Abstract
The extracellular matrix protein Fibulin-1 (Fbln1) has been shown to be involved in numerous processes including cardiovascular and lung development. Here we have examined the role of Fbln1 in bone formation. Alizarin red staining of skulls from Fbln1 deficient mice showed reduced mineralization of both membranous and endochondral bones. Micro CT (μCT) analysis of the calvarial bones (i.e., frontal, parietal and interparietal bones collectively) indicated that bone volume in Fbln1 nulls at neonatal stage P0 were reduced by 22% (p = 0.015). Similarly, Fbln1 null frontal bones showed a 16% (p = 0.035) decrease in bone volume, with a reduction in the interfrontal bone, and a discontinuity in the leading edge of the frontal bone. To determine whether Fbln1 played a role in osteoblast differentiation during bone formation, qPCR was used to measure the effects of Fbln1 deficiency on the expression of Osterix (Osx), a transcription factor essential for osteoblast differentiation. This analysis demonstrated that Osx mRNA was significantly reduced in Fbln1-deficient calvarial bones at developmental stages E16.5 (p = 0.049) and E17.5 (p = 0.022). Furthermore, the ability of BMP-2 to induce Osx expression was significantly diminished in Fbln1-deficient mouse embryo fibroblasts. Together, these findings indicate that Fbln1 is a new positive modulator of the formation of membranous bone and endochondral bone in the skull, acting as a positive regulator of BMP signaling.
Keywords: Fibulin-1, Extracellular matrix protein, Bmp-2, Osterix, Osteoblast differentiation
Introduction
Bone formation is a complex process that involves the mineralization of extracellular matrix (ECM) by osteoblasts to form bone [1] and the erosion of bone by osteoclasts to allow for growth and remodeling [2]. The formation and erosion of the mineralized matrix must be carefully coordinated and an imbalance in osteoblast and/or osteoclast activity has been shown to occur in several pathological conditions (e.g., osteoporosis, osteopetrosis) [3,4]. The balance between osteoblast and osteoclast formation is controlled in part by ECM proteins. Bone morphogenetic protein-2 (Bmp-2) is an ECM growth factor in the TGFβ super-family [5] that stimulates mesenchymal cells to differentiate into osteoblasts [6,7] and promotes differentiation of osteoclast precursors into mature osteoclasts [8,9]. A relatively large number of ECM proteins negatively regulate Bmp-2-mediated osteoblast differentiation including Noggin (Nog) [10,11], Twisted gastrulation (Twsg1) [10,12], Connective tissue growth factor (CCN2/Ctgf) [13], Nephroblastoma overexpressed (CCN3/Nov) [14], and Fibrillin-2 (Fbn2) [15]. By contrast, a small number of ECM proteins have been defined as positive modulators of Bmp-2 signaling in bone formation (e.g., CCN4/Wisp1) [16]. Bmp-2 controls osteoblast differentiation by upregulating the osteogenic transcription factor Osterix (Osx) [7]. The importance of Osterix in osteoblast differentiation is evidenced by the absence of bone synthesis in Osterix deficient mice [17].
Fibulin-1 (Fbln1) is an ECM protein [18,19] known to be expressed in adult bone marrow [20] and osteoblasts derived from adult bone [21]. In Fbln1 deficient mice, which die perinatally, bone size and ossification is reduced in the skull, indicating a role for Fbln1 in bone formation [22]. Here we have performed a more detailed characterization of the consequence of Fbln1 deficiency on skull bones, investigating mechanisms whereby Fbln1 could impact the process of bone formation.
Methods
Fbln1 deficient mice
The studies employed two Fbln1-deficient mouse strains that have overlapping phenotypes including 1) a Fbln1 gene trap mutant [22], and 2) a strain containing targeted deletion of Fbln1 exon 1 [23]. All procedures and protocols were done in accordance with a Medical University of South Carolina IACUC approved protocol.
Histology and immunohistochemistry
P0 neonate skulls were fixed in 1× phosphate buffered saline (PBS) containing 4% paraformaldehyde for 2 h. After fixation, skulls were embedded in Optimal Cutting Temperature (OCT) compound and sectioned at 10 μm thickness. Immunohistochemical staining was performed on cryosections with rabbit anti-fibulin-1 [24]. Primary antibody was detected with Alexa-Flour Dye conjugated secondary antibodies (Life Technologies, Carlsbad, CA). Nuclei were stained using either propidium iodide (Life Technologies) or DRAQ5 (Cell Signaling, Danvers, MA) using the manufacturer’s instructions. To detect Fbln1 in whole mount tissue segments of P0, were permeabilized in PBS containing 1% Triton X-100 for 15 min. Skull segments were washed in PBS and then blocked in 1% fatty acid free BSA (Sigma) for 1 h. After blocking, skull segments were incubated overnight with rabbit anti-fibulin-1 antiserum at 4 °C [24] and then washed in PBS containing 0.5% Triton X-100 for 4 h at room temperature. Primary antibody was detected by incubating samples for 2 h with Alexa-Flour Dye conjugated secondary antibody at room temperature. Samples were then washed for 2 h in PBS containing 0.5% Triton X-100, equilibrated in 100% glycerol, and subjected to confocal microscopic optical sectioning on a Leica SP2. Specificity of the Fbln1 antibody was confirmed by incubating Fbln1 null sections with anti-Fbln1 IgG, demonstrating the absence of immunoreactivity (Supplemental Fig. 2).
In vivo proliferation analysis
Timed pregnant females from Fbln1 heterozygous matings were given an intraperitoneal (IP) injection of 5-bromo-2′-deoxyuridine (BrdU) (Sigma, St. Louis, MO) at E17.5. Embryos were isolated 6 h after injection and fixed in 1× PBS containing 4% paraformaldehyde. BrdU incorporation into DNA was detected by immunohistochemistry using a BrdU IHC kit (BD Biosciences, San Jose, CA) following the manufacturer’s recommendations. Specificity of the BrdU immunodetection was confirmed using appropriate negative controls, including 1) skull sections obtained from timed pregnant females not injected with BrdU, and 2) skull sections treated without anti-BrdU antibody (Supplemental Fig. 3). Proliferation indices were determined by counting BrdU positive cells in the dermis that were located adjacent to and within the osteogenic front of the frontal bone; this was performed using a series of sections through the supraorbital ridge from wild-type (n = 3) and Fbln1 null (n = 3) skulls. BrdU positive cells are reported as a percentage of the total number of hematoxylin stained nuclei within the dermis or osteogenic front of the frontal bone in wild-type and Fbln1 null skulls. Fisher’s exact test was used to determine the statistically significant differences between the wild-type and Fbln1 null skulls.
RNA purification and RT-QPCR
Calvarial tissue from wild-type and Fbln1 null embryos was collected at various stages and stored in RNAlater RNA Stabilization Reagent (Life Technologies, Carlsbad, CA). Total RNA was isolated from stored tissues using TRIzol (Life Technologies) and a Qiagen RNeasy Mini Kit (Qiagen, Valencia, CA). cDNA was prepared from total RNA using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA) according to the manufacturer’s specifications. cDNA preparations were diluted to 25–50 μl, and 2 μl was used in 10-μl quantitative PCR (qPCR) reactions with the SsoFast EvaGreen Supermix reagent (Bio-Rad). Oligonucleotide primers (Integrated DNA Technologies, Coralville, IA) used in qPCR reactions were Osx 5′ TGAGGAAGAAGCCCATTCAC 3′ (forward), 5′ ACTTCTTCTCCCGGGTGTG 3′ (reverse) [25], Alkaline phosphatase, (Alpl) 5′ AACAACCTGACTGACCCTTCGC 3′ (forward), 5′ ATTTTCCCGT TCACCGTCC 3′ (reverse) [26], Runx2 5′ ATGATGACACTGCCACCTCT GAC 3′ (forward), 5′ AACTGCCTGGGGTCTGAAAAAGG 3′ (reverse) [27], and Osteocalcin 5′ GAACAGACTCCGGCGCTA 3′ (forward), 5′ AGGG AGGATCAAGTCCCG 3′ (reverse) [28]. Thermal cycling was performed using a Bio-Rad CFX96 Real-Time PCR Detection System (Bio-Rad); all samples were amplified in duplicate. Resulting data were analyzed with the PCR Miner Web tool [29] to calculate reaction efficiencies and cycle thresholds. Starting fluorescence values (mRNA levels) were calculated using the method of Liu and Saint [30]. mRNA levels for genes of interest (Runx2, Osterix, Osteocalcin and Alkaline phosphatase) were standardized in relation to the reference gene Hprt.
NanoString nCounter gene expression profiling
Expression of 248 genes in E17.5 wild-type and Fbln1 null calvarial total RNA samples was interrogated using a NanoString nCounter (NanoString Technologies) gene expression system [31]. Analysis was performed with a custom NanoString nCounter code set consisting of 248 probes that included 6 housekeeping controls (Eif4a2, GusB, Oaz1, Stk36, Tceb1, and Tubb4a). RNA samples were hybridized with sequence-specific capture and reporter probes (50 bp each) for each transcript of interest according to the manufacturer’s instructions. Resulting counts were normalized and analyzed for statistical significance using NanoStriDE (http://nanostride.soe.ucsc.edu) [32].
Micro-computed tomography (microCT) analysis
P0 neonatal skulls evaluated by microCT (μCT) analysis were scanned at a resolution of 6 μm using a Scanco μCT40 ex vivo scanner in the MUSC Center for Oral Health Research. Data analysis and 3D bone reconstruction imaging was performed using Scanco 3D analysis software version V6.1. Skull length measurements were obtained for 3D reconstructions of P0 controls and Fbln1 null skulls; these measurements represent the combined lengths of the nasal, frontal and parietal bones.
Tartrate-resistant acid phosphatase (TRAP) staining and TUNEL analysis
TRAP staining was performed in tissue sections of E17.5 wild-type (n = 3) and Fbln1 null (n = 3) skulls using a leukocyte acid phosphatase kit (Sigma). After TRAP staining, slides were counterstained with hematoxylin. Osteoclasts were defined as multinucleated TRAP-positive cells in contact with bone surfaces. Bone area was calculated using the threshold color function in Image J (NIH software) [33]. Apoptotic cells in the skulls of E17.5 wild-type (n = 3) and Fbln1 null (n = 3) embryos were identified using a TUNEL detection kit (Millipore, Billerica, MA). Nicked DNA in apoptotic cells was detected by biotin-16-dUTP labeling using terminal deoxynucleotidyl transferase according to the manufacturer’s instructions (Millipore). Negative control sections from E17.5 wild-type and Fbln1 null embryos were processed as recommended with the exception that the terminal deoxynucleotidyl transferase step was omitted.
Alizarin red staining of bone
P0 mouse skulls were fixed in 4% paraformaldehyde for 12 h. After fixation the skulls were placed in 1% potassium hydroxide for 2 h and then transferred to a solution of 0.03% alizarin red and 1% potassium hydroxide for 24 h. Skulls were subsequently placed in a solution of 2% potassium hydroxide and 10% glycerol for 48 h.
Generation of Fbln1 depleted serum
An anti-Fbln1 IgG column was generated by coupling the monoclonal antibody 3A11 to Sepharose [19]. Fbln1 was depleted from fetal bovine serum (FBS) by passing the serum over the monoclonal anti-Fbln1 3A11 IgG Sepharose column six times. Depletion of Fbln1 protein was verified by ELISA or immunoblotting.
Generation of mouse embryonic fibroblasts (MEFs) and treatment with Bmp-2
Embryos (E12.5) were isolated from timed pregnant Fbln1 heterozygous females. A segment of embryonic tissue was collected for genotyping and the remaining tissue was dispersed by trituration using 0.05% trypsin and a glass Pasteur pipette. Isolated wild-type and Fbln1 null MEFs were separately plated on fibronectin-coated plates in serum free DMEM-H. After 2 h, the serum free media was removed and replaced with DMEM-H media containing 10% FBS depleted of Fbln1. Cells were allowed to adhere for 12 h then treated with 300 ng/ml Bmp-2 (R&D Systems) or vehicle. Following 18 h of treatment, RNA was isolated from the MEF cultures and used for qPCR analysis as described above. Bmp-2 experiments were repeated three times using two independent batches of wild-type and Fbln1 null MEFs.
Fbln1-Sepharose binding assay
Fibulin-1 was purified from extracts of human placenta by immunoaffinity chromatography using mouse monoclonal 3A11 anti-fibulin-1 IgG-Sepharose as previously described [18,24]. Purified Fbln1 was cross-linked to CNBr-activated Sepharose 4B (GE Healthcare Life Sciences, Piscataway, NJ) according to the manufacturer’s instructions. Binding of Fbln1 to Bmp-2 was assessed by an in vitro pull down binding assay. Briefly, Bmp-2 (200 ng/ml, R&D Systems, Minneapolis, MN) and BSA (500 ng/ml fatty acid-free, Sigma) were added to 300 μl of binding buffer (20 mM Tris, pH 7.0, 137 mM NaCl, 1% IGEPAL detergent, 10% glycerol) [34] in 1.5 ml microcentrifuge tubes. Samples were pre-cleared by incubation with Sepharose 4B beads resuspended in binding buffer. After pre-clearing, Fbln1 Sepharose 4B beads or plain Sepharose 4B beads were added to each sample. Total bead volume was normalized in the Fbln1 Sepharose 4B samples by adding additional plain Sepharose 4B beads. Samples were rocked end over end for 8 h at 4 °C. Beads were then washed 4 times with 500 μl of 20 mM Tris, pH 7.0, 137 mM NaCl, 1% IGEPAL detergent, 10% glycerol followed by two 500 μl washes with PBS. Bound proteins were eluted from the beads by boiling in reducing sample buffer and analyzed by Western blotting [35] using antibodies to Bmp-2 (R&D Systems) or Ccn3 (Abcam, Cambridge, MA). As a positive control, the in vitro pull-down binding assay was performed with Fbln1 Sepharose 4B beads and Ccn3 (200 ng/ml, Abcam), which has been shown to bind to Fbln1 [36].
Statistical analysis
Fisher’s exact test, Student’s t-test (unpaired, two tailed variants) or one-way ANOVA with Tukey’s post hoc test was used to determine the statistical significance of experimental results using Prism version 6.01 software (GraphPad, La Jolla, CA). Differences were considered significant at p < 0.05.
Results
Fibulin-1-deficient skulls display reduced alizarin red staining and bone
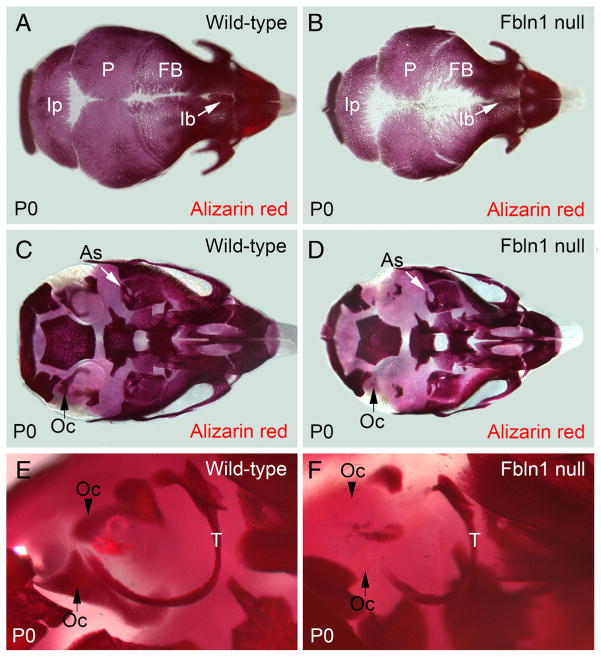
Previous studies of mice homozygous for a gene trap insertion in the fibulin-1 gene (designated hereafter as Fbln1 nulls) have shown that Fbln1 plays a role in skull bone formation [22]. To extend on those initial findings we examined skulls from P0 wild-type and Fbln1 null neonates (P0 neonates represent the latest developmental stage that Fbln1 null mice can be obtained before lethality) using alizarin red to stain mineralized extracellular matrix. Fbln1 null mice had less alizarin red stain incorporated into the membranous bones of the calvarium (i.e., frontal (F), parietal (P) and interparietal bones (Ip)) (Fig. 1). Fbln1 null skulls also displayed decreased alizarin red staining along the midline and showed little to no interfrontal bone (Ib) at P0 (Fig. 1). Examination of Fbln1 null bones of the calvarium revealed increased bone porosity and a pronounced irregular edge as compared to wild-type (Fig. 1B).
Fig. 1.
Fbln1 null neonates have reduced alizarin red staining of membranous bones and endochondral bones in the cranium. A and B show that alizarin red staining in the membranous frontal, parietal and interparietal bones from a P0 Fbln1 null neonate (B) is reduced as compared to P0 wild-type (A). Arrow in B points to the interfrontal bone in P0 Fbln1 null that is reduced compared to P0 wild-type. C and D show reduced alizarin red staining of the otic capsule from P0 Fbln1 null (D) as compared to P0 wild-type (C). Arrow (black) in C points to a bone in the otic capsule that is absent in the P0 Fbln1 null (black arrow in D). Arrow (white) shows alisphenoid bone in P0 Fbln1 null that has reduced alizarin red staining. E and F are higher magnification views of the otic region in P0 wild-type and P0 Fbln1 null skull showing reduced ossification of the otic capsule and tympanic ring bone (T) in P0 Fbln1 null. Arrowhead in E points to a bone in P0 wild-type otic capsule that is absent in the P0 Fbln1 null otic capsule. As, alisphenoid bone; FB, frontal bone; Ib, interfrontal bone; Ip, interparietal bone; Oc, otic capsule; P, parietal bone; T, tympanic ring bone.
The consequence of Fbln1 deficiency on skull bones was evident in both membranous and endochondral bones. Fbln1 nulls at P0 showed reduced alizarin red staining in both bone types at the cranial base (i.e., alisphenoid bone (As), tympanic ring (T), and the bones that form the otic capsule (Oc)) (Fig. 1). Reduced levels of alizarin red staining were also observed in the frontal, parietal and interparietal bones of an alternate model of Fbln1 deficiency designated as the Fbln1 exon 1 null model (Supplemental Fig. 1).
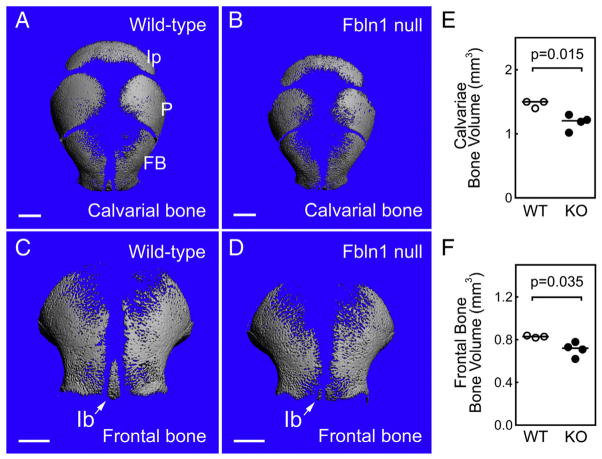
μCT was used to quantify the size and amount of bone (i.e., frontal, parietal and interparietal bones) in Fbln1 null and control skulls at P0 (Fig. 2) (See Table 1). This analysis showed that Fbln1 deficient skulls were smaller in size overall with a 14% decrease in the combined length of the nasal, frontal and parietal bones as compared to controls (mean length of Fbln1 null skulls = 668 mm (n = 4); mean length of control skulls = 764 mm (n = 3), p = 0.046). μCT analysis also revealed that bone volume (BV) was reduced by 22% in Fbln1 null calvariae and 16% in Fbln1 null frontal bones compared to controls (p = 0.035; n = 3 controls; n = 4 Fbln1 nulls).
Fig. 2.
Membranous bones in Fbln1 nulls have reduced bone volume and bone size. A and B show μCT reconstructions of wild-type and Fbln1 null calvariae from neonates (P0). C and D show μCT reconstructions of wild-type and Fbln1 null frontal bones from neonates (P0). Note that the interfrontal bone (Ib) (arrow) in the Fbln1 null (D) is poorly formed compared to wild-type (C). Graphs in E and F, show bone volumes (BV) of calvariae and frontal bones from control and Fbln1 null neonates (P0). Horizontal lines in the graphs indicate mean values for each data group. Scale bars in A–D are 1 mm. Indicated p values were determined using Student’s t-test. Ip, interparietal bone; P, parietal bone; FB, frontal bone; Ib, interfrontal bone.
Table 1.
Measurements obtained from Micro-CT analysis of Control and Fbln1 null P0 skulls.
| Control b | Fbln1 null c | P value d | % difference e | |
|---|---|---|---|---|
| Skull length (cm)a | 763.7 ± 12.5 | 668 ± 61 | 0.046 | 14 |
| Calvarial bone volume (mm3)f | 1.5 ± 0.06 | 1.2 ± 0.13 | 0.011 | 22 |
| Frontal bone volume (mm3) | 0.8 ± 0.01 | 0.7 ± 0.07 | 0.035 | 16 |
Measurements are the combined lengths of the nasal, frontal and parietal P0 bones of wild type as compared to Fbln1 nulls.
n = 3 (2 wild-type P0 neonate skulls and 1 Fbln1 heterozygous P0 neonate skull).
n = 4
P value was calculated using Student’s t-test.
Percent difference calculated from the mean of wild type measurements and the mean of Fbln1 null measurements.
Includes frontal, parietal and interparietal bones.
Together, these results suggest that Fbln1 is critical for regulating cranial bone BV, a measure of ossification. Similar to what was observed in the Fbln1 nulls, the frontal, parietal and interparietal bone areas were reduced in Fbln1 exon 1 nulls (Supplemental Fig. 1).
Fibulin-1 is deposited around condensed mesenchymal cells, osteoblasts and osteocytes in the ECM of the frontal bone, with prominent detection in blood vessels
To establish the spatial expression pattern of Fbln1 in the frontal bone we performed Fbln1 immunolabeling on wild-type frontal bone from P0 neonates. The frontal bone segments were scanned en face using a Leica SP2 confocal microscope, and the collected z-series images were collapsed to generate a single optical section for analysis (Fig. 3A). In this imaging study, Fbln1 immunolabeling was most prominent in the ECM around clusters of condensed mesenchymal cells (Cmc) within the frontal bone (Fig. 3A). Fbln1 immunolabeling was also observed around osteoblasts (defined as cells adjacent to mineralized regions) in the frontal bone at P0. Less prominent Fbln1 immunolabeling was observed around bone-encased osteocytes in the frontal bone (Fig. 3A). Finally, Fbln1 immunolabeling was present around blood vessels (V) in the frontal bone at P0 (Fig. 3A).
Fig. 3.
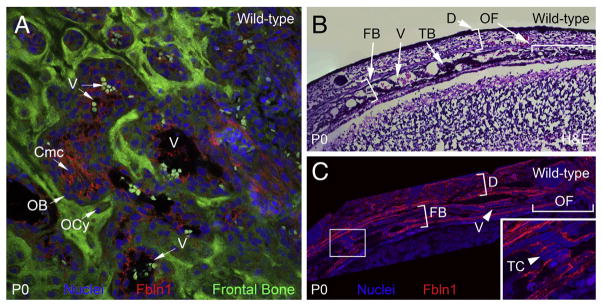
Fibulin-1 is deposited in the extracellular matrix surrounding osteoblasts in the frontal bone. Confocal microscopy in A, shows Fbln1 deposited in the ECM in wild-type mouse frontal bone (P0) (red). Panel A is derived from a collapsed z-series of multiple optical sections. Section of wild-type neonate (P0) skull in B, (containing frontal bone) is stained with H&E. Section of wild-type neonate (P0) skull C, is immunolabeled with antibodies to Fbln1 (red). The inset panel in C shows a higher magnification of the boxed region. Note that Fbln1 is deposited in the ECM around osteoblasts (arrowheads) located in the frontal bone. Nuclei in A and C were stained using Draq5 (blue). V, blood vessel; Cmc, condensed mesenchymal cells; D, dermis; FB, frontal bone; OB, osteoblast; OCy, osteocytes; OF, osteogenic front; TB, trabecular bone; TC, cell adjacent to trabecular bone.
We also performed H&E staining (Fig. 3B) and anti-Fbln1 immunolabeling (Fig. 3C) on a series of sections made through the supraorbital ridge from a P0 wild-type skull. H&E staining showed trabecular bone (TB) formation within the frontal bone. Fbln1 immunolabeling of the P0 wild-type skull (Fig. 3C) revealed that Fbln1 is deposited in the ECM 1) of the dermis (D) that covers the frontal bone (FB), 2) around cells adjacent to the trabecular bone (TC) within the frontal bone, 3) around cells of the osteogenic front (OF), and 4) around blood vessels (V) within the frontal bone.
Fibulin-1 deficient mice display discontinuous frontal bone formation
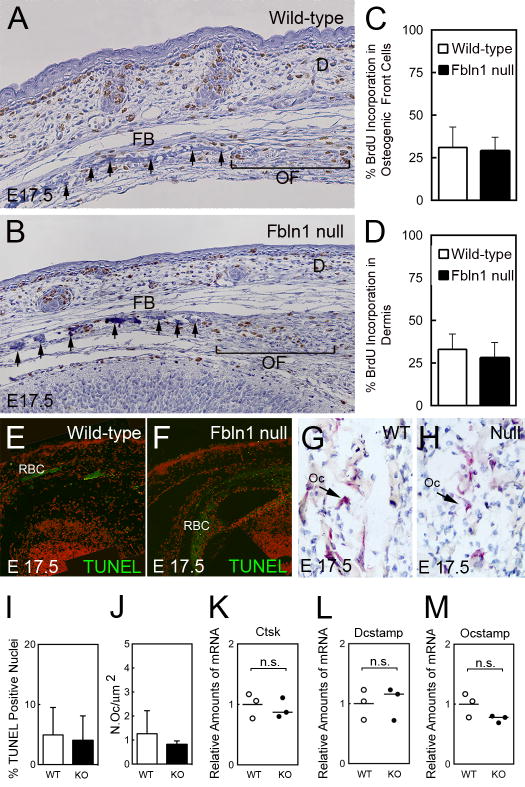
The effect of Fbln1 deficiency on frontal bone morphology was examined using hematoxylin staining of wild-type and Fbln1 null E17.5 skulls. These studies revealed that the leading edge of the frontal bone in the Fbln1 nulls was discontinuous as compared to frontal bones in wild-type E17.5 skulls (Figs. 4A and B, compare arrows in the frontal bone regions).
Fig. 4.
Fibulin-1 deficiency does not alter the proliferation or survival of osteoprogenitors or increase osteoclast activity in the frontal bone of Fbln1 nulls. A and B show BrdU incorporation into DNA by immunohistological analysis of frontal bone sections from wild-type and Fbln1 null E17.5 embryos using an antibody to BrdU. Note that the Fbln1 null shows a discontinuous frontal bone compared to wild-type. Graphs in C and D, show % BrdU positive nuclei determined in the osteogenic front (brackets in A and B) and in the dermis adjacent to the osteogenic front. BrdU positive nuclei were counted in multiple sections from three E17.5 skulls per genotype. Error bars represent standard deviation of the mean; significance was determined by Fisher’s Exact test. E and F shows TUNEL analysis of wild-type and Fbln1 null E17.5 frontal bone sections. G and H show histological TRAP staining for osteoclasts in the trabecular region of frontal bone in wild-type and Fbln1 null E17.5 skulls. Graph in I, shows % TUNEL positive nuclei in the osteogenic front and trabecular bone region of the frontal bone in wild-type (n = 3) and Fbln1 null (n = 3) E17.5 skulls. Error bars represent standard deviation of the mean and significance determined by Fisher Exact test. Graph in J, shows the number of TRAP positive osteoclasts determined from the trabecular bone region of the frontal bone in wild-type (n = 3) and Fbln1 null (n = 3) E17.5 skulls. Error bars represent standard deviation of the mean and significance determined by Students t-test. Graphs in K, L, and M, show levels of Ctsk, Dcstamp and Ocstamp mRNA determined by nanostring analysis of RNA isolated from E17.5 wild-type (WT) and Fbln1 null (KO) calvariae. The graphed values show mean relative amounts of mRNA based of WT (n = 3) and KO (n = 3). P values were calculated using NanoStriDE. D, dermis; FB, frontal bone; OF, osteogenic front; Oc, osteoclast; RBC, red blood cells; Cathepsin K, Ctsk.
We sought to determine whether the observed discontinuity in Fbln1 null frontal bones was due to deficient cell proliferation. BrdU labeling of wild-type and Fbln1 null embryos (E17.5) revealed no significant difference in cell proliferation in the osteogenic front of the frontal bone through the supraorbital ridge for Fbln1 nulls versus wild-types (Fig. 4C). Moreover, the incorporation of BrdU in the mesenchyme of the adjacent dermis was not significantly different between these genotypes (Fig. 4D).
To evaluate whether the reduced bone formation in the Fbln1 null frontal bones resulted from increased cell death we performed TUNEL analysis. Examination of wild-type and Fbln1 null E17.5 frontal bone sections through the supraorbital ridge region showed only scattered TUNEL positive cells in the osteogenic front and the trabecular bone (Figs. 4E and F). Quantification of TUNEL positive cells within these regions showed no significant difference in the number of apoptotic cells between these genotypes (Fig. 4I). Taken together, these data suggest that it is unlikely that the reduced BV seen in Fbln1 null calvariae results from decreased osteoblast proliferation or reduced osteoblast survival.
The observations of multiple perforations within the calvariae of P0 Fbln1 nulls suggested a possible increase in osteocytic activity (Fig. 1B). To determine if the reduced bone volume seen in Fbln1 null skulls resulted from increased osteoclast numbers we performed TRAP staining on E17.5 wild-type and Fbln1 null skull sections. The patterns of TRAP staining observed in both genotypes were overtly similar (Figs. 4G and H). Quantification of TRAP positive osteoclasts within the trabecular bone region of the frontal bone revealed no significant difference between E17.5 wild-type and Fbln1 nulls (Fig. 4J). We also performed gene expression analysis on RNA isolated from E17.5 wild-type and Fbln1 null calvariae using the NanoString nCounter system to measure expression of Dcstamp and Ocstamp, two genes involved in osteoclast differentiation, and Cathepsin K, which is expressed by mature osteoclasts. Dcstamp, Ocstamp and Cathepsin K expression was not significantly different between these genotypes, suggesting that the changes in BV in Fbln1 nulls was not due to an increase in osteoclasts.
We then considered the possibility that reduced bone formation in Fbln1 null skulls was the result of defective vascularization of bone. This possibility was suggested in part by the prior finding that Fbln1 deficient mice display blood vessel abnormalities [22,23]. Immunohistological analysis of sections from the cranial base of E17.5 wild-type and Fbln1 nulls using a PECAM antibody to detect endothelial cells, showed that both genotypes had endothelial cells in the bone marrow cavities of endochondral bones. However, the PECAM staining pattern in Fbln1 nulls was less well defined compared to wild-type (Supplemental Fig. 4).
Fibulin-1 deficient calvariae displayed reduced osterix expression
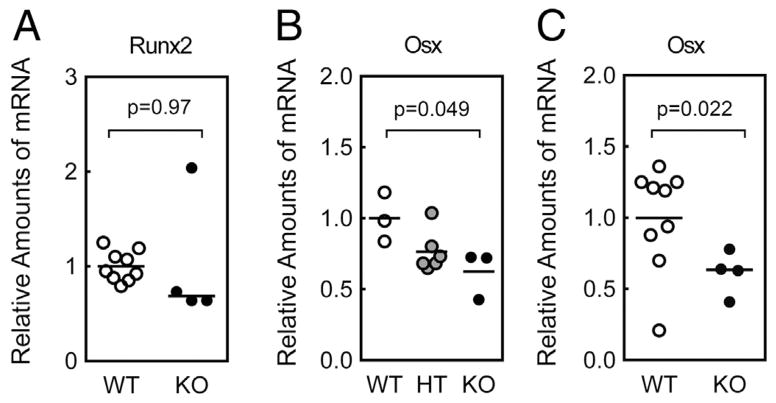
We next examined whether the reduced bone volume and bone discontinuity seen in Fbln1 null frontal bones were related to reduced osteoblast differentiation. The transcription factors Runx2 and Osterix (Osx) are essential for bone formation [17,37]. Runx2 is required to differentiate mesenchymal cells along an osteoblast lineage [38]. To determine if Fbln1 deficiency impacted osteoblast differentiation we measured Runx2 expression in E17.5 calvariae from wild-type and Fbln1 null embryos. However, qPCR analysis of Runx2 mRNA expression in this bone tissue detected no significant difference between wild-type and Fbln1 nulls (Fig. 5A).
Fig. 5.
Fibulin-1 null calvariae express reduced levels of Osterix mRNA. A shows Runx2 levels determined by qPCR analysis of mRNA isolated from E17.5 wild-type (WT) and Fbln1 null (KO) calvariae. B shows levels of the Osterix (Osx) mRNA determined by qPCR analysis of RNA isolated from E16.5 wild-type (WT), Fbln1 heterozygous (HT) and Fbln1 null (KO) calvariae. C shows Osx levels determined by qPCR analysis of mRNA isolated from E17.5 wild-type (WT) and Fbln1 null (KO) calvariae. The graphed values show mean relative amounts of mRNA based on WT (n = 3), HT (n = 6), and KO (n = 3) for E16.5 calvariae and WT (n = 9) and KO (n = 4) for E17.5 calvariae. P value was calculated using one-way ANOVA with Tukey’s post hoc test for B and a Students t-test for A and C.
We next examined the expression of Osx mRNA in Fbln1 null embryos. qPCR analysis of RNA from E16.5 and E17.5 calvariae showed that Fbln1 nulls exhibited a significant decrease in Osx mRNA levels compared to wild-type embryos (p = 0.049, E16.5 Fbln1 null calvariae; p = 0.022, E17.5 Fbln1 null calvariae) (Figs. 5B and C). We also examined the mRNA expression of Osteocalcin and Osteopontin, two proteins synthesized by mature osteoblasts. Surprisingly, neither gene was detected as differentially expressed in the Fbln1 null samples (Supplemental Fig. 5). However, on the whole, these results indicate that Fbln1 influences Osx expression during calvarial bone formation.
Fibulin-1 binds Bmp-2 to promote induction of Osterix and Alkaline phosphatase
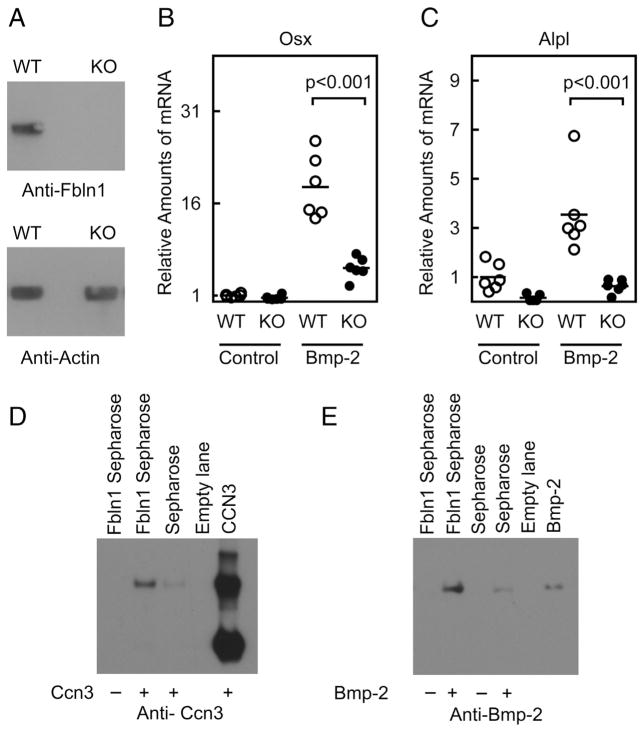
To gain further insights into the relationship between Fbln1 and Osx, we examined whether Fbln1 augmented Osx via a mechanism involving Bmp-2, which is a known upstream regulator of Osx. Using mouse embryo fibroblasts (MEFs) isolated from wild-type and Fbln1 null embryos, we evaluated how Bmp-2 treatment affected Osx mRNA expression. qPCR analysis of Bmp-2 treated cultures revealed that the induction of Osx mRNA expression by Bmp-2 was significantly reduced in Fbln1 null MEFs compared to wildtype MEFs (p < 0.001; n = 6) (Fig. 6B). Similarly, the expression of Alkaline phosphatase (Alpl) mRNA in response to Bmp-2 treatment was significantly reduced in Fbln1 null MEFs compared to wild-type MEFs (p < 0.001; n = 6) (Fig. 6C). These findings indicate that Fbln1 is required to mediate Bmp-2-induced expression of Osx and Alpl.
Fig. 6.
Fbln1 potentiates Bmp-2 induced expression of Osx and Alpl. Immunoblot analysis in A, using anti-Fbln1 to demonstrate loss of Fbln1 expression in Fbln1 null MEFs. Anti-actin immunoblotting was performed to demonstrate equal loading of protein extracts. B and C show Osterix (Osx) and Alkaline phosphatase (Alpl) mRNA expression measured by qPCR analysis of RNA isolated from wild-type (WT) and Fbln1 null MEFs (KO) treated for 18 h with vehicle or Bmp-2 (300 ng/ml). Plotted values show mean relative amounts of mRNA normalized to Hprt mRNA. Statistical significance between WT and Fbln1 KO MEFs in response to Bmp-2 treatment was calculated using one-way ANOVA with Tukey’s post hoc test. Results of a CCN3 pull-down assay in D, performed with either Fbln1-Sepharose beads or plain Sepharose beads. Results of a Bmp-2 pull-down assay in E, performed using either Fbln1-Sepharose beads or plain Sepharose beads in the presence or absence of Bmp-2.
Given the positive effect of Fbln1 on Bmp-2 signaling, we investigated the possibility that Fbln1 interacted physically with Bmp-2. Using Sepharose beads conjugated to purified Fbln1 (Fbln1-Sepharose), pull-down assays were conducted in vitro with CCN3 (as a positive control) and Bmp-2 protein. Western blot analysis of eluates from Fbln1-Sepharose and CCN3 pull-downs confirmed that Fbln1 interacted with CCN3, as has been previously shown [36] (Fig. 6D). Importantly, western analysis of eluates from Fbln1-Sepharose pull-downs with Bmp-2 revealed that Fbln1 interacted with Bmp-2 (Fig. 6E). Densitometric analysis of immunodetected bands showed that Fbln1-Sepharose pulled down Bmp-2 at a substantially higher level (9.5-fold increase) than a Sepharose negative control (Fig. 6E). These findings illustrate that Fbln1 can interact with Bmp-2 in vitro, supporting the possibility that this interaction may also occur in vivo. Such an interaction offers a potential mechanistic basis for how Fbln1 modulates Bmp-2 induction of osteogenic gene.
Discussion
ECM proteins in bone have been shown to promote osteoprogenitor survival, proliferation and differentiation, yet these processes are poorly understood. Here we have investigated how the ECM protein Fbln1 influences bone formation, focusing on the developing skull, which displays reduced bone size and BV in Fbln1 deficient mice. The results presented here show that Fbln1 promotes formation of both the membranous and endochondral bones in the skull. Furthermore, during development, Fbln1 was present in the ECM at sites of bone formation, in proximity to osteoprogenitors and osteoblasts, therefore suggesting several possible mechanisms as to how Fbln1 may influence the bone formation process.
One possible mechanistic explanation for reduced skull bone volume in Fbln1 nulls was that Fbln1 is necessary for proliferation of osteogenic progenitors. Fbln1 null skulls display a discontinuity in the leading edge of the frontal bones at E17.5. Similar discontinuities have been reported in other mouse genetic deficiency models, e.g., Engrailed 1, Msh homeobox 2, and Distal-less homeobox 5. In these other models, the discontinuity has been linked to disruption of osteoprogenitor proliferation [39,40]. However in vivo BrdU analysis of Fbln1 nulls detected no abnormality in osteoprogenitor proliferation in the osteogenic front of frontal bones at E17.5. Thus the discontinuity in the leading edge of Fbln1 null frontal bones is not apparently due to abnormal osteoprogenitor proliferation. Additionally, we did not observe a reduction in osteoblast survival in Fbln1 nulls indicating that apoptosis is unlikely the cause of reduced bone volume. The porous nature of the membranous calvariae of Fbln1 nulls suggested the possibility of increased osteoclast activity as seen in Engrailed deficient mice [39]. However, TRAP staining and mRNA analysis of genes involved in osteoclastogenesis did not indicate an increase in osteoclast activity.
Vascularization is important for normal bone formation, and is especially important for the formation of endochondral bones [41–43]. Previous studies have shown that Fbln1 deficient mice exhibit vascular defects characterized by disruptions in the capillary endothelial layer [23] and leakage of red blood cells from the vasculature into surrounding tissues [22,23]. Deficient vascularization could potentially contribute to the reductions in skull bone volume we report here. Our analysis of endothelial cells in endochondral bones revealed that the bones did contain blood vessels, and thus vascular invasion does occur in Fbln1 deficient skull bone. However, the staining pattern of PECAM, used to detect the endothelial cells, appeared to be more diffuse in Fbln1 deficient bones. Therefore, despite the observed vascularization of Fbln1 null bone marrow, we cannot rule out the possibility that the vascular integrity or structure is compromised, causing the bone marrow to be undersupplied. It is interesting to note that Osx has been reported to positively regulate VEGF expression during bone formation [44] and that mice deficient in VEGF expression display defects in bone formation attributed to defects in vascular formation [41]. Therefore the reduced Osx expression that we observed in Fbln1 null calvariae may have a dual effect in the Fbln1 nulls, causing reductions in osteoblast differentiation as well as blood vessel formation.
Our studies did not detect any alterations in osteoprogenitor proliferation or osteoblast survival in the skulls of Fbln1 nulls. These finding suggest that the activity of two ECM growth factors fgf 8 (required for osteoprogenitor survival) [45] and fgf 18 (required for osteoprogenitor proliferation) [46,47] are unlikely responsible for the reduced bone volume seen if Fbln1 nulls. However, our observation that Fbln1 in wildtype mice was deposited in the ECM in close association with osteoblasts suggested the possibility that Fbln1 supported osteoblast differentiation. The transcription factor Osx has been shown to be essential for osteoblast differentiation and subsequent bone formation. Thus our finding that Osx mRNA expression was significantly reduced in Fbln1 null bones supported the idea that Fbln1 contributes to osteoblast differentiation, and further suggested the possibility that Fbln1 influences the signaling pathway that regulates Osx expression. Osx is regulated by Bmp-2 through signaling mechanisms that include the Smad 1/ 5/8 signaling pathway and the p38 MAPK pathway [7,48]. Our analysis of MEFs confirmed that Fbln1 potentiated the osteogenic signaling activity of Bmp-2, affecting the induction of Osx as well as the early marker of osteoblast differentiation, Alpl. Furthermore, our pull-down experimentation with purified Fbln1 and Bmp-2 revealed that these two proteins can interact directly.
It remains to be established how Fbln1 potentiates Bmp-2 mediated induction of Osx. An interaction between Fbln1 and Bmp-2 may affect how Bmp-2 interacts with the type I and II Bmp receptors. Alternatively, an interaction between Fbln1 and Bmp-2 may influence how Bmp-2 interacts with other regulatory proteins. CCN2 and CCN3 are two proteins that bind Bmp-2 and inhibit Bmp-2-mediated osteoblast differentiation [13,16,49]. Interestingly, CCN2 and CCN3 also bind to Fbln1 [36]. Another member of the CCN family, CCN4, binds Bmp-2 and promotes Bmp-2-mediated bone formation [16]. Thus there is the potential that Fbln1 interacts with Bmp-2 in a manner that influences the association of Bmp-2 with stimulatory CCN4 versus inhibitory CCN2 and CCN3.
Supplementary Material
Supplemental Fig. 1 Reduced ossification of skull bones in Fbln1 exon 1 nulls. A and B, μCT scans of wild-type (Fbln1 +/+) and Fbln1 exon 1 null (Fbln1 −/−) neonatal (P0) skulls. Note reduced ossification of the frontal bone (FB), parietal bone (P) and interparietal (Ip) bone in Fbln1 exon 1 nulls as compared to WT. C and D show alizarin red stained skulls from wild-type and Fbln1 exon 1 null neonates (P0). Note the increased bone porosity in the parietal and interparietal bones of the Fbln1 exon 1 null as compared to wild-type.
Supplemental Fig. 2 Fbln1 nulls have no immunologically detectable Fbln1. A and B, anti-Fbln1 (green) immunolabeling of wild-type and Fbln1 null E16.5 skull tissue sections. Nuclei were stained using propidium iodide (red). The inset panel in A shows a higher magnification of the boxed region. Note that Fbln1 is deposited in the ECM around osteoblasts (arrow) located in the frontal bone. The inset panel in B shows a higher magnification of the boxed region. Note that Fbln1 is not deposited in the ECM in the frontal bone. Red blood cells (RBC) were detected in both sections in the green fluorescence channel due to autofluorescence.
Supplemental Fig. 3 Analysis of BrdU incorporation in the dermis of E17.5 wild-type and Fbln1 null. A and C show BrdU incorporation into DNA by immunohistological analysis of skull bone sections from wild-type and Fbln1 null E17.5 embryos using an antibody to BrdU. B and D are negative controls that show the absence of BrdU nuclear staining in wild-type and Fbln1 null E17.5 skull sections in the absence of a BrdU antibody. E is a negative control showing the absence of BrdU nuclear staining in a wild-type E17.5 embryo not exposed to BrdU.
Supplemental Fig. 4 PECAM staining of blood vessels in the endochondral bones from E17.5 wild-type and Fbln1 nulls. A and B, show anti-PECAM (green) immunolabeling of tissue sections from the cranial base of wild-type and Fbln1 null E17.5 skulls. The inset panel in A shows a higher magnification of the boxed region in the wild-type and the inset panel in B shows a higher magnification of the boxed region in the Fbln1 null. Note that PECAM staining in the Fbln1 null sample (B) appears more diffused compared to the cell surface localization in the wild-type sample (A). Red blood cells (RBC, arrowheads) were detected in both sections in the green fluorescence channel due to autofluorescence.
Supplemental Fig. 5 Osteocalcin and Osteopontin mRNA are not significantly different in Fibulin-1 null calvariae. A shows Osteocalcin (Ocn) levels determined by qPCR analysis of mRNA isolated from E17.5 wild-type (WT) and Fbln1 null (KO) calvariae. B shows levels of the Osteopontin (Opn) mRNA determined by nanostring analysis of RNA isolated from E17.5 wild-type (WT) and Fbln1 null (KO) calvariae. The graphed values show mean relative amounts of mRNA based on WT (n = 9) and KO (n = 4) for E17.5 calvariae in A and WT (n = 3) and KO (n = 3) for E17.5 calvariae in B. P values were calculated using Students t-test for A and using NanoStriDE for B.
Highlights.
Fbln1 is deposited in the ECM around bone producing cells during skull development.
Membranous and endochondral skull bones are reduced in Fbln1 nulls.
Fbln1 deficient calvariae have reduced bone volume and reduced Osterix expression.
Fbln1 binds Bmp-2 in in vitro binding assays.
Fbln1 is a positive modulator of Bmp-2-mediated induction of Osterix.
Acknowledgments
Research reported in this publication was supported in part by pilot funding from the National Institutes of Health (NIH) grants (5P20RR017696 and P30GM10331). This work was also supported by NIH grants R01HL095067, 5R21AG043718 to WSA. JLB was supported by NIH grants P30GM103342 and P20GM103499. This study used the services of the Morphology, Imaging and Instrumentation (MMI) Core, which is supported by NIH-NIGMSP30GM103342 to the South Carolina COBRE for Developmentally Based Cardiovascular Diseases. This work was conducted in a facility constructed with support from the National Institutes of Health, Grant Number C06 RR015455 from the Extramural Research Facilities Program of the National Center for Research Resources. The authors wish to thank Johannes Aartun in the Center for Oral Health Research (Charleston, SC, USA) for his expert assistance with the MicroCT analysis and 3D-bone reconstruction.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.bone.2014.07.038.
References
- 1.Titorencu I, Pruna V, Jinga VV, Simionescu M. Osteoblast ontogeny and implications for bone pathology: an overview. Cell Tissue Res. 355:23–33. doi: 10.1007/s00441-013-1750-3. [DOI] [PubMed] [Google Scholar]
- 2.Weitzmann MN. The role of inflammatory cytokines, the RANKL/OPG Axis, and the immunoskeletal interface in physiological bone turnover and osteoporosis. Scientifica (Cairo) 2013:125705. doi: 10.1155/2013/125705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rachner TD, Khosla S, Hofbauer LC. Osteoporosis: now and the future. Lancet. 377:1276–1287. doi: 10.1016/S0140-6736(10)62349-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Michou L, Brown JP. Genetics of bone diseases: Paget’s disease, fibrous dysplasia, osteopetrosis, and osteogenesis imperfecta. Joint Bone Spine. 78:252–258. doi: 10.1016/j.jbspin.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 5.Lin SJ, Lerch TF, Cook RW, Jardetzky TS, Woodruff TK. The structural basis of TGF-beta, bone morphogenetic protein, and activin ligand binding. Reproduction. 2006;132:179–90. doi: 10.1530/rep.1.01072. [DOI] [PubMed] [Google Scholar]
- 6.Hassan MQ, Tare RS, Lee SH, Mandeville M, Morasso MI, Javed A, van Wijnen AJ, Stein JL, Stein GS, Lian JB. BMP2 commitment to the osteogenic lineage involves activation of Runx2 by DLX3 and a homeodomain transcriptional network. J Biol Chem. 2006;281:40515–26. doi: 10.1074/jbc.M604508200. [DOI] [PubMed] [Google Scholar]
- 7.Matsubara T, Kida K, Yamaguchi A, Hata K, Ichida F, Meguro H, Aburatani H, Nishimura R, Yoneda T. BMP2 regulates Osterix through Msx2 and Runx2 during osteoblast differentiation. J Biol Chem. 2008;283:29119–25. doi: 10.1074/jbc.M801774200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pham L, Beyer K, Jensen ED, Rodriguez JS, Davydova J, Yamamoto M, Petryk A, Gopalakrishnan R, Mansky KC. Bone morphogenetic protein 2 signaling in osteoclasts is negatively regulated by the BMP antagonist, twisted gastrulation. J Cell Biochem. 112:793–803. doi: 10.1002/jcb.23003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jensen ED, Pham L, Billington CJ, Jr, Espe K, Carlson AE, Westendorf JJ, Petryk A, Gopalakrishnan R, Mansky K. Bone morphogenic protein 2 directly enhances differentiation of murine osteoclast precursors. J Cell Biochem. 109:672–682. doi: 10.1002/jcb.22462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Canalis E, Economides AN, Gazzerro E. Bone morphogenetic proteins, their antagonists, and the skeleton. Endocr Rev. 2003;24:218–35. doi: 10.1210/er.2002-0023. [DOI] [PubMed] [Google Scholar]
- 11.Wan DC, Pomerantz JH, Brunet LJ, Kim JB, Chou YF, Wu BM, Harland R, Blau HM, Longaker MT. Noggin suppression enhances in vitro osteogenesis and accelerates in vivo bone formation. J Biol Chem. 2007;282:26450–9. doi: 10.1074/jbc.M703282200. [DOI] [PubMed] [Google Scholar]
- 12.Gazzerro E, Deregowski V, Vaira S, Canalis E. Overexpression of twisted gastrulation inhibits bone morphogenetic protein action and prevents osteoblast cell differentiation in vitro. Endocrinology. 2005;146:3875–82. doi: 10.1210/en.2005-0053. [DOI] [PubMed] [Google Scholar]
- 13.Mundy C, Gannon M, Popoff SN. Connective tissue growth factor (CTGF/CCN2) negatively regulates BMP-2 induced osteoblast differentiation and signaling. J Cell Physiol. 229:672–681. doi: 10.1002/jcp.24491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsushita Y, Sakamoto K, Tamamura Y, Shibata Y, Minamizato T, Kihara T, Ito M, Katsube K, Hiraoka S, Koseki H, Harada K, Yamaguchi A. CCN3 protein participates in bone regeneration as an inhibitory factor. J Biol Chem. 288:19973–19985. doi: 10.1074/jbc.M113.454652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nistala H, Lee-Arteaga S, Smaldone S, Siciliano G, Carta L, Ono RN, Sengle G, Arteaga-Solis E, Levasseur R, Ducy P, Sakai LY, Karsenty G, Ramirez F. Fibrillin-1 and -2 differentially modulate endogenous TGF-beta and BMP bioavailability during bone formation. J Cell Biol. 190:1107–1121. doi: 10.1083/jcb.201003089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ono M, Inkson CA, Kilts TM, Young MF. WISP-1/CCN4 regulates osteogenesis by enhancing BMP-2 activity. J Bone Miner Res. 26:193–208. doi: 10.1002/jbmr.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, de Crombrugghe B. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- 18.Godyna S, Mann DM, Argraves WS. A quantitative analysis of the incorporation of fibulin-1 into extracellular matrix indicates that fibronectin assembly is required. Matrix Biol. 1995;14:467–77. doi: 10.1016/0945-053x(95)90004-7. [DOI] [PubMed] [Google Scholar]
- 19.Tran H, VanDusen WJ, Argraves WS. The self-association and fibronectin-binding sites of fibulin-1 map to calcium-binding epidermal growth factor-like domains. J Biol Chem. 1997;272:22600–6. doi: 10.1074/jbc.272.36.22600. [DOI] [PubMed] [Google Scholar]
- 20.Gu YC, Nilsson K, Eng H, Ekblom M. Association of extracellular matrix proteins fibulin-1 and fibulin-2 with fibronectin in bone marrow stroma. Br J Haematol. 2000;109:305–13. doi: 10.1046/j.1365-2141.2000.02011.x. [DOI] [PubMed] [Google Scholar]
- 21.Hergeth SP, Aicher WK, Essl M, Schreiber TD, Sasaki T, Klein G. Characterization and functional analysis of osteoblast-derived fibulins in the human hematopoietic stem cell niche. Exp Hematol. 2008;36:1022–34. doi: 10.1016/j.exphem.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 22.Cooley MA, Kern CB, Fresco VM, Wessels A, Thompson RP, McQuinn TC, Twal WO, Mjaatvedt CH, Drake CJ, Argraves WS. Fibulin-1 is required for morphogenesis of neural crest-derived structures. Dev Biol. 2008;319:336–45. doi: 10.1016/j.ydbio.2008.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kostka G, Giltay R, Bloch W, Addicks K, Timpl R, Fassler R, Chu ML. Perinatal lethality and endothelial cell abnormalities in several vessel compartments of fibulin-1-deficient mice. Mol Cell Biol. 2001;21:7025–34. doi: 10.1128/MCB.21.20.7025-7034.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Argraves WS, Tran H, Burgess WH, Dickerson K. Fibulin is an extracellular matrix and plasma glycoprotein with repeated domain structure. J Cell Biol. 1990;111:3155–64. doi: 10.1083/jcb.111.6.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X, Kua HY, Hu Y, Guo K, Zeng Q, Wu Q, Ng HH, Karsenty G, de Crombrugghe B, Yeh J, Li B. p53 functions as a negative regulator of osteoblastogenesis, osteoblast-dependent osteoclastogenesis, and bone remodeling. J Cell Biol. 2006;172:115–25. doi: 10.1083/jcb.200507106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nissen-Meyer LS, Jemtland R, Gautvik VT, Pedersen ME, Paro R, Fortunati D, Pierroz DD, Stadelmann VA, Reppe S, Reinholt FP, Del Fattore A, Rucci N, Teti A, Ferrari S, Gautvik KM. Osteopenia, decreased bone formation and impaired osteoblast development in Sox4 heterozygous mice. J Cell Sci. 2007;120:2785–95. doi: 10.1242/jcs.003855. [DOI] [PubMed] [Google Scholar]
- 27.Kodama N, Nagata M, Tabata Y, Ozeki M, Ninomiya T, Takagi R. A local bone anabolic effect of rhFGF2-impregnated gelatin hydrogel by promoting cell proliferation and coordinating osteoblastic differentiation. Bone. 2009;44:699–707. doi: 10.1016/j.bone.2008.12.017. [DOI] [PubMed] [Google Scholar]
- 28.Beier EE, Maher JR, Sheu TJ, Cory-Slechta DA, Berger AJ, Zuscik MJ, Puzas JE. Heavy metal lead exposure, osteoporotic-like phenotype in an animal model, and depression of Wnt signaling. Environ Health Perspect. 121:97–104. doi: 10.1289/ehp.1205374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao S, Fernald RD. Comprehensive algorithm for quantitative real-time polymerase chain reaction. J Comput Biol. 2005;12:1047–64. doi: 10.1089/cmb.2005.12.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu W, Saint DA. Validation of a quantitative method for real time PCR kinetics. Biochem Biophys Res Commun. 2002;294:347–53. doi: 10.1016/S0006-291X(02)00478-3. [DOI] [PubMed] [Google Scholar]
- 31.Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, Fell HP, Ferree S, George RD, Grogan T, James JJ, Maysuria M, Mitton JD, Oliveri P, Osborn JL, Peng T, Ratcliffe AL, Webster PJ, Davidson EH, Hood L, Dimitrov K. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26:317–25. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 32.Brumbaugh CD, Kim HJ, Giovacchini M, Pourmand N. NanoStriDE: normalization and differential expression analysis of NanoString nCounter data. BMC Bioinformatics. 12:479. doi: 10.1186/1471-2105-12-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cooley MA, Broome JM, Ohngemach C, Romer LH, Schaller MD. Paxillin binding is not the sole determinant of focal adhesion localization or dominant-negative activity of focal adhesion kinase/focal adhesion kinase-related nonkinase. Mol Biol Cell. 2000;11:3247–63. doi: 10.1091/mbc.11.9.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–5. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 36.Perbal B, Martinerie C, Sainson R, Werner M, He B, Roizman B. The C-terminal domain of the regulatory protein NOVH is sufficient to promote interaction with fibulin 1C: a clue for a role of NOVH in cell-adhesion signaling. Proc Natl Acad Sci U S A. 1999;96:869–74. doi: 10.1073/pnas.96.3.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–64. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 38.Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen BR, Selby PB, Owen MJ. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–71. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- 39.Deckelbaum RA, Majithia A, Booker T, Henderson JE, Loomis CA. The homeoprotein engrailed 1 has pleiotropic functions in calvarial intramembranous bone formation and remodeling. Development. 2006;133:63–74. doi: 10.1242/dev.02171. [DOI] [PubMed] [Google Scholar]
- 40.Chung IH, Han J, Iwata J, Chai Y. Msx1 and Dlx5 function synergistically to regulate frontal bone development. Genesis. 48:645–655. doi: 10.1002/dvg.20671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maes C, Carmeliet P, Moermans K, Stockmans I, Smets N, Collen D, Bouillon R, Carmeliet G. Impaired angiogenesis and endochondral bone formation in mice lacking the vascular endothelial growth factor isoforms VEGF164 and VEGF188. Mech Dev. 2002;111:61–73. doi: 10.1016/s0925-4773(01)00601-3. [DOI] [PubMed] [Google Scholar]
- 42.Ishijima M, Suzuki N, Hozumi K, Matsunobu T, Kosaki K, Kaneko H, Hassell JR, Arikawa-Hirasawa E, Yamada Y. Perlecan modulates VEGF signaling and is essential for vascularization in endochondral bone formation. Matrix Biol. 31:234–245. doi: 10.1016/j.matbio.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stickens D, Behonick DJ, Ortega N, Heyer B, Hartenstein B, Yu Y, Fosang AJ, Schorpp-Kistner M, Angel P, Werb Z. Altered endochondral bone development in matrix metalloproteinase 13-deficient mice. Development. 2004;131:5883–95. doi: 10.1242/dev.01461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang W, Yang F, Li Y, de Crombrugghe B, Jiao H, Xiao G, Zhang C. Transcriptional regulation of Vascular Endothelial Growth Factor (VEGF) by osteoblast-specific transcription factor Osterix (Osx) in osteoblasts. J Biol Chem. 287:1671–1678. doi: 10.1074/jbc.M111.288472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abu-Issa R, Smyth G, Smoak I, Yamamura K, Meyers EN. Fgf8 is required for pharyngeal arch and cardiovascular development in the mouse. Development. 2002;129:4613–25. doi: 10.1242/dev.129.19.4613. [DOI] [PubMed] [Google Scholar]
- 46.Ohbayashi N, Shibayama M, Kurotaki Y, Imanishi M, Fujimori T, Itoh N, Takada S. FGF18 is required for normal cell proliferation and differentiation during osteogenesis and chondrogenesis. Genes Dev. 2002;16:870–9. doi: 10.1101/gad.965702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guntur AR, Reinhold MI, Cuellar J, Jr, Naski MC. Conditional ablation of Pten in osteoprogenitors stimulates FGF signaling. Development. 138:1433–1444. doi: 10.1242/dev.058016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Celil AB, Campbell PG. BMP-2 and insulin-like growth factor-I mediate Osterix (Osx) expression in human mesenchymal stem cells via the MAPK and protein kinase D signaling pathways. J Biol Chem. 2005;280:31353–9. doi: 10.1074/jbc.M503845200. [DOI] [PubMed] [Google Scholar]
- 49.Rydziel S, Stadmeyer L, Zanotti S, Durant D, Smerdel-Ramoya A, Canalis E. Nephroblastoma overexpressed (Nov) inhibits osteoblastogenesis and causes osteopenia. J Biol Chem. 2007;282:19762–72. doi: 10.1074/jbc.M700212200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Fig. 1 Reduced ossification of skull bones in Fbln1 exon 1 nulls. A and B, μCT scans of wild-type (Fbln1 +/+) and Fbln1 exon 1 null (Fbln1 −/−) neonatal (P0) skulls. Note reduced ossification of the frontal bone (FB), parietal bone (P) and interparietal (Ip) bone in Fbln1 exon 1 nulls as compared to WT. C and D show alizarin red stained skulls from wild-type and Fbln1 exon 1 null neonates (P0). Note the increased bone porosity in the parietal and interparietal bones of the Fbln1 exon 1 null as compared to wild-type.
Supplemental Fig. 2 Fbln1 nulls have no immunologically detectable Fbln1. A and B, anti-Fbln1 (green) immunolabeling of wild-type and Fbln1 null E16.5 skull tissue sections. Nuclei were stained using propidium iodide (red). The inset panel in A shows a higher magnification of the boxed region. Note that Fbln1 is deposited in the ECM around osteoblasts (arrow) located in the frontal bone. The inset panel in B shows a higher magnification of the boxed region. Note that Fbln1 is not deposited in the ECM in the frontal bone. Red blood cells (RBC) were detected in both sections in the green fluorescence channel due to autofluorescence.
Supplemental Fig. 3 Analysis of BrdU incorporation in the dermis of E17.5 wild-type and Fbln1 null. A and C show BrdU incorporation into DNA by immunohistological analysis of skull bone sections from wild-type and Fbln1 null E17.5 embryos using an antibody to BrdU. B and D are negative controls that show the absence of BrdU nuclear staining in wild-type and Fbln1 null E17.5 skull sections in the absence of a BrdU antibody. E is a negative control showing the absence of BrdU nuclear staining in a wild-type E17.5 embryo not exposed to BrdU.
Supplemental Fig. 4 PECAM staining of blood vessels in the endochondral bones from E17.5 wild-type and Fbln1 nulls. A and B, show anti-PECAM (green) immunolabeling of tissue sections from the cranial base of wild-type and Fbln1 null E17.5 skulls. The inset panel in A shows a higher magnification of the boxed region in the wild-type and the inset panel in B shows a higher magnification of the boxed region in the Fbln1 null. Note that PECAM staining in the Fbln1 null sample (B) appears more diffused compared to the cell surface localization in the wild-type sample (A). Red blood cells (RBC, arrowheads) were detected in both sections in the green fluorescence channel due to autofluorescence.
Supplemental Fig. 5 Osteocalcin and Osteopontin mRNA are not significantly different in Fibulin-1 null calvariae. A shows Osteocalcin (Ocn) levels determined by qPCR analysis of mRNA isolated from E17.5 wild-type (WT) and Fbln1 null (KO) calvariae. B shows levels of the Osteopontin (Opn) mRNA determined by nanostring analysis of RNA isolated from E17.5 wild-type (WT) and Fbln1 null (KO) calvariae. The graphed values show mean relative amounts of mRNA based on WT (n = 9) and KO (n = 4) for E17.5 calvariae in A and WT (n = 3) and KO (n = 3) for E17.5 calvariae in B. P values were calculated using Students t-test for A and using NanoStriDE for B.