) was assessed. In a liquid culture medium supplemented with 5 μM of copper sulphate after 5 days of incubation, Nigrospora sp. presented the highest Lcc activity (25.2 U·L−1). The effect of copper on Lcc gene expression was evaluated by reverse transcriptase polymerase chain reaction. Nigrospora sp. showed the highest gene expression of Lcc under the same conditions of Lcc synthesis. The highest Lcc expression by the Arthopyrenia sp. was detected at 96 h of incubation in absence of copper. Molecular approaches allowed the detection of Lcc isozymes and suggest the presence of at least two undescribed putative genes. Additionally, Lcc sequences from the both fungal strains clustered with other Lcc sequences from other fungi that inhabit marine environments.
Keywords: Copper sulphate, Gene expression, Laccase, Marine fungi
Introduction
Marine environments host a huge diversity of microorganisms. Fungi constitute a large part of this microbiota and are also diverse and are important from ecological and biotechnological points of view (Panno et al. 2013). Sponge-derived fungi have repeatedly been shown as interesting sources of novel bioactive metabolites previously not found from terrestrial strains of the same species (Subramani et al. 2013), because they are adapted, amongst others factors, to the harsh marine environment (e.g., high pressure, low temperature, oligotrophic nutrients, high salinity) (Chen et al. 2011). Among the metabolites produced by marine fungi stand out enzymes, that represent enormous potential for the production of pharmaceutical compounds, food, beverages, aromas, fragrances, agrochemicals, and fine chemicals (Rocha et al. 2012).
Laccases (Lcc, EC 1.10.3.2) are found across kingdoms of life (e.g., plants, insects, bacteria and fungi) and, in the Nature act preferably on phenolic compounds. They belong to the class of oxidoreductases and to the family multicopper oxidases as well as ferroxidases, bilirubin oxidases and ascorbate oxidases (Ramos et al. 2011). Coding genes from this family appear to be redundant in fungal genomes, probably due to the different physiological roles played by their coding products and their regulation depending on environmental conditions (Ramos et al. 2011).
The widespread occurrence of Lcc in fungi and their versatility, especially in white-rot fungi, contribute to further research to obtain new sources of improved enzymes (Haibo et al. 2009). Although the copper catalytic centres are similar for all fungal Lcc, significant differences are observed on thermodynamic and kinetic properties depending on the microorganism (de Oliveira et al. 2009). The fungal Lcc often occur as multiple isoenzymes expressed under different growth conditions. Various strategies have been employed to improve the production of Lcc, such as medium optimization, isolation and breeding of high-producing strains, the utilization of inducers, and the heterologous expression of Lcc genes (Wang et al. 2013). Furthermore, most of Lcc are extracellular inducible enzymes, their rates of synthesis and activity being strongly dependent on the presence of suitable inductor which plays an important role in increasing their production (Kocyigit et al. 2012).
Reports investigating enzymes produced by marine-derived fungi, and their potential applications are very few. Recently, Chen et al. (2011) reported the production of laccases by a Pestalotiopsis strain. These authors obtained highest level of Lcc activity (2.0 U·l−1) under submerged growth using the untreated sugarcane bagasse. Bonugli-Santos et al. (2010) identified Lcc, manganese peroxidase (MnP) and lignin peroxidase (LiP) activities from marine-derived fungi such as Aspergillus esclerotiorum, Cladosporium cladosporioides and Mucor racemosus. Lcc are of particular interest with regard to potential industrial applications due to their ability to oxidize a wide range of toxic and polluting substrates, including polycyclic aromatic hydrocarbons (PAH) derived from petroleum, textile dyes and pesticides.
Fungal Lcc production is highly regulated by the media composition. Thus, medium optimization has become one of the main methods to enhance Lcc production. Carbon sources and copper ions are the two most critical factors in improving or stimulating Lcc (Wang et al. 2014). Different studies have shown that Lcc production are regulated by metal ions such as Cu2+ and Fe3+ by gene expression induction or through translational or post-translational regulation (Fonseca et al. 2010). Recently, Manavalan et al. (2013) described significantly increased (1.5 U ml−1) for Lcc production by Ganoderma lucidum when the culture medium was amended with 0.4 mM CuSO4. Nakade et al. (2013) using Pycnoporus cinnabarinus assessed the effect of different inducers (anicidine, catechol, guaiyacol, 2,5-xylidine, ferulic acid, ethanol ,H2O2, Cu+2, Mn+2, Fe+2) in different concentrations for Lcc activity. These authors concluded that the most effective inducer was CuSO4 (0.25 mM) where the Lcc activity detected (34.6 U·ml−1) was 20 times higher than in the absence of this inducer. In work by Lorenzo et al. (2006) the addition of copper to the growth medium stimulated Lcc production by Trametes versicolor. The cultures treated with 3.5 mM copper sulphate showed the highest Lcc activities of approximately 8 U·ml−1. This represented an increase of more than 12-fold in relation to the control culture and was nearly 25% higher than those obtained in the cultures with copper sulphate at 2 mM.
The fungal strains investigated in the present paper (Nigrospora sp. CBMAI 1328 and Arthopyrenia sp. CBMAI 1330) were isolated from the marine sponge Dragmacidon reticulatum and previously selected because of their capacity to synthesise great levels of Lcc (7.7 and 6.5 U·l−1), respectively (Passarini 2012). In the present work, the effect of copper sulphate on laccase synthesis by the strains was investigated to further enhance activity. In addition, the laccase gene diversity was determined.
Materials and methods
Microorganisms
The marine-derived fungi Nigrospora sp. and Arthopyrenia sp. were isolated from the sponge Dragmacidon reticulatum and identified according to Passarini et al. (2013). Both strains were deposited at the Brazilian culture collection “CBMAI/Coleção Brasileira de Micro-organismos de Ambiente e Indústria” (CPQBA/UNICAMP) under the accession number CBMAI 1328 and CBMAI 1330, respectively.
For the assays related to the present work the strains were maintained on Malt Extract Agar (MEA, Oxoid malt extract 20 g∙l−1, Sigma glucose monohydrate 20 g∙l−1, Oxoid bacto peptone1 g∙l−1, Oxoid agar-3 15 g∙l−1) at 4°C and subcultured every month.
Culture conditions
Influence of copper sulphate on laccase gene expression
Lcc synthesis assays were performed in 250 ml Erlenmeyer flasks containing 100 ml of liquid culture medium (LCM: Oxoid malt extract 20 g l−1, Panreac NaCl 30 g l−1, Sigma copper sulphate 1–10 μM). Five plug disks of 8-mm diameter collected from the 7 days old colony of the fungi Nigrospora sp. CBMAI 1328 and Arthopyrenia sp. CBMAI 1330, previously growth on pre-adaption medium (PAM: LCM added by Oxoid agar-3 15 g∙l−1), were used as inocula. The submerged cultures were incubated in a Certomat rotary shaker (150 rpm) for 7 days at 28°C. Controls were carried out under identical conditions without copper sulphate.
Molecular analyses
Genomic DNA isolation and amplification
Five 8-mm diameter plugs were cut as described above and used to inoculate 250 ml Erlenmeyer flasks with 100 ml GYP (Oxoid malt extract 3 g·l−1, Panreac glucose 10 g·l−1, Oxoid yeast extract 3 g·l−1 and Oxoid peptone 5 g·l−1). The cultures were incubated on a Certomat rotary shaker for 7 days at 28°C and 150 rpm. Mycelia from the Nigrospora sp. CBMAI 1328 and Arthopyrenia sp. CBMAI 1330 were frozen at −80°C and later used for genomic DNA extraction as previously described by Raeder and Broda (1985).
The degenerate primer pair LAC2FOR (5′-GGIACIWIITGGTAYCAYWSICA-3′) and LAC3REV (5′-CCRTGIWKRTGIAWIGGRTGIGG-3′) (Invitrogen) were used for amplifying two of the four copper binding regions (designated II and III) (Lyons et al. 2003). The housekeeping gene β-tubulin from the white rot fungus Trametes versicolor was used as reference (GenBank accession no. AY944858.1: 5′-CGGTGAGAGGCGTCGGACAC-3′). DNA amplification was performed using a thermocycler (Bio-Rad, MyCycler) with an initial denaturation of 3 min at 94°C; followed by 35 cycles of 0.5 min at 94°C, 0.5 min at x°C (x = Lcc: 48; β-tubulin = 47), 2 min at 72°C; final extension of 10 min at 72°C and cooled to 4°C. PCR reactions were done in a 50 μl final volume containing: 0.5 μl of DNA (97 ng μl−1), 1 μl of 200 mM deoxynucleotide triphosphates (Promega), 3 μl of MgCl2 solution (25 mM), 10 μl of GoTaq Flexi buffer (10×), 1 μl of each pair of primers (10 mM), 0.25 μl of enzyme GoTAQ Hot Start Polymerase (Promega) and MilliQ water.
Amplified products were visualized in 1.2% agarose gels stained with ethidium bromide. The bands of the expected sizes were removed and purified with GFX PCR DNA and Gel Band Purification Kit (GE Healthcare).
Cloning, sequencing and sequence analysis
PCR products were cloned into pGEM-T Easy Vector (Promega) according to the manufacturer’s instructions and transformed into E. coli JM109 competent cells (Promega). About 15 clones per insert were sequenced with primers M13f (5′-CGCCAGGGTTTTCCCAGTCACGAC-3′) and M13r (5′-TTTCACACAGGAAACAGCTATGAC-3′). Amplified products were purified using GFX PCR DNA and Gel Band Purification Kit (GE Healthcare) for subsequent sequencing with DYEnamic ET Dye Terminator Cycler Sequencing Kit in an automated MegaBace DNA Analysis System 1000 (GE Healthcare) according to the manufacturer’s instructions.
The Phred/Phrap/CONSED software was used to assemble the sequences into a contig. Sequences were identified using BALSTn and BLASTx search (with Lcc gene references in the GenBank database) and aligned using ClustalX (Thompson et al. 1997). The determination of the intron was done according to Bonugli-Santos et al. (2010). For that end, the target sequences were aligned with known Lcc cDNA (e.g., Neurospora crassa AAA33591 or Trametes versicolor U44431) and were manually corrected with BIOEDIT 7 (Hall, 1999) and ClustalW (Thompson et al. 1997). The introns were discarded and the deduced protein sequences were determined before uploading the alignment into phylogenetic programmes. A distance approach using the Kimura 2-parameter model (Kimura 1980) as implemented in MEGA software version 5.0 (Tamura et al. 2011) was used as a substitution model.
Sequences were also used to create a picture of Lcc gene structures using FancyGene v1.4 (Rambaldi and Ciccarelli 2009).
RNA extraction and RT-PCR
As described above five 8-mm diameter plugs of the Nigrospora sp. CBMAI 1328 and Arthopyrenia sp. CBMAI 1330 grown in PAM were inoculated in a 250 ml Erlenmeyer flask with 100 ml GYP and incubated on a Certomat rotary shaker for 5 days at 28°C and 150 rpm. For each fungus, biomass was then retrieved and carefully washed by vacuum filtration under sterile conditions for three times with 150 ml sterile water. From that biomass, approximately 1 g was transferred into 3 different sets of four Erlenmeyer flasks, each containing 100 ml of the LCM with the following conditions: copper sulphate 1 μM, 5 μM and control without the inducer. The twelve Erlenmeyer flasks were then incubated for a period of 48, 72, 96 and 120 h, at 150 rpm and 28°C. For each condition, total RNA was extracted according Chomczynski and Sacchi (1987).
For the cDNA syntheses were used the SuperScript™ III Reverse Transcriptase kit (Invitrogen). PCR amplifications were performed as previously described in Section 2.3.1 (x = Lcc: 48; β-tubulin = 47). PCR reactions were done in a final volume of 50 μl, containing: 2 μl of cDNA of those samples, 1 μl of 200 mM deoxynucleotide triphosphates (Promega), 3 μl of MgCl2 solution (25 mM), 10 μl of GoTaq Flexi buffer (10×), 1 μl of each pair of primers (10 mM), 0.25 μl of enzyme GoTAQ Hot Start Polymerase (Promega) and MilliQ water. PCR products were separated by electrophoresis. Amplified products were visualized by using Gel Doc XR System (Bio-Rad). The quantification of expression levels of the gene was performed by densitometry. Documented images of the amplicons were treated with the support of the program ImageJ 1.44f (http://imagej.net/). Results were normalized by densitometry according to the constitutive gene expression of β-tubulin. Thus, it was possible to establish the ratio between the Lcc gene expressions relative to β-tubulin under different conditions.
Analytical methods
Biomass
For the dry weight biomass measurement the samples were filtered on filter paper n° 41 (45-mm of diameter) under vacuum and kept at 105°C for 8 hours. Biomass was calculated by subtracting the initial weight from the final weight.
Enzymatic assays
The enzymatic activities of Lcc (ε525nm = 65000 M−1 cm−1) and proteases (ε440nm = 4600 M−1 cm−1) were determined as described by Martins et al. (2003). For each enzymatic activity assay, the same reaction mixtures containing boiled supernatant samples were used as control. One unit (U) of enzyme activity was defined as the amount of the enzyme for changing the absorbance by 0.01 per minute. Enzyme activities of all the samples were expressed as U · l−1.
Determination of total proteins
Total proteins were determined by Bradford method using Coomassie Protein Assay Kit (Thermo Scientific Pierce) according to the manufacturer. A standard calibration curve was constructed with three replicates. Total proteins content were presented as μg·ml−1.
Results
Effect of copper sulphate on laccase synthesis
Results showed the efficiency of these two fungal strains to synthesize Lcc in a LCM supplemented with different copper sulphate concentrations (Table 1). Nigrospora sp. CBMAI 1328 displayed the greatest amount of Lcc activity (25.2 U·l−1) when the medium was supplemented with copper sulphate at 5 μM. In the presence of the highest copper sulphate concentrations (10 μM) there was a decrease in the Lcc activity, indicating a possible enzymatic inhibition. In contrast, Arthopyrenia sp. CBMAI 1330 presented highest (3.9 to 7.7 U·l−1) Lcc activities without copper sulphate (the control). The Lcc production by this fungus in the presence of copper sulphate (1–10 μM) ranged from 1.7 to 5.5 U·l−1. In fact, Lcc activity decreased with increased copper sulphate (Table 1).
Table 1.
Data related to the biomass, total protein and enzymatic activity by the selected marine fungi after incubation in LCM supplemented with different [CuSO 4 ] during 7 days at 28°C and 150 rpm
| Fungal strain | Time (d) | Control* | [CuSO 4 ] 1 μM | [CuSO 4 ] 5 μM | [CuSO 4 ] 10 μM | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Biomass** (g·l −1 ) | Total protein (μg·ml −1 ) | Lcc (U·l −1 ) | Biomass (g·l −1 ) | Total protein (μg·ml −1 ) | Lcc (U·l −1 ) | Biomass (g·l −1 ) | Total protein (μg·ml −1 ) | Lcc (U·l −1 ) | Biomass (g·l −1 ) | Total protein (μg·ml −1 ) | Lcc (U·l −1 ) | ||
| Nigrospora sp. CBMAI 1328 | 1 | 0.3 | 6.0 | 1.2 | 0.1 | 5.6 | 2.7 | 1.4 | 6.4 | 5.5 | 0.1 | 5.1 | 1.4 |
| 3 | 1.6 | 5.2 | 1.8 | 2.7 | 5.8 | 2.8 | 3.8 | 6.5 | 5.6 | 3.6 | 5.2 | 4.3 | |
| 5 | 1.8 | 5.7 | 6.5 | 3.9 | 5.4 | 9.5 | 4.0 | 7.0 | 25.2 | 4.0 | 5.6 | 8.5 | |
| 7 | 2.6 | 8.5 | 5.3 | 4.2 | 6.6 | 6.8 | 4.5 | 7.0 | 22.5 | 4.5 | 6.4 | 7.0 | |
| Arthopyrenia sp . CBMAI 1330 | 1 | 0.6 | 2.6 | 3.9 | 0.7 | 3.0 | 4.4 | 0.5 | 4.8 | 1.8 | 0.7 | 4.8 | 1.7 |
| 3 | 1.6 | 2.6 | 7.5 | 0.9 | 4.6 | 5.2 | 0.7 | 5.4 | 3.0 | 0.9 | 5.0 | 2.1 | |
| 5 | 2.1 | 3.3 | 7.7 | 2.9 | 5.5 | 5.1 | 2.9 | 5.5 | 3.3 | 2.5 | 5.1 | 2.8 | |
| 7 | 2.0 | 3.4 | 6.0 | 2.8 | 5.4 | 4.4 | 2.3 | 6.5 | 3.5 | 2.2 | 5.2 | 2.7 | |
*Control = the same culture conditions without CuSO4.
**Biomass = dry matter.
The relationship between copper induction, biomass and total protein was evaluated in the present study (Table 1). Arthopyrenia sp. CBMAI 1330 presented no significant variation in the biomass production for the assays with and without CuSO4. However, an increasing in total protein was observed for all concentrations of CuSO4 in comparison to the control. In contrast, the Nigrospora sp. CBMAI 1328 produced more biomass in the presence of CuSO4. In the culture conditions where the best results of laccase activities were achieved by this fungus (5 μM of CuSO4), the total protein and biomass obtained were highest than that one from the other assays, with the exception of the total protein produced in the control after 7 days and the biomass produced in the presence of CuSO4 10 μM after 5 and 7 days.
Laccase genes expression
By using the RT-PCR approach, the levels of Lcc gene expression by the Nigrospora sp. CBMAI 1328 and Arthopyrenia sp. CBMAI 1330 were determined. Laccase gene expression was detected in the presence and absence of CuSO4 (Figure 1). These results allow the inference that these fungi the enzyme is constitutively and inductively expressed. The highest rate of Lcc gene expression by Nigrospora sp. CBMAI 1328 was achieved in the presence of 5 μM CuSO4 after 120 h of incubation in LCM. In these same conditions this fungus produced the higher level of laccase (25.2 U·L−1), as showed in Table 1. For Arthopyrenia sp. CBMAI 1330 higher Lcc gene expression was detected in LCM without supplementation of CuSO4 (control) after 96 h of incubation (Figure 1b). Coincidently, the higher rates of Lcc activity by this fungus were also observed in submerged LMC without CuSO4 after 96 and 120 h of incubation (Table 1).
Figure 1.
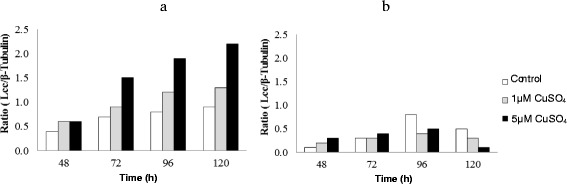
Detection of Lcc gene expression by Nigrospora sp. CBMAI 1328 (a) and Arthopyrenia sp. CBMAI 1330 (b) in different conditions by using RT-PCR. The Lcc gene expression was determined by the relative intensity based on the ratio between the Lcc gene expressions relative to β-tubulin.
Laccase genes characterization
Lcc genes from Nigrospora sp. CBMAI 1328 and Arthopyrenia sp. CBMAI 1330 were characterized based on cloning and sequencing analysis. An amount of 17 and 20 clone sequences were recovered from the fungi Nigrospora and Arthopyrenia, respectively. Amongst all of them, only tree clones presented similarity with sequences from fungal Lcc genes (Table 2): (i) two (C9 and F9) from Arthopyrenia sp. CBMAI 1330 which showed high similarity (between 70 to 100%) with a Lcc gene from Clavariopsis aquatica, a marine ascomycete fungus representative of order Microascales and, (ii) one (A1) from Nigrospora sp. CBMAI 1328 that showed 45% similarity with the fungus Stagonospora sp., a representative of order Pleosporales that can be found in marine environments such as swamps (Lyons et al. 2003).
Table 2.
Similarity of clones recovered by PCR of laccase gene from strains Nigrospora sp. CBMAI 1328 and Arthopyrenia sp. CBMAI 1330
Data derived from phylogenetic analyses showed that only the sequence of clone F9 formed a cluster with the Lcc gene from fungus Clavariopsis aquatica. The other two sequences (from clone C9 and A1) clustered together and separated from the sequences recovered from GenBank database (Figure 2).
Figure 2.
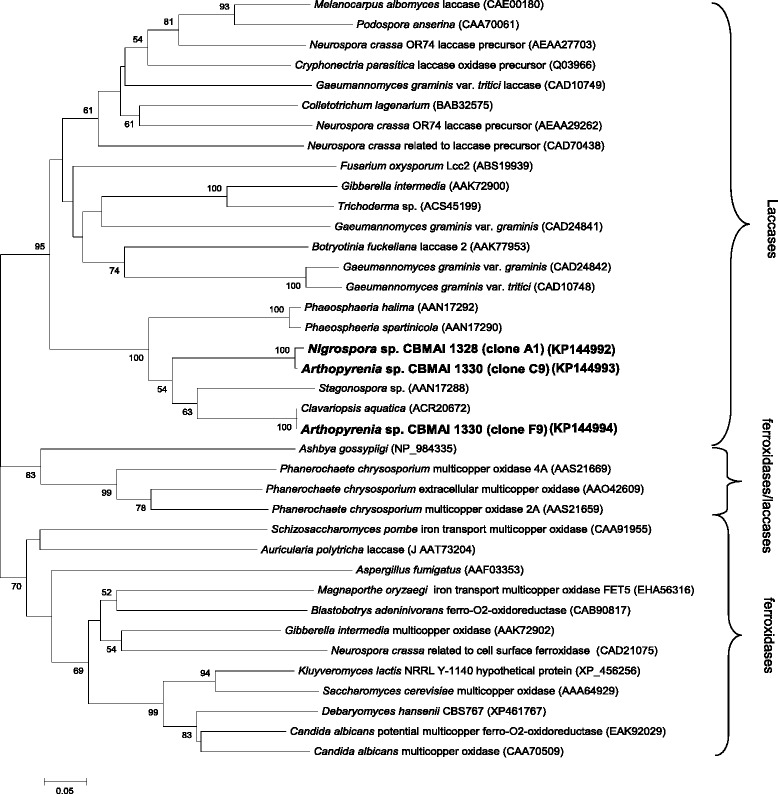
Phylogenetic tree constructed based on the amino acid alignment of protein sequences of fungal Lcc genes. Bootstrap with 1000 replicates was performed by analysis of P-distance. Values greater than 50% are indicated at the nodes of the branches. The horizontal bar represents a distance of 0.05 amino acid substitutions per site.
Molecular characterisation of Lcc genes from the Nigrospora sp. CBMAI 1328 and Arthopyrenia sp. CBMAI 1330 are showed in Figure 3. Comparative schematics of Lcc genes from both marine-derived fungi and Clavariopsis aquatica are presented in Figure 4.
Figure 3.
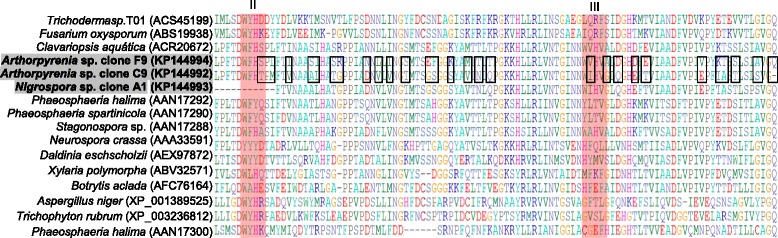
Lcc gene sequences from Nigrospora sp. CBMAI 1328 (clone A1) and Arthopyrenia sp. CBMAI 1330 (clones F9 and C9) aligned together with sequences recovered from GenBank. The red boxes show the possible II and III regions of binding of copper ions and the black rectangles show the differences between the amino acids sequences from clones F9 and C9.
Figure 4.

Comparative schematics of Lcc genes from Nigrospora sp. CBMAI 1328 (clone A1), Arthopyrenia sp. CBMAI 1330 (clones F9 and C9) and Clavariopsis aquatic (ACR20672). The red boxes show the possible copper binding regions (I and II).
Discussion
Copper is an essential micronutrient for most living organisms, and requirements by fungi are usually satisfied by very low concentrations of the metal, in the order of 1–10 μM. Copper present in higher concentrations of its free, cupric form is extremely toxic to fungal cells (Galhaup and Haltrich 2001). The mechanism of the Lcc induction by copper is associated with its role in the Lcc active centre, and its participation in the regulation of Lcc genes transcription and post-transcription modifications; whereas, copper toxicity is attributed to the interaction of copper ions with proteins, enzymes, nucleic acids, and metabolites associated with cell functions and viability, and due to oxidative (Kannaiyan et al. 2012).
Copper is also often a strong inducer of Lcc gene transcription, and this may be related to a defence mechanism against oxidative stress caused by free copper ions (Viswanath et al., 2008). In a recent study, Pezzella et al. (2013) performed a transcriptional analysis of nine Pleurotus ostreatus Lcc genes by RT-PCR in different growth conditions. The authors concluded that the addition of copper to the culture medium resulted in strong induction of lcc9/lcc10 and lcc2 genes, and a lesser induction of other lcc genes. In some cases these responses were dependent on the time of growth. Santo et al. (2012) evaluated the degradation of polyethylene by polyethylene-degrading Rhodococcus ruber and concluded that copper markedly affected the induction and activity of Lcc, resulting in polyethylene degradation. The mRNA quantification by RT-PCR revealed a 13-fold increasing in Lcc mRNA levels from copper-treated cultures in comparison with the untreated control. Additionally, the authors emphasise that the addition of copper to R. ruber cultures containing polyethylene, enhanced by 75% the biodegradation of this compound.
According to Kannaiyan et al. (2012), the induction of Dichomitus squalens Lcc activity required copper sulphate addition as low as 0.06 mM. Palanisami and Lakshmanan (2011) using the marine filamentous non-heterocystous cyanobacterium Phormidium valderianum observed a negative effect of Lcc activity when the concentration of copper was greater than 10 μM and, consequently, a gradual decrease in the rate of Poly-R478 dye decolourisation. Singhal et al. (2009) reported a seven fold enhancement of Lcc production by Cryptococcus albidus after the optimizing of the growth media, which contained 2 mmol·l−1 of copper sulphate. However, for C. albidus, Lcc production was inhibited with increasing copper sulphate concentration. Cordi et al. (2007) showed that the addition of 0.07-0.1 mM copper sulphate during the cultivation of Trametes versicolor promoted higher Lcc activity, reaching a maximum value of 40.44 U·l−1 on day 12 of incubation. In contrast, in the absence of this inductor the values detected for Lcc were considered insignificant. In Sun et al. (2009) the relationship between CuSO4 and fungal biomass was discussed. The authors observed that at high levels of copper sulphate (0.8 mM) the activity of crude Lcc was significantly inhibited possibly due to the weak fungal growth.
Data derived from phylogenetic analyses may be explained by the absence of sequences close to the Nigrospora sp. CBMAI 1328 and Arthopyrenia sp. CBMAI 1330 Lcc genes or may illustrate the recovery of potential new Lcc genes from fungi associated with marine sponges. In a previous work carried out by our research group, three putative new Lcc genes were detected in a marine-derived basidiomycete (Bonugli-Santos et al. 2010). In addition, Lyons et al. (2003) identified 15 distinct sequences of Lcc from ascomycetes isolated from salt marshes in the southeastern U.S.
A study by Hoegger et al. (2006) reported that the composition of multicopper oxidases in fungal species can be very different and some species appear to encode only one type of enzyme such as, ferroxidases, as well as other species can produce other types of enzymes belonging to this family. Based on this report, representative sequences of Lcc, ferroxidase and “ferroxidases/Lcc” (enzymes with strong ferroxidase activity and weak Lcc activity) were recovered and aligned with Nigrospora sp. CBMAI 1328 and Arthopyrenia sp. CBMAI 1330 Lcc protein sequences (Figure 2). Results from phylogenetic analyses showed that Lcc genes from the studied marine fungi formed a cluster with protein sequences that encoding strictly Lcc enzymes. In addition, all Lcc sequences from Nigrospora sp. CBMAI 1328 and Arthorpyrenia sp. CBMAI 1330 grouped with Lcc protein sequences from ascomycetes derived from marine environments.
Since the primer set used was designed to be specific for fungal Lcc and targeted conserved sequences around two pairs of histidine involved in two of the four copper binding regions designated II and III, the alignment of protein sequences from Arthopyrenia clones F9 and C9 and from Nigrospora clone A1 may be related to regions II and III of binding copper ions (Figures 3 and 4).
The comparison between Arthopyrenia clones F9 and C9 protein sequences (Figure 3) and gene sequences alignment (data not shown) revealed that, five different nucleotides and 29 different amino acids were found despite these two Lcc genes being very similar. These results confirm the hypothesis of divergent copies of Lcc genes within the genome of this fungus, where more than one isoform can be expressed under specific experimental conditions.
The presence of multiple Lcc genes in the fungal genome has been discussed previously. According to Ramos et al. (2011), this is probably due to the different physiological roles played by their products and their regulation by environmental conditions. Susla et al. (2007) reported that the diversity of genes can be attributed to post-transcriptional modifications of enzymes within the cell. In conclusion, the use of Lcc gene libraries from filamentous fungi has been a powerful technology for the characterization of this gene (D’Souza et al. 1996; Dedeyan et al. 2000; Dubé et al. 2008).
The detection of two different Lcc genes in the genome of Arthopyrenia sp. CBMAI 1330, suggests the presence of two isoforms. For this fungus, the expression of Lcc was higher when the copper ion was absent in the culture medium. However, in the screening experiments (data not shown) Lcc activity in the range of 15 U·l−1 was achieved when 1 mM of CuSO4 was added to de medium. It is reasonable to speculate that a different isoform may have been expressed by the use of copper ion.
The present work revealed how Lcc activity from two marine-derived fungal strains, identified as belonging to the genera Nigrospora and Arthopyrenia could be increased by the addition of copper sulphate. Molecular and phylogenetic analyses allow the classification of these enzymes and suggest the presence of putative new enzymes. In addition, Lcc from the Nigrospora sp. CBMAI 1328 and Arthopyrenia sp. CBMAI 1330 clustered with other Lcc from fungi that inhabit marine environments.
Fungi from marine environments can produce different enzymes from those produced by the same terrestrial species since they are adapted to oceans where in some cases includes extremes of pH and salinity. The two marine-derived fungal strains studied could be considered strategic for the biotechnology, since they showed to be potential genetic resources, which could be applied in saline environments and/or technological processes.
Acknowledgments
M. Passarini was supported by Ph.D. grant from FAPESP (2008/06720-7), São Paulo, Brazil. The authors thank FAPESP for financial support (BIOTA-FAPESP grant 2010/50190-2 and FAPESP grant 2013/19486-0) and Roberto G.S. Berlinck and CEBIMAR for the support related to samples collecting. L.D. Sette thanks CNPq for Productivity Fellowships 304103/2013-6.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Contributor Information
Michel Rodrigo Zambrano Passarini, Email: michelpassarini@gmail.com.
Cristiane Angelica Ottoni, Email: c.a.ottoni@hotmail.com.
Cledir Santos, Email: cledir.santos@deb.uminho.pt.
Nelson Lima, Email: nelson@ie.uminho.pt.
Lara Durães Sette, Email: larasette@rc.unesp.br.
References
- Bonugli-Santos RC, Durrant LR, Da Silva M, Sette LD. Production of laccase, manganese peroxidase and lignin peroxidase by Brazilian marine-derived fungi. Enzym Microbiol Technol. 2010;46:32–37. doi: 10.1016/j.enzmictec.2009.07.014. [DOI] [Google Scholar]
- Chen H-Y, Xue D-S, Feng X-Y, Yao S-J. Screening and production of ligninolytic enzyme by a marine-derived fungal Pestalotiopsis sp. J63. Appl Biochem Biotechnol. 2011;165:1754–1769. doi: 10.1007/s12010-011-9392-y. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1016/0003-2697(87)90021-2. [DOI] [PubMed] [Google Scholar]
- Cordi L, Minussi RC, Freire RS, Durán N. Fungal laccase: copper induction, semi-purification, immobilization, phenolic effluent treatment and electrochemical measurement. Afr J Biotechnol. 2007;6:1255–1259. [Google Scholar]
- D’Souza TM, Boominathan K, Reddy CA. Isolation of laccase gene-specific sequences from white rot and brown rot fungi by PCR. Appl Environ Microbiol. 1996;62:3739–3744. doi: 10.1128/aem.62.10.3739-3744.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira IRWZ, Fatibello-Filho O, Fernandes SC, Vieira IC. Imobilização da lacase em micropartículas de quitosana obtidas por spray drying e usadas na construção de biossensores. Quim Nova. 2009;32:1195–1201. doi: 10.1590/S0100-40422009000500021. [DOI] [Google Scholar]
- Dedeyan B, Klonowska A, Tagger S, Tron T, Iacazio G, Gil G, Le Petit J. Biochemical and molecular characterization of a laccase from Marasmius quercophilus. Appl Environ Microbiol. 2000;66:925–929. doi: 10.1128/AEM.66.3.925-929.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubé E, Shareck F, Hurtubise Y, Daneault C, Beauregard M. Homologous cloning, expression, and characterization of a laccase from Streptomyces coelicolor and enzymatic decolourisation of an indigo dye. Appl Microbiol Biotechnol. 2008;79:597–603. doi: 10.1007/s00253-008-1475-5. [DOI] [PubMed] [Google Scholar]
- Fonseca MI, Shimizu E, Zapata PD, Villalba LL. Copper inducing effect on laccase production of white rot fungi native from Misiones (Argentina) Enzyme Microb Tech. 2010;46:534–539. doi: 10.1016/j.enzmictec.2009.12.017. [DOI] [PubMed] [Google Scholar]
- Galhaup C, Haltrich D. Enhanced formation of laccase activity by the white-rot fungus Trametes pubescens in the presence of copper. Appl Microbiol Biotechnol. 2001;56:225–232. doi: 10.1007/s002530100636. [DOI] [PubMed] [Google Scholar]
- Haibo Z, Yinglong Z, Feng H, Peiji G, Jiachuan C. Purification and characterization of a thermostable laccase with unique oxidative characteristics from Trametes hirsute. Biotechnol Lett. 2009;31:837–843. doi: 10.1007/s10529-009-9945-0. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Hoegger PJ, Kilaru S, James TY, Thacker JR, Rües U. Phylogenetic comparison and classification of laccase and related multicopper oxidase protein sequences. EBS J. 2006;273:2308–2326. doi: 10.1111/j.1742-4658.2006.05247.x. [DOI] [PubMed] [Google Scholar]
- Kannaiyan R, Mahinpey N, Mani T, Martinuzzi RJ, Kostenko V. Enhancement of Dichomitus squalens tolerance to copper and copper-associated laccase activity by carbon and nitrogen sources. Biochem Eng J. 2012;67:140–147. doi: 10.1016/j.bej.2012.06.007. [DOI] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequence. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Kocyigit A, Pazarbasi MB, Yasa Y, Ozdemir G, Karaboz I. Production of laccase from Trametes trogii TEM H2: a newly isolated white-rot fungus by air sampling. J Basic Microbiol. 2012;52:1–9. doi: 10.1002/jobm.201100341. [DOI] [PubMed] [Google Scholar]
- Lorenzo M, Moldes D, Sanromán MÁ. Effect of heavy metals on the production of several laccase isoenzymes by Trametes versicolor and on their ability to decolourise dyes. Chemosphere. 2006;63:912–917. doi: 10.1016/j.chemosphere.2005.09.046. [DOI] [PubMed] [Google Scholar]
- Lyons JI, Newell SY, Buchan A, Moran MA. Diversity of ascomycete laccase gene sequences in a Southeastern US salt. Microb Ecol. 2003;45:270–281. doi: 10.1007/s00248-002-1055-7. [DOI] [PubMed] [Google Scholar]
- Manavalan T, Manavalan A, Thangavelu KP, Heese K. Characterization of optimized production, purification and application of laccase from Ganoderma lucidum. Biochem Eng J. 2013;70:106–114. doi: 10.1016/j.bej.2012.10.007. [DOI] [Google Scholar]
- Martins MAM, Lima N, Silvestre AJD, Queiroz MJ. Comparative studies of fungal degradation of single or mixed bioaccessible reactive azo dyes. Chemosphere. 2003;52:967–973. doi: 10.1016/S0045-6535(03)00286-8. [DOI] [PubMed] [Google Scholar]
- Nakade K, Nakagawa Y, Yano A, Konno N, Sato T, Sakamoto Y. Effective induction of pblac1 laccase by copper ion in Polyporus brumalis ibrc05015. Fungal Biol. 2013;117:52–61. doi: 10.1016/j.funbio.2012.11.005. [DOI] [PubMed] [Google Scholar]
- Palanisami S, Lakshmanan U. Role of copper in poly R-478 decolorization by the marine cyanobacterium Phormidium valderianum BDU140441. World J Microbiol Biotechnol. 2011;27:669–677. doi: 10.1007/s11274-010-0505-5. [DOI] [Google Scholar]
- Panno L, Bruno M, Voyron S, Anastasi A, Gnavi G, Miserere L, Varese GC. Diversity, ecological role and potential biotechnological applications of marine fungi associated to the seagrass Posidonia oceanica. N Biotechnol. 2013;30:686–694. doi: 10.1016/j.nbt.2013.01.010. [DOI] [PubMed] [Google Scholar]
- Passarini MRZ. Caracterização da diversidade de fungos filamentosos associados a esponjas marinhas e avaliação da produção de lacase. Thesis: Universidade Estadual de Campinas; 2012. [Google Scholar]
- Passarini MRZ, Santos C, Lima N, Berlinck RGS, Sette LD. Filamentous fungi from the Atlantic marine sponge Dragmacidon reticulatum. Arch Microbiol. 2013;195:99–111. doi: 10.1007/s00203-012-0854-6. [DOI] [PubMed] [Google Scholar]
- Pezzella C, Lettera V, Piscitelli A, Giardina P, Sannia G. Transcriptional analysis of Pleurotus ostreatus laccase genes. Appl Microbiol Biotechnol. 2013;97:705–717. doi: 10.1007/s00253-012-3980-9. [DOI] [PubMed] [Google Scholar]
- Raeder U, Broda P. Rapid preparation of DNA from filamentous fungi. Lett Appl Microbiol. 1985;1:17–20. doi: 10.1111/j.1472-765X.1985.tb01479.x. [DOI] [Google Scholar]
- Rambaldi D, Ciccarelli FD. FancyGene: dynamic visualization of gene structures and protein domain architectures on genomic loci. Bioinformatics. 2009;17:2281–2282. doi: 10.1093/bioinformatics/btp381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos JAT, Barends S, Verhaert RMD, de Graaff LHT. The Aspergillus niger multicopper oxidase family: analysis and overexpression of laccase-like encoding genes. Microb Cell Fact. 2011;10:1–11. doi: 10.1186/1475-2859-10-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha LC, Luiz RF, Rosset IG, Raminelli C, Seleghim MHR, Sette LD, Porto ALM. Bioconversion of Iodoacetophenones by Marine Fungi. Mar Biotechnol. 2012;14:396–401. doi: 10.1007/s10126-012-9463-2. [DOI] [PubMed] [Google Scholar]
- Santo M, Weitsman R, Sivan A. The role of the copper-binding enzyme e laccase e in the biodegradation of polyethylene by the actinomycete Rhodococcus ruber. Int Biodeter Biodegr. 2012;84:204–210. doi: 10.1016/j.ibiod.2012.03.001. [DOI] [Google Scholar]
- Singhal A, Choudhary G, Thakur IS. Optimization of growth media for enhanced production of laccase by Cryptococcus albidus and its application for bioremediation of chemicals. Can J Civ Eng. 2009;36:1253–1264. doi: 10.1139/L09-050. [DOI] [Google Scholar]
- Subramani R, Kumar R, Prasad P, Aalbersberg W. Cytotoxic and antibacterial substances against multi-drug resistant pathogens from marine sponge symbiont: Citrinin, a secondary metabolite of Penicillium sp. Asian Pac J Trop Biomed. 2013;3:291–296. doi: 10.1016/S2221-1691(13)60065-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun QY, Hong YZ, Xiao YZ, Fang W, Fang J. Decolorization of textile reactive dyes by the crude laccase produced from solid-state fermentation of agro-byproducts. World J Microbiol Biotechnol. 2009;25:1153–1160. doi: 10.1007/s11274-009-9994-5. [DOI] [Google Scholar]
- Susla M, Novotn C, Svobodová K. The implication of Dichomitus squalens laccase isoenzymes in dye decolorization by immobilized fungal cultures. Bioresour Technol. 2007;98:2109–2115. doi: 10.1016/j.biortech.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5-molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmou-gin F, Higgins DG. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanath B, Chandra MS, Kumar KP, Pallavi H, Reddy BR. Fungal laccases and their biotechnological applications with special reference to bioremediation. Dyn Biochem Process Biotech Mol Biol. 2008;2:1–13. [Google Scholar]
- Wang F, Guo C, Liu C-Z. Immobilization of Trametes versicolor cultures for improving laccase production in bubble column reactor intensified by sonication. J Ind Microbiol Biotechnol. 2013;40:141–150. doi: 10.1007/s10295-012-1214-x. [DOI] [PubMed] [Google Scholar]
- Wang J, Zheng X, Lin S, Lin J, Guo L, Chen X, Chen Q. Identification of differentially expressed genes involved in laccase production in tropical white-rot fungus Polyporus sp. PG15. J Basic Microbiol. 2014;54:142–151. doi: 10.1002/jobm.201200310. [DOI] [PubMed] [Google Scholar]


