Abstract
Background and Objective
Nowadays, the presence of extended-spectrum β-lactamases (ESBLs) producing strains in Serratia genus causes the emergence of resistance to many antibiotics. So, the lack of proper diagnosis of ESBLs strains can lead to failure in the treatment. The objective of the present study was to investigate ESBLs production in Serratia strains isolated from the clinical blood samples in Shiraz, Iran.
Materials and Methods
In this study, 39 Serratia strains isolated from the patients referred to Namazi Hospital, during a 2 year period were tested. The antimicrobial resistance of the isolates to 21 antibiotics was evaluated using Kirby-Bauer disk diffusion method. Combination disk method was used to determine the ESBL phenotype among the isolates. PCR was performed to investigate the presence of ESBL genes of SHV, OXA and TEM types.
Results
The lowest antibiotic resistance rates belonged to meropenem (7.69%) and imipenem (5.12%). Overall, positive ESBL phenotype was identified in 69% (n = 27) of the isolates, 70.37% (n = 19) for S. marcescens and 29.62% (n = 8) for S. liquefaciens. Results obtained by PCR showed that only 20.51% carried OXA gene and 15.38% carried SHV-1 gene. TEM gene was detected in none of the isolates.
Conclusion
This study showed a high prevalence of the emerging ESBL producing strains among clinical isolates of Serratia that could lead to an increase in antibiotic resistance. However, ESBLs genes other than those tested here may be more responsible for the emergence of ESBL phenotype among Serratia clinical isolates in our region.
Keywords: Serratia, Extended-spectrum β-lactamases, OXA, SHV, TEM
INTRODUCTION
Extended spectrum β-lactamases (ESBLs) emerged among Enterobacteriaceae causes a major problem in the treatment of nosocomial infections, thanks to rendered resistance to oxyimino-cephalosporins (e.g. ceftazidime and cefepime) and monobactams (e.g. aztreonam) (1). ESBLs are plasmid-mediated enzymes, mostly found in Klebsiella pneumonia and Escherichia coli, but also described in bacteria of Citrobacter, Enterobacter, Salmonella, Shigella, Proteus, Serratia, and Pseudomonas aeroginosa (2-4). Non-ESBL producing organisms can be changed to ESBL-producing organisms mostly through mutations in parent β-lactamases, particularly TEM-1, TEM-2 and SHV-1. There are various genes of ESBLs worldwide, with TEM, OXA, CTX-M and SHV as the most common ones. The SHV, TEM and CTX-M enzymes are of Ambler molecular class A and Bush group 2be, but OXA- type β-lactamase belongs to class D and group 2d (5). First reports on ESBL-producing organisms were published in Germany in 1983 (6). In the next decade, the majority of reports came from France, and then ESBL-producing organisms spread across the world (7). In recent years, nosocomial infections caused by Serratia strains have become a growing concern. The ability of this opportunistic pathogen to acquire resistance to a broad spectrum of antibiotics has made the effective treatment more difficult. ESBL-producing enzymes rose up among Serratia genus and have increased multidrug resistance (8). Such β-lactamases are quickly transferable and have become a significant issue for human health conditions. The objectives of this study were to evaluate ESBL production and identify the types of such enzymes among Serratia strains, isolated from the blood samples of the patients in Shiraz Namazi Hospital, southern Iran.
MATERIALS AND METHODS
Serratia isolates
This study was conducted at Professor Alborzi Clinical Microbiology Research Center (PACMRC) in Namazi Hospital, Shiraz, Iran, on 39 Serratia strains. The strains were isolated from BACTEC bottles of blood samples of the patients referred to the lab during a period of 2 years from March 2010 to March 2012. All isolates were identified based on the biochemical-microbial tests and then speciated using diagnostic Microgene Bioproducts kit (GN-ID A Panel U.K).
Determination of antimicrobial resistance
Antimicrobial resistance of the strains against 21 antibiotics was determined using standard Kirby-Bauer agar disk diffusion test, according to the CLSI protocol (Mast Co, U.K antimicrobial disk) (9). Escherichia coli ATCC 25922, Klebsiella pneumonia ATCC 700603 and Pseudomonas aeruginosa ATCC 27853 were used as control strains. The antibiotics and their respective amounts were as follows: gentamicin (10 μg), cephalexin (30 μg), ciprofloxacin (5 μg), chloramphenicol (30 μg), meropenem (10 μg), cefuroxime (30 μg), ceftriaxone (30 μg), cefotaxime (30 μg), imipenem (10 μg), ampicillin (10 μg), cefepime (30 μg), ceftazidime (30 μg), tetracycline (30 μg), amikacin (30 μg), ticarcillin (75 μg), ceftizoxime (30 μg), tobramycin (10 μg), piperacillin (100 μg), piperacillin/tazobactam (110 μg), augmentin (30 μg) and aztreonam (30 μg) (Mast Co, U.K.).
Phenotypic confirmatory test for ESBL
For phenotypic confirmation of the presence of ESBLs among the isolates, Combination-Disk Synergistic Test (CDST) was used as recommended by the Clinical and Laboratory Standard Institute (10). Antibiotic disks used in this method included ceftazidime (30 μg), ceftazidime/clavulanate (40 μg), cefotaxime (30 μg) and cefotaxime/clavulanate (40 μg) (Mast Co, UK). A difference of ≥ 5 mm in the zone of growth inhibition diameter between cephalosporin disks and their respective cephalosporin/clavulanate disk confirmed ESBL phenotype among the isolates.
PCR amplification for the detection of ESBL genes
To amplify ESBL genes by PCR method, initially bacterial genome was extracted using boiling water bath for 10 minutes. Briefly, a bacterial suspension in 1.5 ml of distilled water was prepared and incubated at 95°C for 10 minutes. The sample was centrifuged at 8000 rpm for 15 min and then, the supernatant was used as DNA template. The presence of bla OXA-1 and blaSHV-1 within bacterial genome was investigated by multiplex PCR and for blaTEM gene by single PCR, using specific primers as shown in Table 1. The cycling conditions for the detection of blaOXA and blaSHV were as follows: initial denaturation at 95°C for 15 min, 30 cycles of 94°C for 30 sec, 62°C for 90 sec and 72°C for 60 sec, and a final extention at 72°C for 10 min. The cycling conditions for the detection of blaTEM were initial denaturation at 94°C for 2 min, 30 cycles of 94°C for 15 sec, 55°C for 30 sec and 72°C for 45 sec, and a final elongation at 72°C for 10 min. Finally, the PCR products were detected by electrophoresis in a 2% agarose gel with 1X TAE buffer. Gel staining was then performed using ethidium bromide (0.5 μg/ml) and photographed under UV lamp (Gel documenter Uvitec, EEC, England).
Table 1.
Primers used for the detection of blaOXA, blaSHV and blaTEM genes.
Escherichia coli ATCC 25922 and previously characterized blaOXA and SHV positive Klebsiella pneumoniae, blaTEM-positive Salmonella enteritidis and Shigella sonnei strains were used as controls for ESBL analysis (3, 4).
Statistical analysis
Phenotypic results and the data obtained from PCR assays were compared using SPSS statistical software, version 15 and Fisher’s exact test. P-value equal or below 0.05 was considered as significant.
RESULTS
Serratia isolates
Using diagnostic Microgene kit, from 39 Serratia clinical isolates, 30 (77%) were identified as Serratia marcescens and 9 (23%) as Serratia liquefaciens.
Antimicrobial susceptibility testing
The data revealed that; 56.41%, 48.71%, 46.15%, 43.58%, 41.02%, 33.33%, 33.33%, 33.33%, 33.33%, 33.33%, 30.76% and 25.64% of the Serratia strains were susceptible to ceftazidime, ceftizoxime, gentamicin, cefepime, aztreonam, chloramphenicol, amikacin, piperacillin, ticarcillin, ceftriaxone, cefotaxime and tobramycin, respectively. Overall, these strains were found to be more susceptible to imipenem (94.87%), meropenem (92.30%), ciprofloxacin (87.17%) and piperacillin/tazobactam (84.61%). The least susceptibility was observed for augmentin (7.69%) and tetracycline, ampicillin, cefuroxime (5.12%) but none of the isolates was susceptible to cephalexin.
Multidrug resistant strains
The strains exhibiting resistance to more than 2 or 3 classes of antibiotics were considered as multidrug resistant (MDR) strains. The most frequent patterns of MDR among the 39 Serratia isolates were as cephalexin-chloramphenicol-cefuroxime-ampicillin-tetracycline-augmentin and gentamicin-cephalexin-chloramphenicol-cefuroxime-ceftriaxone-cefotaxime-ampicillin-cefepime-ceftazidime-tetracycline-amikacin-ticarcillin-ceftizoxime-tobramycin-piperacillin-augmentin-aztreonam with frequencies of 15.38% and 12.82%, respectively (Table 2).
Table 2.
Frequency of MDR patterns among the Serratia isolates.
| Pattern of MDR | % of MDR |
|---|---|
| CFX-C-CXM-AP-T-AUG | 15.38% |
| GM-CFX-C-CXM-CRO-CTX-AP-CPM-CAZ-T-AK-TC-CZX-TN-PRL-AUG-ATM | 12.82% |
| GM-CFX-C-CXM-CRO-CTX-AP-CPM-CAZ-T-AK-TC-CZX-TN-PRL-PTZ-AUG-ATM | 7.69% |
CFX, cephalexin; C, chloramphenicol; CXM, cefuroxime; AP, ampicillin; T, tetracycline; AUG, augmentin; GM, gentamicin; CRO, ceftriaxone; CTX, cefotaxime; CPM, cefepime; CAZ, ceftazidime; AK, amikacin; TC, ticarcillin; CZX, ceftizoxime; TN, tobramycin; PRL, piperacillin; ATM, aztreonam; PTZ, piperacillin/tazobactam.
Determination of ESBL phenotype
The results of combination disk assay showed that from all the strains 69% (n = 27) were identified as positive ESBL phenotype. Moreover, it was also indicated that a high percentage of Serratia samples were fully resistant (R = 100%) to cefotaxime (CTX), but none was completely resistant to ceftazidime (CAZ).
Detection of blaOXA, blaSHV and blaTEM genes by PCR
Of 39 Serratia isolates, 10 samples (25.64%) demonstrated the presence of either OXA or SHV or both genes at the same time (Fig. 1a). The OXA-1 gene was observed in 20.51% (n = 8) of the strains but SHV gene in 15.38% (n = 6). Moreover, blaTEM gene was not detected in any of the strains (Fig. 1b).
Fig. 1.
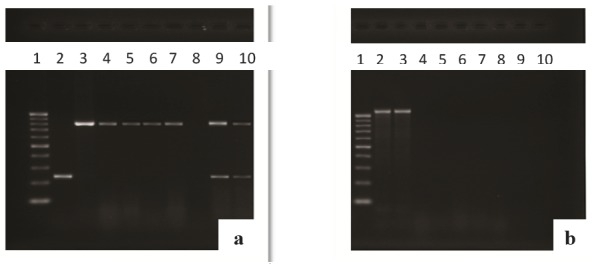
(a): Multiplex PCR assay for blaOXA and blaSHV. Lane 1, 100-bp DNA ladder; lane 2, positive control for SHV gene (237 bp); lane 3, positive control for OXA gene (813 bp); lanes 4,5,6,7 clinical isolates of ESBL positive (blaOXA); lanes 9,10, clinical isolates of ESBL positive for both blaSHV and blaOXA. (b): Single PCR assay for blaTEM. Lane 1, 100-bp DNA ladder; lanes 2,3, positive control for TEM gene (1080 bp); lanes 4-10, clinical isolates of ESBL negative (blaTEM).
Statistical analysis
To determine the relationship between the presence of ESBL gene and ESBL phenotype among the Serratia isolates, Fisher’s exact test was performed and p-value was determined for each of the genes. P-value represents the reliability of the target gene for displaying the ESBL phenotype. The Statistical analysis showed that p-value for blaOXA and blaSHV genes were 0.042 and 0.151, respectively. However, due to the absence of blaTEM among the examined isolates, statistical analysis was not performed for this gene. The results were reliable for the blaOXA gene but it was not for SHV gene.
DISCUSSION
The Serratia strains are opportunistic pathogens and responsible for 2% of nosocomial infections that are often caused by S. marcescens species, however, Serratia liquefaciens infections are rare and mostly due to poor hygiene. These organisms are involved in the development of some diseases such as meningitis, urinary tract infections, sepsis, bacteremia and endocarditis. Serratia strains are resistant to a range of antibiotics used to treat bacterial infections. ESBL genes are highly frequent in the majority of bacteria, especially Enterobacteriaceae family. Their presence among the bacteria makes them ESBL expressing organisms. Such bacteria become resistant to third and fourth generation cephalosporins and monobactams, in addition to the majority of antibiotics (14). ESBL producing organisms often colonized in the patients hospitalized for a long time, especially those who use urinary catheters or central venous lines. Different reports indicated the presence of ESBL producing strains among the Serratia genus (15,16). The treatment of diseases caused by ESBL organisms must be based on the results of new antibiogram tests but not empirical therapy because they have potential resistance to multiple antibiotics (17,18). In this study, phenotypic evaluation by combination disk method was indicative of a prevalence of 69% for ESBL producing strains in Serratia species. We diagnosed only two species of marcescens and liquefaciens among the Serratia isolates in our study. The prevalence of positive ESBL among these strains was 70.37% (n = 19) and 29.62% (n = 8), respectively. However, the rate of ESBL production in the Serratia strains was consistent with that in a previous study by Wu LT, et al in 2004 (65% positive ESBL phenotype) (15). Based on the results of disk diffusion method, the isolates showed a high degree of resistance to the third generation cephalosporins (cefotaxime, 69.23%; ceftriaxone, 66.66%; ceftizoxime, 51.28%; ceftazidime, 43.58%) and the fourth generation cephalosporins (cefepime, 56.41%). The findings also indicate that due to the emergence of ESBL strains, the third and fourth generation cephalosporins could not be reliable for the treatment of the diseases caused by Serratia strains. In this study, the effects of ciprofloxacin, one of the fluoroquinolines, on the Serratia isolates were examined and resistance of 12.82% was observed.
However, it has been demonstrated that from 21 antibiotics tested, imipenem (5.12%) and meropenem (7.69) can be considered as the most effective drug for the treatment of infection caused by ESBL strains of Serratia, followed by fluoroquinolines (19). Among the strains under our study 94.87% were MDR. This is probably due to the presence of mobile resistance elements carrying resistance genes to multiple antibiotics among the isolates. The PCR results obtained from this study showed that of the three genes under investigation for ESBL producing strains, blaOXA gene (20.51%) was the most frequent one among Serratia isolates. The frequency of blaSHV gene was 15.38%, but none of the isolates had blaTEM gene. Overall, the frequency of ESBL strains based on PCR results was 25.64% but 69.23% by phenotypic evaluation. The exact prevalence of ESBL genes among the bacteria is not clear, maybe due to the lack of sufficient studies in this field or improper identification of the genes. However, it is clear that ESBL organisms are widely spread worldwide among Enterobacteriaceae family. The most frequent types of ESBL enzymes include SHV, TEM, OXA and CTX-M, but there are other ESBL enzymes with different frequenies among the bacteria (VEB, PER, GES) (20, 2). The differences between the results of this study and the others could be because of the presence of other ESBL genes within bacterial total genome that were not considered here. In two previous studies, blaSHV and blaCTX-M-3 were identified as the most common ESBL genes in Serratia marcescens (21, 22). However, it should be considered that the frequency and type of ESBL genes vary in different geographical areas and even from region to region. The results of statistical analysis indicated a significant relationship between the presence of blaOXA gene and ESBL phenotype (p = 0.042), but there was no such a relationship between SHV and ESBL phenotype (p = 0.151). This might be due to the limited number of samples in this study or the lack of SHV betalactamase in these ESBL strains.
In conclusion, these data showed that carbapenems followed by ciprofloxacin and piperacillin/tazobactam are the most effective antibiotics against the Serratia strains isolated in our region, while cephalexin showed no effects on them. Moreover, according to the present phenotypic and genotypic data it seems that about 44% of the present ESBL producing strains carry β-lactamases genes other than the genes under investigation in this study. More genetic studies for detection of other ESBL genes are being pursued in our lab. Regarding the important role of Serratia as a causative agent in nosocomial infections and widespread presence of ESBL strains among them, developing and performing tests for the detection of ESBL producing Serratia strains as a routine test in clinical laboratories is recommended.
Acknowledgments
This study has been financially supported by grand#90-25, provided by Prof. Alborzi Clinical Microbiology Research Center, Shiraz University of Medical Sciences. We are grateful to Sareh Roosta in the Center for Development of Clinical Research of Namazi Hospital for her assistance in data analysis and Mrs Marziyeh Hosseini in Professor Alborzi Clinical Microbiology Research Center for her critical assistance in performing the molecular diagnostic methods. Our thanks are also due to Hassan Khajehei, Ph.D, for copy editing of the manuscript.
References
- 1.Karen B, George AJ, Antone AM. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;39:1211–33. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.David LP, Robert AB. Extended-spectrum β-lactamases: a clinical update. Clin Microbiol Rev. 2005;18:657–686. doi: 10.1128/CMR.18.4.657-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ranjbar R, Ghazi FM, Farshad S, Giammanco GM, Aleo A, Owlia P, et al. The occurrence of extended-spectrum β-lactamase producing Shigella spp. in Tehran, Iran. Iran J Microbiol. 2013;5:108–12. [PMC free article] [PubMed] [Google Scholar]
- 4.Ranjbar R, Giammanco GM, Aleo A, Plano MR, Naghoni A, Owlia P, et al. Characterization of the first extended-spectrum beta-lactamase-producing nontyphoidal Salmonella strains isolated in Tehran, Iran. Foodborne Pathog Dis. 2010;7:91–5. doi: 10.1089/fpd.2009.0382. [DOI] [PubMed] [Google Scholar]
- 5.Karen B. New β-lactamases in Gram-negative bacteria: diversity and impact on the selection of antimicrobial therapy. Clin Infect Dis. 2001;32:1085–1089. doi: 10.1086/319610. [DOI] [PubMed] [Google Scholar]
- 6.Du Bois SK, Marriott MS, Amyes SG. TEM- and SHV- derived extended-spectrum beta-lactamases: relationship between selection, structure and function. J Antimicrob Chemother. 1995;35:7–22. doi: 10.1093/jac/35.1.7. [DOI] [PubMed] [Google Scholar]
- 7.George AJ. Extended-spectrum beta-lactamases. Antimicrob Agents Chemother. 1989;33:1131–1136. doi: 10.1128/aac.33.8.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu WL, Lin CW, Wang DY. Serratia marcescens bacteremia: clinical features and antimicrobial susceptibilities of isolates. J Microbiol Immunol Infect. 1998;31:171–179. [PubMed] [Google Scholar]
- 9.National Committee for Clinical Laboratory Standards (NCCLS) Performance standards for antimicrobial susceptibility testing. Disk diffusion, 15th Informational supplement. M100-S15. Wayne (PA): 2005. [Google Scholar]
- 10.Clinical and Laboratory Standards Institute (CLSI) Performance Standards for Antimicrobial Susceptibility Testing, 19th informational supplement. M100-S19. Wayne (PA): 2009. [Google Scholar]
- 11.Dong L, Min Y, Jianying H, Jing Z, Ruyin L, Xin G, et al. Antibiotic-resistance profile in environmental bacteria isolated from penicillin production wastewater treatment plant and the receiving river. Environ Microbiol. 2009;11:1506–1517. doi: 10.1111/j.1462-2920.2009.01878.x. [DOI] [PubMed] [Google Scholar]
- 12.Hong F, Christina L, Barbro OL, Goran H, Emma L, Asa R, et al. Molecular epidemiological analysis of Escherichia coli isolates producing extended-spectrum beta-lactamases for identification of nosocomial outbreaks in Stockholm Sweden. J Clin Microbiol. 2004;42:5917–5920. doi: 10.1128/JCM.42.12.5917-5920.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marc O, Luc B, Paul HR. Precise insertion of antibiotic resistance determinants into Tn21-like transposons: nucleotide sequence of the OXA-1 beta-lactamases gene. Proc Natl Acad Sci USA. 1987;84:7378–7382. doi: 10.1073/pnas.84.21.7378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patricia AB. Extended-spectrum β-lactamases in the 21 st century: characterization, epidemiology, and detection of this important resistance threat. Clin Microbiol Rev. 2001;14:933–951. doi: 10.1128/CMR.14.4.933-951.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lii Tzu W, Mei Fen T, Hwa Jene W, Hui En C, Yin Ching C, Wen Liang Y. Survey of CTX-M-3 extended-spectrum β-lactamase (ESBL) among cefotaxime-resistant Serratia marcescens at a medical center in middle Taiwan. Diagn Microbiol Infect Dis. 2004;49:125–129. doi: 10.1016/j.diagmicrobio.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 16.Wen Liang Y, Lii Tzu W, Michael AP, Patricia LW, Ronald NJ. Confirmation of extended-spectrum β-lactamase-producing Serratia marcescens: preliminary report from Taiwan. Diagn Microbiol Infect Dis. 2003;45:221–224. doi: 10.1016/s0732-8893(02)00539-4. [DOI] [PubMed] [Google Scholar]
- 17.David LP, Wen Chien K, Anne Von G, Jose Maria C, Lutfiye M, Keith PK, et al. Outcome of cephalosporin treatment for serious infections due to apparently susceptible organisms producing extended-spectrum β-lactamases: implications for the clinical microbiology laboratory. J Clin Microbiol. 2001;39:2206–2212. doi: 10.1128/JCM.39.6.2206-2212.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.David LP, Wen Chien K, Anne Von G, Sunita M, Jose Maria C, Herman G, et al. Antibiotic therapy for Klebsiella pneumonia bacteremia: implications of production of extended-spectrum β-lactamases. Clin Infect Dis. 2004;39:31–37. doi: 10.1086/420816. [DOI] [PubMed] [Google Scholar]
- 19.Mario T, Michela S, Enrico Maria T, Fiammetta L, Marianna R, Barbara F, et al. Bloodstream infections caused by extended-spectrum-beta-lactamase- producing Escherichia coli: risk factors for inadequate initial antimicrobial therapy. Antimicrob Agents Chemother. 2008;52:3244–3252. doi: 10.1128/AAC.00063-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marek G. Evolution and epidemiology of extended-spectrum β-lactamases (ESBLs) and ESBL-producing microorganisms. Clin Microbiol Infect. 2001;7:597–608. doi: 10.1046/j.1198-743x.2001.00330.x. [DOI] [PubMed] [Google Scholar]
- 21.Elvira GG, Sandra IMI, Jorge MLD, Gloria MG. Molecular characterization and antimicrobial susceptibility of extended-spectrum {beta}-lactamase-producing Enterobacteriaceae isolates at a tertiary-care center in Monterrey Mexico. J Med Microbiol. 2011;60:84–90. doi: 10.1099/jmm.0.022970-0. [DOI] [PubMed] [Google Scholar]
- 22.Kuo Chen C, Yin Ching C, Lii Tzu W, Guan Cheng H, Wen Liang Y. Clinical experiences of the infections caused by extended-spectrum β-lactamase-producing Serratia marcescens at a medical center in Taiwan. Jpn J Infect Dis. 2006;59:147–152. [PubMed] [Google Scholar]


