Abstract
Background and Objectives
Annual incidence of infection with S. Typhi is estimated to be about 17 million cases worldwide. A systematic search among actinomycete isolates from soil of Iran aimed at finding active actinomycetes against the causative agent of typhoid fever, Salmonella Typhi was carried out during this study.
Materials and Methods
Our anti-Salmonella screening program resulted in nine highly active actinomycete isolates. All nine antibiotic producing strains showed broad-spectrum antibacterial activity, as five strains showed antifungal activity as well. Based on microscopic morphology and cell wall analysis, all nine active actinomycete strains were representatives of the genus Streptomyces. Three of the producing strains including the isolates HG87, HG116 and HG443 with inhibition zone of >20 mm, were selected for further identification and investigation of cytotoxic effects.
Results and Conclusion
None of the producing strains showed cytotoxicity on HEK and USSC cell lines, while strain HG116 showed excellent antitumor activity on T47D cancer cell lines. Isolates HG87, HG116 and HG443 can be distinguished from the related species by some phenotypic and biochemical characteristics. Our results demonstrate the broad-range biological activity exhibited by bioactive compounds of soil actinomycetes from Iran.
Keywords: Actinomycetes, Antibiotic, Cytotoxic activity, Salmonella enterica serovar Typhi
INTRODUCTION
Worldwide mortality due to infection with S. Typhi is estimated at about 600,000 annually (1). Typhoid is common in less-industrialized countries, principally owing to the problem of unsafe drinking-water, inadequate sewage disposal. Multidrug-resistant Salmonella enterica (MDR) serovar Typhi was first isolated in 1980s which showed resistance to chloramphenicol, ampicillin, and cotrimoxazole and currently its treatment is limited to fluoroquinolone, mostly ciprofloxacin (2). When fluoroquinolone resistance is observed, extended-spectrum cephalosporins and macrolide antibiotics are administered as appropriate alternatives (3). However, extended-spectrum β-lactamase (ESBL)-producing Salmonella spp. are reported in Iran and neighboring countries (4-5).
Actinomycetes, especially Streptomycetes, have proven their potential in discovering new antibiotics (6). One of the effective ways in confronting multidrug-resistant pathogens and to discover novel antibiotics is screening of microorganisms from unexplored regions. Iran has diverse climates and geological structures while it is located in one of the biodiversity hotspot of the world. On the other hand Iran’s natural resources are not highly explored for bioactive microorganisms. Recently some new species of actinomycetes having antimicrobial activities (7-10, 11) and new antibiotic compounds (12) have been reported from Iran. However, few screening efforts were done to find actinomycetes active against Gram negative pathogenic bacteria (13, 14). In this work, a screening program on isolation and selection of actinomycetes active against S. Typhi is carried out.
MATERIALS AND METHODS
Microorganisms
Salmonella enterica serovar Typhi NCTC 5761, Bacillus subtilis ATCC 6633, methicillin resistant Staphylococcus aureus UTMC 01401, Klebsiella pneumoniae UTMC 01405, Escherichia coli NCTC 11954, Pseudomonas aeruginosa ATCC 9027, Candida albicans and Aspergillus niger ATCC 16404 were used as test strains in antimicrobial assays.
Sample collection and isolation of actinomycetes
Thirty seven soil samples were collected from different regions of Iran since 2008 to 2009. The habitats were the rhizosphere of plants, river sediments and various virgin soils including desert, mountain and forest soils. The soil samples were collected from 10-15 cm depth, air-dried for one week at room temperature, grinded and eventually sieved. Enrichment of actinomycetes was done by addition of 10% CaCO3 to 1 gram of air dried soil sample. UV pretreatment at 257 nm for 5 minutes was also used to isolate actinomycete strains and to minimize the growth of undesired microorganisms (15). Serial dilutions up to 10-3 were prepared using sterile physiological serum. An aliquot of 100 μl of each dilution was taken and spread evenly over the surface of two selective culture media including glucose asparagine agar and glycerol arginine agar (16). The plates were incubated at 28°C for 14 days. Actinomycete isolates were purified on Yeast Extract-Malt Extract Glucose (ISP2) agar medium and incubated at 28°C for two weeks.
Anti-S. Typhi activity screening from liquid production media
Well-agar diffusion method was employed for screening of anti-S. Typhi activity. A loopful spore of each actinomycete isolate was inoculated into a 100 ml flask containing 20 ml of ISP2 broth. The flasks were incubated at 220 rpm, 28°C, for 120 hours. The fermentation broths were then centrifuged at 4000 rpm for 10 minutes. Antimicrobial activity of the supernatant was examined using well (8 mm in diameter) agar diffusion assay against selected test strains on Muller-Hinton agar plates. After incubation for 18 hrs at 37°C, the diameter of inhibition zone of active broth was measured in millimeters using a slide caliper with an accuracy of 0.1 mm. The activity was assessed in triplicates for positive isolates and the un-reproducible isolates were removed from the study.
Initial extraction of bioactive principles from anti-S. Typhi activity strains
For confirmation of anti-S. Typhi activity, detection was done using disk-agar diffusion method. The amount of 2% (v/v) of ISP2 broth seeding medium was used to inoculate the fermentation medium of 200 ml ISP2 in 1000 ml Erlenmeyer flasks. The cell-free supernatant was extracted by equal volume of ethyl acetate and stirred vigorously for 1 hour. This procedure was repeated twice. The organic phase was separated and evaporated to dryness under reduced pressure and temperature. Sterile filter paper discs (6 mm in diameter) were impregnated with extracts, dried and placed onto Muller Hinton agar freshly seeded with S. Typhi. In order to prevent losing highly polar agents, bioactivity of 100 μl of aqueous phase was also examined by disk-agar diffusion test. The biological activity assays were performed using NCCLS (National Committee for Clinical Laboratory Standards) protocols (17).
Effect of incubation period on antibiotic production
Effect of fermentation period was assessed in 250 ml flasks containing 50 ml ISP2 broth. Samples were taken at 24 hrs interval and the biological activity of the cell-free supernatant against S. Typhi and other test microorganisms was determined by well-agar diffusion method (wells of 6 mm in diameter).
MTT colorimetric cytotoxicity assay
The cytotoxic effect of the anti-S. Typhi activity crude extracts were evaluated against three human cell lines including normal kidney cells (HEK 260), unrestricted somatic stem cells (USSC) and breast cancer cells (T47D). To prepare, 600 ml of the supernatant was extracted with ethyl acetate and the weight of dry residue was determined. Afterwards, it was dissolved in 10 μl of DMSO solvent and 0.5 μl of the solution was added to 2500 μl of cells culture medium. The used ratio of the solvent to medium (2 micromolar) has been determined to be non-toxic on cell lines. First the cells were trypsinized to dissociate from flasks and inoculated into complete culture medium of Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% of fetal calf serum at a density of 103 cells per well. The cells were incubated at 37°C in a humidified atmosphere with 5% CO2 for 24 hrs. Then the medium was removed and replaced with the medium containing crude extract. Thiazole tetrazolium (MTT) colorimetric assay was carried out to assess the cell proliferation or inhibition (18). Medium and the DMSO solvent were used as control. The absorbance was measured at 580 nm using ELISA plate reader. Cell cytotoxic activity was expressed as the mean concentration of the extract in μl required to inhibit 50% of the cell population (IC50 value). The cell inhibitory rate was calculated according to the following formula:
Statistical and data analysis
All of the assays were calculated as an average of at least three independent experiments. One-way analysis of variance (ANOVA) was used at p value <0.05. The rate of microbial cell growth (rx = dx /dt), the rate of antibiotic production (rp = dp /dt), maximum specific growth rate (mmax =rx /x̄), maximum specific production rate (qp max = rx /x̄) and yield of product to biomass (Yp / x = ∆p /∆x) were calculated, where t is time, x is biomass concentration and p is product concentration. The diameter of the inhibition zone (mm) which is indirectly indicative of antibiotic production was considered as p. The estimated time was calculated between t0 and tmax.
Phenotypic characterizations
Morphological features of all of the producing strains and the diaminopimelic acid (DAP) type, were determined. Selected strains were characterized morphologically and physiologically using the instructions of the International Streptomyces Project (ISP) (19) and Bergey’s Manual of Systematic Bacteriology (20).
Molecular identification
Genomic DNA was extracted as described previously (21). Amplification of the 16S rRNA gene of the selected strains was performed using two universal primers 27F: 5′-AGAGTTTGATCCTGGCTCAG-3′ (22) and 1541R: 5’-AAGGAGGTGATCCAGCC-3’ (23). The sequenced 16S rRNA genes were blasted via EzTaxon GenBank database to estimate the degree of similarity. The phylogenetic tree was constructed with the neighbor-joining (NJ) method of MEGA 5.0 software for analysis of evolutionary relationships (24).
Culture preservation
The selected strains were deposited in the University of Tehran Microorganisms Collection (UTMC) and maintained using deep freezing (at -70°C) method.
RESULTS
Anti-S. typhi activity and range of antimicrobial effectiveness
From 37 soil samples collected from different regions of Iran, 522 colonies were isolated and screened for their bioactivity against S. Typhi. Nine of the isolated strains had potent antibiotic activity against S. Typhi (Fig. 1). The activity of these isolates was also assayed against other Gram negative and Gram positive representatives. All of the nine active strains exhibited broad spectrum antibacterial activities. Activity against S. Typhi and other Gram negative test strains was less than that of against Gram positive test strains (Table 1). The morphological characteristics and the type of the diaminopimelic acid (L-DAP) of all of the bioactive strains were consistent with their classification in the genus Streptomyces (20).
Fig. 1.
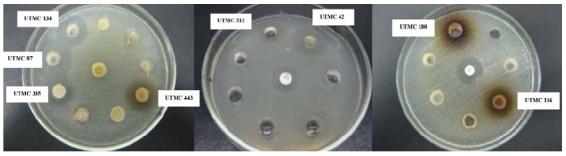
Anti-S. typhi activity of some actinomycetes isolates; the numbers of potent strains are presented on the images
Table 1.
The antimicrobial activity of the active actinomycete strains against test microorganisms expressed as inhibition zone diameter (data accuracy is 0.1 mm).
| Strains | Test microorganisms |
||||||
|---|---|---|---|---|---|---|---|
| S. Typhi | B. subtilis | MRSA | E. coli | P. aeruginosa | C. albicans | A. niger | |
| HG42 | 19 | 23 | 20 | 16 | 12 | 13 | 13 |
| HG87 | 21 | 24 | 23 | 18 | 18 | 0 | 0 |
| HG116 | 16 | 40 | 38 | 12 | 14 | 0 | 0 |
| HG134 | 17 | 16 | 20 | 14 | 15 | 12 | 12 |
| HG180 | 14 | 17 | 15 | 12 | 12 | 20 | 17 |
| HG335 | 16 | 33 | 30 | 0 | 0 | 0 | 0 |
| HG442 | 21 | 21 | 18 | 13 | 0 | 26 | 23 |
| HG443 | 21 | 20 | 16 | 13 | 0 | 25 | 22 |
| HG512 | 16 | 25 | 20 | 12 | 0 | 0 | 0 |
The bioactivity of the crude extracts was evaluated by disk-agar diffusion assay. The crude extracts of all of the nine bioactive strains mentioned in Table 1, showed activity against S. Typhi. The isolates HG87, HG116 and HG443 which showed inhibition zone of >20 mm against S. Typhi in disk or well diffusion methods, were selected for further analysis.
Time course of antibiotic production of the selected strains against S. Typhi was studied. The maximum antibiotic production obtained by the selected strains HG87, HG116 and HG443 were after 192 hrs, 120 hrs and 48 hrs, respectively. The fluctuations in concentration of antibiotic and biomass and pH value are exhibited by Fig. 2. These parameters are the important parameters to monitor the antibiotic production.
Fig. 2.
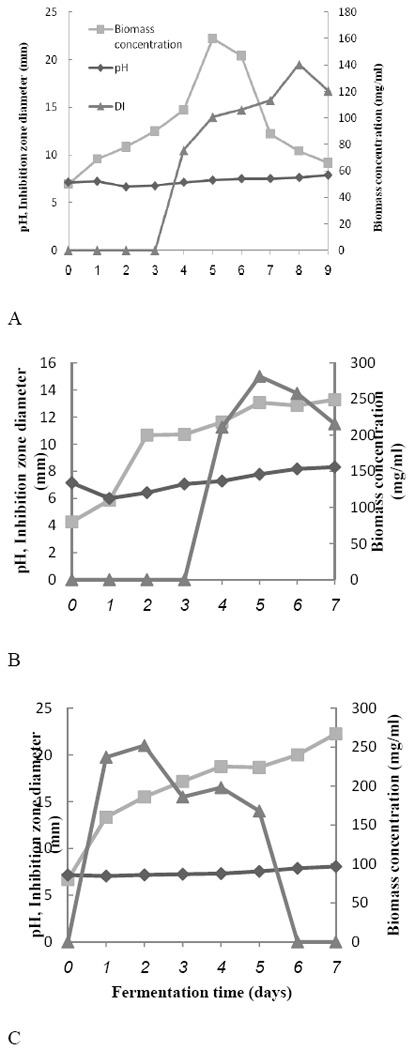
Growth and antibiotic production relationship of the selected strains A, HG87 B, HG116 and C, HG443 against S. Typhi
To find a relation between pH value, concentration of biomass and antibiotic production (in order to use them as rapid indicator of the process), the regression line equations between them were calculated. The coefficient (R2) between pH and antibiotic for HG87, HG116 and HG 742 were 0.71, 0.66 and 0.44, respectively. The coefficient (R2) between pH and biomass for HG87, HG116 and HG 742 were 0.007, 0.44 and 0.55, respectively. As seen, there is no significant correlation (p < 0.05) between the pH and biomass and antibiotic concentration. The related parameters to the rate and yield of product formation and biomass concentration are calculated and presented in Table 2.
Table 2.
Data analysis of growth and production parameters of antibiotic producing actinomycete isolates.
| Strains | Parameters |
||||
|---|---|---|---|---|---|
| rxmax | μmax | rpmax | qpmax | Yp/x | |
| HG87 | 1.87 | 0.019 | 2.6 | 0.026 | 1.4 |
| HG116 | 29 | 0.262 | 3.2 | 0.029 | 0.11 |
| HG443 | 22 | 0.212 | 10 | 0.097 | 2.09 |
Study of cytotoxic activity
The produced antibiotics were studied for their cytotoxic activities. In the issue of cancer, breast cancer has been estimated to be the second most common death cause among women. In proportion, kidney cancer encompasses lower prevalence and mortality rate, with more incidence cases among men and children (25). Tumor cell lines associated with the two mentioned cancers were subjected for evaluation of the cytotoxicity of the anti-S. Typhi active agents produced by the selected strains. Table 3 represents inhibitory rate of the cultured cells due to the crude extracts of the selected strains. The selected strain HG116, exhibited cytotoxic activity towards T47D cell lines at crude extract concentration of 1 μg/ml.
Table 3.
Cell inhibitory rate of the extracted bioactive metabolites against normal and tumor cell lines.
| Strains | Cell lines |
||
|---|---|---|---|
| HEK | USSC | T47D | |
| HG87 | 4.96% | 4.83% | 4.53% |
| HG116 | 20% | 13.23% | 87/97% |
| HG443 | 0.621% | 12.72% | 12.34% |
Taxonomical characterization of selected isolates
Comparison of an almost complete 16S rRNA sequence of the selected strains with that of Streptomyces species deposited in GenBank and EzTaxon database was done. The 16S rRNA sequence analysis of HG87 revealed 99.92% similarity to S. xinghaiensis. The other similar species to HG87 were S. feralitis and S. tateyamensis sharing 97.91% identity to the strain. HG116 shared 100% resemblance to S. griseoincarnatus, S. labedae, S. variabilis and S. erythrogriseus and HG443 was found to be most similar to S. toxytricini and S. globosus with a sequence identity of 100%. The selected strains HG87, HG116 and HG443 were deposited as Streptomyces sp. UTMC 01390 (GenBank accession number JX291534), Streptomyces sp. UTMC 01392 (GenBank accession number JX291533) and Streptomyces sp. UTMC 01397 (GenBank accession number JX291535), respectively and are accessible in University of Tehran Microorganisms Collection. The phylogenetic trees of the selected strains depict the evolutionary relationships between different Streptomyces species and the selected strains of this screening program (Fig. 3).
Fig. 3.
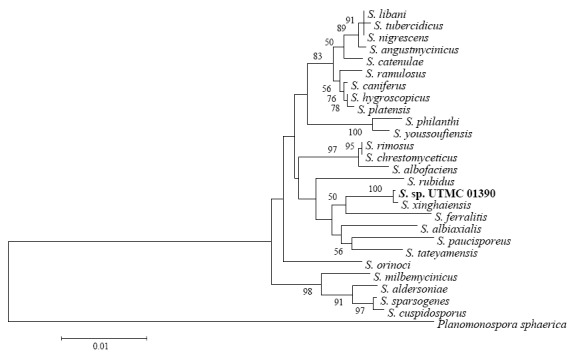
Phylogenetic trees of the 16S rRNA sequences of the selected strains A, Streptomyces sp. UTMC 01390, B, Streptomyces sp. UTMC 01392 and C, Streptomyces sp. UTMC 01397 (Bootstrap values under 50 are not shown)
Streptomyces is the largest bacterial genus (26). 16S rDNA-based similarity levels of more than 99% and even 100%, has been described between different Streptomyces species (27-28). Despite the high degree of 16S rRNA identity, the selected strains showed some distinctions with respect to their morphological, physiological and biochemical properties to their similar strains which increase the probability of being novel species. The comparison of the cultural, morphological and physiological features of the selected bioactive strains and the close Streptomyces species are summarized in Tables 4, 5 and 6.
Table 4.
Comparison of the cultural and morphological characteristics of strains Streptomyces sp. UTMC 01390 (HG87), Streptomyces sp. UTMC 01392 (HG116) and Streptomyces sp. UTMC 01397 (HG443) with their closest related species.
| Characteristic | HG87 | S. xinghaiensis | HG116 | S. labedae | S. variabilis | S. erythrogriseus | S. griseoincarnatus | HG443 | S. toxytricini | S. globosus |
|---|---|---|---|---|---|---|---|---|---|---|
| Color on ISP2 agar | White | White | Gray | Gray | Gray | Gray | Gray | White | Red | Gray |
| Spore chain morphology | Sa | RFc | S | RF | S or RAe | S | S or RA | RF | RF | RF |
| Spore Surface | Smooth | Smooth | Spiny | Smooth | Spiny | Spiny | Spiny | Smooth | Smooth | Smooth |
| Soluble pigments | b− | − | d+ | − | − | + or − | + | + | − | + |
| Melanoid pigments | + | − | − | − | − | − | − | + | + | + |
Spiral,
Negative,
Recti-flexibilis,
Positive,
Retina-culaperti
Table 5.
Comparison of the physiological properties of strains Streptomyces sp. UTMC 01390 (HG87), Streptomyces sp. UTMC 01392 (HG116) and Streptomyces sp. UTMC 01397 (HG443) with their closest related species.
| Characteristic | HG87 | S. xinghaiensis | HG116 | S. labedae | S. variabilis | S. erythrogriseus | S. griseoincarnatus | HG443 | S. toxytricini | S. globosus |
|---|---|---|---|---|---|---|---|---|---|---|
| Hydrolysis of starch | + | − | + | NA | NA | NA | + | + | ++ | NA |
| Degradation of tween 80 | + | NAb | + | + | d | NA | NA | + | d | NA |
| Caseinase | + | + | + | NA | NA | NA | + | + | + | NA |
| Chitinase | + | NA | − | NA | NA | NA | NA | + | NA | NA |
| Urease | + | NA | + | + | NA | − | + | + | NA | − |
| Lecithinase activity | − | NA | − | dc | − | NA | NA | + | + | NA |
| Liquefaction of gelatin | + | + | + | + | NA | + | d++ | + | we | w |
| Nitrate reduction | + | NA | − | d | d | NA | + | − | d | − |
| H2S production | NDa | NA | − | + | + | − | − | + | d | − |
| Tyrosinase reaction | − | NA | + | NA | NA | NA | W | + | NA | NA |
| Blood hemolysis | − | NA | + | NA | NA | NA | NA | + | NA | NA |
| Growth temperature | C°25-40 | C°10-45 | C°15-50 | NA | NA | NA | C°Growth at 45 | C°20-40 | NA | NA |
| pH range | 7-12 | 7-9 | 6-12 | NA | NA | NA | NA | 7-10 | NA | NA |
| NaCl tolerance | 0-7.5% | 0-9% | 0-9% | 0-7% | 0-7% | 0-5% | 10%~7 0- | 0-4% | NA | 0-5% |
Not detected,
Not available,
11-99% of strains are positive,
Strong activity,
Weak activity
Table 6.
Carbon source utilization patterns.
| Carbon source | HG87 | S. xinghaiensis | HG116 | S. labedae | S. variabilis | S. erythrogriseus | S. griseoincarnatus | HG443 | S. toxytricini | S. globosus |
|---|---|---|---|---|---|---|---|---|---|---|
| Glucose | − | + | + | + | + | + | + | + | + | + |
| Inositol | − | − | ++ | d | + | + | + | − | d | w/+ |
| Cellulose | a± | NA | ± | + | NA | − | − | − | NA | − |
| Mannitol | + | + | ++ | d | + | + | + | − | − | − |
| Raffinose | − | − | − | d | d | − | − | − | − | w/+ |
| Galactose | − | + | ++ | + | NA | NA | + | − | + | + |
| Rhamnose | − | + | ++ | d | + | + | d | − | d | − |
| Inuline | − | NA | + | NA | NA | NA | NA | ++ | NA | NA |
| Arabinose | ++ | − | ++ | + | + | + | + | − | + | + |
| Lactose | + | NA | ++ | NA | NA | NA | NA | − | NA | NA |
| Ribose | − | + | ± | NA | NA | NA | NA | − | NA | NA |
| Sorbitol | − | − | − | NA | NA | NA | NA | − | NA | NA |
| Sucrose | ++ | + | − | d | d | − | d | + | d | + |
| Glycerol | − | NA | + | NA | NA | NA | NA | + | NA | NA |
| Fructose | ++ | + | ++ | + | + | + | + | ± | − | w/− |
Doubtful utilization
The strain Streptomyces sp. UTMC 01390, produced white aerial spore mass and yellow substrate mycelium with no distinct diffusible pigments as for S. xinghaiensis. Colony margins were ruffled in S. xinghaiensis, while the macroscopical characteristic for Streptomyces sp. UTMC 01390 was smooth. This strain produced melanoid pigments on ISP6 medium whereas S. xinghaiensis did not. The aerial mycelium of the isolate Streptomyces sp. UTMC 01390 formed short spiral hyphae (Fig. 4A) whereas S. xinghaiensis formed long straight or flexuous hyphae. S. xinhaiensis failed to use arabinose sugar as its sole carbon source whereas Streptomyces sp. UTMC 01390 strongly utilized it. The other next obvious difference was the range of the temperature for growth. S. xinghaiensis had the ability to grow at 10°C but Streptomyces sp. UTMC 01390 could hardly grow at 20°C (29).
Fig. 4.
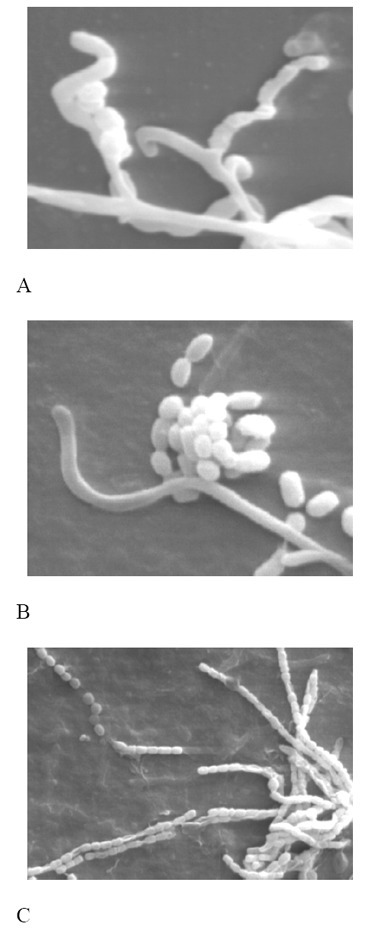
Scanning electron micrographs of A, Streptomyces sp. UTMC 01390 strain grown on ISP4 agar, B, Streptomyces sp. UTMC 01392 and C, Streptomyces sp. UTMC 01397 on ISP2 agar medium after incubation at 28°C for 14 days
The next selected strain, Streptomyces sp. UTMC 01392 developed spiral gray aerial mycelium (Fig. 4B). However, the available phenotypic characteristics of the similar species show clear distinctions with Streptomyces sp. UTMC 01392. From available data, it was evident that S. erythrogriseus differed in urease activity with Streptomyces sp. UTMC 01392. The representative strains of the related species of S. labedae and S. variabilis were positive for H2S production, but Streptomyces sp. UTMC 01392 gave negative result for the reaction. Moreover, positive response to production of diffusible pigments was not observed in ISP2 culture of S. variabilis. Finally, considering the last most similar species, S. griseoincarnatus, the ability to reduce nitrate possessed, whereas Streptomyces sp. UTMC 01392 was negative for this important biochemical test (30-31).
Having white aerial spore-mass, the other next selected strain Streptomyces sp. UTMC 01397, was mainly distinguishable from its related species (Fig. 4C). Negative lipolytic activity and absence of diffusible pigments in S. toxytricini, further differentiated it from Streptomyces sp. UTMC 01397. The latter characteristic is regarded as one of the key diagnostic characters in Streptomyces classification (32). Arabinose consumption and absence of H2S production by S. globosus were among distinctive phenotypic properties which discriminated these similar strains (20, 33-35).
DISCUSSION
In the present study, 1.7% of the isolates produced active compounds against S. Typhi. The isolates Streptomyces sp. UTMC 01390, Streptomyces sp. UTMC 01392 and Streptomyces sp. UTMC 01397 were found to have greater potency in inhibition of our pathogenic test strain; S. Typhi.
Assessing using well diffusion method, Streptomyces sp. UTMC 01390 was the most active strain against S. Typhi (the inhibition zone of 21 mm after 192 hrs) and showed the highest activity towards P. aeruginosa and K. pneumonia amongst the three potent strains. After extraction with the organic solvent, the aqueous phase of the strain Streptomyces sp. UTMC 01390 was able to suppress the growth of S. Typhi with the diameter of the inhibition zone of 15 mm. The crude extract of Streptomyces sp. UTMC 01392 showed the most inhibition activity against S. Typhi by disk method (21 mm). That can be ascribable to solubility of produced bioactive metabolite in ethyl acetate.
Higher specific growth rate was seen as the strain Streptomyces sp. UTMC 01392 subjected to antibiotic production assessment. The parameter was estimated to be the least for the strain Streptomyces sp. UTMC 01390. The less the specific growth rate, the easier would be the process of product recovery in large scale production programs at future. However, more yield of product to biomass and specific rate of antibiotic production were observed for Streptomyces sp. UTMC 01397 (Yp/x = 2.09, q pmax = 0.097). The strain showed specific potential in antibiotic production reaching to its maximum level after 48 hrs (r pmax = 10).
Usually, higher growth rate and productivity are the major characteristics in selection of industrial strains (36), therefore Streptomyces sp. UTMC 01397 can be considered as a promising candidate for further research.
The level of antibiotic production after 24 hrs was about 30% higher than the level reached after 72 hrs. The high production rate is an industrially valuable factor that might make the production economically efficient in the next stages.
Inhibition activity against S. Typhi is reported in some recent researches in bioassays accomplished in solid media (37-40). But these screenings were done alongside with other Gram positive and Gram negative pathogenic bacteria and were not solely directed towards S. Typhi. This target directed approach increase the chance of finding more potent strains and maybe helpful in dereplication of already known broad range antibiotics that are frequently dispersed among actinomyctes. To our best knowledge, this is the first literature in which S. Typhi has solely been the focus of a screening program of antibiotic producing actinomycetes.
Taxonomic identification proposed all of 3 selected anti-S. Typhi active strains as new isolates of Streptomyces genus. Phenotypic differences were more prominent for the strain Streptomyces sp. UTMC 01390. Streptomyces sp. UTMC 01392, as compared to its closest producing strains (41-42), could inhibit the growth of Gram negative pathogenic bacteria. Streptomyces sp. UTMC 01397 had broader spectrum of activity and stronger effect on pathogenic fungi than the producing strains of the assigned species; S. toxytricini and S. globosus (35). The differences in antimicrobial spectra can be indicative of involvement of different compounds.
The produced metabolite by the selected strains exhibit no cytotoxic activity on human normal cell lines and had inhibition rate of <50% (42). The strain Streptomyces sp. UTMC 01392, exposed high cytotoxic activity towards T47D cell lines (Inhibition rate of 87% against T47D cell lines). Consistent with the American National Cancer Institute (NCI) rules, the criteria for cytotoxic activity of crude extracts is an IC50 value of ≤30 μg/ml, hence based on the evidences the effective concentration was in accordance with the standard range.
Considering the mentioned results, all of the produced bioactive metabolites by the selected strains could be considered as candidates in antibiotic discovery for typhoid. The results lend support to the notion that in screening programs, the potential of Streptomycetes as a source of antibiotic could be still worthwhile (6). The findings suggest that screening unexplored habitats could lead to isolation of safer and broader spectrum antibiotic producing actinomycetes with high yield of production not only against existing and newly emerging drug resistant pathogens, but for application in treatment of various life-threatening diseases. More studies are needed to define the chemical characterization of compound(s) and genetic of the microorganisms.
References
- 1.Rupali P, Abraham O, Jesudason M, John T, Zachariah A. Treatment failure in typhoid fever with ciprofloxacin susceptible Salmonella enterica Serotype Typhi. Diagn Microbiol Infect Dis. 2004;49:1–3. doi: 10.1016/j.diagmicrobio.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 2.Rowe B, Ward LR, Threlfall EJ. Multidrug-resistant Salmonella Typhi: a worldwide epidemic. Clin Infect Dis. 1997;24:106–109. doi: 10.1093/clinids/24.supplement_1.s106. [DOI] [PubMed] [Google Scholar]
- 3.Morita M, Takai N, Terajima J, Watanabe H, Kurokawa M, Sagara H, Ohnishi K, Izumiya H. Plasmid-mediated resistance to cephalosporins in Salmonella enterica serovar Typhi. AntimicrobAgents Chemother. 2010;54:3991–3992. doi: 10.1128/AAC.00225-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Irajian G, Ranjbar R, Moghadas A. Detection of extended spectrum beta lactamase producing Salmonella spp. and multidrug resistance pattern. Iran J Pathol. 2009;4:128–132. [Google Scholar]
- 5.Jabeen K, Zafar A, Irfan S, Khan E, Mehraj V, Hasan R. Increase in isolation of extended spectrum beta lactamase producing multidrug resistant non typhoidal Salmonellae in Pakistan. BMC Infect Dis. 2010;10:101. doi: 10.1186/1471-2334-10-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Watve MG, Tickoo R, Jog MM, Bhole BD. How many antibiotics are produced by the genus Strepto-myces? Arch Microbiol. 2001;176:386–390. doi: 10.1007/s002030100345. [DOI] [PubMed] [Google Scholar]
- 7.Hamedi J, Mohammadipanah F, Klenk HP. Streptomyces iranensis sp. nov., isolated from soil. Int J Syst Evol Microbiol. 2010a;60:1504–1509. doi: 10.1099/ijs.0.015339-0. [DOI] [PubMed] [Google Scholar]
- 8.Hamedi J, Mohammadipanah F, Klenk HP, Pötter G, Schumann P, Kroppensted RM. Nocardiopsis sinuspersici sp.nov.,isolated from the sandy rhizospheric soil. Int J Syst Evol Microbiol. 2010b;60:2346–2352. doi: 10.1099/ijs.0.018366-0. [DOI] [PubMed] [Google Scholar]
- 9.Hamedi J, Mohammadipanah F, Pötter G, Spröer C, Schumann P, Göker M, Klenk HP. Nocardiopsis arvandica sp. nov., isolated from the banks of the Arvand river in Iran. Int J Syst Evol Microbiol. 2010c;61:1466–5026. doi: 10.1099/ijs.0.022756-0. [DOI] [PubMed] [Google Scholar]
- 10.Hamedi J, Mohammadipanah F, Maghsoudi N, Khodagholi F, Klenk HP. Inhibition of oxidative stress-induced amyloid β formation in NT2 neurons by culture filtrate of a strain of Streptomyces antibioticus. Appl Microbiol Biotechnol. 2010d;86:1805–1811. doi: 10.1007/s00253-010-2456-z. [DOI] [PubMed] [Google Scholar]
- 11.Hamedi J, Mohammadipanah F, Klenk H, Pötter G, Schumann P, Spröer C, Klenk HP. Nocardiopsis arvandica sp nov, isolated from the sandy soil of Iran. Int J Syst Evol Microbiol. 2011;61:1189–1194. doi: 10.1099/ijs.0.022756-0. [DOI] [PubMed] [Google Scholar]
- 12.Mohammadipanah F, Matasyoh J, Hamedi J, Klenk HP, Laatsch H, Persipeptides A. Persipeptides A and B, Two Cyclic Peptides from Streptomyces sp. UTMC 1154. Bioorg Medicinal Chem. 2012;20:335–339. doi: 10.1016/j.bmc.2011.10.076. [DOI] [PubMed] [Google Scholar]
- 13.Chatterjee S, Nadkarni SR, Vijayakumar EKS, Patel MV, Ganguli BN. Napsamycins, new Pseudomonas active antibiotics of the mureidomycin family from Streptomyces sp. HIL Y-82,11372. J Antibiot. 1994;47:595–598. doi: 10.7164/antibiotics.47.595. [DOI] [PubMed] [Google Scholar]
- 14.Saadoun I, Gharaibeh R. The Streptomyces flora of Jordan and it’s potential as a source of antibiotics active against antibiotic-resistant Gram-negative bacteria. World J Microbiol Biotechnol. 2002;18:465–470. [Google Scholar]
- 15.Galatenko OA, Trekhova LP. Isolation of antibiotic producing actinomycetes from soil samples exposed to UV light. Antibiot Khimioter. 1990;35:6–8. [PubMed] [Google Scholar]
- 16.EL-Nakeeb MA, Lechevalier HA. Selective isolation of aerobic actinomycetes. Appl Microbiol. 1963;11:75–77. doi: 10.1128/am.11.2.75-77.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lorian V. Antibiotics in laboratory medicine. Lippincott Williams and Wilkins; Baltimore: 2005. pp. 1–90. [Google Scholar]
- 18.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 19.Shirling EB, Gottlieb D. Methods for characterization of Streptomyces species. Int J Syst Bacteriol. 1966;16:313–340. [Google Scholar]
- 20.Bergey DH, Williams ST, Sharpe ME, Holt JG. Bergey’s manual of systematic bacteriology. Lippincott Williams and Wilkins; Baltimore: 1989. pp. 2452–2492. [Google Scholar]
- 21.Rabah FL, Elshafei A, Saker M, Cheikh B, Hocine H. Screening, isolation and characterization of a novel antimicrobial producing actinomycete, strain RAF10. Asian J Biotechnol. 2007;6:489–495. [Google Scholar]
- 22.Boudjella H, Bouti K, Zitouni A, Mathieu F, Lebrihi A, Sabaou N. Taxonomy and chemical characterization of antibiotics of Streptosporangium Sg 10 isolated from a Saharan soil. Microbiol Res. 2006;161:288–298. doi: 10.1016/j.micres.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 23.Kawasaki H, Koyama K, Kurokawa S, Watanabe K, Nakazawa M, Izawa K, Nakamatsu T. Production of (R)-3-amino-3-phenylpropionic acid and (S)-3-amino-3-phenylpropionic acid from (R,S)-N-acetyl-3-amino-3-phenylpropionic acid using microorganisms having enantiomer-specific amidohydrolyzing activity. Biosci Biotechnol Biochem. 2006;70:99–106. doi: 10.1271/bbb.70.99. [DOI] [PubMed] [Google Scholar]
- 24.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 25.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 26.Ramakrishnan J, Shunmugasundaram M, Narayanan M. Streptomyces sp. SCBT isolated from rhizosphere soil of medicinal plants is antagonistic to pathogenic bacteria. Iran J Biotechnol. 2009;7:75–81. [Google Scholar]
- 27.Arunachalam R, Wesely EG, George J, Annadurai G. Novel approaches for identification of Streptomyces noboritoensis TBG-V20 with cellulase production. Curr Res Bacteriol. 2010;3:15–26. [Google Scholar]
- 28.Rong X, Huang Y. Taxonomic evaluation of the Streptomyces griseus clade using multilocus sequence analysis and DNA–DNA hybridization, with proposal to combine 29 species and three subspecies as 11 genomic species. Int J Syst Evol Microbiol. 2010;60:696–703. doi: 10.1099/ijs.0.012419-0. [DOI] [PubMed] [Google Scholar]
- 29.Zhao XQ, Li WJ, Jiao WC, Li Y. Streptomyces xinghaiensis sp. nov., isolated from marine sediment. Int J Syst Evol Microbiol. 2009;59:2870–2874. doi: 10.1099/ijs.0.009878-0. [DOI] [PubMed] [Google Scholar]
- 30.Lacey J. Nomenclature of Saccharopolyspora erythraea Labeda and Streptomyces erythraeus (Waksman 1923) Waksman and Henrici 1948, and proposals for the alternative epithet Streptomyces labedae sp. nov. Int J Syst Bacteriol. 1987;37:458. [Google Scholar]
- 31.Palleroni NJ, Reichelt KE, Mueller D, Epps R, Tabenkin B. Production of a novel red pigment, rubrolone, by Streptomyces echinoruber sp. nov. J Antibiot (Tokyo) 1978;31:1218–1225. doi: 10.7164/antibiotics.31.1218. [DOI] [PubMed] [Google Scholar]
- 32.Kuster E. Simple working key for the classification and identification of named taxa included in the international Streptomyces project. J Syst Bacteriol. 1972;22:139–148. [Google Scholar]
- 33.Luo Y, Xiao J, Wang Y, Xu J, Xie S, Xu J. Streptomyces indicus sp. nov., a novel actinomycete isolated from deep-sea sediment of the Indian Ocean. Int J Syst Evol Microbiol. 2010;61:2712–2716. doi: 10.1099/ijs.0.029389-0. [DOI] [PubMed] [Google Scholar]
- 34.Shirling EB, Gottlieb D. Cooperative description of type cultures of Streptomyces V. Additional descriptions. Int J Syst Bacteriol. 1972;22:265–394. [Google Scholar]
- 35.Wu RY, Chen MH. Identification of the Streptomyces strain KS3-5. Bot Bull Acad Sin. 1995;36:201–205. [Google Scholar]
- 36.Waites MJ, Morgan NL, Rockey JS, Higton G. Industrial Microbiology: An Introduction. Wiley-Blackwell; 2001. pp. 75–85. [Google Scholar]
- 37.Baskaran R, Vijayakumar R, Mohan PM. Enrichment method for the isolation of bioactive actinomycetes from mangrove sediments of Andaman Islands India. Malaysia J Microbiol. 2011;7:26–32. [Google Scholar]
- 38.Hozzein WN, Rabie W, Ali MIA. Screening the Egyptian desert actinomycetes as candidates for new antimicrobial compounds and identification of a new desert Streptomyces strain. Afr J Biotechnol. 2011;10:2295–2301. [Google Scholar]
- 39.Remya M, Vijayakumar R. Isolation and characterization of marine antagonistic actinomycetes from west coast of India. Facta Univ Ser Med Biol. 2008;15:13–19. [Google Scholar]
- 40.Satheeja SV, Jebakumar SRD. Phylogenetic analysis and antimicrobial activities of Streptomyces isolates from mangrove sediment. J Basic Microbiol. 2011;51:71–79. doi: 10.1002/jobm.201000107. [DOI] [PubMed] [Google Scholar]
- 41.Hayakawa Y, Iwakiri T, Imamura K, Seto H, Otake N. Studies on the isotetracenone antibiotics III. A new isotetracenone antibiotic, grincamycin. J Antibiot (Tokyo) 1987;40:1785–1787. doi: 10.7164/antibiotics.40.1785. [DOI] [PubMed] [Google Scholar]
- 42.Sajid I, Shaaban KA, Hasnain S. Antitumour compounds from a saline soil isolate, Streptomyces griseoincarnatus CTF15. Nat Prod Res. 2011;25:549–559. doi: 10.1080/14786419.2010.534993. [DOI] [PubMed] [Google Scholar]
- 43.Sui X, Yin J, Ren X. Antiviral effect of diammonium glycyrrhizinate and lithium chloride on cell infection by pseudorabies. Antivir Res. 2010;85:346–353. doi: 10.1016/j.antiviral.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]


