Abstract
Neuronal cells are highly dependent on mitochondria, and mitochondrial dysfunction is associated with neurodegenerative diseases. As perturbed mitochondrial function renders neurons extremely sensitive to a wide variety of insults, such as oxidative stress and bioenergetic defects, mitochondrial defects can profoundly affect neuronal fate. Several studies have linked ALS with mitochondrial dysfunction, stemming from observations of mitochondrial abnormalities, both in patients and in cellular and mouse models of familial forms of ALS. Mitochondrial changes have been thoroughly investigated in mutants of superoxide dismutase 1 (SOD1), one of the most common causes of familial ALS, for which excellent cellular and animal models are available, but recently evidence is emerging also in other forms of ALS, both familial and sporadic. Mitochondrial defects in ALS involve many critical physiopathological processes, from defective bioenergetics to abnormal calcium homeostasis, to altered morphology and impaired trafficking. In this review, we summarize established evidence of mitochondrial dysfunction in ALS, especially in SOD1 mutant models of familial ALS. The main focus of the review is on defective mitochondrial quality control (MQC) in ALS. MQC operates at multiple levels to clear damaged proteins through proteostasis and to eliminate irreparably damaged organelles through mitophagy. However, since ALS motor neurons progressively accumulate damaged mitochondria, it is plausible that the MQC is ineffective or overwhelmed by excessive workload imposed by the chronic and extensive mitochondrial damage.
Keywords: ALS, mitochondria, mitophagy, SOD1, parkin, p62
Introduction
Amyotrophic Lateral Sclerosis (ALS) is the most prevalent, adult-onset, motor neuron disease, characterized by the degeneration of upper and lower motor neurons, progressive muscle weakness and atrophy, leading to muscle paralysis. (Cleveland and Rothstein, 2001; Sabatelli et al., 2013). The incidence is 2 in 100,000 individuals per year (Hirtz et al., 2007). Patients die of respiratory failure in 2 to 5 years, after clinical symptoms develop. There is no cure for ALS, and only one drug, Riluzole, has been approved for treatment (Bensimon et al., 1994; Lacomblez et al., 1996; Pandya et al., 2013). Unfortunately, Riluzole only prolongs average patients’ survival by a few months. This dramatic lack of effective treatments prompts the need for a better understanding of disease pathogenesis to identify appropriate therapeutic targets and to develop disease biomarkers, which could help stratifying patients and select appropriate therapies.
Approximately, 90% of the patients develop ALS with unknown etiology, classified as sporadic ALS (sALS), while the remaining 10% are familial ALS cases (fALS) due to genetic defects, which are directly linked to the pathogenesis of the disease. An increasing number of genes are being linked to ALS (Andersen and Al-Chalabi, 2011; Renton et al., 2014). Superoxide dismutase 1 (SOD1) was the first gene discovered as causative for fALS in 1993 (Rosen, 1993) and since then more than 160 pathogenic mutations in SOD1 have been described (Andersen, 2006; Moreira et al., 2013). Mutations in SOD1 account for 20% of the fALS cases, and they have rarely been described in idiopathic cases (Tortelli et al., 2013).
In the last decade, new breakthroughs in the genetics of ALS emerged from the discovery that mutations in several DNA/RNA binding proteins, such as TAR DNA-binding protein (TARDBP, TDP-43) (Sreedharan et al., 2008) and fused in sarcoma (FUS) (Kwiatkowski et al., 2009; Vance et al., 2009), are associated with fALS and frontotemporal dementia (FTD), pointing to new pathological mechanisms related to RNA metabolism (Lattante et al., 2013; Ling et al., 2013; van Blitterswijk and Landers, 2010). Other fALS genes, like valosin-containing protein (VCP)(Johnson et al., 2010), ubiquilin 2 (UBQLN2) (Deng et al., 2011), and sequestosome 1 (SQSTM1) (Fecto et al., 2011), highlighted the importance of proteostatic pathways in neuronal cells (Fecto and Siddique, 2012). Profilin 1 (PFN1) (Wu et al., 2012) and dynactin (DCTN1) (Puls et al., 2003) mutations in fALS indicate that also cytoskeletal and axonal transport abnormalities are involved in disease pathogenesis. Recently, the largest proportion of fALS cases (40%) have been linked to intronic hexanucleotide repeat expansions in C9ORF72 (DeJesus-Hernandez et al., 2011; Renton et al., 2011), a gene with still unknown function, opening a new field of research that connects ALS/FTD to DNA repeat expansion (Majounie et al., 2012).
Several molecular mechanisms have been proposed to explain the neuronal degeneration in ALS. It is noteworthy that many of them emerge from studies of mutant SOD1, owing to the fact that many cellular and animal models have been developed and studied. The proposed mechanisms include oxidative stress, toxic gain of function of misfolded and aggregated proteins, endoplasmic reticulum stress, mitochondrial dysfunction, and axonal disorganization, including organelle transport defects(Cozzolino and Carri, 2011; Ferraiuolo et al., 2011; Kawamata and Manfredi, 2010b; Magrane and Manfredi, 2009; Pasinelli and Brown, 2006).
In this review, we will summarize established evidence of the involvement of mitochondrial dysfunction in ALS, especially in SOD1 mutant models of fALS; we will then focus on mitochondrial quality control (MQC) mechanisms. Mutations in critical components of MQC have been associated with a number of neurodegenerative diseases, including parkin and PINK1 (Phosphatase and tensin homolog (PTEN)-induced putative kinase 1) links to familial Parkinson disease (PD) (Scarffe et al., 2014), but also in familial ALS/FLTD (Frontotemporal Lobar Degeneration). However, despite evidence that damaged mitochondria accumulate in ALS, the role of MQC has not been fully elucidated yet.
Mitochondrial damage and dysfunction in ALS
Mitochondria are essential organelles for a wide variety of cellular processes, including cell intermediate metabolism, calcium homeostasis, bioenergetics, and intrinsic cell death processes. Mitochondrial dysfunction has long been associated with neurodegenerative diseases, because of the dependence on mitochondrial function of neuronal cells (Schon and Przedborski, 2011). Many studies have linked SOD1-fALS with mitochondrial morphological abnormalities, both in patients’ biopsies and postmortem tissues (Sasaki and Iwata, 1996; Sasaki and Iwata, 2007; Sasaki, 2010) and in cellular and mouse models of fALS (Magrane et al., 2009; Magrane et al., 2014; Vinsant et al., 2013a; Vinsant et al., 2013b).
Although SOD1 is considered to be a cytosolic enzyme, a fraction of the protein resides in mitochondria. The majority of the SOD1 in mitochondria resides in the intermembrane space (IMS) (Jaarsma et al., 2001; Mattiazzi et al., 2002). Although it is not entirely clear which role the enzyme could be performing in mammalian mitochondria, a protective antioxidant effect was suggested (Okado-Matsumoto and Fridovich, 2001). This concept is supported by studies performed in mice lacking SOD1, in which the expression of a wild type SOD1 targeted to the mitochondria was able to rescue motor neuron loss (Fischer et al., 2011).,
Mitochondria are targets of mutant SOD1 toxicity (Cozzolino et al., 2013; Hervias et al., 2006; Higgins et al., 2002; Martin, 2011). Mutant SOD1 mice develop progressive bioenergetic abnormalities in the CNS, characterized by decreased mitochondrial respiratory chain activity and impaired ATP production (Mattiazzi et al., 2002), and defective calcium uptake (Damiano et al., 2006; Kim et al., 2012; Parone et al., 2013). These mitochondrial deficits could directly contribute to disease pathogenesis, because they compromise fundamental functions in neuronal cells.
In mitochondria, mutant SOD1 misfolds and aggregates, causing oxidative stress (Carri and Cozzolino, 2011). Mitochondrial damage can also ensue from the aberrant interaction of mutant SOD1 with proteins of the outer membrane (OM), such as VDAC (Voltage-dependent anion channel) (Israelson et al., 2010) and Bcl2 (B-cell lymphoma 2) (Pasinelli et al., 2004; Pedrini et al., 2010), and from the accumulation of misfolded mutant SOD1 on the OM (Vande Velde et al., 2011), in the IMS (Kawamata and Manfredi, 2010a) and on the inner membrane (IM) (Vijayvergiya et al., 2005).
In mutant SOD1 neurons, early studies reported deficits in axonal transport (Williamson and Cleveland, 1999; Zhang et al., 1997) resulting in defective mitochondrial transport (De Vos et al., 2007). Direct interactions between some axonal motor complexes (essentially dynein, the major motor protein required for retrograde axonal transport) and mutant SOD1 (Ligon et al., 2005; Shi et al., 2010; Zhang et al., 2007) are required for the formation of mutant SOD1 aggregates and have been proposed as the cause for the disruption of axonal transport. Thus, either formation of mutant SOD1-containing aggregates or sequestration of essential components, such as the dynein motor complex,) for axonal transport, together with reduced binding of cargoes, probably as a consequence of altered mitochondrial outer membrane structure, result in a deficit on mitochondrial transport (Magrane et al., 2009; Marinkovic et al., 2012). In addition, mitochondrial fusion and fission are imbalanced in SOD1 mutant neurons (Magrane et al., 2012). As a result, mutant SOD1 motor neurons accumulate mitochondria with abnormal morphology (Vande Velde et al., 2011), ultrastructure (Gould et al., 2006), and bioenergetics (Damiano et al., 2006; Kawamata and Manfredi, 2010a; Mattiazzi et al., 2002). Evidence of progressive accumulation of abnormal mitochondria with impaired axonal transport was also found in vivo in both mutant SOD1 and TDP43 transgenic mice, starting early on in the course of the disease, before onset of symptoms and motor neuron death (Magrane et al., 2014). Moreover, mitochondrial structural and dynamics abnormalities have been linked to physical association of mutant TDP-43 with mitochondria in cultured cells (Wang et al., 2013b). Although it is still unknown in which mitochondrial compartment TDP-43 resides, the authors showed that interactions with mitofusin 2 (Mfn2) on the OM might play a pathogenic role. Indeed, it was recently shown that TDP-43 perturbs the ER (endoplasmic reticulum)-mitochondria contact sites (or MAMs), by disrupting the interaction between VAPB (vesicle-associated membrane protein-associated protein-B) and the mitochondrial protein tyrosine phosphatase-interacting protein 51 (PTPIP51), resulting in dysregulation of intracellular calcium homeostasis (Stoica et al., 2014). Thus, abnormal TDP-43 expression could interfere with mitochondrial function.
Accumulation of damaged mitochondria may actively cause neuronal toxicity by excessive free radical production, leakage of pro-apoptotic factors, and clogging of the proteostatic and autophagic machineries. The toxicity from damaged mitochondria could act synergistically with impaired bioenergetics, leading to synaptic dysfunction and neuronal degeneration. Intriguingly, genetic ablation of cyclophilin D, a positive regulator of calcium-dependent mitochondrial permeability transition, increased calcium capacity in the CNS mitochondria of SOD1 mutant mice and the number of surviving motor neurons cell bodies, while decreasing the amount of SOD1 inclusions. However, it did not improve muscle denervation (Parone et al., 2013), and in one study it decreased survival in female SOD1 mutant mice (Kim et al., 2012). These results suggest that simply increasing mitochondrial calcium capacity by eliminating a highly conserved modulator, such as cyclophilin D, is not a viable strategy to protect motor neuron function.
Mitochondrial quality control mechanisms (MQC) and their implications for ALS
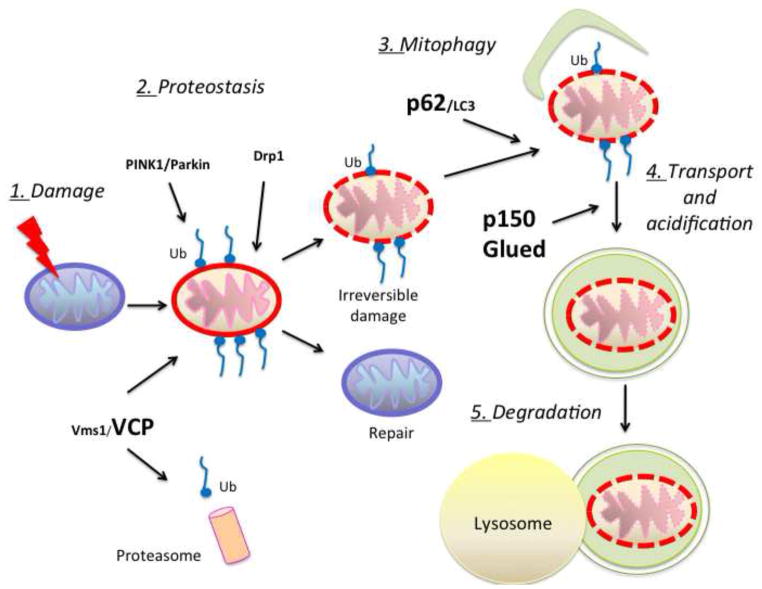
Different mechanisms have evolved to ensure proper mitochondrial homeostasis: new mitochondria are generated by highly regulated biogenesis and damaged mitochondria are subjected to quality control processes. Why ALS motor neurons are unable to repair or eliminate damaged mitochondria and maintain homeostasis is unclear. Mitochondrial quality control systems (MQC) operate at multiple levels to clear damaged proteins through proteostasis and to eliminate irreparably damaged organelles through selective removal of damaged mitochondrial components or selective removal of organelles through mitophagy (Ashrafi and Schwarz, 2013; Cherra et al., 2010). Figure 1 schematically summarizes the putative major steps of MCQ in cells that express mutant SOD1 or other similar mitochondrial stressors. The figure highlights the MCQ components that have been linked to ALS. Since mutant SOD1 motor neurons accumulate damaged mitochondria, it is plausible that their MQC is ineffective. This could be the result of mutant SOD1 interference with the MQC machinery or excessive load imposed on the MQC by constitutive and extensive mitochondrial damage.
Figure 1. Impaired pathways of mitochondria quality control (MQC) could be involved in ALS.
Mitochondrial homeostasis is maintained through repair or degradation of damaged mitochondria. In response to mitochondrial stress, as for example accumulation of mutant and misfolded fALS proteins (SOD1 and TDP-43), proteostasis mechanisms act to repair damaged mitochondria. In some instances, excessive overloading damage requires the complete elimination of mitochondria, in a process defined as mitophagy. The various steps (numbered 1–5 in this scheme) involved in these mechanisms require proteins such as VCP, p62 and p150Glued (in bold), which have been associated with fALS, suggesting that ineffective MQC participates in motor neuron degeneration.
Mitochondrial proteostasis protects against mitochondrial damage
Mitochondria with loss of membrane potential or subject to protein oxidation and misfolding become targets of MQC. There are three main, partially interconnected, pathways of MQC: protein degradation, vesicular degradation, and mitophagy.
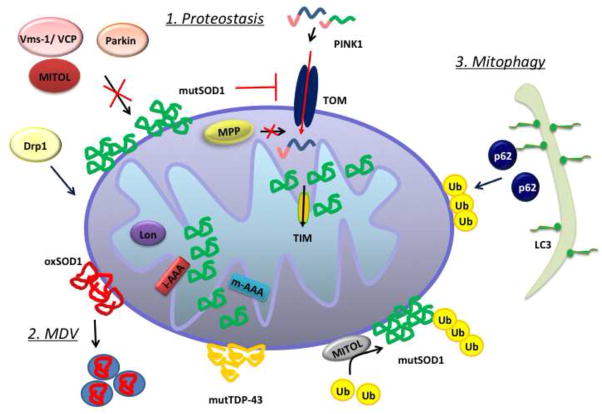
The first line of defense to maintain a pool of healthy mitochondria consists in mitochondrial proteostasis. This mechanism ensures the proteolysis and subsequent degradation of misfolded, oxidized, and damaged proteins by proteases residing in different compartments of the mitochondria (Anand et al., 2013). Figure 2 illustrates schematically various components of the proteostatic machinery dedicated to tagging and removal of damaged proteins in mitochondria.
Figure 2. Proteostasis and autophagy pathways in mitochondria.
The elimination of misfolded or oxidized proteins in mitochondria proceeds through different mechanisms, depending on the nature of the protein and its localization in mitochondria. 1. Proteostasis: ubiquitin ligases, such as Parkin, MITOL, and Mulan are responsible for the proteasomal degradation of components of the OM; mitochondrial proteases (Lon, i-AAA, m-AAA) degrade proteins of the IM and the matrix. 2: MDV: Elimination of oxidized components can also occur through turnover of mitochondria-derived vesicles (MDVs). The accumulation of damaged proteins in the mitochondria could interfere with protein import, causing matrix protease (MPP) malfunction. 3. Mitophagy: if the damage is too extensive to be dealt with by proteostasis alone, mitophagy pathways could be activated for the elimination of whole organelles, mediated by ubiquitination of specific proteins of the OM and their interaction with the mitophagy adaptor p62 and autophagosomes, upon LC3 modification.
Mitochondria have internal proteases, such as the AAA-protease complex (ATPases Associated with diverse cellular Activities) of the IM (Gerdes et al., 2012) and the Lon protease of the matrix (Matsushima and Kaguni, 2012). Mitochondria are also endowed with their own unfolded protein response (UPR), which is activated when misfolded proteins accumulate in the matrix (Pellegrino et al., 2013) and in the IMS (Papa and Germain, 2011). Although there is no direct link between mitochondrial protein degradation and ALS yet, it could be hypothesized that the accumulation of misfolded and aggregated proteins in different sub-compartments of the mitochondria would activate proteolytic mechanisms. Excessive accumulation of misfolded and oxidized proteins may slow down the system and, in the worst-case scenario, block it.
Less studied, in terms of quality control mechanisms, are the mitochondrial processing peptidases (MPP), which play a crucial role in the import of proteins into mitochondria. However, defective mitochondrial protein import has been reported in spinal cord-purified mitochondria from transgenic mutant SOD1 rats (Li et al., 2010), suggesting that there could be a dysfunction in the processing of the imported peptides.
Mitochondria rely on the cytosolic ubiquitin-proteasome system (UPS) to eliminate damaged proteins destined to the OM or before they engage in the mitochondrial import pathway (Karbowski and Neutzner, 2012; Radke et al., 2008). Ubiquitin ligases, such as Parkin, ubiquitinate oxidized or misfolded OM proteins (Heo and Rutter, 2011). Parkin recruitment to the OM of damaged mitochondria that have lost membrane potential has been ascribed to PINK1, following its incomplete processing and import across the OM of depolarized mitochondria (Matsuda et al., 2010; Narendra et al., 2008; Narendra et al., 2010b; Vives-Bauza et al., 2010). Parkin-mediated OM protein ubiquitination can recruit valosin-containing protein (VCP or p97/Cdc48) to mitochondria (Kim et al., 2013b). VCP is an AAA+-ATPase, whose segregase activity extracts ubiquitinated proteins from the OM, as well as other membranes and organelles, and targets them for proteasomal degradation (Xu et al., 2011; Ye et al., 2005). Mutations in VCP are associated with multisystem disorders (Watts et al., 2004), including IBMPFD (Inclusion body myopathy with early-onset Paget disease and frontotemporal dementia) and ALS (Nalbandian et al., 2011). VCP mutations cause mitochondrial structural changes in transgenic mice, and loss of VCP impairs the clearance of damaged mitochondria (Kim et al., 2013b). Selective VCP translocation to damaged mitochondria depends on Vms1 (VCP/Cdc48-associated Mitochondrial Stress responsive 1), a cytosolic protein that senses local mitochondrial stress (Heo et al., 2010; Heo et al., 2013) and binds VCP allowing its translocation to damaged mitochondria. Ubiquitination of OM proteins can be operated by the PINK1/Parkin system, but also by other ligases, such as the MITOchondrial ubiquitin Ligase MITOL (or MARCH5), an E3-ubiquitin ligase resident in the OM (Nagashima et al., 2014), which ubiquitinates OM proteins involved in mitochondrial fusion (Mfn1 and Mfn2) (Park et al., 2010; Sugiura et al., 2013) and fission (Drp1) (Karbowski et al., 2007; Nakamura et al., 2006; Yonashiro et al., 2006). Interestingly, MITOL ubiquitinates and increases the turnover of mutant SOD1 on the OM (Yonashiro et al., 2009). Mitochondrial Ubiquitin Ligase Activator of NF-kB (MULAN) is another, less characterized, ubiquitin-ligase, which has also been implicated in the ubiquitination of Drp1 (Li et al., 2008b).
Degradation of oxidized components by mitochondrial-derived vesicles
It is unclear what drives the switch from proteostasis to mitophagy, but the extent of mitochondrial damage is likely a discriminating factor: when proteostasis cannot repair mitochondria, mitophagy ensues. In some cases, however, a newly discovered form of communication between mitochondria and lysosomes, involving limited sections of mitochondrial membranes (mitochondria-derived vesicles, MDV) containing oxidized proteins, could be sufficient to repair the damage, prevent full-blown mitophagy and maintain mitochondria with adequate membrane potential (Soubannier et al., 2012a).
Besides the continuous generation of MDV at basal levels, an increase in several pools of MDV (in terms of cargo selectivity) can be detected in response to increased levels of reactive oxygen species (ROS), in a Drp-1-independent fashion. The majority of MDVs reaches the lysosomes for degradation. However, this process is completely independent of canonical macroautophagy, as cells non-competent for autophagy maintain basal levels of MDV formation and respond to ROS challenge by increasing the number of MDV that are released from mitochondria. In vitro reconstitution of MDVs further determined the content of these vesicles (Soubannier et al., 2012b), which is selective based upon the nature of the ROS source applied, and established that MDVs are enriched in oxidized mitochondrial proteins. Recently, it has been proposed that PINK1-parkin activity on mitochondria is indispensable for the formation of a subtype of MDVs (OM Tom20 negative and matrix proteins positive) as a fast response to intramitochondrial ROS insults (McLelland et al., 2014), preceding mitophagic degradation of the whole organelle. No direct evidences point to the participation of MDVs in the pathology of ALS yet, but a crosstalk between oxidized SOD1 and MDVs could be hypothesized as a mechanism for the elimination of oxidized cargo in lysosomes. This mechanism may emerge as a first line of defense, when the damage to mitochondria is moderate, to protect against accumulation of oxidized and misfolded SOD1. However, this pathway may become insufficient, and mitophagy could be required to degrade extremely damaged mitochondria
Mitophagy as a pathway to clear damaged mitochondria
The property which best describes mitophagy is specificity, as only those mitochondria that are properly tagged, in a controlled cascade of events, are degraded. Mitophagy plays a role in the selective clearance of mitochondria during development in certain cell types, but its involvement in the clearance of damaged mitochondria in neurons and in neurodegenerative diseases is still largely unknown.
As mentioned above, the molecular mechanism that couples the irreversible loss of mitochondrial membrane potential to mitophagy is the accumulation of PINK1 in the OM, phosphorylation of various targets and recruitment of the E3-ubiquitin ligase parkin. In some cases, parkin phosphorylation by PINK1 is necessary for its further activity (Birsa et al., 2014; Kondapalli et al., 2012).
Ubiquitination of mitochondrial proteins is part of the quality control system outlined above, but also one of the best-studied signals for the activation of mitophagy. Ubiquitination and degradation of Mfn1 and Mfn2 initiate mitochondrial fission (Poole et al., 2010; Tanaka et al., 2010; Ziviani et al., 2010), while degradation of the tubulin cargo adaptor Miro1 results in the arrest of mitochondrial transport (Wang et al., 2011). Fragmentation of the mitochondrial network and immobilization facilitate the engulfment of damaged mitochondria in autophagic vesicles. Ubiquitination of other OM proteins, such as VDAC1 (Geisler et al., 2010), can also trigger the initiation of mitophagy, since Parkin-mediated mitophagy can take place in the absence of mitofusins (Chan et al., 2011).
Mitophagy requires an ubiquitin-binding adaptor that recruits mitochondria to the autophagosome by binding to the lipidated form of LC3 (microtubule associated protein 1 light chain 3). p62/SQSTM1 is one of the adaptors, but others must exist, since mitophagy can occur in a p62-independent manner (Narendra et al., 2010a; Okatsu et al., 2010). Autophagosomes containing mitochondria are transported to lysosomes for the formation of autolysosomes, where mitochondrial components are degraded. In neurons, fully active lysosomes are mostly localized in the soma, and autophagosomes containing mitochondria from axons and dendrites must be retro-transported to the soma. Thus, retrograde axonal transport and mitophagy are intimately interconnected processes. Recently, the presence of mature lysosomes was reported in the axons, where lysosomes are capable of fusion with autophagosomes. Retrograde transport of the autolysosomes to the soma is needed to achieve complete degradation of cargoes (Maday et al., 2012). Further studies from Ashrafi and colleges, described degradation of autophagosomes containing mitochondria, upon fusion with mature lysosomes, in the axon. It was suggested that immobilization of individual mitochondria and engulfment by autophagosomes follows PINK1/parkin-mediated Miro1 degradation and consequent mitochondrial arrest (Ashrafi et al., 2014). However, excessive and localized damage to mitochondria in distal axons, as it occurs in ALS, could result in sequestration and depletion of essential mitophagy components.
Mutant SOD1 impairs mitochondrial retrograde axonal transport (Magrane et al., 2014). Transport changes are accompanied by mitochondrial fragmentation (Vande Velde et al., 2011), especially in the distal portions of the motor axons (Magrane et al., 2012). These observations suggest that mitophagy takes place in mutant SOD1 neurons, since mitochondrial motility arrest (Wang et al., 2011) and increased fission (Twig et al., 2008) are associated with mitophagy. They may also suggest that mitophagy fluxes are delayed in ALS neurons. Therefore, delayed mitophagy fluxes in mutant SOD1 neurons could be due to both defective retrograde transport of autophagosomes and exhaustion of rate-limiting components.
The existence of the “canonical” PINK1/Parkin mitophagy pathway in neurons is somehow controversial (Grenier et al., 2013). Some investigators have detected PINK1 and Parkin recruitment after treatment with the potent uncoupler CCCP (Cai et al., 2012; Joselin et al., 2012; Koyano et al., 2013; Seibler et al., 2011), while others have not (Van Laar et al., 2011). In neurons, this event appears to be highly dependent on conditions, since the presence of antioxidants in the medium prevents mitophagy (Joselin et al., 2012), possibly by protecting mitochondria from oxidative damage. Mitophagy following PINK1-Parkin translocation requires longer time in neurons than in other cell types (Cai et al., 2012), and it involves only a subset of mitochondria, even when cells are treated with CCCP. This suggests that limited MQC may be taking place in neurons, with the goal of achieving a steady state of functional mitochondria through selective organellar degradation and recycling. This is of essence, since neurons cannot survive with exclusive glycolytic metabolism, and thus cannot dispense of their whole mitochondrial complement at once. It is plausible that in neurons both Parkin-dependent and independent MQC mechanisms coexist and that Parkin and other ubiquitin-ligases are involved in both proteostasis of OM proteins (i.e., mitochondrial repair) and mitophagy (mitochondrial 1elimination), depending on the severity of the damage. For example, MITOL or Mulan could compensate for loss of Parkin. In addition, cardiolipin exposure in the OM was found in neurons subjected to mitochondrial stress and has been proposed as an alternative mechanism for signaling mitophagy to the canonical PINK1/parkin (Chu et al., 2013). In this case, cardiolipin interacts directly with LC3 II and serves as a receptor for the fusion with the autophagosome.
Some reports have suggested interactions between key players in mitophagy and TDP-43. A decrease in RNA and protein levels of parkin was detected in cellular and animal models of TDP-43 and FUS depletion and in sALS spinal cord samples (Lagier-Tourenne et al., 2012; Polymenidou et al., 2011), which could eventually lead to increase vulnerability to mitochondrial dysfunction and cell death. Accordingly, parkin overexpression was capable of counteracting the deleterious effects of TDP-43 overexpression, through mechanisms involving ubiquitination and subsequent TDP-43 subcellular re-localization (Hebron et al., 2014; Hebron et al., 2013). This role of TDP-43 in cell survival was ascribed to its nuclear functions, without any implications of TDP-43 or FUS aggregates in the cytoplasm or in mitochondria. However, in ALS, disturbances in mitophagy mechanisms could also arise from excessive deposition of mutant and misfolded proteins, such as SOD1 and TDP43, on mitochondria, which might interfere with the normal PINK1/parkin pathway. In support of this hypothesis, it was demonstrated that overexpression of mutant TDP-43 results in a depletion of Parkin from the brain (Stribl et al., 2014).
Increased motor neuron-specific localization of the autophagic marker LC3 II has been reported in transgenic SOD1 mice (Li et al., 2008a; Morimoto et al., 2007). Others have reported the accumulation of autophagosomes and autophagolysosomes in human spinal cord ALS samples (Sasaki, 2011), together with p62 aggregates in degenerating motor neurons. Interestingly, many of the autolysosomes contained mitochondria, a clear indication of active mitophagy. Moreover, increased mitophagy was observed in NSC34 motor neuronal cell lines transfected with TDP-43 constructs (Hong et al., 2012). Altogether, biochemical and histological evidences point to an up regulation of macroautophagy in ALS, and mitophagy specifically.
Progressive accumulation of p62 occurs in the spinal cord of G93A mutant SOD1 transgenic mice (Gal et al., 2007) and LC3-positive vacuoles are increased in motor neurons (Li et al., 2008a). However, the interpretation of these data is complex, because mutant SOD1 can bind p62 and LC3 directly, indicating that SOD1 could interfere with the p62-LC3 complex (Gal et al., 2009). Although it is still unclear whether these markers indicate increased or impaired autophagy in SOD1-fALS the role of p62 in ALS is supported by several lines of evidence. Gal and coworkers found that p62 enables the sequestration of mutant SOD1 into cytoplasmic inclusions, independent of ubiquitination, and delivers these aggregates to autophagosomes, through direct interaction with LC3 (Gal et al., 2007; Gal et al., 2009). However, mitochondrial degradation was not specifically investigated, and autophagic degradation of SOD1 aggregates could be a process distinct from mitophagy.
A partner of p62 in the recognition of ubiquitinated mitochondria is HDAC6 (Histone Deacetylase 6) (Yan et al., 2013), a cytosolic member of the family of histone deacetylases (Hubbert et al., 2002). This protein works as an adaptor between ubiquitinated proteins and dynein motors, enabling the transport of cargoes retrogradely along microtubules to form the aggresome. Recently, it was proposed that HDAC6 contributes to the formation of large inclusions of SOD1 (Gal et al., 2013). Interactions between HDAC6 and ALS proteins, such as TDP43 and SOD1, have emerged as functionally relevant in ALS (Fiesel et al., 2010; Taes et al., 2013). HDAC6 also functions as a regulator of the fusion between autophagosomes and lysosomes, since it is capable of promoting the remodeling of F-actin cytoskeleton, in a mechanism dependent of cortactin-recruitment (Lee et al., 2010). In this context, the retrograde transport of mitophagic vacuoles to reach the soma and fuse with the lysosomes is an essential step in mitophagy (Maday et al., 2012; Maday and Holzbaur, 2014) and mutant SOD1 impairs retrograde axonal transport in vivo and in vitro (Magrane et al., 2012; Magrane et al., 2014).
Dynein and dynactin form a complex that mediates interactions between cargoes and microtubules and is responsible for the retrograde transport. Perturbations in this complex result in motor neuron degeneration (Ikenaka et al., 2013). Mutations in the dynactin subunit p150 Glued have been linked to familial motor neuron disease (Levy et al., 2006). This raises the possibility that impairment of vesicular transport necessary for MQC is a pathological mechanism, especially in the context of the extremely large axons of the motor neurons, in which transport of autophagosome-engulfed mitochondria and components of the mitophagy machinery is essential for completing the mitophagy program and to ensure a proper turnover of damaged organelles.
Interestingly, genetic deletion of HDAC6 extended the survival of mutant SOD1 mice and maintained motor axon integrity. This protective effect was associated with increased 3-tubulin acetylation (Taes et al., 2013). Mitochondrial transport was not directly investigated in this study, but it is possible that increased 3-tubulin acetylation results in increased axonal transport and contributes to ameliorating the defect of mitochondrial dynamics in mutant SOD1 mice.
Therapeutic perspectives
Autophagy is emerging as a process that can be targeted for therapeutic interventions for neurological diseases. Pharmacological stimulation of autophagy was shown to be beneficial in a mouse model of mitochondrial encephalopathy (Johnson et al., 2013). In ALS, approaches to increase autophagy could lead to prospective therapeutic interventions aimed at potentiate the elimination of damaged components of the cell, such as dysfunctional mitochondria.
Several studies have investigated autophagy modulation in mutant SOD1 mice with varying outcomes. Down regulation of X-box-binding protein-1 (XBP-1) stimulated autophagy in mutant SOD1 mice, resulting in motor neuron protection and disease improvement (Hetz et al., 2009). Initial reports showed contradictory results upon treatment with general autophagy enhancers, such as lithium (Fornai et al., 2008; Pizzasegola et al., 2009) and rapamycin (Zhang et al., 2011), but several recent studies have demonstrated beneficial effects of enhancing autophagy in mutant SOD1 mice and other mouse models (Castillo et al., 2013; Kim et al., 2013a; Wang et al., 2012; Wang et al., 2013a; Zhang et al., 2013; Zhang et al., 2014). The therapies tested so far in ALS have been aimed at potentiating general macroautophagy mechanisms and not mitophagy specifically. Currently, there are no viable approaches to selectively enhance mitophagy or any other components of MQC pathways. A first step in this direction could be made by genetically targeting these pathways, especially in cellular and mouse models of ALS, where there is a clear mitochondrial involvement. Modulating the expression of genes that encode for components of the MQC could help determining the significance of maintaining appropriate mitochondrial homeostasis in ALS and could reveal unsuspected commonalities with different forms of neurodegeneration, such as familial Parkinson disease, where impairment of the MQC has been shown to play a determining role.
Highlights.
Cells are endowed with mitochondria quality control (MQC) systems, such as proteostasis and mitophagy.
Mitochondrial dysfunction and degeneration are associated with familial and sporadic forms of ALS.
MQC systems fail to control mitochondrial damage in ALS motor neurons.
This review discusses mitochondrial damage and the involvement of MQC in ALS.
Acknowledgments
funding: NIH grants NS051419 and NS062055, The Packard Center for ALS Research, The Muscular Dystrophy Association.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anand R, et al. Proteolytic control of mitochondrial function and morphogenesis. Biochim Biophys Acta. 2013;1833:195–204. doi: 10.1016/j.bbamcr.2012.06.025. [DOI] [PubMed] [Google Scholar]
- Andersen PM. Amyotrophic lateral sclerosis associated with mutations in the CuZn superoxide dismutase gene. Curr Neurol Neurosci Rep. 2006;6:37–46. doi: 10.1007/s11910-996-0008-9. [DOI] [PubMed] [Google Scholar]
- Andersen PM, Al-Chalabi A. Clinical genetics of amyotrophic lateral sclerosis: what do we really know? Nat Rev Neurol. 2011;7:603–15. doi: 10.1038/nrneurol.2011.150. [DOI] [PubMed] [Google Scholar]
- Ashrafi G, Schwarz TL. The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ. 2013;20:31–42. doi: 10.1038/cdd.2012.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashrafi G, et al. Mitophagy of damaged mitochondria occurs locally in distal neuronal axons and requires PINK1 and Parkin. J Cell Biol. 2014;206:655–70. doi: 10.1083/jcb.201401070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensimon G, et al. A controlled trial of riluzole in amyotrophic lateral sclerosis. ALS/Riluzole Study Group. N Engl J Med. 1994;330:585–91. doi: 10.1056/NEJM199403033300901. [DOI] [PubMed] [Google Scholar]
- Birsa N, et al. K27 ubiquitination of the mitochondrial transport protein Miro is dependent on serine 65 of the Parkin ubiquitin ligase. J Biol Chem. 2014 doi: 10.1074/jbc.M114.563031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q, et al. Spatial parkin translocation and degradation of damaged mitochondria via mitophagy in live cortical neurons. Curr Biol. 2012;22:545–52. doi: 10.1016/j.cub.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carri MT, Cozzolino M. SOD1 and mitochondria in ALS: a dangerous liaison. J Bioenerg Biomembr. 2011;43:593–9. doi: 10.1007/s10863-011-9394-z. [DOI] [PubMed] [Google Scholar]
- Castillo K, et al. Trehalose delays the progression of amyotrophic lateral sclerosis by enhancing autophagy in motoneurons. Autophagy. 2013;9:1308–20. doi: 10.4161/auto.25188. [DOI] [PubMed] [Google Scholar]
- Chan NC, et al. Broad activation of the ubiquitin-proteasome system by Parkin is critical for mitophagy. Hum Mol Genet. 2011;20:1726–37. doi: 10.1093/hmg/ddr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherra SJ, 3rd, et al. Review: autophagy and neurodegeneration: survival at a cost? Neuropathol Appl Neurobiol. 2010;36:125–32. doi: 10.1111/j.1365-2990.2010.01062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu CT, et al. Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nat Cell Biol. 2013;15:1197–205. doi: 10.1038/ncb2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland DW, Rothstein JD. From Charcot to Lou Gehrig: deciphering selective motor neuron death in ALS. Nat Rev Neurosci. 2001;2:806–19. doi: 10.1038/35097565. [DOI] [PubMed] [Google Scholar]
- Cozzolino M, Carri MT. Mitochondrial dysfunction in ALS. Prog Neurobiol. 2011 doi: 10.1016/j.pneurobio.2011.06.003. [DOI] [PubMed] [Google Scholar]
- Cozzolino M, et al. Mitochondria and ALS: implications from novel genes and pathways. Mol Cell Neurosci. 2013;55:44–9. doi: 10.1016/j.mcn.2012.06.001. [DOI] [PubMed] [Google Scholar]
- Damiano M, et al. Neural mitochondrial Ca2+ capacity impairment precedes the onset of motor symptoms in G93A Cu/Zn-superoxide dismutase mutant mice. J Neurochem. 2006;96:1349–61. doi: 10.1111/j.1471-4159.2006.03619.x. [DOI] [PubMed] [Google Scholar]
- De Vos KJ, et al. Familial amyotrophic lateral sclerosis-linked SOD1 mutants perturb fast axonal transport to reduce axonal mitochondria content. Hum Mol Genet. 2007;16:2720–8. doi: 10.1093/hmg/ddm226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–56. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng HX, et al. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature. 2011;477:211–5. doi: 10.1038/nature10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fecto F, et al. SQSTM1 mutations in familial and sporadic amyotrophic lateral sclerosis. Arch Neurol. 2011;68:1440–6. doi: 10.1001/archneurol.2011.250. [DOI] [PubMed] [Google Scholar]
- Fecto F, Siddique T. UBQLN2/P62 cellular recycling pathways in amyotrophic lateral sclerosis and frontotemporal dementia. Muscle Nerve. 2012;45:157–62. doi: 10.1002/mus.23278. [DOI] [PubMed] [Google Scholar]
- Ferraiuolo L, et al. Molecular pathways of motor neuron injury in amyotrophic lateral sclerosis. Nat Rev Neurol. 2011;7:616–30. doi: 10.1038/nrneurol.2011.152. [DOI] [PubMed] [Google Scholar]
- Fiesel FC, et al. Knockdown of transactive response DNA-binding protein (TDP-43) downregulates histone deacetylase 6. EMBO J. 2010;29:209–21. doi: 10.1038/emboj.2009.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer LR, et al. SOD1 targeted to the mitochondrial intermembrane space prevents motor neuropathy in the Sod1 knockout mouse. Brain. 2011;134:196–209. doi: 10.1093/brain/awq314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornai F, et al. Lithium delays progression of amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A. 2008;105:2052–7. doi: 10.1073/pnas.0708022105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal J, et al. p62 accumulates and enhances aggregate formation in model systems of familial amyotrophic lateral sclerosis. J Biol Chem. 2007;282:11068–77. doi: 10.1074/jbc.M608787200. [DOI] [PubMed] [Google Scholar]
- Gal J, et al. Sequestosome 1/p62 links familial ALS mutant SOD1 to LC3 via an ubiquitin-independent mechanism. J Neurochem. 2009;111:1062–73. doi: 10.1111/j.1471-4159.2009.06388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal J, et al. HDAC6 regulates mutant SOD1 aggregation through two SMIR motifs and tubulin acetylation. J Biol Chem. 2013;288:15035–45. doi: 10.1074/jbc.M112.431957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–31. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- Gerdes F, et al. Mitochondrial AAA proteases--towards a molecular understanding of membrane-bound proteolytic machines. Biochim Biophys Acta. 2012;1823:49–55. doi: 10.1016/j.bbamcr.2011.09.015. [DOI] [PubMed] [Google Scholar]
- Gould TW, et al. Complete dissociation of motor neuron death from motor dysfunction by Bax deletion in a mouse model of ALS. J Neurosci. 2006;26:8774–86. doi: 10.1523/JNEUROSCI.2315-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenier K, et al. Parkin- and PINK1-Dependent Mitophagy in Neurons: Will the Real Pathway Please Stand Up? Front Neurol. 2013;4:100. doi: 10.3389/fneur.2013.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebron M, et al. Parkin reverses TDP-43-induced cell death and failure of amino acid homeostasis. J Neurochem. 2014;129:350–61. doi: 10.1111/jnc.12630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebron ML, et al. Parkin ubiquitinates Tar-DNA binding protein-43 (TDP-43) and promotes its cytosolic accumulation via interaction with histone deacetylase 6 (HDAC6) J Biol Chem. 2013;288:4103–15. doi: 10.1074/jbc.M112.419945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo JM, et al. A stress-responsive system for mitochondrial protein degradation. Mol Cell. 2010;40:465–80. doi: 10.1016/j.molcel.2010.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo JM, Rutter J. Ubiquitin-dependent mitochondrial protein degradation. Int J Biochem Cell Biol. 2011;43:1422–6. doi: 10.1016/j.biocel.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo JM, et al. Intramolecular interactions control Vms1 translocation to damaged mitochondria. Mol Biol Cell. 2013;24:1263–73. doi: 10.1091/mbc.E13-02-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervias I, et al. Mitochondrial dysfunction and amyotrophic lateral sclerosis. Muscle Nerve. 2006;33:598–608. doi: 10.1002/mus.20489. [DOI] [PubMed] [Google Scholar]
- Hetz C, et al. XBP-1 deficiency in the nervous system protects against amyotrophic lateral sclerosis by increasing autophagy. Genes Dev. 2009;23:2294–306. doi: 10.1101/gad.1830709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins CM, et al. Mutant Cu, Zn superoxide dismutase that causes motoneuron degeneration is present in mitochondria in the CNS. J Neurosci. 2002;22:RC215. doi: 10.1523/JNEUROSCI.22-06-j0001.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirtz D, et al. How common are the “common” neurologic disorders? Neurology. 2007;68:326–37. doi: 10.1212/01.wnl.0000252807.38124.a3. [DOI] [PubMed] [Google Scholar]
- Hong K, et al. Full-length TDP-43 and its C-terminal fragments activate mitophagy in NSC34 cell line. Neurosci Lett. 2012;530:144–9. doi: 10.1016/j.neulet.2012.10.003. [DOI] [PubMed] [Google Scholar]
- Hubbert C, et al. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417:455–8. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- Ikenaka K, et al. dnc-1/dynactin 1 knockdown disrupts transport of autophagosomes and induces motor neuron degeneration. PLoS One. 2013;8:e54511. doi: 10.1371/journal.pone.0054511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israelson A, et al. Misfolded mutant SOD1 directly inhibits VDAC1 conductance in a mouse model of inherited ALS. Neuron. 2010;67:575–87. doi: 10.1016/j.neuron.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaarsma D, et al. CuZn superoxide dismutase (SOD1) accumulates in vacuolated mitochondria in transgenic mice expressing amyotrophic lateral sclerosis-linked SOD1 mutations. Acta Neuropathol. 2001;102:293–305. doi: 10.1007/s004010100399. [DOI] [PubMed] [Google Scholar]
- Johnson JO, et al. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2010;68:857–64. doi: 10.1016/j.neuron.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SC, et al. mTOR inhibition alleviates mitochondrial disease in a mouse model of Leigh syndrome. Science. 2013;342:1524–8. doi: 10.1126/science.1244360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joselin AP, et al. ROS-dependent regulation of Parkin and DJ-1 localization during oxidative stress in neurons. Hum Mol Genet. 2012;21:4888–903. doi: 10.1093/hmg/dds325. [DOI] [PubMed] [Google Scholar]
- Karbowski M, et al. The mitochondrial E3 ubiquitin ligase MARCH5 is required for Drp1 dependent mitochondrial division. J Cell Biol. 2007;178:71–84. doi: 10.1083/jcb.200611064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbowski M, Neutzner A. Neurodegeneration as a consequence of failed mitochondrial maintenance. Acta Neuropathol. 2012;123:157–71. doi: 10.1007/s00401-011-0921-0. [DOI] [PubMed] [Google Scholar]
- Kawamata H, Manfredi G. Import, maturation, and function of SOD1 and its copper chaperone CCS in the mitochondrial intermembrane space. Antioxid Redox Signal. 2010a;13:1375–84. doi: 10.1089/ars.2010.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamata H, Manfredi G. Mitochondrial dysfunction and intracellular calcium dysregulation in ALS. Mech Ageing Dev. 2010b;131:517–26. doi: 10.1016/j.mad.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, et al. The mitochondrial calcium regulator cyclophilin D is an essential component of oestrogen-mediated neuroprotection in amyotrophic lateral sclerosis. Brain. 2012;135:2865–74. doi: 10.1093/brain/aws208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, et al. Autophagy activation and neuroprotection by progesterone in the G93A-SOD1 transgenic mouse model of amyotrophic lateral sclerosis. Neurobiol Dis. 2013a;59:80–5. doi: 10.1016/j.nbd.2013.07.011. [DOI] [PubMed] [Google Scholar]
- Kim NC, et al. VCP is essential for mitochondrial quality control by PINK1/Parkin and this function is impaired by VCP mutations. Neuron. 2013b;78:65–80. doi: 10.1016/j.neuron.2013.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondapalli C, et al. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biol. 2012;2:120080. doi: 10.1098/rsob.120080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyano F, et al. The principal PINK1 and Parkin cellular events triggered in response to dissipation of mitochondrial membrane potential occur in primary neurons. Genes Cells. 2013;18:672–81. doi: 10.1111/gtc.12066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski TJ, Jr, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–8. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- Lacomblez L, et al. Dose-ranging study of riluzole in amyotrophic lateral sclerosis. Amyotrophic Lateral Sclerosis/Riluzole Study Group II. Lancet. 1996;347:1425–31. doi: 10.1016/s0140-6736(96)91680-3. [DOI] [PubMed] [Google Scholar]
- Lagier-Tourenne C, et al. Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long pre-mRNAs. Nat Neurosci. 2012;15:1488–97. doi: 10.1038/nn.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lattante S, et al. TARDBP and FUS mutations associated with amyotrophic lateral sclerosis: summary and update. Hum Mutat. 2013;34:812–26. doi: 10.1002/humu.22319. [DOI] [PubMed] [Google Scholar]
- Lee JY, et al. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. EMBO J. 2010;29:969–80. doi: 10.1038/emboj.2009.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy JR, et al. A motor neuron disease-associated mutation in p150Glued perturbs dynactin function and induces protein aggregation. J Cell Biol. 2006;172:733–45. doi: 10.1083/jcb.200511068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, et al. Altered macroautophagy in the spinal cord of SOD1 mutant mice. Autophagy. 2008a;4:290–3. doi: 10.4161/auto.5524. [DOI] [PubMed] [Google Scholar]
- Li Q, et al. ALS-linked mutant superoxide dismutase 1 (SOD1) alters mitochondrial protein composition and decreases protein import. Proc Natl Acad Sci U S A. 2010;107:21146–51. doi: 10.1073/pnas.1014862107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, et al. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle’s dynamics and signaling. PLoS One. 2008b;3:e1487. doi: 10.1371/journal.pone.0001487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligon LA, et al. Mutant superoxide dismutase disrupts cytoplasmic dynein in motor neurons. Neuroreport. 2005;16:533–6. doi: 10.1097/00001756-200504250-00002. [DOI] [PubMed] [Google Scholar]
- Ling SC, et al. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron. 2013;79:416–38. doi: 10.1016/j.neuron.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maday S, et al. Autophagosomes initiate distally and mature during transport toward the cell soma in primary neurons. J Cell Biol. 2012;196:407–17. doi: 10.1083/jcb.201106120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maday S, Holzbaur EL. Autophagosome biogenesis in primary neurons follows an ordered and spatially regulated pathway. Dev Cell. 2014;30:71–85. doi: 10.1016/j.devcel.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magrane J, et al. Mutant SOD1 in neuronal mitochondria causes toxicity and mitochondrial dynamics abnormalities. Hum Mol Genet. 2009;18:4552–64. doi: 10.1093/hmg/ddp421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magrane J, Manfredi G. Mitochondrial function, morphology, and axonal transport in amyotrophic lateral sclerosis. Antioxid Redox Signal. 2009;11:1615–26. doi: 10.1089/ars.2009.2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magrane J, et al. Mitochondrial dynamics and bioenergetic dysfunction is associated with synaptic alterations in mutant SOD1 motor neurons. J Neurosci. 2012;32:229–42. doi: 10.1523/JNEUROSCI.1233-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magrane J, et al. Abnormal mitochondrial transport and morphology are common pathological denominators in SOD1 and TDP43 ALS mouse models. Hum Mol Genet. 2014;23:1413–24. doi: 10.1093/hmg/ddt528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majounie E, et al. Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. Lancet Neurol. 2012;11:323–30. doi: 10.1016/S1474-4422(12)70043-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinkovic P, et al. Axonal transport deficits and degeneration can evolve independently in mouse models of amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A. 2012;109:4296–301. doi: 10.1073/pnas.1200658109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin LJ. Mitochondrial pathobiology in ALS. J Bioenerg Biomembr. 2011;43:569–79. doi: 10.1007/s10863-011-9395-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda N, et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol. 2010;189:211–21. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima Y, Kaguni LS. Matrix proteases in mitochondrial DNA function. Biochim Biophys Acta. 2012;1819:1080–7. doi: 10.1016/j.bbagrm.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattiazzi M, et al. Mutated human SOD1 causes dysfunction of oxidative phosphorylation in mitochondria of transgenic mice. J Biol Chem. 2002;277:29626–33. doi: 10.1074/jbc.M203065200. [DOI] [PubMed] [Google Scholar]
- McLelland GL, et al. Parkin and PINK1 function in a vesicular trafficking pathway regulating mitochondrial quality control. EMBO J. 2014;33:282–95. doi: 10.1002/embj.201385902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira LG, et al. Structural and functional analysis of human SOD1 in amyotrophic lateral sclerosis. PLoS One. 2013;8:e81979. doi: 10.1371/journal.pone.0081979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto N, et al. Increased autophagy in transgenic mice with a G93A mutant SOD1 gene. Brain Res. 2007;1167:112–7. doi: 10.1016/j.brainres.2007.06.045. [DOI] [PubMed] [Google Scholar]
- Nagashima S, et al. Roles of mitochondrial ubiquitin ligase MITOL/MARCH5 in mitochondrial dynamics and diseases. J Biochem. 2014 doi: 10.1093/jb/mvu016. [DOI] [PubMed] [Google Scholar]
- Nakamura N, et al. MARCH-V is a novel mitofusin 2- and Drp1-binding protein able to change mitochondrial morphology. EMBO Rep. 2006;7:1019–22. doi: 10.1038/sj.embor.7400790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalbandian A, et al. The multiple faces of valosin-containing protein-associated diseases: inclusion body myopathy with Paget’s disease of bone, frontotemporal dementia, and amyotrophic lateral sclerosis. J Mol Neurosci. 2011;45:522–31. doi: 10.1007/s12031-011-9627-y. [DOI] [PubMed] [Google Scholar]
- Narendra D, et al. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D, et al. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy. 2010a;6:1090–106. doi: 10.4161/auto.6.8.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra DP, et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010b;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okado-Matsumoto A, Fridovich I. Subcellular distribution of superoxide dismutases (SOD) in rat liver: Cu,Zn-SOD in mitochondria. J Biol Chem. 2001;276:38388–93. doi: 10.1074/jbc.M105395200. [DOI] [PubMed] [Google Scholar]
- Okatsu K, et al. p62/SQSTM1 cooperates with Parkin for perinuclear clustering of depolarized mitochondria. Genes Cells. 2010;15:887–900. doi: 10.1111/j.1365-2443.2010.01426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandya RS, et al. Therapeutic neuroprotective agents for amyotrophic lateral sclerosis. Cell Mol Life Sci. 2013;70:4729–45. doi: 10.1007/s00018-013-1415-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa L, Germain D. Estrogen receptor mediates a distinct mitochondrial unfolded protein response. J Cell Sci. 2011;124:1396–402. doi: 10.1242/jcs.078220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YY, et al. Loss of MARCH5 mitochondrial E3 ubiquitin ligase induces cellular senescence through dynamin-related protein 1 and mitofusin 1. J Cell Sci. 2010;123:619–26. doi: 10.1242/jcs.061481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parone PA, et al. Enhancing mitochondrial calcium buffering capacity reduces aggregation of misfolded SOD1 and motor neuron cell death without extending survival in mouse models of inherited amyotrophic lateral sclerosis. J Neurosci. 2013;33:4657–71. doi: 10.1523/JNEUROSCI.1119-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasinelli P, et al. Amyotrophic lateral sclerosis-associated SOD1 mutant proteins bind and aggregate with Bcl-2 in spinal cord mitochondria. Neuron. 2004;43:19–30. doi: 10.1016/j.neuron.2004.06.021. [DOI] [PubMed] [Google Scholar]
- Pasinelli P, Brown RH. Molecular biology of amyotrophic lateral sclerosis: insights from genetics. Nat Rev Neurosci. 2006;7:710–23. doi: 10.1038/nrn1971. [DOI] [PubMed] [Google Scholar]
- Pedrini S, et al. ALS-linked mutant SOD1 damages mitochondria by promoting conformational changes in Bcl-2. Hum Mol Genet. 2010;19:2974–86. doi: 10.1093/hmg/ddq202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrino MW, et al. Signaling the mitochondrial unfolded protein response. Biochim Biophys Acta. 2013;1833:410–6. doi: 10.1016/j.bbamcr.2012.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzasegola C, et al. Treatment with lithium carbonate does not improve disease progression in two different strains of SOD1 mutant mice. Amyotroph Lateral Scler. 2009;10:221–8. doi: 10.1080/17482960902803440. [DOI] [PubMed] [Google Scholar]
- Polymenidou M, et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci. 2011;14:459–68. doi: 10.1038/nn.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole AC, et al. The mitochondrial fusion-promoting factor mitofusin is a substrate of the PINK1/parkin pathway. PLoS One. 2010;5:e10054. doi: 10.1371/journal.pone.0010054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puls I, et al. Mutant dynactin in motor neuron disease. Nat Genet. 2003;33:455–6. doi: 10.1038/ng1123. [DOI] [PubMed] [Google Scholar]
- Radke S, et al. Mitochondrial protein quality control by the proteasome involves ubiquitination and the protease Omi. J Biol Chem. 2008;283:12681–5. doi: 10.1074/jbc.C800036200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton AE, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–68. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton AE, et al. State of play in amyotrophic lateral sclerosis genetics. Nat Neurosci. 2014;17:17–23. doi: 10.1038/nn.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen DR. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;364:362. doi: 10.1038/364362c0. [DOI] [PubMed] [Google Scholar]
- Sabatelli M, et al. Clinical and genetic heterogeneity of amyotrophic lateral sclerosis. Clin Genet. 2013;83:408–16. doi: 10.1111/cge.12117. [DOI] [PubMed] [Google Scholar]
- Sasaki S, Iwata M. Dendritic synapses of anterior horn neurons in amyotrophic lateral sclerosis: an ultrastructural study. Acta Neuropathol. 1996;91:278–83. doi: 10.1007/s004010050426. [DOI] [PubMed] [Google Scholar]
- Sasaki S, Iwata M. Mitochondrial alterations in the spinal cord of patients with sporadic amyotrophic lateral sclerosis. J Neuropathol Exp Neurol. 2007;66:10–6. doi: 10.1097/nen.0b013e31802c396b. [DOI] [PubMed] [Google Scholar]
- Sasaki S. Determination of altered mitochondria ultrastructure by electron microscopy. Methods Mol Biol. 2010;648:279–90. doi: 10.1007/978-1-60761-756-3_19. [DOI] [PubMed] [Google Scholar]
- Sasaki S. Autophagy in spinal cord motor neurons in sporadic amyotrophic lateral sclerosis. J Neuropathol Exp Neurol. 2011;70:349–59. doi: 10.1097/NEN.0b013e3182160690. [DOI] [PubMed] [Google Scholar]
- Scarffe LA, et al. Parkin and PINK1: much more than mitophagy. Trends Neurosci. 2014 doi: 10.1016/j.tins.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schon EA, Przedborski S. Mitochondria: the next (neurode)generation. Neuron. 2011;70:1033–53. doi: 10.1016/j.neuron.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibler P, et al. Mitochondrial Parkin recruitment is impaired in neurons derived from mutant PINK1 induced pluripotent stem cells. J Neurosci. 2011;31:5970–6. doi: 10.1523/JNEUROSCI.4441-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi P, et al. Effects of ALS-related SOD1 mutants on dynein- and KIF5-mediated retrograde and anterograde axonal transport. Biochim Biophys Acta. 2010;1802:707–16. doi: 10.1016/j.bbadis.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soubannier V, et al. A vesicular transport pathway shuttles cargo from mitochondria to lysosomes. Curr Biol. 2012a;22:135–41. doi: 10.1016/j.cub.2011.11.057. [DOI] [PubMed] [Google Scholar]
- Soubannier V, et al. Reconstitution of mitochondria derived vesicle formation demonstrates selective enrichment of oxidized cargo. PLoS One. 2012b;7:e52830. doi: 10.1371/journal.pone.0052830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreedharan J, et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–72. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoica R, et al. ER-mitochondria associations are regulated by the VAPB-PTPIP51 interaction and are disrupted by ALS/FTD-associated TDP-43. Nat Commun. 2014;5:3996. doi: 10.1038/ncomms4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stribl C, et al. Mitochondrial dysfunction and decrease in body weight of a transgenic knock-in mouse model for TDP-43. J Biol Chem. 2014;289:10769–84. doi: 10.1074/jbc.M113.515940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura A, et al. MITOL regulates endoplasmic reticulum-mitochondria contacts via Mitofusin2. Mol Cell. 2013;51:20–34. doi: 10.1016/j.molcel.2013.04.023. [DOI] [PubMed] [Google Scholar]
- Taes I, et al. Hdac6 deletion delays disease progression in the SOD1G93A mouse model of ALS. Hum Mol Genet. 2013;22:1783–90. doi: 10.1093/hmg/ddt028. [DOI] [PubMed] [Google Scholar]
- Tanaka A, et al. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J Cell Biol. 2010;191:1367–80. doi: 10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortelli R, et al. Amyotrophic lateral sclerosis: a new missense mutation in the SOD1 gene. Neurobiol Aging. 2013;34:1709 e3–5. doi: 10.1016/j.neurobiolaging.2012.10.027. [DOI] [PubMed] [Google Scholar]
- Twig G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–46. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Blitterswijk M, Landers JE. RNA processing pathways in amyotrophic lateral sclerosis. Neurogenetics. 2010;11:275–90. doi: 10.1007/s10048-010-0239-4. [DOI] [PubMed] [Google Scholar]
- Van Laar VS, et al. Bioenergetics of neurons inhibit the translocation response of Parkin following rapid mitochondrial depolarization. Hum Mol Genet. 2011;20:927–40. doi: 10.1093/hmg/ddq531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance C, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–11. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vande Velde C, et al. Misfolded SOD1 associated with motor neuron mitochondria alters mitochondrial shape and distribution prior to clinical onset. PLoS One. 2011;6:e22031. doi: 10.1371/journal.pone.0022031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayvergiya C, et al. Mutant superoxide dismutase 1 forms aggregates in the brain mitochondrial matrix of amyotrophic lateral sclerosis mice. J Neurosci. 2005;25:2463–70. doi: 10.1523/JNEUROSCI.4385-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinsant S, et al. Characterization of early pathogenesis in the SOD1(G93A) mouse model of ALS: part I, background and methods. Brain Behav. 2013a;3:335–50. doi: 10.1002/brb3.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinsant S, et al. Characterization of early pathogenesis in the SOD1(G93A) mouse model of ALS: part II, results and discussion. Brain Behav. 2013b;3:431–57. doi: 10.1002/brb3.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vives-Bauza C, et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci U S A. 2010;107:378–83. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang IF, et al. Autophagy activators rescue and alleviate pathogenesis of a mouse model with proteinopathies of the TAR DNA-binding protein 43. Proc Natl Acad Sci U S A. 2012;109:15024–9. doi: 10.1073/pnas.1206362109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang IF, et al. Autophagy activation ameliorates neuronal pathogenesis of FTLD-U mice: a new light for treatment of TARDBP/TDP-43 proteinopathies. Autophagy. 2013a;9:239–40. doi: 10.4161/auto.22526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, et al. The ALS disease-associated mutant TDP-43 impairs mitochondrial dynamics and function in motor neurons. Hum Mol Genet. 2013b;22:4706–19. doi: 10.1093/hmg/ddt319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, et al. PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell. 2011;147:893–906. doi: 10.1016/j.cell.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts GD, et al. Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat Genet. 2004;36:377–81. doi: 10.1038/ng1332. [DOI] [PubMed] [Google Scholar]
- Williamson TL, Cleveland DW. Slowing of axonal transport is a very early event in the toxicity of ALS-linked SOD1 mutants to motor neurons. Nat Neurosci. 1999;2:50–6. doi: 10.1038/4553. [DOI] [PubMed] [Google Scholar]
- Wu CH, et al. Mutations in the profilin 1 gene cause familial amyotrophic lateral sclerosis. Nature. 2012;488:499–503. doi: 10.1038/nature11280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, et al. The AAA-ATPase p97 is essential for outer mitochondrial membrane protein turnover. Mol Biol Cell. 2011;22:291–300. doi: 10.1091/mbc.E10-09-0748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, et al. SQSTM1/p62 interacts with HDAC6 and regulates deacetylase activity. PLoS One. 2013;8:e76016. doi: 10.1371/journal.pone.0076016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, et al. Recruitment of the p97 ATPase and ubiquitin ligases to the site of retrotranslocation at the endoplasmic reticulum membrane. Proc Natl Acad Sci U S A. 2005;102:14132–8. doi: 10.1073/pnas.0505006102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonashiro R, et al. A novel mitochondrial ubiquitin ligase plays a critical role in mitochondrial dynamics. EMBO J. 2006;25:3618–26. doi: 10.1038/sj.emboj.7601249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonashiro R, et al. Mitochondrial ubiquitin ligase MITOL ubiquitinates mutant SOD1 and attenuates mutant SOD1-induced reactive oxygen species generation. Mol Biol Cell. 2009;20:4524–30. doi: 10.1091/mbc.E09-02-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, et al. Neurofilaments and orthograde transport are reduced in ventral root axons of transgenic mice that express human SOD1 with a G93A mutation. J Cell Biol. 1997;139:1307–15. doi: 10.1083/jcb.139.5.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, et al. Interaction between familial amyotrophic lateral sclerosis (ALS)-linked SOD1 mutants and the dynein complex. J Biol Chem. 2007;282:16691–9. doi: 10.1074/jbc.M609743200. [DOI] [PubMed] [Google Scholar]
- Zhang K, et al. Food restriction-induced autophagy modulates degradation of mutant SOD1 in an amyotrophic lateral sclerosis mouse model. Brain Res. 2013;1519:112–9. doi: 10.1016/j.brainres.2013.04.050. [DOI] [PubMed] [Google Scholar]
- Zhang X, et al. Rapamycin treatment augments motor neuron degeneration in SOD1(G93A) mouse model of amyotrophic lateral sclerosis. Autophagy. 2011;7:412–25. doi: 10.4161/auto.7.4.14541. [DOI] [PubMed] [Google Scholar]
- Zhang X, et al. MTOR-independent, autophagic enhancer trehalose prolongs motor neuron survival and ameliorates the autophagic flux defect in a mouse model of amyotrophic lateral sclerosis. Autophagy. 2014:10. doi: 10.4161/auto.27710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziviani E, et al. Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin. Proc Natl Acad Sci U S A. 2010;107:5018–23. doi: 10.1073/pnas.0913485107. [DOI] [PMC free article] [PubMed] [Google Scholar]