Abstract
Three isolates of a bacterium recovered from human endometrium using conventional culture methods were characterized biochemically and subjected to 16S rRNA gene sequencing and phylogenetic analysis. Isolates were non-motile, obligately anaerobic, non-spore forming, asaccharolytic, non-cellulolytic, indole positive, Gram positive rods. Cell wall fatty acid profiling revealed C14:0, C16:0, C18:2 ω6, 9c, C18:1 ω9c and C18:0 to be the major fatty acid composition. The DNA mol % G+C was determined to be 44.2%. 16S rRNA gene sequence analysis revealed only 91% sequence similarity with the closest cultivated bacterial isolate, Saccharofermentans acetigenes. Based on genotypic and phenotypic data, all three isolates are considered to be members of the same species and data suggest it represents a novel genus and species in the order Clostridiales with an association with Clostridium rRNA cluster III within the family Ruminococcaceae. We propose the name, Mageeibacillus indolicus gen. nov., sp. nov. The type strain is BAA-2120T and CCUG 59143T.
Keywords: bacterial vaginosis associated bacterium 3; Mageeibacillus indolicus gen. nov., sp. nov.; taxonomy; genital tract
1. Introduction
Bacterial vaginosis (BV) is a common polymicrobial syndrome affecting women of reproductive age. In women with BV, the vaginal microbiome is characterized by a higher abundance and greater diversity of both facultative and anaerobic organisms and a decrease in Lactobacillus species. BV is associated with an increased risk of acquiring and transmitting sexually transmitted infections including human immunodeficiency virus type 1 (HIV) and herpes simplex virus type 2 [1, 2, 3]. Despite treatment, one in four women fails antibiotic therapy and the proportion of women who develop recurrent BV increases over time [4]. Failure to describe all of the etiologic agents through conventional culture-based methods has limited our current understanding of the syndrome as well as the identification of alternative treatment regimens.
Advances in molecular approaches, specifically the sequencing and phylogenetic analysis of the 16S rRNA gene has enhanced the understanding of the complex bacterial biota of the human vagina. Fredricks et al. reported the use of molecular methods for detecting fastidious and non-cultivable organisms highly specific for BV, including three novel species designated BV-associated bacterium 1, 2, and 3 (BVAB1, BVAB2 and BVAB3) in the order Clostridiales [5, 6, 7]. Although clinical criteria or the Nugent scoring method to detect altered vaginal flora were not implemented in their study, Zhou et al. also found novel species in the order Clostridiales in vaginal communities not dominated by Lactobacillus species [8].
In this study, we characterized three bacterial isolates (0009-5T, 0004-9, 0019-D) that were indistinct from each other, and were classified as “anaerobic Gram positive rods” by initial biochemical testing. Further characterization of this group using phenotypic and genotypic approaches suggested that these isolates represent a novel genus and species that we here designate Mageeibacillus indolicus gen. nov., sp. nov.
2. Methods
2.1. Isolation and cultivation
The isolates were obtained from endometrial biopsy samples from women being evaluated for pelvic inflammatory disease (PID) in a protocol approved by the University of Pittsburgh Institutional Review Board. Briefly, a Pipelle® endometrial sampling device was inserted through the cervix, into the uterine cavity and the biopsy was aspirated by suction. The biopsy was initially inoculated onto Brucella agar supplemented with 5% sheep blood (bioMerieux, Durham, NC) and incubated anaerobically at 37°C for four to seven days. After the initial isolation and identification, all isolates were frozen at −80°C in litmus milk (Becton Dickinson, Franklin Lakes, NJ). Immediately prior to advanced testing, isolates were thawed, inoculated onto the same media and incubated as described above. Isolates were subcultured at least twice before testing to ensure purity.
Primary examination of cell morphology, Gram reaction and motility by dispersion in chopped meat broth (Becton Dickinson, prepared in-house) were examined with light microscopy at 1000X and 400X, respectively. Cell wall structure was also assessed using the rapid potassium hydroxide (KOH) test previously performed by Halebian et al. [9] Colony morphology was described at 20X with a dissecting microscope. Aerotolerance was tested by subculturing onto a Brucella agar plate, supplemented with 5% sheep’s blood (bioMerieux) and incubated separately in 5–7% CO2 and an AnaeroPack® system containing a Pack-MicroAero sachet and jar (Mitsubishi Gas Company, Tokyo, Japan) at 37°C for 48 and 96 hours, respectively. Initial biochemical tests performed included catalase (3%) (Fisher Scientific, Pittsburgh, PA), spot indole (prepared in-house), urease (Becton Dickinson), oxidase (Becton Dickinson) and susceptibility to the special potency ID discs; vancomycin, 5μg and colistin, 10μg (Becton Dickinson). Lipase and lecithinase activity on Egg Yolk agar (bioMerieux), carbohydrate fermentation testing using peptone-yeast (PY) broth with the separate addition of glucose and ribose (prepared in-house), as well as PRAS esculin and gelatin hydrolysis (Anaerobe Systems, Morgan Hill, CA) were also tested.
Due to poor growth in broth, growth stimulation testing was performed and included the separate addition of 0.5% hemin (Sigma-Aldrich, St. Louis, Missouri), 0.5% arginine (Sigma-Aldrich), 0.001% sodium bicarbonate (Fisher Scientific), 0.5% Tween 80 (Beckton Dickinson), 5% horse serum (Life Technologies, Carlsbad, CA) and 1% glucose (Beckton Dickinson) to chopped meat carbohydrate (CMC) broth and PY broth. All initial biochemical testing and growth stimulation testing was performed according to Summanen et al. [10].
Cellulose degradation was assessed using a plate assay developed by Kasana et al. [11]. Briefly, inoculation on carboxymethylcellulose agar (prepared in-house) was performed and plates were incubated anaerobically at 37°C for 96 hours. Gram’s iodine was then flooded onto the plate to assess for zones of hydrolysis (agar without the uptake of Gram’s iodine) around the colonies suggesting cellulase production by the organism.
Optimal temperature for growth was determined by inoculating and incubating the isolates anaerobically as described above at varying temperatures. Minimum and maximum temperatures for growth were 35°C and 42°C, respectively.
Demonstration of spores was performed according to Johnson, Summanen and Finegold [12]. Briefly, a turbid suspension was made in CMC broth supplemented with 5% horse serum and incubated anaerobically at 37°C for one week followed by heat and ethanol treatments.
Additional biochemical testing was performed using API rapid ID 32A and API ZYM enzyme detection panels (bioMerieux) according to the manufacturer’s instructions.
2.2. Antimicrobial Susceptibility Testing
Antimicrobial susceptibility testing using the agar dilution method was performed to test susceptibility of the three strains to metronidazole, clindamycin, cefoxitin and doxycycline according to the Clinical Laboratory Standards Institute guidelines (CLSI) for anaerobic bacteria, 5th edition [13].
2.3. Fatty Acid Methyl Extraction Analysis
Fatty acid methyl extraction (FAME) was performed to determine the cell wall fatty acid composition. Because of the poor growth observed in PY broth, the standard growth medium used for FAME for comparison to their standard libraries at Microbial ID, the following recommendation by personnel at Microbial ID for processing the organisms for FAME was performed; isolates were cultured on Brucella agar with 5% sheep blood as previously described and colonies were harvested at two days growth, used to make a turbid suspension in DNAse free water and pelleted by centrifugation at 12 000 r.p.m for two minutes. The supernatant was discarded and the pellet was frozen at −80°C. The pellets were sent on dry ice overnight to Microbial ID (Newark, DE) where fatty acid analysis was performed by gas chromatography using a standard protocol, the MIDI Sherlock MIS system [14].
2.4. Electron Microscopy
For transmission electron microscopy, cells were fixed in 2.5% gluteraldehyde and negative staining was performed on copper grids with a formver membrane. Micrographs were recorded on a Jeol JEM-1011 electron microscope operating at 80kV. For scanning electron microscopy, cells were fixed in 2.5% gluteraldehyde and 1% osmium tetroxide (OsO4). Specimens were dehydrated in ethanol, coated with 3.5 nm of gold/palladium and analyzed with a JEM-6335F field emission SEM at 3kV.
2.5. DNA mol% G+C content determination
0009-5T was submitted to the J. Craig Venter Institute for whole genomic sequencing as part of the Human Microbiome Project. The DNA mol% G+C content was determined by analyzing the complete genomic sequence using Molecular Evolutionary Genetics Analysis (MEGA4) software [15].
2.6. DNA isolation, 16S rRNA gene sequencing and phylogenetic analysis
Whole genomic DNA was extracted as previously described in Antonio et al. [16]. DNA from the three isolates was PCR-amplified using broad-range primers 27F (5´-AGAGTTTGATCMTGGCTCAG-3´) (Frank et al.) [17] and 1541R (5´-AAGGAGGTGATCCAGCCGCA-3´) (Löeffler et al.) [18] to amplify nearly complete 16S rRNA genes. Each 50 μl PCR reaction contained 1X Pfu Buffer, 0.8 mM deoxynucleotide triphosphate mixture, 1.5 U PfuTurbo Hot Start DNA Polymerase (Stratagene-Agilent, Santa Clara, CA), 0.2 μM forward and reverse primers. The PCR conditions included a pre-melt at 95 °C for 10 min and then 40 cycles of 95°C for 30 seconds (melt), 55°C for 30 seconds (annealing) and 72°C for 30 seconds (extension), followed by a final extension step at 72 °C for 7 min. The PCR products were visualized after electrophoresis using 1.5% agarose gels and staining with Gel-Red (Biotium, Hayward, CA). The PCR products were sequenced (Big Dye Version 3, Life Technologies) using a panel of broad-range 16S rRNA gene-directed sequencing primers to ensure that every base was read at least 2 times. The sequencing primers included 27F, 1541R, 338F (5´-ACTCCTRCGGGAGGCAGCAG-3´), 1060F (5´-TGTCGWCAGCTCGTGYYGTGA-3´), 806R (5´-GGACTACCAGGGTATCTAAT-3´), 338R (5´-CTGCTGCCTCCCGYAGGAGT-3´) and 1407F (5´-TGYACWCACCGCCCGTC-3´) (Loy et al.) [19]. Raw sequence data were edited and assembled using Sequencher (Gene Codes, Ann Arbor, MI). A multiple sequence alignment was created from the consensus sequence of the novel isolates and type strains of close relatives obtained from NCBI GenBank and Ez Taxon databases [20]. Evolutionary analyses were conducted in MEGA6 [21]. A phylogenetic tree was constructed with 26 sequences using the Neighbor-Joining method to infer evolutionary history [22]. Evolutionary distances were computed using the Jukes-Cantor method [23] and bootstrap analysis (1000 replicates) was performed to determine the reliability of the inferred tree [24]. All positions containing gaps and missing data were eliminated and a total of 1203 positions were included in the final dataset.
2.7. GenBank Accession Numbers
The partial 16S rRNA gene sequences from the isolates were submitted to GenBank and the Accession numbers for bacterial isolates 0004-9, 0009-5T, and 0019-D are GQ900631, GQ900632 and GQ900633, respectively. The whole genomic sequence for 0009-5T was submitted to GenBank and the Accession number is CP001850.
3. Results and Discussion
All three isolates were non-motile, vancomycin sensitive, colistin resistant, obligately anaerobic rods that stained Gram negative with blunt or tapered ends and were asaccharolytic, non-cellulolytic, catalase negative, indole positive, urease negative, oxidase negative and were negative for esculin and gelatin hydrolysis, and lecithinase and lipase activity. A negative rapid KOH test (absence of stringing reaction with the addition of bacteria to a 3% KOH solution) validated disk susceptibility results and confirmed the bacteria as a Gram positive rod. There was no demonstration of spores following heat and ethanol treatments. The optimal temperature for growth ranged from 35–42°C. The DNA mol% G+C content was determined to be 44.2%. Table 1 shows phenotypic and genotypic data which are useful for differentiating Mageeibacillus indolicus from other taxa in the Clostridium rRNA cluster III and related organisms.
Table 1.
Characteristics for differentiating related organisms within Clostridium rRNA cluster III and cluster IV a, b
| Mageeibacillus indolicus |
Saccharofermentans acetigenes |
Fastidiosipila sangunis |
Clostridium caenicola |
Clostridium clariflavum |
Clostridium sufflavum |
Flavonifractor plautii |
Ruminococcus albus |
|
|---|---|---|---|---|---|---|---|---|
| % similarity | 91% | 88% | 88% | 90% | 89% | 86% | 84% | |
| Characteristics | ||||||||
| Cluster | III | III | III | III | III | III | IV | IV |
| Cellular Morphology | rod | coccobacillary | cocci | rod | rod | rod | rod | cocci |
| Spores | neg | pos | neg | pos | pos | pos | neg | neg |
| Gram stain | neg | pos | pos | neg | variable | neg | variable | pos |
| Motility | neg | neg | neg | pos | NO | pos | pos | neg |
| Flagella | neg | neg | neg | PB | P | P | P | neg |
| Cellulose degradation | neg | neg | neg | pos | pos | pos | neg | neg |
| Growth at 60°C | neg | neg | neg | pos | pos | neg | neg | neg |
| Mol % G+C | 44.2 | 55.6 | 33 | 51.1 | 36.9 | 40.7 | 58–61.6 | 42.6–45.8 |
| Predominant Fatty Acids (>5%) | C14:0, C16:0, C18:2ω6, 9c, C18:1ω9c, C18:0 | iso-C15:0, anteiso-C15:0, iso-C14:0, 3-OH | C14:0, C16:0, C18:2ω6, 9c, C18:1ω9c, C18:0 | iso-C15:0, anteiso-C15:0, C16:0, iso-C17:0, anteiso-C17:0 | C16:0, iso-C16:0, C16:0 DMA, iso-C17:0 | iso-C15:0, iso-C14:0, C16:0 DMA, | Unknown compound between C12:0 and C13:0, C14:0, Unknown compound between anteiso-C15:0 and C15:0, C16:0, C17:1ω8c, C18:0 | iso-C14:0, Unknown compoud between iso-C14:0 and C14:0, C14:0, anteiso-C15:0, C15:0, iso-C16:0, C16:0 |
Characteristics are described as (neg) negative, (pos) positive, (ND) not determined, (NO) not observed, (PB) polar bundle, (P) peritrichous.
Following five days of anaerobic incubation at 37°C on Brucella agar supplemented with 5% sheep’s blood (bioMerieux), colony morphology revealed a one millimeter, entire, flat fried egg with a transparent pink edge and slight iridescent cream center. Longer incubation resulted in a slight spreading of colonies and larger colony.
Growth was not supported in PY or CMC broth alone. Increased growth was not observed in either broth when supplemented as described above, however the initial inoculum was maintained in PY supplemented with glucose, arginine, sodium bicarbonate, hemin and horse serum. Growth was also maintained in CMC supplemented with horse serum.
The same positive reactions to twelve substrates were observed for all three strains following API ID 32A and API ZYM testing and include the following; indole, arginine arylamidase, phenylalanine arylamidase, leucine arylamidase, tyrosine arylamidase, glycine arylamidase, α-fucosidase, histidine arylamidase, serine arylamidase, valine arylamidase, acid phosphatase, and napthol-AS-BI-phosphohydrolase suggesting amino acids are utilized as a primary source of carbon. Enzymatic activity was observed for alanine arylamidase for two of the three strains tested (0019-D and 0004-9). Reactions to substrates were rated as positive regardless if color change was weak or robust.
Electron micrograph images (available as supplementary data, S1 and S2) taken from the strains grown on Brucella agar revealed rods approximately 1.25 μm in length and 588 nm in diameter occurring in chains or in small aggregates. S1 shows a Gram positive cell wall with a relatively thin layer of peptidoglycan and may explain why the organism stains Gram negative.
Predominant fatty acids (> 5%) observed among all strains were of the unbranched saturated and unsaturated forms; C14:0 (4.83–11.09%), C16:0 (30.13–39.31%), C18:2 ω6, 9c (7.62–10.18%), C18:1 ω9c (25.52–32.75%) and C18:0 (5.69–10.89%). Minor fatty acids present consisted of unbranched and branched iso- and anteiso- forms; C12:0 (1.18–1.95%), C15:0 (1.85–3.05%), iso-C15:0 (0–0.41%), anteiso-C15:0 (0–0.66%), C16:0 aldehyde (0–0.58%), C16:1 ω7c (1.23–1.56%), C16:0 DMA (0–0.93%), anteiso-C17:0 (0.49–0.61%), C17:0 (1.56–2.56%). The Sherlock MIS software used to analyze fatty acid composition profiles did not recognize the pattern generated for comparison to known cultivated and tested bacteria, further supporting the novelty of this bacterium.
Sexually Transmitted Diseases Treatment guidelines recommend treatment of BV with metronidazole or clindamycin and PID with cefoxitin in combination with doxycycline with or without metronidazole [33]. These antibiotics were chosen for susceptibility testing because these novel strains were found to be highly associated with BV and these microorganisms were recovered from the upper genital tract of women being evaluated for pelvic inflammatory disease. Breakpoints of resistance for metronidazole, clindamycin and cefoxitin are ≥ 32 μg ml−1, ≥ 8 μg ml−1, and ≥64 μg ml−1, respectively. CLIS guidelines do not list breakpoints for doxycycline for anaerobes, so susceptibility could not be determined. The range of minimal inhibitory concentrations observed for the three isolates for doxycycline however was (4–16 μg ml−1). Although only three isolates were tested, no resistance to metronidazole, clindamycin or cefoxitin was observed among the strains and the ranges of minimal inhibitory concentrations for those drugs were (0.25–0.5 μg ml−1), (0.125 μg ml−1) and (4–8 μg ml−1), respectively.
Comparing our novel isolate sequences with sequences in GenBank using the NCBI BLAST tool revealed that the 16S rRNA gene sequences were most closely related (≥ 98%) to the sequences from uncultivated bacterial clones detected by PCR and designated as BVAB3 [5], a vaginal clone (AY995273) in the study by Zhou et al., [8] and skin clones (GQ043025, GQ038494) reported by Grice et al., [34]. The novel isolate sequences were also distinct from sequence types obtained from clones previously described as BVAB1 (sequence similarity, 82%) and BVAB2 (sequence similarity, 89%) from the human vagina [5].
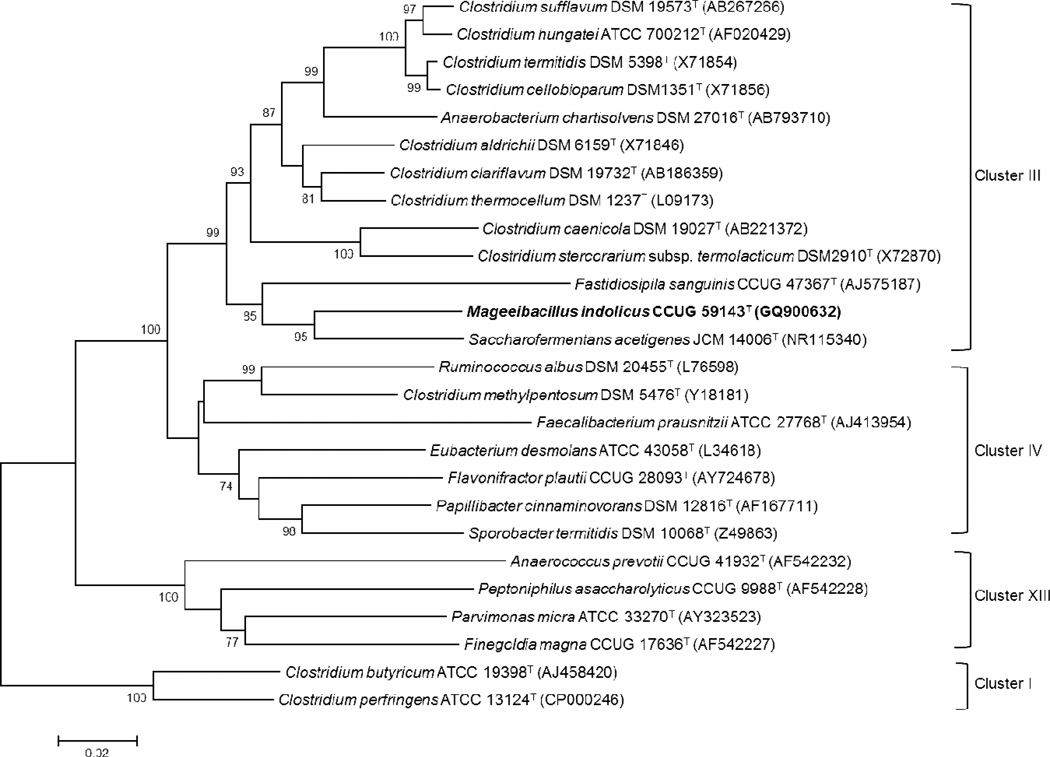
The closest bacterial isolate Saccharofermentans acetigenes described by Chen et al. [25] had a 91% sequence similarity. Phylogenetic analysis revealed that the novel bacterium formed deep branches with S. acetigenes and Fastidiosipila sanguinis described by Falsen et al. [26], both of which are associated with Clostridium rRNA cluster III (as defined by Collins et al.) [35] and are now included in the family Ruminococcaceae [36]. These associations are supported by high bootstrap values as shown in Figure 1. The high level of sequence divergence suggests that strain 0009-5T could represent a novel species of a new genus in this cluster.
Figure 1.
Neighbor-joining phylogenetic dendrogram showing the phylogenetic position of Mageeibacillus indolicus CCUG 59143T among closely related species using 16S rRNA sequences. The tree was rooted with C. butyricum ATCC 19398T and C. perfringes ATCC 13124T. Numbers at branch points represent bootstrap support (%) based on an analysis of 1000 resampled datasets. GenBank accession numbers of the 16S rRNA gene sequences are provided in parentheses. Bar, 2% sequence divergence.
This study resulted in identification of our novel isolate as the fastidious uncultivated clone previously designated as BVAB3 [5]. Numerous studies have been conducted regarding BV-associated bacteria, specifically BVAB3, and its role in the etiology of BV and resulting sequelae remain unclear. While it has been associated with BV in some studies [5, 6], others have reported that this organism was linked to BV in univariate but not multivariate models [37]. It remains to be determined whether BVAB3 is critical to the development of BV in some women. In addition to the vagina and endometrium, this organism has also been detected from anal swabs of women having BV, suggesting that extragenital reservoirs for this organism can occur [38]. However, this organism was never recovered from the oral cavity and was only in 4% of anal swab samples [38]. Foxman et al. evaluated the role of several BV-associated bacteria with preterm birth and reported that pregnant women having this organism had a decreased risk of preterm delivery [39]. BV has been linked with increased shedding of HIV [1]. Among HIV infected women, those having BVAB3 were more likely to shed HIV even when on effective antiretroviral therapy [40]. Further research will be required to elucidate the role of this organism in BV and reproductive tract sequelae.
Description of Mageeibacillus gen. nov.
Mageeibacillus: Ma.gee.i.ba.cil’lus. N.L. n. Mageea, named after Magee; L. masc. n. bacillus, a rod; N.L. masc. n. Mageeibacillus, a rod isolated at Magee-Womens Research Institute, and where biochemical identification was performed. Cells are Gram positive, catalase negative, oxidase negative, non-motile, asaccharolytic, obligately anaerobic, non-spore forming rods. Cellulose is not degraded. Optimal temperature range for growth is 35–42°C. Predominant fatty acids observed are unbranched saturated and unsaturated forms. The type species is Mageeibacillus indolicus.
Description of Mageeibacillus indolicus sp. nov.
Mageeibacillus indolicus (in.dol’icus. N.L. masc. adj. indolicus for pertaining to the ability of the organism to produce indole).
Colonies are entire, 2mm, flat fried egg with a transparent pink edge and slight iridescent cream center at eight days growth on Brucella agar supplemented with 5% sheep blood incubated anaerobically at 37°C. Cells are Gram positive but stain Gram negative, are vancomycin sensitive, colistin resistant, non-motile, catalase, oxidase, urease, lipase and lecithinase negative, non-cellulolytic and are indole positive. Rods are approximately 1.25 μm in length and 588 nm in diameter occurring in chains or in small aggregates when grown as previously mentioned. Optimal temperature for growth is 37°C. Strains are asaccharolytic but enzymatic activities are present for arginine arylamidase, phenylalanine arylamidase, leucine arylamidase, tyrosine arylamidase, glycine arylamidase, α-fucosidase, histidine arylamidase, serine arylamidase, valine arylamidase, acid phosphatase and napthol-AS-BI-phosphohydrolase. Susceptibility testing to metronidazole, clindamycin and cefoxitin was performed with no resistance observed, however only three isolates were tested. Predominant fatty acids observed were C14:0, C16:0, C18:2 ω6, 9c, C18:1 ω9c and C18:0. The DNA base composition is 44.2 mol% G+C.
The type stain is BAA-2120T and CCUG 59143T, isolated from the human genital tract.
Supplementary Material
Highlights.
Three bacteria were isolated from the endometrium with conventional culture methods.
Isolates are anaerobic, asporulous, asaccharolytic, indole positive, positive rods.
Sequencing revealed only 91% similarity to cultivated organisms in GenBank.
All data suggest a novel genus and species within the Clostridium rRNA cluster III.
Acknowledgements
The authors are grateful to Donna Beer Stolz, Associate Director for the Center for Biologic Imaging and Associate Professor, Cell Biology and Physiology, University of Pittsburgh Medical School as well as Doris Clay and Jonathan Franks, Center for Biologic Imaging, for performing TEM and SEM and technical assistance.
The authors are also grateful to Kerin Tyrrell, Senior Research Associate at the R.M. Alden Research Laboratory for all of her advice and technical assistance.
This research was supported by grant AI 41624.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The GenBank Accession Numbers for 16S rRNA gene sequences and whole genomic sequence: GQ900631-GQ900633 and CP001850, respectively.
References
- 1.Cohen CR, Lingappa JR, Baeten JM, Ngayo MO, Spiegel CA, Hong T, et al. Bacterial vaginosis associated with increased risk of female-to-male HIV-1 transmission: A prospective cohort analysis among African couples. PLoS Med. 2012;9(6):e1001251. doi: 10.1371/journal.pmed.1001251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atashili J, Poole C, Ndumbe PM, Adimora AA, Smith JS. Bacterial vaginosis and HIV acquisition: A meta-analysis of published studies. AIDS. 2008;22(12):1493–1501. doi: 10.1097/QAD.0b013e3283021a37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cherpes TL, Meyn LA, Krohn MA, Hillier SL. Risk factors for infection with herpes simplex virus type 2. Sex Transm Dis. 2003;30(5):405–410. doi: 10.1097/00007435-200305000-00006. [DOI] [PubMed] [Google Scholar]
- 4.Bradshaw CS, Morton AN, Hocking J, Garland SM, Morris MB, Moss LM, et al. High recurrence rates of bacterial vaginosis over the course of 12 months after oral metronidazole therapy. J Infect Dis. 2006;193:1478–1486. doi: 10.1086/503780. [DOI] [PubMed] [Google Scholar]
- 5.Fredricks DN, Fiedler TL, Marrazzo JM. Molecular identification of bacteria associated with bacterial vaginosis. N Engl J Med. 2005;353:1899–1911. doi: 10.1056/NEJMoa043802. [DOI] [PubMed] [Google Scholar]
- 6.Fredricks DN, Fiedler TL, Thomas KK, Oakley BB, Marazzo JM. Targeted PCR for detection of vaginal bacteria associated with bacterial vaginosis. J Clin Microbiol. 2007;45(10):3270–3276. doi: 10.1128/JCM.01272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fredricks DN, Fiedler TL, Thomas KK, Mitchell CM, Marazzo JM. Changes in vaginal bacterial concentrations with intravaginal metronidazole therapy for bacterial vaginosis as assessed by quantitative PCR. J Clin Microbiol. 2009;47(3):721–726. doi: 10.1128/JCM.01384-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou X, Brown CJ, Abdo Z, Davis CC, Hansmann MA, Joyce P, et al. Differences in the composition of vaginal microbial communities found in healthy Caucasian and black women. Inter Soc Microb Ecol. 2007;1(2):121–133. doi: 10.1038/ismej.2007.12. [DOI] [PubMed] [Google Scholar]
- 9.Halebian S, Harris B, Finegold SM, Rolfe RD. Rapid method that aids in distinguishing Gram positive from Gram negative anaerobic bacteria. J Clin Microbiol. 1981;13(3):444–448. doi: 10.1128/jcm.13.3.444-448.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Summanen P, Baron E, Citron DM, Strong CA, Wexler HM, Finegold SM. Wadsworth Anaerobic Bacteriology Manual. 5th edn. Belmont, CA: Veterans Administration Wadsworth Medical Center, and Departments of Medicine, and Microbiology and Immunology, UCLA School of Medicine; 1993. [Google Scholar]
- 11.Kasana RC, Salwan R, Dhar H, Dutt S, Gulati A. A rapid and easy method for the detection of microbial cellulases on agar plates using Gram’s iodine. Curr Microbiol. 2008;57:503–507. doi: 10.1007/s00284-008-9276-8. [DOI] [PubMed] [Google Scholar]
- 12.Johnson EA, Summanen P, Finegold SM. Clostridium. In: Murray PR, editor. Manual of Clinical Microbiology. 9th edn. Washington, D.C.: ASM Press; 2007. pp. 889–910. [Google Scholar]
- 13.Clinical Laboratory Standards Institute. Methods for antimicrobial susceptibility testing of anaerobic bacteria; Approved standard, 5th edn. CLSI document M11-A5. Wayne PA: Clinical Laboratory Standards Institute; 2001. [Google Scholar]
- 14.Sasser M. Identification of bacteria by gas chromatography of cellular fatty acids (technical note no. 101) Newark, DE: Microbial ID Inc.; 1990. ( http://www.microbialid.com/pages/publications_fa.html). [Google Scholar]
- 15.Tamura K, Dudley J, Nei M, Kumar S. Mega4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24(8):1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 16.Antonio MA, Hawes SE, Hillier SL. The identification of vaginal Lactobacillus species and the demographic and microbiologic characteristics of women colonized by these species. J Infect Dis. 1999;180:1950–1956. doi: 10.1086/315109. [DOI] [PubMed] [Google Scholar]
- 17.Frank JA, Reich CI, Sharma S, Weisbaum JS, Wilson BA, Olsen GJ. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl Environ Microbiol. 2008;4:2461–2470. doi: 10.1128/AEM.02272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Löffler FE, Sun Q, Li J, Tiedje JM. 16S rRNA gene-based detection of tetrachloroethene-dechlorinating Desulfuromonas and Dehalococcoides species. Appl Environ Microbiol. 2008;66:1369–1374. doi: 10.1128/aem.66.4.1369-1374.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loy A, Maixner F, Wagner M, Horn M. probeBase- an online resource for rRNA targeted oligonucleotide probes: new features 2007. Nucleic Acids Res. 2007;35(Database issue):D800–D804. doi: 10.1093/nar/gkl856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chun J, Lee LH, Jung Y, Kim M, Kim BK, Lim YW. EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int J Syst Evol Microbiol. 2007;57(10):2259–2261. doi: 10.1099/ijs.0.64915-0. [DOI] [PubMed] [Google Scholar]
- 21.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. Mega6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Bio Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 23.Jukes TH, Cantor CR. Evolution of protein molecules. In: Munro NH, editor. Mammalian Protein Metabolism. New York: Academic Press; 1969. pp. 21–132. [Google Scholar]
- 24.Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 25.Chen S, Niu L, Zhang Y. Saccharofermentans acetigenes gen. nov., sp. nov., an anaerobic bacterium isolated from sludge treating brewery wastewater. Int J Syst Evol Microbiol. 2010;60:2735–2738. doi: 10.1099/ijs.0.017590-0. [DOI] [PubMed] [Google Scholar]
- 26.Falsen E, Collins MD, Welinder-Olsson C, Song Y, Finegold SM, Lawson PA. Fastidiosipila sanguinis gen. nov., sp. nov., a new Gram positive, coccus-shaped organism from human blood. Int J Syst Evol Microbiol. 2005;55:853–858. doi: 10.1099/ijs.0.63327-0. [DOI] [PubMed] [Google Scholar]
- 27.Moore ERB. [Date of access, 10/29/14]; www.ccug.se/default.cfm?page=search_record.cfm&ccugno=47711 Culture Collection University of Goteborg, 5/17/2006. Web.
- 28.Shiratori H, Sasaya K, Ohiwa H, Ikeno H, Ayame S, Kataoka N, et al. Clostridium clariflavum sp. nov. and Clostridium caenicola sp. nov., moderately thermophilic, cellulose-/cellobiose-digesting bacteria isolated from methanogenic sludge. Int J Syst Evol Microbiol. 2009;59:1764–1770. doi: 10.1099/ijs.0.003483-0. [DOI] [PubMed] [Google Scholar]
- 29.Nishiyama T, Ueki A, Kaku N, Ueki K. Clostridium sufflavum sp. nov., isolated from a methanogenic reactor treating cattle waste. Int J Syst Evol Microbiol. 2009;59:981–986. doi: 10.1099/ijs.0.001719-0. [DOI] [PubMed] [Google Scholar]
- 30.Carlier JP, Bedora-Faure M, K’ouas G, Alauzet C, Mory F. Proposal to unify Clostridium orbiscindens Winter et al. 1991 and Eubacterium plautii (Seguin 1928) Hofstad and Aasjord 1982, with description of Flavonifractor plautii gen. nov., comb. nov., and reassignment of Bacteroides capillosus to Pseudoflavonifractor capillosus gen. nov., comb. nov. Int J Syst Evol Microbiol. 2010;60:585–590. doi: 10.1099/ijs.0.016725-0. [DOI] [PubMed] [Google Scholar]
- 31.Ezaki T. Genus I. Ruminococcus Sijpesteijn 1948, 152AL (Effective publication: Hungate 1957 307AL) In: DeVos P, Garrity GM, Jones D, Krieg NR, Ludwig W, Rainey FA, et al., editors. Bergey’s Manual of Systematic Bacteriology. 2nd edn. Vol. 3. New York: Springer; 2009. pp. 1016–1017. [Google Scholar]
- 32.Ifkovits RW, Ragheb HS. Cellular fatty acid composition and identification of rumen bacteria. Appl Microbiol. 1968;16(9):1406–1413. doi: 10.1128/am.16.9.1406-1413.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Centers for Disease Control. Sexually transmitted diseases treatment guidelines. Morbidity Mortality Weekly Report. 2010;59:64–66. [Google Scholar]
- 34.Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young MA, et al. Topographical and temporal diversity of the skin microbiome. Science. 2009;324:1190–1192. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Collins MD, Lawson PA, Willems A, Cordoba JJ, Fernandez-Garayzabal J, Garcia P, et al. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int J Syst Bacteriol. 1994;44:812–826. doi: 10.1099/00207713-44-4-812. [DOI] [PubMed] [Google Scholar]
- 36.Rainey FA. Family VIII: Ruminococcaceae fam. nov. In: DeVos P, Garrity GM, Jones D, et al., editors. Bergey’s Manual of Systematic Bacteriology. 2nd edn. Vol. 3. New York: Springer; 2009. pp. 1016–1018. [Google Scholar]
- 37.Fethers K, Twin J, Fairley CK, Fowkes FJI, Garland SM, Fehler G, et al. Bacterial vaginosis (BV) candidate bacteria: Associations with BV and behavioural practices in sexually-experienced and inexperienced women. PLoS ONE. 2012;7(2):e30633. doi: 10.1371/journal.pone.0030633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marrazzo JM, Fiedler TL, Srinivasan S, Thomas KK, Liu C, Ko D, et al. Extravaginal reservoirs of vaginal bacteria as risk factors for incident bacterial vaginosis. J Infect Dis. 2012;205:1580–1588. doi: 10.1093/infdis/jis242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Foxman B, Wen A, Srinivasan U, Goldberg D, Marrs CF, Owen J, et al. Mycoplasma, bacterial vaginosis-associated bacteria BVAB3, race, and risk of preterm birth in a high-risk cohort. Am J Obstet Gynecol. 2014;210(226):e1–e7. doi: 10.1016/j.ajog.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mitchell C, Balkus JE, Fredricks D, Liu C, McKernan-Mullin J, Frenkel LM, et al. Interaction between lactobacilli, bacterial vaginosis-associated bacteria, and HIV type 1 RNA and DNA genital shedding in U.S. and Kenyan women. AIDS Res Hum Retroviruses. 2013;29(1):13–19. doi: 10.1089/aid.2012.0187. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.