Abstract
Cytoplasmic dynein associates with dynactin to drive cargo movement on microtubules, but the structure of the dynein-dynactin complex is unknown. Using electron microscopy, we determined the organization of native bovine dynein, dynactin, and the dynein-dynactin-microtubule quaternary complex. In the microtubule-bound complex, the dynein motor domains are positioned for processive unidirectional movement and the cargo binding domains of both dynein and dynactin are accessible.
Cytoplasmic dynein-1 (hereafter dynein) interacts with its cofactor dynactin to transport a variety of cellular cargoes toward microtubule (MT) minus ends, supporting events critical for cell division and survival1. Despite elucidation of the structure and mechanical behavior of simplified versions of its catalytic motor domain2,3, the mechanism by which the dimeric dynein holoenzyme powers movement of intracellular cargoes over long distances remains undefined, primarily due to lack of structural information for the dynein-dynactin-MT complex.
Dynein is a dimeric multisubunit complex comprising a pair of ≈500 kDa heavy chains (HC) that contain the motor domain. Each HC also binds a light intermediate chain (LIC) and intermediate chain (IC) that is complexed with three light chains (LCs; LC7, LC8 and Tctex); together, these components form the tail domain that binds cargo4. We performed negative stain EM studies of vertebrate dynein, which can be observed as “V”-shaped dimers (Supplementary Fig. 1). Traditional 2D analysis of whole particles yielded averages with limited resolvable structural detail due to the extreme conformational flexibility of the head-tail link5–7 (Supplementary Fig. 1b). To overcome this challenge, we developed a “divide and conquer” image processing approach (see Online Methods and Supplementary Fig. 2) to reveal the organization of the dynein holoenzyme in unprecedented detail.
In the class averages two tail-dimerization sites are visible: one at the distal end of the tail, and a second positioned about 13 nm closer to the motor domains. The distal mass has long been ascribed to the three LCs8. The second mass, which has never been described previously, is positioned between donut-shaped lobes corresponding to the IC beta-propeller domains9 (Fig. 1). The organization of the three LCs on the IC polypeptide has been established biochemically10, and the proximity of this second dimerization density to the WD40 domains suggests it corresponds to the LC7 dimer (Fig. 1). Focused 2D analysis of the region extending beyond the putative LC7 dimer shows two densities that likely correspond to the LC8 and Tctex dimers that bind the IC N-terminus. This mobile IC-LC domain does not appear to interact with any portion of the HC. These data lead to a refined and novel model of dynein tail organization in which the distal portion exclusively comprises the HC, consistent with a recent report20.
Fig. 1. Proposed architecture of native vertebrate cytoplasmic dynein.
(A and B) Negative stain class average of the purified dynein dimer. Middle: Atomic coordinates for known dynein subunits (head domain, yellow, PDBid: 3VKH; LIC, orange, PDBid: 3W7G; LC7, green, PDBid: 3L9K) and a 7-bladed beta propeller (blue) were filtered to simulate low resolution EM density and overlaid on the class averages. Right: Cartoon representation of the proposed subunit architecture in the class average (see text for discussion). The locations of the HC dimerization domain and IC WD40 domain are established20. (C) IC-associated densities were identified through focused 2D analysis of the region surrounding the putative LC7 dimer density (white dotted circle). Densities proposed to correspond to the LC8 and Tctex dimers project away from the LC7 dimer at a range of orientations relative to the dynein tail, and do not appear to make any contact with the HC. (D) Crystal structures for LC7 (green, PDBid: 3L9K), LC8, and Tctex (magenta and orange, respectively, PDBid: 2PG1) are superimposed on the 2D averages in (C); LC positions are based on the literature. Density corresponding to the IC N-terminus (IC N-term) is also visible at the far end of the structure. (E) Cartoon representation of the proposed organization of dynein subunits. Scale bars correspond to 10nm.
Beyond the IC WD40 domains, a small crescent-shaped density is bound to each HC. We attribute these to the light intermediate chains (LICs) whose HC binding site (residues 650–800 11), lies between the IC binding site and motor domain. Beyond the LIC, the HC exhibits a dramatic kink that has not been previously described5–7,12. The function of the kink, which is observed in 100% of dynein averages (Supplementary Fig. 2b, bottom row), is unclear but its location between the tail and heads suggests it may serve as a hinge that allows motions associated with the mechanochemical cycle of the head to occur without disrupting tail-cofactor-cargo interactions13.
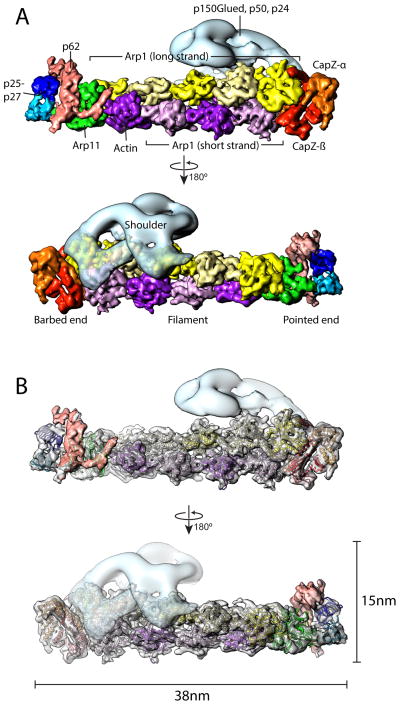
Dynein-based movement of cellular cargoes depends upon dynactin, a multi-protein assembly containing distinct domains that bind dynein, MTs, and cargoes14. The dynactin assembly consists of an actin-like polymer of the actin-related protein, Arp1, with distinct protein complexes attached at either end15. A large structure (“shoulder”) containing a dimeric assembly of p24, p50, and p150Glued projects from the side of the Arp filament near its barbed end. The pointed end of the Arp filament binds a complex of Arp11, p62, p27 and p25, subunits that contribute to dynactin complex stability and cargo binding16,17. Although models exist for dynactin subunit organization based on biochemical analysis and low-resolution EM18,19, the detailed architecture of this complex has only recently been described 20.
We determined the structure of native dynactin purified from bovine brain4. The Arp filament is resolvable to 6.5Å by cryoEM, but the shoulder readily detaches during vitrification and is not present in the reconstruction. We determined the organization of the entire vertebrate dynactin complex by fitting the 6.5Å cryoEM structure of the filament into a 24Å resolution negative stain structure of the intact complex (Fig. 2, Supplementary Figs. 3, 4). The two-stranded helical organization of the Arp polymer (four copies in one strand, five in the other) is obvious in the structures (Fig. 2). Dynactin’s single actin protomer14 lies at pointed end of the four-strand polymer 20. At the barbed end of the filament, helices corresponding to the α- and β- “tentacles” of CapZα-β are seen bound to the first Arp1 in each filament strand (Supplementary Fig. 4c). Arp11 contacts protomers of both filament strands at the opposite end. The remainder of the pointed end domain is not as well resolved owing to intrinsic flexibility, but densities corresponding to the p25-p27 subunits are discernible as two paralleloid prisms (Fig. 2b). The remaining unaccounted density, which lies between Arp11 and the p25-p27 dimer, is attributed to the p62 subunit.
Fig. 2. Subunit organization of vertebrate dynactin.
(A) The 6.5Å resolution cryoEM structure of the dynactin filament including the barbed and pointed end complexes (solid surface representation) was combined with a 24Å resolution negative stain structure of the shoulder domain (transparent surface representation, light blue). Each subunit of the dynactin is colored individually. The Arp filament contains a long strand (5 subunits, yellow and light yellow) and a short strand (4 subunits, purple and light purple). The protomer at the pointed end of the short strand is actin 20; all others are Arp1. The CapZ heterodimer (orange and red) caps the filament at the barbed end, and the pointed end complex (Arp11, p62, p27, p25) cap the filament at the pointed end. (B) The atomic model for CapZ (PDBid: 1IZN), homology models for Arp1 & Arp11, and two copies of p27 (PDBid: 3TV0) were fit into the uncolored transparent cryoEM density. (A corresponding atomic model is not available for p62.)
The p150Glued dimer, a component of the shoulder, contains an N-terminal coiled-coil domain that is absent in the 3D reconstruction due to its flexibility. Focused 2D analysis of the shoulder region reveals a mobile coiled-coil extension with a globular density at its end (Supplementary Fig. 3c). The globular structure we observe is much larger than what would be expected for the MT binding CAP-Gly domain located at the p150Glued N-terminus14. Furthermore, the position of the density 20nm from the shoulder is incompatible with the predicted 50nm length of the p150Glued coiled-coil. This domain likely corresponds to the recently described p150Glued ≈40 kDa intercoil domain20. The globular density may contribute to dynactin regulation in heretofore undefined ways, perhaps governing the mobility of the coiled-coil region to accommodate and facilitate simultaneous interactions of dynactin with both MTs and dynein.
Vertebrate dynein is not processive in the absence of dynactin, but instead undergoes diffusive, bi-directional movement21. Dynactin binding converts this behavior into longer range, unidirectional motion22, particularly when the N-terminal fragment of cofactor Bicaudal D homolog 2 (BicD2N) is added12,21. Attempts by other groups to determine the structural arrangement of the dynein-dynactin-BicD2N (DDB) complex were inconclusive, presumably due to the conformational heterogeneity of dynein12,21.
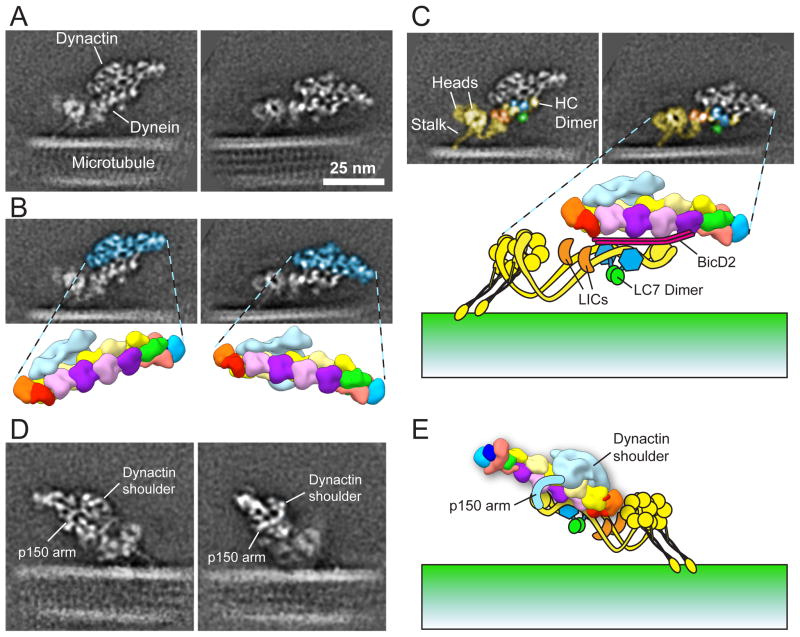
Because MT binding is expected to limit the conformational flexibility of dynein and the DDB complex, we prepared native dynein-dynactin complexes stabilized with BicD2N and bound to MTs (DDB-MT). In negative stain micrographs the DDB complexes are discernible as structures ~45nm in length attached at a range of angles to the MT surface, but tilted in a single direction, consistent with directional movement (Supplementary Fig. 5c). Detailed class averages of the DDB-MT complex were achieved in the same focused classification strategy used for dynein (Supplementary Fig. 5d,e).
In the DDB-MT complex, dynein is aligned between dynactin and the MT with the distal part of the dynein tail near the dynactin pointed end. The dynein tail associates with the short strand of the dynactin Arp filament, on the side opposite the shoulder (Fig. 3), consistent with the recently determined structure of the isolated dynein tail-dynactin-BicD2N complex20. Densities that cannot be attributed to either dynactin or dynein likely correspond to the BicD2N coiled-coil. Notably, the centers of the dynein heads are approximately 17 nm away from the MT and the MT-binding stalks are oriented at an acute angle (Supplementary Fig. 5d,e). Unlike the motor domains in free DDB complexes12,21, which exhibit a range of orientations and separation, those in DDB-MT complexes are in close proximity to one another (about 7 nm apart) but are not locked into a single orientation relative to each other (Supplementary Fig. 5e,g). The adjacent HC kink may provide a flexible “shock absorber” that allows the motor domains to undergo the structural changes that underlie stepping without interfering with dynactin and cargo interactions. The organization of dynein and dynactin in the DDB-MT complex also elucidates the arrangement of cargo-binding domains. Using the isolated dynein class averages for comparison, densities corresponding to the IC-LC dimerization domain and the LICs (Fig. 3b, green and orange, respectively) are apparently exposed in the DDB-MT complex. Given the importance of these regions for interactions with dynactin and other binding partners, it makes sense that these parts of the tail remain accessible.
Fig. 3. Organization of the DDB-MT complex.
(A) Two class averages of MT-bound DDB complexes, attached at slightly different angles relative to the MT. (B) 2D averages in the same orientation as (A) overlaid with a low-pass filtered 3D dynactin model (from Fig. 2) colored blue. (C) Dynein components in the 2D averages are assigned based on information determined from the isolated dynein class averages (Fig. 1). HCs are yellow, LICs are orange, the IC C-term WD40 domains are blue, and the putative LC7 dimer is green. (D) Two class averages of DDB-MT complexes showing the location of the shoulder and an extension (labeled “p150 arm”) that wraps around the Arp1 filament and contacts dynein. We propose this corresponds to the p150Glued coiled-coil. (E) A model of the DDB complex in the same orientation as the averages in (D), with the path of the putative p150Glued coiled-coil extension (“p150 arm”) traced in light blue. Scale bar in (A) corresponds to 25 nm, and all EM images are at the same scale.
The dynactin shoulder position on the opposite side of the DDB complex from the MT is intriguing given that the projecting p150Glued arm contains a MT-interacting domain. Although we did not observe a MT-associated density that might correspond to this domain, a thin filamentous extension emerging from the shoulder was seen in certain averages (Fig. 3d). The extension curves around the dynactin Arp filament and appears to make contact with the IC dimerization domain, but cannot be traced further. We propose that this extension is the C-terminal portion of the p150Glued coiled-coil, which may engage with dynein to help secure it to dynactin during processive movement.
This analysis provides the first visualization of the dynein-dynactin complex bound to microtubules, revealing an organization in which the HCs are oriented with the motor domains aligned to favor unidirectional movement. At the same time, the complex is structured in a way that favors cargo recruitment (Fig. 3c). The insights provided here will serve as paradigm for understanding the dynein-dynactin interactions that are essential for a wide range of minus-end directed motile phenomena. Although more detailed structural information will certainly improve understanding of the mechanism of dynein-dynactin-dependent movement, high-resolution determination of this entire structure will be extremely challenging due to the large size of the DDB complex and the heterogeneity of its link to the MT surface. The organizational framework of the DDB complex presented here provides the foundation for future work that supplies a new structural context for interpreting decades of biophysical and biochemical studies aimed at deciphering the mechanics of this fundamental and ancient cellular transport system.
ONLINE METHODS
Purification of dynein and dynactin and analysis of MT binding
Dynein and dynactin were purified from bovine brain by combining sucrose gradient centrifugation and ion-exchange chromatography as described in reference 23 with the following changes. Dynein and dynactin were eluted from the MonoQ 10/100 column using a linear salt gradient. Then each pool (identified by SDS-PAGE) was rechromatographed separately on a MonoQ 5/50 column and eluted with a salt bump. Between 2 and 5 ml of the peak dynein and dynactin fractions were supplemented with 1 mM TCEP and microconcentrated (without desalting or dilution) to ≈0.5 ml to yield a final concentration of 1–3 mg/ml. Proteins were kept on ice until use. The appearance of these complexes in negative stain EM was indistinguishable from those isolated using AMP-PNP-MT binding from chick embryo brain as described previously24. AMP-PNP-dependent binding of bovine dynein to MTs (Supplementary Fig. 5a) was analyzed using the method described previously24 except purified dynein was used and nucleotide conditions in the MT-co-pelleting step were varied.
Expression and purification of BicD2N
A plasmid encoding mouse BicD2 AA 25–400 (BicDNSh; hereafter BicD2N) in pET28a (Kanamycin resistant) with an N-terminal 6X-His and strepII-superfold GFP tag 21, (generously provided by R. McKenney), was transformed into BL21(DE3) cells containing a pACYCDuet-1 (Chloramphenicol resistant) plasmid with rare tRNAs for codon-usage in E. coli. Cells were then grown at 30°C in 2X LB media 25 containing 35 μg/ml Chloramphenicol and 50 μg/ml Kanamycin, till cell density reached O.D600nm ~0.4. The temperature was then dropped to 18°C and induced with 1mM IPTG (Sigma Aldrich). Cells were grown overnight at 18°C, harvested, and the cell pellet was flash frozen in liquid nitrogen and stored at −80°C for later purification.
The frozen cell pellet was thawed on ice and resuspended in lysis buffer containing 50mM NaH2PO4 (pH 8.0), 300mM NaCl, 10mM Imidazole, and 1X EDTA-free protease inhibitor (G Biosciences). After the cells were lysed by ultrasonication, the lysate was cleared of insoluble cell debris by centrifuging at 35,000 × g for 45 min, passed through a 0.2 μm syringe filter and incubated with HisPur™ Ni-NTA Resin (Fisher Scientific) for two hours at 4°C. The Ni resin was washed extensively with 50mM NaH2PO4 (pH 8.0), 300mM NaCl, 20mM imidazole and BicD2N was eluted using the same buffer containing 250mM imidazole. The His-tag purified BicD2N was concentrated and passed over Superose 6 gel-filtration column (GE Healthcare), equilibrated with 50mM NaH2PO4 (pH 8.0), 300mM NaCl and 1mM TCEP. Pure BicD2N fractions identified using by SDS-PAGE were pooled and glycerol was added to final concentration of 5% (w/v). The pooled fractions were then concentrated to 18μM and flash frozen in liquid nitrogen and stored at −80°C for later use.
Purification of dynein-dynactin-BicD2N complex bound to MT from mouse brain
Five g of fresh mouse brains were washed with ice cold PMEE buffer (35 mM Pipes, pH 7.2, 5 mM MgSO4, 1 mM EGTA, 0.5 mM EDTA, 6.8% (w/v) glycerol), supplemented with 1X protease inhibitor and 1mM TCEP, then homogenized in an ice cold Dounce homogenizer. The homogenate was centrifuged at 36,000 × g for 15mins at 2°C and the supernatant centrifuged at 90,000 × g for 30mins at 2°C. BicD2N was added to the final supernatant at 500 nM and the mixture was incubated on ice for 2 hrs with gentle swirling every 20 mins to mix. 4mM MgSO4, 1mM GTP (Sigma Aldrich) and 4mM AMP-PNP (Sigma Aldrich) was added and the mixture incubated for 10 min at 37°C. AMP-PNP was used to promote tight binding of the dynein motor to the MT surface (Supplementary Fig. 5a) The mixture was supplemented with 20μM taxol (Cytoskeleton Inc.) and further incubated for 15 mins at 37°C. The polymerized MTs, along with bound DDB particles, were pelleted by centrifuging at 21,000× g for 30 mins at 30°C. The pellet was washed by resuspension in PMEE buffer containing 4mM MgSO4, 1mM GTP, 4mM AMP-PNP and 20μM taxol and the MTs were pelleted as above. The final pellet was re-suspended in ten times its volume of PMEE buffer containing 4mM MgSO4, 1mM GTP, 4mM AMP-PNP and 20μM taxol at room temperature. The presence of DDB-MT complexes in the suspension was confirmed by SDS-PAGE and negative stain EM (Supplementary Fig. 5b,c).
Sample preparation for negative stain EM analysis
Grids for negative stain analysis of isolated dynein or dynactin were prepared in a similar fashion. Dynein and dynactin samples were diluted to 20 and 50μg/ml, respectively, in a buffer containing 35mM Tris, pH 7.2, 5mM MgSO4, 150mM KCl, and 1mM TCEP. A 4μl aliquot was applied to freshly-cleaned 400 mesh Cu-Rh maxtaform grids (Electron Microscopy Sciences) that had been coated with a thin layer of carbon. After incubating for 1min, excess protein was wicked off with a filter paper and the grid was immediately inverted and placed on 50μl droplet of 2% (w/v) uranyl formate solution. After 30s, excess stain was wicked off from the grid by touching the edge with filter paper. This staining step was repeated three times for thorough embedding of the sample, and the grids were air dried after the last blotting step.
In case of isolated dynactin, a majority of the particles exhibited a preferential orientation on the carbon support. To overcome this issue, we pretreated the plasma cleaned carbon surface on grids with 5μl of 0.1 % (w/v) poly L-lysine hydrobromide (Polysciences) for 90s, followed by two washes with 10μl drops of water. After the grids dried, the dynactin sample was applied and stained as described above. This treatment enabled us to obtain several additional views of dynactin that were missing earlier without this pre-treatment.
A similar staining strategy was used to prepare grids for analysis of the MT-DDB complex. The final MT-DDB suspension was diluted 15 fold in PMEE buffer supplemented with 4mM MgSO4, 1mM GTP, 4mM AMP-PNP and 20μM taxol. The staining steps were performed in the same fashion as for the isolated dynein complex.
Sample preparation for cryoEM analysis
Purified dynactin was applied to freshly glow-discharged 400 mesh C-Flat™ grids (Protochips) containing 2μm diameter holes spaced 2μm apart. Immediately before applying protein sample on grid, 0.025% (w/v) Amphipol A8-35 (Anatrace) was mixed with the sample to aid in dispersing dynactin particles across the holes in the carbon. 4μl of the dynactin sample was applied to the grid and excess sample was manually blotted using filter paper for ~5–7s and immediately vitrified by plunge freezing in liquid ethane slurry at −179°C. The entire procedure was carried out at 4°C and 98% humidity.
Data acquisition
All negative stain and cryoEM data were acquired using the Leginon automated data acquisition system 26. Data acquisition for negative stained samples was performed on a Tecnai Spirit (FEI) transmission electron microscope operating at 120keV. Images were collected at a nominal magnification of 52,000X on an F416 CMOS 4Kx4K camera (TVIPS) with a pixel size of 2.05 Å/pixel at specimen level. All micrographs were collected using an electron dose of 20 electrons/Å2 with a defocus range from 0.3 μm to 1.5 μm.
CryoEM data for dynactin were collected on Titan Krios (FEI) transmission electron microscope operating at 300keV, using a Gatan K2 Summit camera operated in counting mode at dose rate of ~10 electrons/pixel/s. Each movie comprised of 30 frames acquired over 6s, with a cumulative does of ~35 electrons/Å2. Imaging was performed at a nominal magnification of 22,500X, with a pixel size of 1.31 Å/pixel at specimen level using a defocus ranging from 0.8μm to 4μm.
Dynein Image Processing
1,200 micrographs of negatively stained dynein sample were collected for 2D analysis (Supplementary Fig. 1). All image preprocessing and initial 2D analysis was performed using the Appion image processing pipeline 27. The contrast transfer function (CTF) of each micrograph was estimated using CTFFindv3 28, and template-based automated particle selection was performed with FindEM 29. Templates for particle selection were generated from 2D class averages from a small set of manually picked particles. Phases for each micrograph were corrected using EMAN 30 and particles were extracted using a 640 X 640 pixel box. The data was binned by a factor of two for faster computation. Individual particles were normalized by eliminating pixels with values above or below 4.5σ of the mean pixel value using the normalization function in the XMIPP package 31.
An initial stack of 120,000 particles were subjected to five rounds of iterative multivariate statistical analysis (MSA) 32 and multi-reference alignment (MRA) in Appion to remove any non-particle features and aggregates that were erroneously picked by automated particle picker. To speed up this process, the particle stack was further binned by a factor of 4 before analysis. Particles belonging to classes that represented non-particles or aggregates were eliminated, resulting in a stack of 100,000 dynein particles. This particle stack was subjected to one generation of reference free 2D alignment and classification using ISAC 33, a module in EMAN2/SPARX software package 34,35, after applying a low pass and high pass filter of 15Å and 650Å, respectively. The stable averages obtained from ISAC had the overall “V” shaped feature of dynein with two circular features at the two free ends (Supplementary Fig. 1b). Due to the conformational heterogeneity of dynein, the class averages lacked structural details. In order to improve the resolvable details, we focused on sub regions of the molecule (Supplementary Fig. 2). For a given class average, soft-edged masks were created to isolate the tail and individual head domains.. These masks were applied to the hundreds of aligned particles belonging to the class average. This resulted in three stacks - one with only the tail region visible, another containing the first head domain, and another containing the second head domain. Each group of particles was individually subjected to three generations of reference-free classification and alignment using ISAC, with pixel error values between 3 and 8 and particles per group ranging from 250 to 100 for different generations. The stable averages obtained from these classifications showed significantly improve structural detail.
In order to generate a representative average of the full dynein molecule, the classification and alignment parameters for each individual classification were applied to the unmasked stack of particles. While the details outside of the focused region of interest were not well resolved after unmasking, structural features such as the kinked HC domain were discernible in many of the tail and head domain averages. These common structural features were used to overlay the head and tail classes in order to reassemble a composite view of the entire dynein molecule. For each set of class averages, the kinked region of one of the head domains was positioned in the center of the image, and the kinked region in the corresponding arm of the tail was also positioned in the center. A rotational and translational search using a limited search radius was used to align the kinked region in both class averages, and the averages were then themselves averaged together. This process was repeated for the second head and tail arm, resulting in a composite class average of the complete dynein molecule. These class averages not only revealed detailed structural features of dynein, but also showed the wide range of conformational heterogeneity of this complex in solution.
A similar masking approach was used to identify the dynein LCs. The unmasked dynein tail averages (Supplementary Fig. 2a) where the LC7 dimer was clearly visible were selected, and a soft mask was applied to the aligned particles around the area next to and including the LC7 dimer (area enclosed within the dotted circle in Fig. 1b). These masked particles were subjected to ISAC 2D alignment and classification, and the resulting class averages were unmasked as described earlier.
Dynactin Image Processing
Preprocessing and initial 2D analysis of the negative stain dynactin dataset was performed in the same manner as described for dynein. From 1,978 micrographs, 88,434 automatically selected particles were extracted with a box size of 480 pixels. After several iterations of particle cleaning by MSA/MRA 2D classification, we obtained 46,734 dynactin particles that included the shoulder domain, which were used for 3D reconstruction. We used a 60Å low pass filtered negative stain reconstruction of dynactin (EMD-2716) 19 as our initial model for 3D refinement by iterative projection matching using EMAN2 and SPARX libraries 34,35. Refinement of the initial model began generating forward projections at an angular increment of 20°, which decreased incrementally to 1°. The refinement would not proceed to the next angular step size until 95% of the particles had converged to a pixel error of less than one pixel. Forward projections of the refined volume showed excellent correlation with reference-free class averages, supporting the quality of reconstruction, free from model bias (Supplementary Fig. 3d). The resolution of the reconstruction was estimated by splitting the particle stack into odd and even halves, and by calculating the Fourier Shell Correlation (FSC) between the resulting volumes. The estimated resolution of the reconstruction at 0.5 FSC was 24Å (Supplementary Fig. 3e).
Due to its flexibility, the p150Glued coiled coil arm was not resolved in the 2D averages or 3D reconstructions of the dynactin complex. Using a soft mask to perform focused analysis on the region surrounding the shoulder domain, we were able to visualize the p150 Glued side arm. A thin filament could be seen extending away from the shoulder domain with a large globular density at the end, ~20nm from shoulder. The class averages show that this domain can undergo a wide range of motion (Supplementary Fig. 3b).
A total of 2,421 cryoEM images of dynactin were collected and analyzed for a higher resolution structure. K2 movie frames were aligned using the GPU frame alignment program described by Li et al. 36, which is incorporated into the Appion pipeline. A frame offset of 5 and B factor of 1000 pixel2 was used for the frame alignment step. CTF parameters were estimated using CTFFindv3, and images reporting a confidence value of less than 90% were discarded. Particles were picked using an automated template picker, as described earlier for negatively stained dynactin data. 133,558 Particles were extracted from the aligned micrographs using a box size of 512 pixels, which were binned by a factor of two for subsequent processing. As described previously, MSA/MRA 2D classification was utilized to discard false or damaged particles. The shoulder domain, which we have observed to readily dissociate from the dynactin complex, was absent in almost all the particles in the cryo dataset.
After removing junk particles, a stack of 59,538 particles remained, whose coordinates were imported into the program Relion 37 for 3D classification and reconstruction. Particles were extracted with a box size of 512 pixels and scaled to 256 pixels, with 2.62 Å/pixel values in order to reduce computational demand. 3D classification into three classes was performed, using the 24Å negative stain reconstruction of dynactin filtered to 60 Å as an initial model. After 25 iterations of 3D classification, 33,305 particles belonging to the most well resolved class average were used for further refinement. Refinement by projection matching in Relion resulted in a 9.5Å resolution (by Gold Standard FSC at a cutoff of 0.143) reconstruction of dynactin, although the pointed end was not resolved at subnanometer resolution due to intrinsic flexibility of this region. A soft 3D mask was generated from the 9.5Å resolution dynactin reconstruction and the pointed end of the complex was removed from the mask. Applying this mask during the refinement improved the alignment of the particles and accordingly improved the resolution of the 3D reconstruction to 7.5Å (by Gold Standard FSC at 0.143). The particle polishing methodology in Relion was then used to improve the resolution of this map to 6.5Å. The local resolution map shown in Supplementary Fig. 4d was calculated from the half volumes using the “blocres” function in the Bsoft package 38.
The individual components that make up the Arp filament of dynactin were clearly discernible, and the map was segmented using the volume tracer tool in UCSF Chimera 39. The cryo EM reconstruction of the Arp filament could be fit with high precision into the 24Å resolution negative stain dynactin reconstruction. By low pass filtering of the cryoEM reconstruction to a comparable resolution and using “diffmap” from the Grigorieff lab, it was possible to isolate density for the shoulder domain from the negative stain structure.
Nine copies of a homology model of Arp1, based on an F-Actin as template (as obtained by 19), were individually fit into the segmented CryoEM map of the filament using UCSF Chimera 39. A homology model for Arp11 was obtained using the Phyre2 40. This model could be docked with moderate confidence into the segmented EM density for Arp11 at the pointed end of dynactin. The overall, shape of the model corresponded to the shape of the density for Arp11. Two copies of the prism-shaped crystal structure of p27 (PDBid: 3TV0) could be docked into two hollow cylindrical densities at the pointed end, suggesting this density to be p25/p27 dimer. As no homology model was available for p62, any density not attributed to Arp11 or the p25/p27 dimer was ascribed to p62. UCSF Chimera 39 was used to generate all surface renderings of the dynactin EM density.
DDB-MT image processing
In order to accumulate sufficient particles for 2D analysis, 24,778 micrographs of negatively stained DDB-bound MTs were collected. 30,758 MT-attached DDB particles were manually selected and extracted from the micrographs with a box size of 480 (Supplementary Fig. 5c). Particles were binned by a factor of 2 for processing. The signal contributed by the MT dominated the 2D reference-free alignment of the particles, resulting in averages of the MTs in which the DDB complex was indistinguishable. To overcome this issue, a soft edged mask was applied to the particles such that the majority of MT was excluded, leaving only the DDB complex and the MT surface. 2D alignment and classification of these particles revealed the overall shape of the complex, which contained an elongated region, corresponding to the dynactin and dynein tail with circular densities closer to the MT surface, corresponding to the dynein motors. However, these averages still lacked sufficient structural detail for interpretation. Similar to the methodology used to examine the isolated dynein dimers, we generated masks to focus on the elongated and circular regions separately, followed by reassembly using correlated common features. To generate the histograms of the angles between the elongated region of DDB and the MT surface (Supplementary Fig. 5f), we used the technique described previously41.
Supplementary Material
Acknowledgments
We thank Rick McKenney (Vale lab, UCSF) for providing the BicD2N construct, and the Encalada lab (Scripps) for providing mouse brains. We are enormously grateful to Ron Milligan for his ongoing support and input into this project. This work was supported by the Damon Runyon Cancer Research Foundation (DFS-#07-13), the Pew Scholars program, the Searle Scholars program, and NIH grant DP2 EB020402-01 to GCL and NIH grant GM44589 to TAS.
Footnotes
ACCESSION CODES:
Density maps of the negative stain and cryoEM dynactin reconstructions have been deposited at the Electron Microscopy Data Bank (EMDB) and assigned reference codes EMDB-6290 and EMDB-6289, respectively.
AUTHOR CONTRIBUTIONS:
S.A.K. prepared the isolated dynein and dynactin. S.C. prepared the MT-DDB complex and performed all electron microscopy, image analysis, and reconstructions. All authors contributed to the experimental design and assembly of the manuscript.
References
- 1.Kardon JR, Vale RD. Regulators of the cytoplasmic dynein motor. Nat Rev Mol Cell Biol. 2009;10:854–865. doi: 10.1038/nrm2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carter AP. Crystal clear insights into how the dynein motor moves. J Cell Sci. 2013;126:705–13. doi: 10.1242/jcs.120725. [DOI] [PubMed] [Google Scholar]
- 3.Roberts AJ, Kon T, Knight PJ, Sutoh K, Burgess SA. Functions and mechanics of dynein motor proteins. Nat Rev Mol Cell Biol. 2013;14:713–26. doi: 10.1038/nrm3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bingham JB, King SJ, Schroer TA. Purification of dynein and dynactin from brain tissue. Meth Enzymol. 1998;298:171–184. doi: 10.1016/s0076-6879(98)98017-x. [DOI] [PubMed] [Google Scholar]
- 5.Vallee RB, Wall JS, Paschal BM, Shpetner HS. Microtubule-associated protein 1C from brain is a two-headed cytosolic dynein. Nature. 1988;332:561–563. doi: 10.1038/332561a0. [DOI] [PubMed] [Google Scholar]
- 6.Amos LA. Brain dynein crossbridges microtubules into bundles. J Cell Sci. 1989;93:19–28. doi: 10.1242/jcs.93.1.19. [DOI] [PubMed] [Google Scholar]
- 7.Steffen W, Hodgkinson JL, Wiche G. Immunogold localisation of the intermediate chain within the protein complex of cytoplasmic dynein. J Struct Biol. 1996;117:227–235. doi: 10.1006/jsbi.1996.0087. [DOI] [PubMed] [Google Scholar]
- 8.Vale RD. The molecular motor toolbox for intracellular transport. Cell. 2003;112:467–480. doi: 10.1016/s0092-8674(03)00111-9. [DOI] [PubMed] [Google Scholar]
- 9.Wilkerson CG, King SM, Koutoulis A, Pazour GJ, Witman GB. The 78,000 M(r) intermediate chain of Chlamydomonas outer arm dynein isa WD-repeat protein required for arm assembly. J Cell Biol. 1995;129:169–78. doi: 10.1083/jcb.129.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hall J, Song Y, Karplus PA, Barbar E. The crystal structure of dynein intermediate chain-light chain roadblock complex gives new insights into dynein assembly. J Biol Chem. 2010;285:22566–75. doi: 10.1074/jbc.M110.103861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tynan SH, Gee MA, Vallee RB. Distinct but overlapping sites within the cytoplasmic dynein heavy chain for dimerization and for intermediate chain and light intermediate chain binding. J Biol Chem. 2000;275:32769–74. doi: 10.1074/jbc.M001537200. [DOI] [PubMed] [Google Scholar]
- 12.Schlager MA, Hoang HT, Urnavicius L, Bullock SL, Carter AP. In vitro reconstitution of a highly processive recombinant human dynein complex. EMBO J. 2014;33:1855–68. doi: 10.15252/embj.201488792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burgess SA, Walker ML, Sakakibara H, Oiwa K, Knight PJ. The structure of dynein-c by negative stain electron microscopy. J Struct Biol. 2004;146:205–216. doi: 10.1016/j.jsb.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 14.Schroer TA. Dynactin. Annu Rev Cell Dev Biol. 2004;20:759–779. doi: 10.1146/annurev.cellbio.20.012103.094623. [DOI] [PubMed] [Google Scholar]
- 15.Eckley DM, et al. Analysis of dynactin subcomplexes reveals a novel actin-related protein associated with the Arp1 filament pointed end. J Cell Biology. 1999;147:307–319. doi: 10.1083/jcb.147.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yeh TY, et al. Dynactin helps target Polo-like kinase 1 to kinetochores via its left-handed beta-helical p27 subunit. EMBO J. 2013;32:1023–35. doi: 10.1038/emboj.2013.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yeh TY, Quintyne NJ, Scipioni BR, Eckley DM, Schroer TA. Dynactin’s pointed-end complex is a cargo-targeting module. Mol Biol Cell. 2012;23:3827–37. doi: 10.1091/mbc.E12-07-0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hodgkinson JL, Peters C, Kuznetsov SA, Steffen W. Three-dimensional reconstruction of the dynactin complex by single-particle image analysis. Proc Natl Acad Sci USA. 2005;102:3667–3672. doi: 10.1073/pnas.0409506102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Imai H, Narita A, Maeda Y, Schroer TA. Dynactin 3D Structure: Implications for Assembly and Dynein Binding. J Mol Biol. 2014;426:3262–71. doi: 10.1016/j.jmb.2014.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Urnavicius L, et al. The structure of the dynactin complex and its interaction with dynein. Science. 2015 doi: 10.1126/science.aaa4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKenney RJ, Huynh W, Tanenbaum ME, Bhabha G, Vale RD. Activation of cytoplasmic dynein motility by dynactin-cargo adapter complexes. Science. 2014;345:337–41. doi: 10.1126/science.1254198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.King SJ, Schroer TA. Dynactin increases the processivity of the cytoplasmic dynein motor. Nat Cell Biol. 2000;2:20–24. doi: 10.1038/71338. [DOI] [PubMed] [Google Scholar]
- 23.Bingham JB, King SJ, Schroer TA. Purification of dynactin and dynein from brain tissue. Methods Enzymol. 1998;298:171–84. doi: 10.1016/s0076-6879(98)98017-x. [DOI] [PubMed] [Google Scholar]
- 24.Schroer TA, Sheetz MP. Two activators of microtubule-based vesicle transport. J Cell Biol. 1991;115:1309–18. doi: 10.1083/jcb.115.5.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bertani G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol. 1951;62:293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suloway C, et al. Automated molecular microscopy: the new Leginon system. J Struct Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 27.Lander GC, et al. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol. 2009;166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–47. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 29.Roseman AM. FindEM--a fast, efficient program for automatic selection of particles from electron micrographs. J Struct Biol. 2004;145:91–9. doi: 10.1016/j.jsb.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 30.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 31.Sorzano CO, et al. XMIPP: a new generation of an open-source image processing package for electron microscopy. J Struct Biol. 2004;148:194–204. doi: 10.1016/j.jsb.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 32.Ogura T, Iwasaki K, Sato C. Topology representing network enables highly accurate classification of protein images taken by cryo electron-microscope without masking. J Struct Biol. 2003;143:185–200. doi: 10.1016/j.jsb.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 33.Yang Z, Fang J, Chittuluru J, Asturias FJ, Penczek PA. Iterative stable alignment and clustering of 2D transmission electron microscope images. Structure. 2012;20:237–47. doi: 10.1016/j.str.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang G, et al. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 35.Hohn M, et al. SPARX, a new environment for Cryo-EM image processing. J Struct Biol. 2007;157:47–55. doi: 10.1016/j.jsb.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 36.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10:584–90. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–30. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heymann JB, Belnap DM. Bsoft: image processing and molecular modeling for electron microscopy. J Struct Biol. 2007;157:3–18. doi: 10.1016/j.jsb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 39.Goddard TD, Huang CC, Ferrin TE. Visualizing density maps with UCSF Chimera. J Struct Biol. 2007;157:281–7. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 40.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–71. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 41.Screpanti E, et al. Direct binding of Cenp-C to the Mis12 complex joins the inner and outer kinetochore. Curr Biol. 2011;21:391–8. doi: 10.1016/j.cub.2010.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.