SUMMARY
Histones and their post-translational modifications influence the regulation of many DNA-dependent processes. Although an essential role for histone-modifying enzymes in these processes is well established, defining the specific contribution of individual histone residues remains a challenge because many histone-modifying enzymes have non-histone targets. This challenge is exacerbated by the paucity of suitable approaches to genetically engineer histone genes in metazoans. Here, we describe a facile platform in Drosophila for generating and analyzing any desired histone genotype, and we use it to test the in vivo function of three histone residues. We demonstrate that H4K20 is neither essential for DNA replication nor for completion of development, unlike conclusions drawn from analyses of H4K20 methyltransferases. We also show that H3K36 is required for viability and H3K27 is essential for maintenance of cellular identity during development. These findings highlight the power of engineering histones to interrogate genome structure and function in animals.
INTRODUCTION
During animal development, a single genome gives rise to a wide diversity of cells. Each cell type differentially regulates genome activity in order to accurately execute a particular program of gene expression, cell cycle progression or DNA replication. Failure of this execution can lead to developmental defects or disease states that reduce organismal fitness. Because the genome sequence is essentially unchanged in most cell types, epigenetic mechanisms have been proposed to bring about cell-type specific regulation of genome activity (Margueron and Reinberg, 2010). Such mechanisms require a substrate that carries regulatory information and a means of propagating this information over time. Histone proteins are particularly attractive candidates as carriers of epigenetic information because they can fulfill both of these criteria. First, histone proteins have the potential to be dynamic regulators of genome activity because they are subject to a broad range of post-translational modifications (PTMs), including phosphorylation, acetylation, and methylation (Rothbart and Strahl, 2014). Histone PTMs are thought to contribute to regulation of genome activity by controlling chromatin packaging (Shogren-Knaak et al., 2006), and by serving as binding sites for protein complexes that control a variety of DNA-dependent processes including transcription, replication, and repair (Lachner et al., 2001). Second, histone proteins provide a potential means of propagating information over time through their partitioning to daughter cells during each cell division (Margueron and Reinberg, 2010).
Whereas critical roles for histone-modifying enzymes in the regulation of genome activity have been clearly demonstrated in a variety of species, the specific contribution of histone residues is less well understood (Henikoff and Shilatifard, 2011). Systematic mutagenesis in the budding yeast S. cerevisiae has identified histone residues essential for viability and for response to environmental challenges (Dai et al., 2008; Nakanishi et al., 2008). However, there are likely to be additional roles for histone residues in multicellular organisms, which exhibit diverse regulation of genome activity across different cell types and developmental stages. In multicellular organisms, the function of histone residues has largely been inferred from phenotypes caused by mutation of histone-modifying enzymes rather than by mutation of histone residues themselves. Examination of phenotypes caused by mutations in histone modifiers is not sufficient to make conclusions regarding causality because many of these enzymes have multiple substrates, including non-histone proteins (Glozak et al., 2005; Huang and Berger, 2008; Sims and Reinberg, 2008). More recently, substitution of methionine for lysine residues in histone proteins has been used to test the function of histone residues in animals (Herz et al., 2014). However these efforts are also insufficient to test causality because the methionine mutants are thought to act by dominantly interfering with histone methyltransferase activity (Lewis et al., 2013), which would likely impact all substrates of the methyltransferases.
A particularly powerful approach for studying the biological function of specific histone PTMs is to change the acceptor residue to an amino acid that cannot be appropriately modified and then to engineer a complete gene replacement for phenotypic analysis. Implementing this strategy in animals is technically challenging because metazoan histones are typically encoded by gene clusters found at multiple chromosomal locations (Marzluff et al., 2008). For example, the human genome has 64 histone genes, clustered at three different loci (Marzluff et al., 2002). In contrast, the Drosophila replication-dependent histone genes are found at a single locus (Lifton et al., 1978). Recently, Herzig and colleagues created a system for complementing deletion of the endogenous histone gene cluster with plasmid-based transgenes (Gunesdogan et al., 2010), allowing for the first analysis of histone residue function in animal development (Hodl and Basler, 2012; Pengelly et al., 2013). However, a minimum of four transgenes was required to rescue the histone locus deletion phenotype, limiting the ease with which this strategy can be used in combination with other genetic tools to study histone gene function in Drosophila.
Here, we present a BAC-based platform that can rescue deletion of the endogenous histone locus with a single transgenic insertion, allowing us to study not only the regulation of histone genes themselves, but also the specific contribution of histones to the regulation of DNA-dependent processes. After demonstrating its in vivo functionality, we used this platform to directly test the function during animal development of three post-translationally modified histone residues: H3K36, H3K27, and H4K20. Unlike results obtained in yeast, we show that H3K36 is required for viability in Drosophila. Consistent with current models, we find that H3K27 is required for the maintenance of Polycomb target gene repression, demonstrating that histone residues can perform an essential function in gene regulation. These results underscore the essential roles played by these two histone residues in gene expression and animal development. Finally, in contrast to current models, we show that a modifiable H4K20 residue is neither required for DNA replication nor for completion of Drosophila development. Together, these studies demonstrate the importance of directly testing the function of individual histone residues in animal development, and highlight the potential of this approach to test the role of histones in metazoan genome structure and function.
RESULTS
A BAC-based platform for histone gene replacement
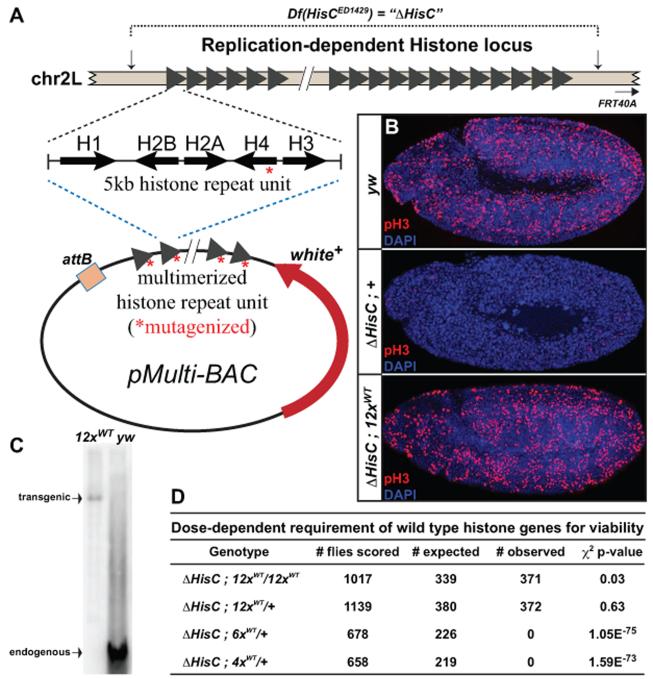
The Drosophila melanogaster replication-dependent histone genes are tandemly arrayed at a single locus on chromosome 2L (Figure 1A). Each 5kb repeat unit contains one copy of each of the four core histone genes (His2A, His2B, His3, His4), plus the linker histone, His1. Using the DrosDel system (Ryder et al., 2004), Herzig and colleagues (Gunesdogan et al., 2010) generated a precise deletion of the histone gene complex, termed Df(2L)HisCED1429 (hereafter ΔHisC). Because zygotic transcription of histone genes is first required during S-phase of cell cycle 15 (Smith et al., 1993; Gunesdogan et al., 2014), ΔHisC homozygotes cannot complete this cycle and die as embryos following depletion of the maternal histone contribution (Figure 1B).
Figure 1. A single histone replacement transgene rescues deletion of the endogenous histone locus.
(A) Schematic of the endogenous replication-dependent histone locus. Breakpoints of the deficiency are indicated by vertical arrows. Each triangle represents a single 5kb histone repeat unit, which was cloned and multimerized into a BAC vector for transgenesis. MCS: Multiple Cloning Site; w+: mini-white cassette; attB: site-specific recombination sequence. (B) Confocal images of cycle 15 embryos stained with antibodies for phospho-histone H3 (red) and DAPI (blue). Genotypes (top to bottom): wild type (yw); homozygous histone deletion (ΔHisC;+); homozygous histone deletion with two copies of the 12× wild type histone transgene (ΔHisC;12×WT). (C) Southern blot of SalI/XhoI digested genomic DNA from 12×WT and wild type (yw) flies, hybridized with an H2A probe. (D) Table of viability tests for various wild type histone arrays. The p-value for the chi-square test is shown. See also Figure S1.
In the Herzig approach, four independent plasmid-based transgenes bearing three copies of the 5kb histone repeat unit were needed to rescue ΔHisC (Gunesdogan et al., 2010). To create a more genetically facile system, we cloned tandem arrays of the native 5kb histone gene repeat unit into a BAC-based vector capable of site-specific transgenesis (Figures 1A and S1). The tandemly repeated organization of the native histone gene repeat sequence in these constructs maintains the cis-regulatory information required for proper histone gene expression, thereby avoiding potential cell toxic effects of histone over-expression or expression outside of S-phase (Gunjan and Verreault, 2003; Singh et al., 2010). Resupplying zygotic histone expression with a BAC-based transgene fully rescues the embryonic cell cycle arrest phenotype, as visualized by phospho-histone H3 staining during cell cycle 15 (Figure 1B), and supports development to adulthood. Thus, the BAC-based, transgenic histone arrays are functional in vivo.
To define the minimal number of transgenic histone genes needed for full rescue of the histone deletion phenotype, we generated tandem arrays with different numbers of repeat units (Figure 1A). These vectors were integrated into the same genomic location, thereby eliminating position-dependent effects on transgene expression. Southern blots of genomic DNA from histone replacement flies propagated for more than 50 generations show that the transgenic histone arrays are stable after integration in the genome (Figures 1C and S1). Viability tests showed that 6 or fewer histone gene repeats are insufficient to rescue lethality of ΔHisC (Figure 1D), whereas 12 or more repeats fully rescues lethality. When homozygous, the 12× transgene supports the propagation of a stable stock lacking all endogenous histone genes. We refer to these genotypes as “12×-Rescue” and “24×-Rescue” strains, respectively. Importantly, we did not observe any developmental delays in 12×-Rescue flies, either in the timing of larval hatching or adult eclosion. We note that the fertility of 24×-Rescue and 12×-Rescue females is somewhat decreased relative to wild type flies; however, the basis for this defect is not known.
The Drosophila genome contains 100 copies of the histone repeat unit
The ability of a single transgene containing 12 histone repeats to support development to adulthood is somewhat surprising, given that original estimates suggested that there are upwards of 100 copies of the histone repeat unit on chromosome 2L (Lifton et al., 1978). In contrast, current genome annotations (FlyBase release version FB_2014_1) list only 23 histone repeats. Thus, the precise number of histone genes in the Drosophila genome remains an open question.
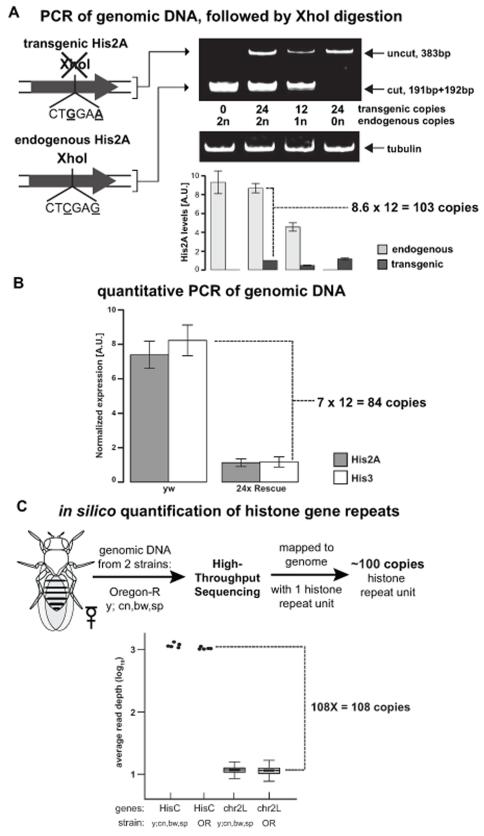
We took two complementary approaches to directly measure the number of endogenous histone genes. First, we reasoned that transgenic histone arrays could be used as in vivo calibrators to accurately measure the endogenous gene copy number by PCR. To discriminate between endogenous and transgenic His2A DNA, we engineered a silent mutation in an XhoI site within the transgenic His2A gene (Figure 2A). Using PCR primers that recognize both endogenous and transgenic templates, we amplified His2A genomic DNA from four genotypes and digested the PCR products with XhoI, cutting the endogenous His2A fragment in two equal halves while leaving the transgenic His2A product intact. Following electrophoresis, quantification of band intensities revealed that the endogenous His2A template is 8-fold more abundant than the transgenic His2A template (Figure 2A). Importantly, semi-quantitative PCR reactions from both the endogenous and transgenic His2A templates are within the linear range, as shown by XhoI digestion assays using genomic DNA from four genotypes with different histone gene copy numbers (Figure 2A). Consistent with our measurements from the XhoI digestion assay, real time PCR indicates that the His2A and His3 genes are 7-fold more abundant in wild type flies than 24×-Rescue flies (Figure 2B). These experiments indicate that the haploid Drosophila genome contains approximately 100 histone repeats.
Figure 2. The haploid Drosophila genome contains 100 copies of the histone repeat unit.
(A) Ethidium bromide-stained gel of XhoI-digested PCR products of endogenous and transgenic His2A genes. For each of the four genotypes, barplots of normalized band intensity are shown below each lane. Error bars represent standard error of the mean (SEM). (B) Barplots of normalized real-time PCR results for wild type (yw) and 24× Rescue genotypes using primers to His2A and His3. Error bars: SEM. (C) Flow chart and plots of in silico quantification of histone gene repeats for two wild type strains (Oregon R (OR), and y;cn,bw,sp). HisC: total read depth for each of the five replication-dependent genes; chr2L: box plots of average read depth for the remaining genes on chromosome 2L. The box represents the inner quartile range (IQR), and whiskers represent 1.5-times IQR. For clarity, outliers were not plotted.
Second, we calculated the histone gene copy number using high-throughput sequencing analysis. We reasoned that the abundance of histone sequences relative to those of other genes on chromosome 2L would reflect the number of copies of histone genes in the genome. To accurately measure their abundance, we sequenced genomic DNA from two different strains and mapped reads to a custom Drosophila genome containing a single histone gene repeat unit (see Experimental Procedures). Comparison of the average read density across the coding sequence of each histone gene to the average read density across coding sequences of the remaining annotated genes on chromosome 2L revealed that the histone genes are ~100-fold more abundant (Figure 2C), consistent with our PCR assays and the original estimates (Lifton et al., 1978).
A histone gene dosage compensation mechanism
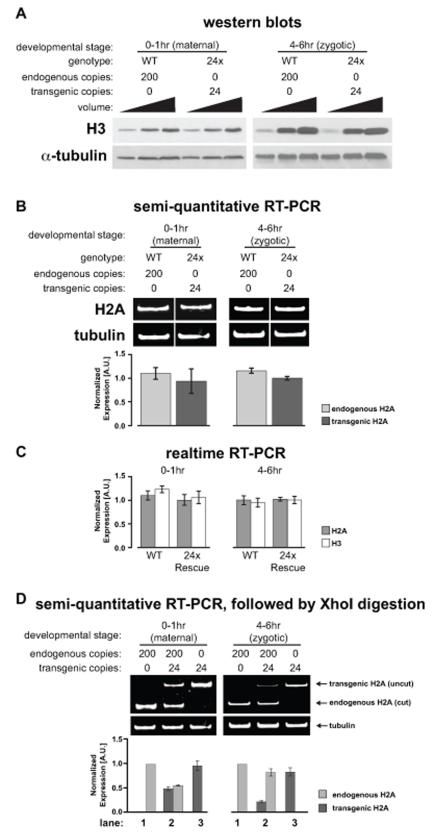
The preceding experiments show that wild type diploid flies contain ~200 copies of the histone repeat unit, and yet a single 12× histone transgene is sufficient to support development of flies lacking all endogenous histone genes. We therefore compared expression levels between the endogenous and transgenic histone genes. Western blot and RT-PCR analysis at two stages of embryogenesis (0-1hr and 4-6hr) showed no significant differences in histone protein or mRNA levels between wild type and 24×-Rescue flies (Figures 3A-C). Because the zygotic histone genes are not active in 0-1hr embryos, histone levels at this time point reflect maternal protein and mRNA derived from the activity of the histone genes during oogenesis. The 4-6hr time point includes cell cycle 15, when zygotic histone gene activity is first required due to destruction of the maternal histone mRNA supply. Despite different demands on histone gene activity between these two stages, the 24× transgenic histone genes produce the same amount of protein and mRNA as 200 copies of the endogenous histone genes (Figure 3A-C). Thus, histone replacement flies express equivalent steady-state levels of histones as wild type flies, despite a ten-fold difference in gene copy number.
Figure 3. Transgenic histone arrays are expressed at levels similar to the endogenous genes.
(A) Western blot of wild type (WT) and 24× Rescue genotypes at 0-1hr and 4-6hrs after egg laying. (B) Ethidium bromide stained gel of RT-PCR products from 0-1hr and 4-6hr wild type (WT) and 24× Rescue embryos. Barplots of normalized band intensity are shown below each lane. Error bars: SEM. (C) Barplots of normalized real-time RT-PCR results for H2A and H3 in 0-1hr and 4-6hr wild type (WT) and 24× Rescue embryos. Error bars: SEM. (D) Ethidium bromide stained gel of XhoI-digested RT-PCR products from 0-1hr and 4-6hr embryos for three genotypes: wild type (lane 1); +/+;12×WT/12×WT (lane 2); 24× Rescue (lane 3). Barplots of normalized band intensity are shown. Error bars: SEM. See also Figure S2.
As both the protein levels and the amino acid sequences of the endogenous and transgenic histones are identical, we infer that the nucleosome and higher-order chromatin organization is similar across the genome in wild type and 24×-Rescue flies. In addition, 12×- and 24×-Rescue flies show no increase in sensitivity to the DNA-replication inhibiting agent hydroxyurea (Figure S2, and data not shown), as we hypothesize would occur if histone production during S-phase was limiting in these animals.
The similar amount of mRNA produced in 24×-Rescue and wild type flies suggests the existence of a histone gene dosage compensation mechanism. To test whether such a mechanism exists, we compared the levels of mRNA in wild type and 24×-Rescue flies to those in flies containing both endogenous and transgenic histone genes (“endogenous + 24×”), discriminating between them using the XhoI digestion assay described above (Figure 3D). Similar to our results from undigested samples (Figure 3B), His2A mRNA levels are the same in wild type and 24×-Rescue embryos (Figure 3D, lane 1 and lane 3). By contrast, His2A mRNA levels originating from both endogenous and ectopic histone genes are reduced in “endogenous + 24×” embryos compared with wild type and 24×-Rescue embryos (Figure 3D, lane 2). Importantly, the sum of endogenous plus ectopic His2A mRNA in “endogenous + 24×” flies equals the levels observed in wild type or 24×-Rescue embryos. Thus, the total amount of histone mRNA at a given stage of embryogenesis is the same for each genotype, suggesting that the steady state level of RNA expressed from individual histone genes is scaled to the total number of histone genes present in the genome.
Transgenic histone gene arrays assemble HLBs that accurately process histone transcripts
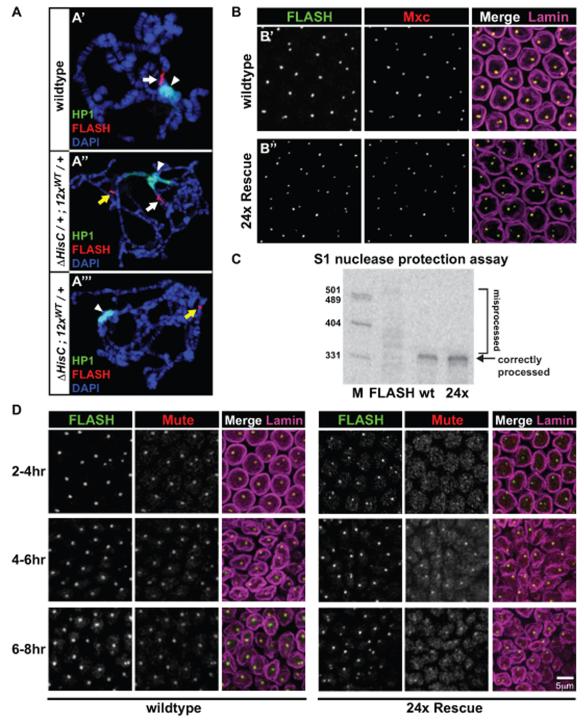
The endogenous histone locus assembles a nuclear sub-compartment termed the Histone Locus Body (HLB), which is thought to facilitate efficient transcription and mRNA processing during S-phase (Liu et al., 2006; Salzler et al., 2013). To test for HLB assembly at our transgenic arrays, we performed immunofluorescence on salivary gland polytene chromosome spreads. In these polyploid cells, the genome is amplified up to 1000-fold, and sister chromatids are aligned in register, allowing for sensitive and high-resolution cytology. Using antibodies to HLB components FLASH, Mxc, and Mute (Figure 4A, and data not shown), we observed a single HLB in wild type polytene chromosomes at the endogenous histone locus on chromosome 2L (Figure 4A’). In “endogenous + 12×” larvae we observed two HLBs: one assembled at the endogenous histone locus and a second (smaller) one at an ectopic location corresponding to the 12× wild-type transgene inserted on chromosome 3L (Figure 4A”). Finally, in 12×-Rescue larvae, which contain no endogenous histone genes, we only detected a single HLB at the ectopic site (Figure 4A’”). We conclude that HLBs assemble at transgenic, ectopic histone loci in salivary gland polytene chromosomes, similar to previous findings (Salzler et al., 2013).
Figure 4. The transgenic histone gene locus assembles an HLB that accurately processes histone transcripts.
(A) Confocal images of salivary gland polytene chromosome squashes stained for FLASH (red), HP1 (green), and DAPI (blue) for the three indicated genotypes. White arrow: endogenous HLB; white arrowhead: chromocenter; yellow arrow: transgenic HLB. (B) Confocal images of blastoderm stage embryos stained for FLASH (green), Mxc (red), and Lamin (magenta) for the two indicated genotypes. (C) Phosphorimager scan of S1 nuclease protection assay performed on total RNA from three genotypes: FLASHPBac/FLASHDf; Oregon R (wt, 4-6hrs); 24× Rescue (24×, 4-6hrs). M: Markers. (D) Confocal images of embryos at 2-4hrs, 4-6hrs, and 6-8hrs stained for FLASH (green), Mute (red) and Lamin (magenta) for wild type and 24× Rescue embryos. For B and D, the maximum projection of four 0.5-micron slices is shown.
To test whether we could also detect HLBs in 24×-Rescue diploid cells, we performed immunofluorescence on post-blastoderm stage embryos. In 24×-Rescue embryos we detected Mxc and FLASH foci in 100% of nuclei (Figure 4B”). Consistent with these cytological results, and with the absence of defects in viability (Figure 1C), S1 nuclease protection assays showed that histone mRNAs were processed normally in 24×-Rescue animals (Figure 4C). However, we note that HLB assembly is not fully recapitulated in histone replacement animals. Localization of Mute reveals diffuse nuclear staining in 2-4hr 24×-Rescue embryos, in contrast to the discrete foci observed when detecting Mute in wild type embryos at this stage (Figure 4D, 2-4hrs). In 24×-Rescue embryos, Mute becomes more concentrated in the HLB as embryogenesis proceeds, and its localization resembles that of wild type embryos by 6-8hrs (Figure 4D), although it never fully achieves the wild type pattern. Previous work has shown that Mute is a repressor of histone gene expression in Drosophila (Bulchand et al., 2010), raising the possibility that Mute’s localization to the HLB in 24×-Rescue embryos helps regulate histone gene transcription.
Together, these data show that we can engineer a functional replacement of the endogenous replication-dependent histone genes, presenting an opportunity to interrogate the function of individual histone residues in animal development. To test this premise, we engineered genotypes that prevent post-translational modification of three different histone residues with proposed roles in well-characterized epigenetic pathways.
H4K20 is dispensable for DNA replication and viability
Histone H4 lysine 20 (H4K20) can be mono-, di-, or tri-methylated, and mono-methylation is cell cycle regulated (Jorgensen et al., 2013). During DNA replication, newly incorporated histones have low levels of H4K20 methylation because of S phase-coupled destruction of PR-Set7/SET8, the enzyme that catalyzes H4K20 mono-methylation (H4K20me1; Havens and Walter, 2011). During G2, H4K20me1 levels rise, allowing for di- and tri-methylation by Suv420H1/H2 enzymes during the subsequent G1-phase. Experimental manipulation of H4K20 methyltransferases suggests that H4K20 methylation contributes to multiple DNA-dependent processes, including replication, maintenance of genomic integrity, and chromatin condensation (Beck et al., 2012b). For example in Drosophila, PR-Set7 mutants display defects in heterochromatin silencing, and PR-Set7 null animals die during larval stages, exhibiting cell proliferation defects (Karachentsev et al., 2005). In mammalian cells, expression of a non-degradable PR-Set7 induces re-replication of DNA due to aberrant licensing of replication origins (Beck et al., 2012b; Tardat et al., 2010). Although these experiments demonstrate the essential role of H4K20 methyltransferases in DNA replication and other processes, the specific function of the H4K20 residue remains untested.
To directly test the requirement for H4K20 in vivo, we generated a histone replacement transgene in which the H4K20 residue is mutated to alanine (12×H4K20A). Expression of H4K20A histones in an otherwise wild type genetic background had no observable phenotype. Given the putative critical role of H4K20 methylation in chromosome duplication and condensation, we were surprised to find that 56% (n=554) of homozygous ΔHisC files with a 12×H4K20A transgene survive to adulthood (hereafter, H4K20A replacement flies). Although H4K20A replacement flies exhibit a significant developmental delay (24-48 hours), their survival is in direct contrast to expectations, as 100% of PR-Set7 null animals die as larvae (Karachentsev et al., 2005).
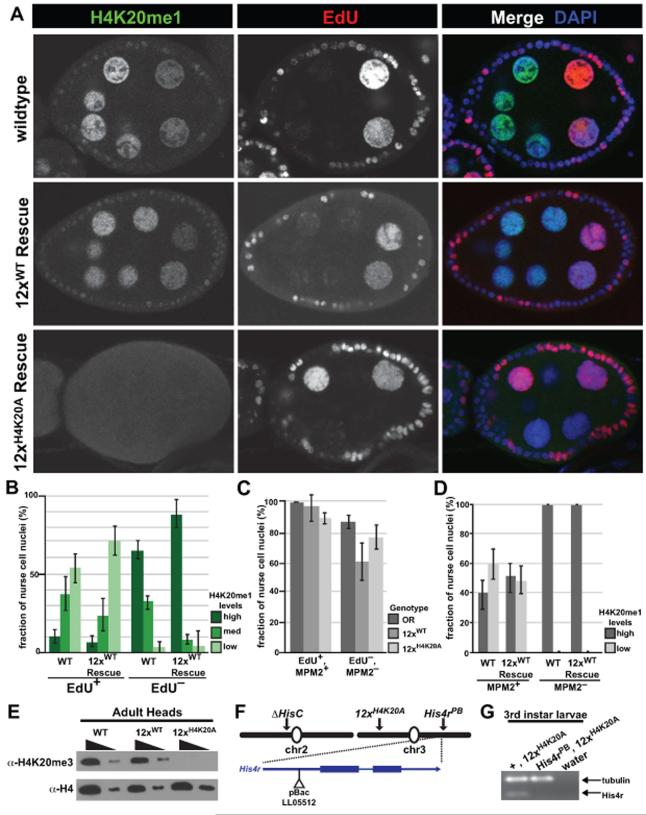
To explicitly test the requirement of H4K20 modification in DNA replication, we performed EdU incorporation studies in combination with H4K20me1 antibody staining of egg chambers from wild type, 12×WT, and 12×H4K20A adult ovaries (Figure 5A, B). The germline nurse cells and somatic follicle cells of Drosophila egg chambers are polyploid, making them amenable to cytological assays. In addition, these cells endoreduplicate asynchronously, allowing for direct comparisons between different stages of the cell cycle within a single egg chamber. We detected robust EdU incorporation in wild type and 12× Rescue nurse and follicle cell nuclei that is inversely correlated with H4K20me1 levels (Figure 5A, B), consistent with previous findings that PR-Set7 activity is lowest during S-phase. We corroborated this result using MPM-2 monoclonal antibody staining of the histone locus body, a readout of CycE-Cdk2 activity that is commonly used as a proxy for active DNA replication (Figure 5C, D, S3A, S3B). In egg chambers from H4K20A replacement flies, there is no detectable H4K20me1 signal in nurse cell or follicle cell nuclei (Figure 5A, S3B). Nevertheless, we detected robust EdU incorporation in these nuclei (Figure 5A, C, S3A), demonstrating that they are capable of DNA replication in the absence of histone H4K20 modification. Because cells lacking detectable H4K20 mono-methylation can actively incorporate EdU during S-phase and progress to gap-phases, we conclude that canonical H4K20 methylation is not essential for viability, cell-cycle progression, or DNA replication.
Figure 5. H4K20 is dispensable for DNA replication and viability.
(A) Confocal images of stage 6-8 egg chambers stained for H4K20me1 (green), EdU (red), and DAPI (blue). (B) Barplot of the fraction of nurse cell nuclei with indicated H4K20me1 levels for each category of EdU incorporation. Error bars: SEM. (C) Barplot of the fraction of nurse cell nuclei with shared EdU and MPM2 status for each genotype. Error bars: SEM. (D) Barplot of the fraction of nurse cell nuclei with the indicated H4K20me1 levels for each category of MPM2 staining. Error bars: SEM (E) Western blots of adult head extracts from 3 genotypes: wild type (WT), 12× Rescue (12×WT), and H4K20A replacement flies (12×H4K20A). (F) Genetic scheme for analysis of His4r function in H4K20A replacement flies. (G) Ethidium bromide stained gel of RT-PCR products from H4K20A replacement flies. Genotypes: (lane 1) ΔHisC/ΔHisC; His4rPB, 12×H4K20A/TM6B. (lane 2) ΔHisC/ΔHisC; His4rPB, 12×H4K20A/ His4rPB. See also Figure S3.
The Drosophila genome contains a single-copy gene encoding a replication-independent version of histone H4 (His4r) that is identical in amino acid sequence to canonical H4 (Akhmanova et al., 1996). Although our immunofluorescence and western blot experiments (Figure 5A, E) could not detect methylation of H4K20, expression of His4r could, in theory, provide a sufficient amount of modifiable H4K20 to compensate for the absence of replication-dependent H4K20. RT-PCR showed that His4r mRNA levels are unchanged in 12×H4K20A replacement flies, arguing against the bulk replacement of canonical H4 with His4r (Figure S3D). To genetically test for the requirement of His4r in 12×H4K20A replacement flies, we obtained a fly line with a Piggy-Bac transposon insertion near the 5′ end of His4r (hereafter His4rPB) (Figure S4B). Sequencing of PCR amplicons from His4rPB flies showed that the transposon is inserted in the first intron or 5′ UTR of His4r, depending on the gene isoform (Figure S3E). His4rPB homozygotes are viable at sub-mendelian ratios, but they are sterile. RT-PCR of whole His4rPB adults or dissected ovaries showed no detectable His4r mRNA (Figure S3F). Thus, His4rPB is a null or strong hypomorphic allele of His4r.
We next took advantage of the facile genetics afforded by our histone replacement platform by recombining the His4r mutant allele with the 12×H4K20A transgene (Figure 5F). 3rd instar larvae homozygous for both His4rPB and ΔHisC covered by 12×H4K20A showed no expression of His4r by RT-PCR (Figure 5G). Despite this fact, ~85% of these flies pupate and 35% develop until late pharate adults (Figure S3G). In one instance, we obtained an overtly healthy adult fly that contained no wild type copies of H4, demonstrating that H4K20 is not essential for completion of development. We note that this experiment may overestimate the importance of H4rK20 because it was performed in the absence of any H4r expression, and thus in the absence of any function of H4r independent of its K20 residue. Together, these findings demonstrate the importance of directly testing the functions ascribed to histone residue PTMs, rather than relying on indirect inferences from mutant phenotypes of the enzymes that catalyze their modification.
H3K27 is required for heritable silencing of Polycomb target genes
The signaling events that specify cell fates during development of multicellular organisms are often transient, yet these decisions need to be remembered over time, through multiple rounds of cell division. Methylation of histone H3 on lysine 27 (H3K27me3) is associated with heritable repression of Polycomb group (PcG) target genes (Cao et al., 2002; Czermin et al., 2002; Muller et al., 2002). Genetic and biochemical studies have identified multiple protein complexes that are required for PcG target gene repression (Klymenko et al., 2006; Kuzmichev et al., 2002; Shao et al., 1999). However, the specific role of H3K27 in PcG target gene repression is less well understood.
We generated histone replacement constructs with either 8 or 12 histone gene repeat units in which H3K27 is mutated to alanine (H3K27A). We recovered transgenic flies with 8 H3K27A tandem gene repeats (8×H3K27A), but did not recover a 12×H3K27A transformant. The presence of 8×H3K27A transgenes in an otherwise wild type genetic background elicited several dominant phenotypes, including decreased viability and diminished fertility. A hemizygous ΔHisC background enhanced these phenotypes, and also resulted in new phenotypes, including aberrant leg and wing morphogenesis (Figure S4, and not shown). The posterior wing phenotype is reminiscent of gain-of-function alleles of Ultrabithorax (Ubx), in which Ubx is ectopically active in cells of the second thoracic segment where it is normally inactive in wild type animals (Lewis, 1978). Ubx is a well-characterized target of PcG complexes in the wing (Christen and Bienz, 1994). To determine whether expression of H3K27A histones results in ectopic expression of Ubx, we performed immunofluorescence experiments on 3rd instar imaginal wing discs. Similar to wild type flies, we detected no Ubx expression in the main epithelium of flies bearing 12×WT transgenes (Figure S4D). By contrast, we detected low-level ectopic expression of Ubx in the posterior compartment of 8×H3K27A 3rd instar imaginal wing discs (Figure S4E), and this ectopic expression of Ubx is enhanced in ΔHisC hemizygous animals (Figure S4F). These experiments are consistent with a dose-dependent requirement for modified H3K27 histones in PcG target gene repression, and indicate that incorporation of non-modifiable H3K27A into chromatin interferes with the ability of PcG complexes to repress target genes.
Because methylation of H3K27 is found at target genes repressed by PcG complexes, we tested for a genetic interaction between a Pc loss of function allele (PcXT) and the 8×H3K27A transgene. The Pc gene is haploinsufficient, and both PcXT heterozygous flies and 8×H3K27A transgenic flies exhibit ectopic sex combs at low frequency (8% and 1%, respectively; Figure S4G). However, 8×H3K27A transgenic flies that are also heterozygous for PcXT exhibit ectopic sex combs at a much higher frequency (68%; Figure S4G). Thus, the presence of H3K27A histones enhances the Pc ectopic sex comb phenotype, consistent with a role for modified H3K27 in PcG target gene silencing.
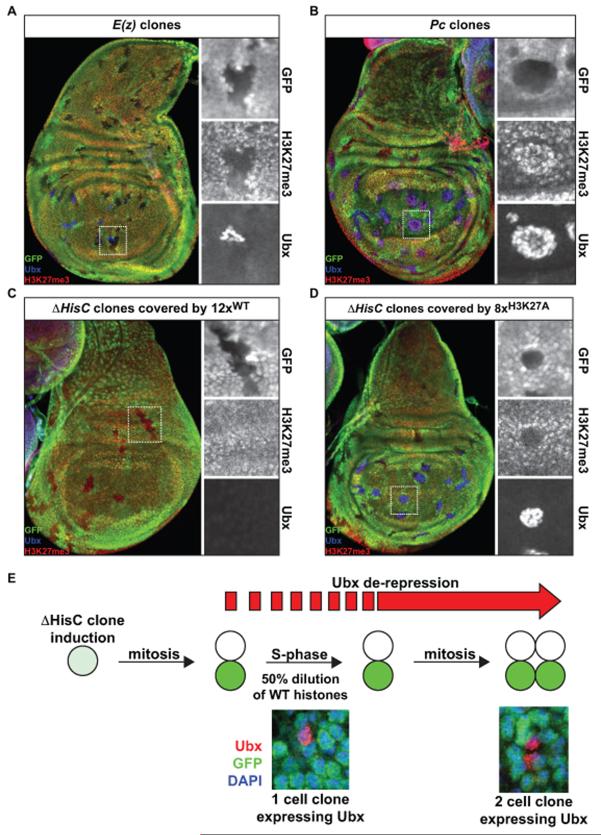
To directly test the requirement of H3K27 in PcG target gene repression, we used the FLP-FRT system (Xu and Rubin, 1993) to generate and compare cell clones that express non-modifiable H3K27A to those that are mutant for E(z), the enzyme that catalyzes methylation of H3K27 (Czermin et al., 2002; Muller et al., 2002), as well as for Pc, which specifically binds to H3K27me3 at PcG target genes (Cao et al., 2002). Clones of E(z) mutant wing imaginal disc cells show ectopic expression of Ubx in the wing pouch near the dorsal-ventral boundary (Figure 6A), consistent with previous findings (Muller et al., 2002). Clones of cells lacking Pc function also ectopically express Ubx in the same spatial pattern as E(z) mutant clones (Figure 6B). Since these clones were made at the same time (48-60 hours after egg laying), the increased Ubx levels in Pc clones relative to E(z) clones may be due to differences in Pc and E(z) protein stability. Strikingly, ΔHisC clones containing an 8×H3K27A transgene (hereafter, H3K27A clones) show robust ectopic expression of Ubx in the same spatial pattern as do E(z) and Pc mutant clones, unlike ΔHisC clones containing a 12×WT or 6×WT transgene that do not express Ubx (Figure 6C, D). H3K27A clones also exhibit ectopic expression of Abd-B and En, two other PcG target genes in wing imaginal discs (not shown). The restriction of ectopic PcG target activity only to clones near the D/V boundary demonstrates that replacement of wild type H3 with H3K27A does not result in widespread de-regulation of gene expression. Instead, there is a specific de-regulation of PcG target genes in the expected cell populations within wing imaginal discs. ΔHisC clones covered by an 8× histone replacement transgene in which H3K27 was mutated to an arginine instead of an alanine (8×H3K27R) de-regulate PcG target genes in the same spatial profile (Figure S4H), demonstrating that loss of a modifiable H3K27 is responsible for the phenotype rather than loss of the positive charge on the lysine side chain, consistent with another recent report (Pengelly et al., 2013). These experiments provide direct evidence that H3K27 is required for regulation of PcG target genes. Activation of PcG target genes in the absence of a modifiable H3K27 residue also demonstrates that other modifications of H3K27 (e.g. acetylation) are not required for expression per se.
Figure 6. H3K27 is required for Polycomb target gene repression.
(A-D) Confocal images of imaginal wing discs of the indicated genotypes with 48-60hr mitotic clones stained for Ubx (blue) and H3K27me3 (red). Mutant and ΔHisC clones are marked by absence of GFP (green). The square outlines the magnified area at the right. (E) Schematic of clone induction and confocal images of 2 different clones induced 24hrs prior to dissection. GFP (green), Ubx (red), DAPI (blue). See also Figure S4.
A hallmark of epigenetic regulators like Polycomb is their ability to propagate gene regulatory states through many cell divisions (Simon and Kingston, 2013). Histone modification levels, chromatin composition, and chromosome structure all change dramatically through the cell cycle (Follmer et al., 2012; Fonseca et al., 2012), raising the possibility that changes in epigenetic states may depend on passage through a particular stage of the cell cycle. We took advantage of the high temporal resolution of mitotic clone analysis to determine when ectopic expression of Ubx is first detectable following removal of wild type histones. Consistent with a cell-doubling time of 12-24 hours in wing discs (Shibutani et al., 2008), we observed many H3K27A clones consisting of only two cells 24-hours after inducing H3K27A clones. Many of these clones show high levels of ectopic Ubx expression (Figure 6E). Remarkably, we also found single cell H3K27A clones with robust ectopic Ubx activity (Figure 6E). Single cell H3K27A clones have yet to divide following induction of the clone. Because the cell cycle-dependent histones are only expressed during S-phase, the observed ectopic expression of Ubx indicates that these clones have completed DNA replication and are in the G2 phase of the cell cycle. Thus, we conclude that cell division is not necessary for ectopic activation of Ubx, and that a 50% dilution of wild type histones by H3K27A histones after a single S-phase results in de-repression of Ubx in the wing.
H3K36 is required for completion of development
Methylation of lysine 36 on histone H3 (H3K36) is a PTM associated with actively transcribed genes (Bannister et al., 2005; Strahl et al., 2002). Studies in the budding yeast S. cerevisiae have demonstrated a role for H3K36 methylation in suppression of spurious transcription initiation in the wake of transcribing RNA polymerase by recruitment of histone deacetylases to gene bodies (Carrozza et al., 2005; Keogh et al., 2005). In animals, depletion of SETD2, the enzyme that catalyzes H3K36me3, is reported to cause dysregulation of alternative exon inclusion, implicating H3K36 methylation in the regulation of pre-mRNA splicing (Luco et al., 2010). A role for H3K36 methylation in regulation of sex chromosome dosage compensation has also been reported (Larschan et al., 2007).
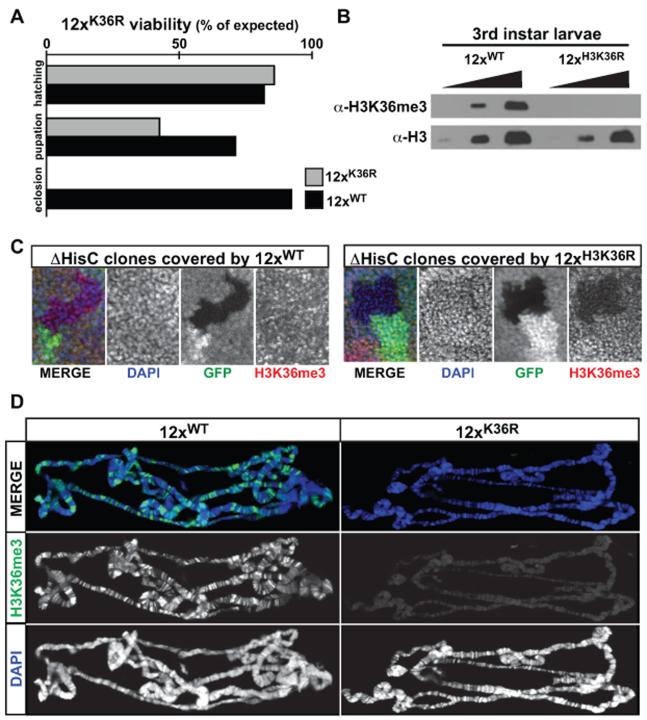
We generated a 12× histone replacement transgene in which H3K36 is mutated to arginine (12×H3K36R). When carried in a wild type or ΔHisC hemizygous background, 12×H3K36R transgenes cause no discernible developmental or fertility defects, demonstrating that expression of H3K36R histones does not result in overtly dominant phenotypes. By contrast, 100% of homozygous ΔHisC animals containing one 12×H3K36R histone replacement transgene (hereafter H3K36R replacement flies) die before the end of pupal development (Figure 7A). Western blots of wild type and H3K36R replacement larvae show that H3K36R histones are expressed and are not recognized by anti-H3K36me3 antibodies (Figure 7B). To examine the cellular basis for the requirement of H3K36 in development, we generated clones of wing imaginal disc cells lacking endogenous histones. Although we observed a marked decrease in H3K36me3 levels in ΔHisC clones covered by a 12×H3K36R transgene (Figure 7C), we did not observe a significant difference in the size of these clones relative to their wild type twins (not shown). We also observed a dramatic decrease in H3K36me3 levels in polytene chromosomes from H3K36R replacement 3rd instar larvae (Figure 7D). Despite this depletion, we detected no apparent defects in chromosome structure. Together, these experiments demonstrate that H3K36 is essential for viability in Drosophila, in contrast to results from budding yeast, where H3K36 mutations show no overt growth defects (Kizer et al., 2005).
Figure 7. H3K36R replacement flies are inviable and lack H3K36me3.
(A) Barplots of the expected fraction of viable flies at each indicated stage. We used the denominator from the previous stage to calculate the percentage observed at each stage. (B) Western blots from wild type (WT) and H3K36R replacement (12×H3K36R) 3rd instar larval nuclei. (C) Confocal images of imaginal wing discs with 48-60hr mitotic clones stained for H3K36me3 (red), and DAPI (blue). ΔHisC clones are marked by absence of GFP (green). (D) Confocal images of salivary gland polytene chromosomes from 12× Rescue (12×WT) and H3K36R replacement flies (12×H3K36R) stained for DAPI (blue) and H3K36me3 (green).
DISCUSSION
A facile genetic platform to study histone function
Distinguishing direct from indirect effects caused by mutations in histone-modifying enzymes can be difficult because histone modifiers can have multiple substrates, including non-histone proteins (Glozak et al., 2005; Huang and Berger, 2008; Sims and Reinberg, 2008). The more direct approach of investigating the role of histone PTMs by mutating residues of interest is intractable in most animal systems. Here we demonstrate functional histone gene replacement in Drosophila with a single BAC-based transgene, improving upon a previously described method requiring multiple transgenic insertions (Gunesdogan et al., 2010). Using our platform, we provide evidence for a mechanism that scales histone expression to compensate for changes in gene number, and demonstrate that the biological function of histone tail residues can be ascertained either in whole animals or specific tissues. Our data reveal that phenotypes caused by mutation of histone-modifying enzymes will not always be recapitulated by mutation of the corresponding histone target residue.
Histone residue mutant phenotypes can differ between yeast and animals
Histone gene replacement strategies in budding yeast have identified multiple histone residues that are essential for viability and responses to environmental challenges (Dai et al., 2008; Nakanishi et al., 2008). Multicellular animals require a greater range of genome regulation to create a diversity of cell types in development, which may result in a greater reliance on post-translationally modified histone residues that affect epigenetic regulation of genome activity. Our data support this hypothesis. We found that H3K36R replacement flies die before completing development, whereas the same mutation in yeast is viable and shows no overt growth defects (Kizer et al., 2005). These findings suggest a greater dependency on H3K36 in the development and survival of multicellular organisms. Thus, our histone replacement platform has the potential to identify additional histone residues that perform essential functions in animals but have not been discovered in previous studies.
The requirement of histone residues in heritable regulation of gene expression
The co-occurrence of H3K27me3 and transcriptional repression by Polycomb proteins has led to the widely held hypothesis that H3K27me3 contributes to repression of PcG target gene expression. However, a direct test for the requirement of H3K27me3 in PcG target gene repression has been lacking. Our data, as well as those of Müller and colleagues (Pengelly et al., 2013), demonstrate that H3K27 performs an essential function in maintaining PcG target gene repression. Thus, at least in certain circumstances, post-translational modification of a histone residue is directly required for regulation of gene expression.
Direct comparison of ΔHisC clones covered by H3K27A and H3K27R transgenes shows the same pattern of PcG target gene de-repression in wing imaginal discs. However, the dominant effects we observe in H3K27A transgenic flies are notably absent from the H3K27R transgenic flies, suggesting that expression of H3K27A histones results in a more severe phenotype. Because both mutations introduce non-modifiable residues, the difference in their phenotypes may result from the charge at this residue, which is maintained as positive by substitution with arginine, but not alanine. The decrease in positive charge in H3K27A histones may decrease the affinity of the interaction between the histone tail and the negatively charged DNA backbone, potentially altering chromatin structure. Polycomb target genes may be particularly sensitive to changes in chromatin structure, as one proposed mode of PcG action is through chromatin compaction (Simon and Kingston, 2013). Thus, the amino acid used to mutate a particular histone residue could distinguish between roles of histone PTMs to serve as binding sites for trans-acting proteins and to act as regulators of chromatin packaging.
H4K20 methylation in metazoans
Our data definitively show that canonical H4K20 is dispensable for DNA replication and Drosophila development. This finding is surprising, given that genetic studies manipulating PR-Set7/Suv420H1/Suv420H2 activity in cell culture and animal models have demonstrated a consistent correlation of deleterious and lethal phenotypes with loss of H4K20 methylation (Beck et al., 2012b). In both mouse and Drosophila a complete loss of the PR-Set7 mono-methyltransferase, which is required for all H4K20 methylation states, results in a failure to complete development (Huen et al., 2008; Karachentsev et al., 2005; Oda et al., 2009). In addition, tethering active, but not catalytically inactive, PR-Set7 to DNA results in the recruitment of components of the ORC and MCM complexes, which are essential DNA replication factors (Beck et al., 2012a; Tardat et al., 2010). Although flies lacking canonical H4K20 are viable, we note that they are not phenotypically wild type. For example, they are sensitive to culture conditions and exhibit a significant developmental delay, even when H4K20A replacement larvae are cultured separately from their wild type siblings. Thus, post-translational modification of H4K20 is likely one of multiple mechanisms that ensures fidelity and robustness to the processes of DNA replication, chromatin condensation, and heterochromatin maintenance.
Several lines of evidence demonstrate that the Drosophila replication-independent H4 gene (His4r) does not compensate for the absence of replication-dependent H4K20. First, we saw no increase in His4r mRNA levels in H4K20A replacement animals. Second, we cannot detect H4K20 PTMs in H4K20A replacement animals by immunofluorescence or by western blot. Third, we show that when covered by 12×H4K20A transgenes, flies homozygous for both His4rPB and ΔHisC can survive until late pharate stages and complete development. We cannot detect His4r mRNA in flies homozygous for His4rPB, demonstrating that this is a strong allele. Any maternally contributed wild type H4 would either be absent or greatly diluted by these late stages of development. Thus maternal H4 is unlikely to have any impact on processes that require a significant concentration of H4K20 PTMs. The decrease in the ability of 12×H4K20A transgenes to rescue ΔHisC in His4rPB homozygotes may be due to a function for His4r that becomes necessary only after mutation of canonical H4K20. His4r rescue experiments will be required to definitively test this possibility.
Altogether, our data demonstrate that H4K20 is neither essential for DNA replication nor for completion of development. One explanation for this finding is that non-histone substrates of PR-Set7/SET8 are essential for DNA replication and cell cycle progression. Comparative phenotypic analysis of mutations in histone modifying enzymes and mutations in their cognate histone residues should provide answers to this question. More generally, our findings raise the important possibility that non-histone proteins can function as carriers of epigenetic information that is required for proper animal development.
Experimental Procedures
Histone locus transgene construction
The pMulti-BAC vector was generated by combining components from pBAC/oriV (Wild et al., 2002) and pattB (Bischof et al., 2007), followed by extensive site-directed mutagenesis. The 5kb histone repeat unit was subcloned into pBluescript prior to multimerization in pMulti-BAC. See Supplemental Information for additional details.
Fly strains and genetic crosses
A list of all fly strains and genetic crosses is included in the Supplemental Information.
Immunofluorescence, clone induction and confocal microscopy
Salivary gland polytene chromosome squashes were performed on wandering 3rd instar larvae, as previously described (Salzler et al., 2013). Embryo stains and mitotic recombination experiments in imaginal discs were performed as previously described (Estella et al., 2008). Egg chamber immunofluorescence was performed as previously described (Deng et al., 2001). EdU incorporation was performed using the Click-iT EdU Alexa Fluor 555 imaging kit (Invitrogen) according to the manufacturer’s instructions. The maximum projection from two adjacent z-slices from stage 6-8 egg chambers was used as representative images for each genotype. For quantification of signal intensities in nurse cell nuclei, each channel was scored independently of the other channels. See Supplemental Information for details and for a list of antibodies used.
In silico quantification of endogenous histone gene repeats
High-throughput genomic DNA sequencing libraries were generated from adult virgin females, as previously described (McKay and Lieb, 2013; see genomic input files under Gene Expression Omnibus accession number GSE38727). The following exceptions were made in order to unambiguously map reads to the histone locus. A custom reference genome was created by removing all replication-dependent histone gene repeat sequences from genome version dm3, and by adding back a single 5kb histone gene repeat unit. An unlimited number of reads were then mapped to this custom genome using bowtie (version 0.12.3) using the options ‘--nomaqround’ and ‘--best’ (Langmead et al., 2009). Coverage values were then calculated for each base in the genome. The average read depth across each gene’s translated sequence was calculated for the five replication-dependent histone genes, and the remaining refSeq genes on chromosome 2L. Data were plotted in R (www.R-project.org).
S1 nuclease protection assay
S1 assays were performed as described in (Salzler et al., 2013) using total RNA isolated from 4-6 hr embryos.
Western blotting
Immunoblot analyses were performed as described (Fuchs et al., 2012) with the following exceptions. Embryos were lysed by bead-beating for 3 minutes in SUTEB buffer (1% SDS, 8M Urea, 10mM Tris pH 6.8, 10mM EDTA, 0.01% bromophenol blue). Lysates were then boiled for 10 minutes and the supernatant was clarified by centrifugation. We could not obtain a reproducible anti-H4K20me1 signal on western blots, so anti-H4K20me3 was used. Adult heads were used for this analysis because they expressed high levels of H4K20me3.
Reverse transcription and PCR assays
For histone gene expression analyses, total RNA was isolated using Trizol, and reverse transcription was performed using random hexamers and Superscript II (Invitrogen), according to the manufacturer’s protocols. RT-PCR was performed using gene-specific primers to His2A, His3, His4r, and α-tubulin. Genomic DNA from 15 adult males was used for histone gene copy number assays. For semi-quantitative analysis of PCR products, amplicons were run on 8% acrylamide gels, and bands were quantified using ImageJ. iTaq Universal SYBR Green Supermix (Bio-Rad) was used for real-time PCRs. Additional details and primer sequences are available in the Supplemental Information.
Supplementary Material
Acknowledgments
The authors would like to thank Bill Marzluff for bringing us all together, and for establishing a fun and collaborative research environment for histone and chromatin biology in Chapel Hill. We also thank A. Herzig for generous gifts of fly lines and for communicating results prior to publication. MPM was supported in part by an NIH predoctoral fellowship, F31-CA177088. MPM, TJP and SLM were also supported in part by NIH predoctoral traineeships, T32-GM007092. SK was supported in part by a National Cancer Institute postdoctoral fellowship, T32 CA009156, and by an NIH diversity supplement to NIH grant R01-GM053034 (to AGM). This work was supported by NIH grants DA036897 (to RJD, AGM & BDS) and GM057859 (to RJD) and by startup funds from the University of North Carolina (to DJM).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Akhmanova A, Miedema K, Hennig W. Identification and characterization of the Drosophila histone H4 replacement gene. FEBS Lett. 1996;388:219–222. doi: 10.1016/0014-5793(96)00551-0. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Schneider R, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J Biol Chem. 2005;280:17732–17736. doi: 10.1074/jbc.M500796200. [DOI] [PubMed] [Google Scholar]
- Beck DB, Burton A, Oda H, Ziegler-Birling C, Torres-Padilla ME, Reinberg D. The role of PR-Set7 in replication licensing depends on Suv4-20h. Genes Dev. 2012a;26:2580–2589. doi: 10.1101/gad.195636.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck DB, Oda H, Shen SS, Reinberg D. PR-Set7 and H4K20me1: at the crossroads of genome integrity, cell cycle, chromosome condensation, and transcription. Genes Dev. 2012b;26:325–337. doi: 10.1101/gad.177444.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci U S A. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulchand S, Menon SD, George SE, Chia W. Muscle wasted: a novel component of the Drosophila histone locus body required for muscle integrity. J Cell Sci. 2010;123:2697–2707. doi: 10.1242/jcs.063172. [DOI] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- Christen B, Bienz M. Imaginal disc silencers from Ultrabithorax: evidence for Polycomb response elements. Mech Dev. 1994;48:255–266. doi: 10.1016/0925-4773(94)90064-7. [DOI] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- Dai J, Hyland EM, Yuan DS, Huang H, Bader JS, Boeke JD. Probing nucleosome function: a highly versatile library of synthetic histone H3 and H4 mutants. Cell. 2008;134:1066–1078. doi: 10.1016/j.cell.2008.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng WM, Althauser C, Ruohola-Baker H. Notch-Delta signaling induces a transition from mitotic cell cycle to endocycle in Drosophila follicle cells. Development. 2001;128:4737–4746. doi: 10.1242/dev.128.23.4737. [DOI] [PubMed] [Google Scholar]
- Estella C, McKay DJ, Mann RS. Molecular integration of wingless, decapentaplegic, and autoregulatory inputs into Distalless during Drosophila leg development. Dev Cell. 2008;14:86–96. doi: 10.1016/j.devcel.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follmer NE, Wani AH, Francis NJ. A polycomb group protein is retained at specific sites on chromatin in mitosis. PLoS Genet. 2012;8:e1003135. doi: 10.1371/journal.pgen.1003135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca JP, Steffen PA, Muller S, Lu J, Sawicka A, Seiser C, Ringrose L. In vivo Polycomb kinetics and mitotic chromatin binding distinguish stem cells from differentiated cells. Genes Dev. 2012;26:857–871. doi: 10.1101/gad.184648.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs SM, Kizer KO, Braberg H, Krogan NJ, Strahl BD. RNA polymerase II carboxyl-terminal domain phosphorylation regulates protein stability of the Set2 methyltransferase and histone H3 di- and trimethylation at lysine 36. J Biol Chem. 2012;287:3249–3256. doi: 10.1074/jbc.M111.273953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glozak MA, Sengupta N, Zhang X, Seto E. Acetylation and deacetylation of non-histone proteins. Gene. 2005;363:15–23. doi: 10.1016/j.gene.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Gunesdogan U, Jackle H, Herzig A. A genetic system to assess in vivo the functions of histones and histone modifications in higher eukaryotes. EMBO Rep. 2010;11:772–776. doi: 10.1038/embor.2010.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunesdogan U, Jackle H, Herzig A. Histone supply regulates S phase timing and cell cycle progression. Elife. 2014;3:e02443. doi: 10.7554/eLife.02443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunjan A, Verreault A. A Rad53 kinase-dependent surveillance mechanism that regulates histone protein levels in S. cerevisiae. Cell. 2003;115:537–549. doi: 10.1016/s0092-8674(03)00896-1. [DOI] [PubMed] [Google Scholar]
- Havens CG, Walter JC. Mechanism of CRL4(Cdt2), a PCNA-dependent E3 ubiquitin ligase. Genes Dev. 2011;25:1568–1582. doi: 10.1101/gad.2068611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Shilatifard A. Histone modification: cause or cog? Trends Genet. 2011;27:389–396. doi: 10.1016/j.tig.2011.06.006. [DOI] [PubMed] [Google Scholar]
- Herz HM, Morgan M, Gao X, Jackson J, Rickels R, Swanson SK, Florens L, Washburn MP, Eissenberg JC, Shilatifard A. Histone H3 lysine-to-methionine mutants as a paradigm to study chromatin signaling. Science. 2014;345:1065–1070. doi: 10.1126/science.1255104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodl M, Basler K. Transcription in the absence of histone H3.2 and H3K4 methylation. Curr Biol. 2012;22:2253–2257. doi: 10.1016/j.cub.2012.10.008. [DOI] [PubMed] [Google Scholar]
- Huang J, Berger SL. The emerging field of dynamic lysine methylation of non-histone proteins. Curr Opin Genet Dev. 2008;18:152–158. doi: 10.1016/j.gde.2008.01.012. [DOI] [PubMed] [Google Scholar]
- Huen MS, Sy SM, van Deursen JM, Chen J. Direct interaction between SET8 and proliferating cell nuclear antigen couples H4-K20 methylation with DNA replication. J Biol Chem. 2008;283:11073–11077. doi: 10.1074/jbc.C700242200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen S, Schotta G, Sorensen CS. Histone H4 lysine 20 methylation: key player in epigenetic regulation of genomic integrity. Nucleic Acids Res. 2013;41:2797–2806. doi: 10.1093/nar/gkt012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karachentsev D, Sarma K, Reinberg D, Steward R. PR-Set7-dependent methylation of histone H4 Lys 20 functions in repression of gene expression and is essential for mitosis. Genes Dev. 2005;19:431–435. doi: 10.1101/gad.1263005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, et al. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- Kizer KO, Phatnani HP, Shibata Y, Hall H, Greenleaf AL, Strahl BD. A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol Cell Biol. 2005;25:3305–3316. doi: 10.1128/MCB.25.8.3305-3316.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klymenko T, Papp B, Fischle W, Kocher T, Schelder M, Fritsch C, Wild B, Wilm M, Muller J. A Polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities. Genes Dev. 2006;20:1110–1122. doi: 10.1101/gad.377406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larschan E, Alekseyenko AA, Gortchakov AA, Peng S, Li B, Yang P, Workman JL, Park PJ, Kuroda MI. MSL complex is attracted to genes marked by H3K36 trimethylation using a sequence-independent mechanism. Mol Cell. 2007;28:121–133. doi: 10.1016/j.molcel.2007.08.011. [DOI] [PubMed] [Google Scholar]
- Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- Lewis PW, Muller MM, Koletsky MS, Cordero F, Lin S, Banaszynski LA, Garcia BA, Muir TW, Becher OJ, Allis CD. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013;340:857–861. doi: 10.1126/science.1232245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifton RP, Goldberg ML, Karp RW, Hogness DS. The organization of the histone genes in Drosophila melanogaster: functional and evolutionary implications. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):1047–1051. doi: 10.1101/sqb.1978.042.01.105. [DOI] [PubMed] [Google Scholar]
- Liu JL, Murphy C, Buszczak M, Clatterbuck S, Goodman R, Gall JG. The Drosophila melanogaster Cajal body. J Cell Biol. 2006;172:875–884. doi: 10.1083/jcb.200511038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T. Regulation of alternative splicing by histone modifications. Science. 2010;327:996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. Chromatin structure and the inheritance of epigenetic information. Nat Rev Genet. 2010;11:285–296. doi: 10.1038/nrg2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff WF, Gongidi P, Woods KR, Jin J, Maltais LJ. The human and mouse replication-dependent histone genes. Genomics. 2002;80:487–498. [PubMed] [Google Scholar]
- Marzluff WF, Wagner EJ, Duronio RJ. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet. 2008;9:843–854. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay DJ, Lieb JD. A common set of DNA regulatory elements shapes Drosophila appendages. Dev Cell. 2013;27:306–318. doi: 10.1016/j.devcel.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O’Connor MB, Kingston RE, Simon JA. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- Nakanishi S, Sanderson BW, Delventhal KM, Bradford WD, Staehling-Hampton K, Shilatifard A. A comprehensive library of histone mutants identifies nucleosomal residues required for H3K4 methylation. Nat Struct Mol Biol. 2008;15:881–888. doi: 10.1038/nsmb.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda H, Okamoto I, Murphy N, Chu J, Price SM, Shen MM, Torres-Padilla ME, Heard E, Reinberg D. Monomethylation of histone H4-lysine 20 is involved in chromosome structure and stability and is essential for mouse development. Mol Cell Biol. 2009;29:2278–2295. doi: 10.1128/MCB.01768-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pengelly AR, Copur O, Jackle H, Herzig A, Muller J. A histone mutant reproduces the phenotype caused by loss of histone-modifying factor Polycomb. Science. 2013;339:698–699. doi: 10.1126/science.1231382. [DOI] [PubMed] [Google Scholar]
- Rothbart SB, Strahl BD. Interpreting the language of histone and DNA modifications. Biochim Biophys Acta. 2014 doi: 10.1016/j.bbagrm.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder E, Blows F, Ashburner M, Bautista-Llacer R, Coulson D, Drummond J, Webster J, Gubb D, Gunton N, Johnson G, et al. The DrosDel collection: a set of P-element insertions for generating custom chromosomal aberrations in Drosophila melanogaster. Genetics. 2004;167:797–813. doi: 10.1534/genetics.104.026658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzler HR, Tatomer DC, Malek PY, McDaniel SL, Orlando AN, Marzluff WF, Duronio RJ. A sequence in the Drosophila H3-H4 Promoter triggers histone locus body assembly and biosynthesis of replication-coupled histone mRNAs. Dev Cell. 2013;24:623–634. doi: 10.1016/j.devcel.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z, Raible F, Mollaaghababa R, Guyon JR, Wu CT, Bender W, Kingston RE. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell. 1999;98:37–46. doi: 10.1016/S0092-8674(00)80604-2. [DOI] [PubMed] [Google Scholar]
- Shibutani ST, de la Cruz AF, Tran V, Turbyfill WJ, 3rd, Reis T, Edgar BA, Duronio RJ. Intrinsic negative cell cycle regulation provided by PIP box- and Cul4Cdt2-mediated destruction of E2f1 during S phase. Dev Cell. 2008;15:890–900. doi: 10.1016/j.devcel.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol Cell. 2013;49:808–824. doi: 10.1016/j.molcel.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ, 3rd, Reinberg D. Is there a code embedded in proteins that is based on post-translational modifications? Nat Rev Mol Cell Biol. 2008;9:815–820. doi: 10.1038/nrm2502. [DOI] [PubMed] [Google Scholar]
- Singh RK, Liang D, Gajjalaiahvari UR, Kabbaj MH, Paik J, Gunjan A. Excess histone levels mediate cytotoxicity via multiple mechanisms. Cell Cycle. 2010;9:4236–4244. doi: 10.4161/cc.9.20.13636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AV, King JA, Orr-Weaver TL. Identification of genomic regions required for DNA replication during Drosophila embryogenesis. Genetics. 1993;135:817–829. doi: 10.1093/genetics/135.3.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Grant PA, Briggs SD, Sun ZW, Bone JR, Caldwell JA, Mollah S, Cook RG, Shabanowitz J, Hunt DF, et al. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol Cell Biol. 2002;22:1298–1306. doi: 10.1128/mcb.22.5.1298-1306.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardat M, Brustel J, Kirsh O, Lefevbre C, Callanan M, Sardet C, Julien E. The histone H4 Lys 20 methyltransferase PR-Set7 regulates replication origins in mammalian cells. Nat Cell Biol. 2010;12:1086–1093. doi: 10.1038/ncb2113. [DOI] [PubMed] [Google Scholar]
- Wild J, Hradecna Z, Szybalski W. Conditionally amplifiable BACs: switching from single-copy to high-copy vectors and genomic clones. Genome Res. 2002;12:1434–1444. doi: 10.1101/gr.130502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1993;117:1223–1237. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.