Abstract
Growth factor signaling regulates tissue-tissue interactions to control organogenesis and tissue homeostasis. Specifically, transforming growth factor beta (TGFβ) signaling plays a crucial role in the development of cranial neural crest (CNC) cell–derived bone, and loss of Tgfbr2 in CNC cells results in craniofacial skeletal malformations. Our recent studies indicate that non-canonical TGFβ signaling is activated whereas canonical TGFβ signaling is compromised in the absence of Tgfbr2 (in Tgfbr2fl/fl;Wnt1-Cre mice). A haploinsufficiency of Tgfbr1 (aka Alk5) (Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+) largely rescues craniofacial deformities in Tgfbr2 mutant mice by reducing ectopic non-canonical TGFβ signaling. However, the relative involvement of canonical and non-canonical TGFβ signaling in regulating specific craniofacial bone formation remains unclear. We compared the size and volume of CNC–derived craniofacial bones (frontal bone, premaxilla, maxilla, palatine bone, and mandible) from E18.5 control, Tgfbr2fl/fl;Wnt1-Cre, and Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ mice. By analyzing three dimensional (3D) micro-computed tomography (microCT) images, we found that different craniofacial bones were restored to different degrees in Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ mice. Our study provides comprehensive information on anatomical landmarks and the size and volume of each craniofacial bone, as well as insights into the extent that canonical and non-canonical TGFβ signaling cascades contribute to the formation of each CNC–derived bone. Our data will serve as an important resource for developmental biologists who are interested in craniofacial morphogenesis.
Keywords: Mouse embryo, craniofacial bone development, TGFβ signaling, micro-computed tomography
Introduction
With recent advancements in mouse genetics, there has been tremendous progress in research in craniofacial developmental biology, particularly with respect to malformations (Chai and Maxson, 2006). Numerous studies demonstrate that there is often a pleiotropic effect on multiple organs following a single gene mutation. In light of such wide-ranging effects, it is becoming clear that analyzing a single tissue or two-dimensional (2D) sections is not adequate to provide a comprehensive view of a mutant mouse embryo. Three-dimensional (3D) volumetric imaging is robust, quantitative, automated and high-throughput. Furthermore, 3D digital data allows custom-designed analysis and viewing (Wong et al., 2012). Micro-computed tomography (microCT) has been used to analyze craniofacial phenotypes in many studies, including ones designed to define anatomical landmarks and perform volumetric analysis for each craniofacial bone in humans and mice (Perrine et al., 2014, Percival et al., 2014, Titiz et al., 2012, Conner et al., 2007). Percival et al. (2014) reported that the patterns of growth and maturation in craniofacial bones formed through endochondral ossification were different than in those formed intramembraneously. However, the regulatory mechanisms of craniofacial bone morphogenesis remain poorly understood.
During craniofacial development, cranial neural crest (CNC) cells give rise to osteoblasts and form most of the craniofacial bones that compose part of the skull (Jiang et al., 2002). The majority of the craniofacial bones, with the exception of the cranial base and parts of the mandible that arise from endochondral ossification, are formed by intramembranous ossification through a mechanism that remains poorly understood (Chai et al., 2003). Transforming growth factor beta (TGFβ) signaling plays a crucial role in craniofacial development, and loss of TGFβ signaling in CNC cells results in cranial skeletal deformities (Ito et al., 2003; Iwata et al., 2011). TGFβ transmits signals through both canonical (SMAD–dependent) and non-canonical (SMAD–independent) signaling pathways, and targets a variety of genes in an embryonic stage-dependent and cell type-specific manner (Ross and Hill, 2008). Mutations in genes for TGFβ receptor type I or type II (TGFBR1 or TGFBR2) are associated with Loeys-Dietz Syndrome (previously called Marfan Syndrome type II) in humans, which can manifest with craniofacial malformations including a small mandible, cleft palate, craniosynostosis, hypertelorism, and vascular defects (Mizuguchi et al., 2004; Loeys et al., 2005).
In order to test the functional significance of TGFβ signaling in regulating the fate of CNC cells, we have generated a mutant animal model in which loss of Tgfbr2 in CNC cells (Tgfbr2fl/fl;Wnt1-Cre) leads to compromised canonical TGFβ signaling, activation of non-canonical TGFβ signaling through the TGFβ RI/RIII complex and an array of craniofacial deformities (Ito et al., 2003). We have recently reported that non-canonical TGFβ signaling is activated whereas canonical TGFβ signaling is compromised in Tgfbr2fl/fl;Wnt1-Cre mice (Iwata et al., 2014; Iwata et al., 2012). In addition, craniofacial deformities in Tgfbr2 mutant mice were largely restored by a haploinsufficiency of Tgfbr1 (Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+), a key receptor that is involved in ectopic non-canonical TGFβ signaling, indicating that altered canonical TGFβ signaling causes craniofacial deformities (Iwata et al., 2012). However, the extent to which canonical and non-canonical TGFβ signaling cascades contribute to CNC–derived craniofacial bone formation is still unknown.
Understanding craniofacial bone development is essential for studying craniofacial anomalies in humans. Defined anatomical landmarks along with 3D microCT images of craniofacial bone morphology combine to produce a powerful method for studying craniofacial anomalies and anatomical defects. MicroCT imaging analyses allow for detailed examination of craniofacial growth disturbances in mutant mouse models (Ford-Hutchinson et al., 2003; Brewer et al., 2004). In the present study, we defined new anatomical landmarks for CNC-derived bones (premaxilla, maxilla, palatine bone, frontal bone, and mandible) that were specifically affected in our mouse models. For the purposes of this study, we excluded non-CNC-derived craniofacial bones such as the occipital and parietal bones. We used previously published landmarks as references to generate new landmark definitions that are necessary for future studies (Perrine et al., 2014; Percival et al., 2014). These new landmark definitions are not available in the literature. We provided these shorthand landmark definitions to simplify the context of the manuscript for the reader. All this information is available through the FaceBase Consortium (https://www.facebase.org/ or http://face.usc.edu/). Furthermore, we validated the TGFβ signaling mechanism during craniofacial bone formation using established mouse models with altered canonical and non-canonical TGFβ signaling. We analyzed 3D microCT images with anatomical landmarks on craniofacial bones from control, Tgfbr2fl/fl;Wnt1-Cre, and Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ mice. We found that canonical TGFβ signaling was specifically required for frontal bone and mandible formation in the proximal region, whereas activation of non-canonical TGFβ signaling adversely affected the growth of the premaxilla, maxilla, proximal region of the mandible, and palatine bone. Thus, canonical and non-canonical TGFβ signaling may act differentially and in concert to regulate craniofacial bone formation.
Materials and Methods
Animals
Wnt1-Cre mice were obtained from the Jackson laboratory and crossed with Tgfbr2fl/fl mice (a gift from Dr. Harold L. Moses, Vanderbilt University, Nashville, Tennessee, USA). To generate Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ mice, we mated Tgfbr2fl/+;Wnt1-Cre;Alk5fl/+ with Tgfbr2fl/fl;Alk5fl/fl mice. Alk5fl/fl mice were obtained from Dr. Vesa M. Kaartinen (University of Michigan, Ann Arbor, Michigan, USA). Genotyping was performed using PCR primers as described previously (Ito et al., 2003; Wang et al., 2006).
MicroCT Imaging and 3D Reconstruction
MicroCT analysis was performed using a SCANCO µCT50 (Scanco V1.28) at the University of Southern California Molecular Imaging Center. The microCT images were acquired from E14.5, E16.5, and E18.5 embryos with the x-ray source at 70 kVp and 250 µA. The data were collected at a resolution of 20 µm. Reconstruction in 3D was achieved using Avizo 7.1 (Visualization Sciences Group). MicroCT scans of five embryos per genotype per stage were studied. All landmarks were determined based on Mouse Development (Eds. J Rossant and P. L. Tam, 2002) and www.getahead.psu.edu.
All bones used in this study were manually segmented. MicroCT scans were uploaded to Avizo as Dicom files. The background noise from these segmentations and bones outside the scope of this study, such as the neck vertebrae, were manually removed using Avizo’s editor tools (Threshold, Contrast, Cropping). The remaining craniofacial bones were isolated and labeled using Avizo’s semiautomatic segmentation editor tool (Magic Wand) as well as manually using pre-scale thresholds that allowed only bones to be labeled (Paintbrush). Volumetric data was then rendered using Avizo’s algorithm tools and a voxel size of 0.01 was used in the calculations in order to produce standard data with the units in millimeters. The mean measurements of the bones were compared across the E18.5 control, mutant, and rescued models.
Statistical Analysis
Two-tailed Student’s t-tests were applied for statistical analysis. For all graphs, error bars represent standard deviations. A p value of less than 0.05 was considered statistically significant.
Results
Defects of Craniofacial Bone Formation in Tgfbr2fl/fl;Wnt1-Cre Mice
Mice with a deletion of Tgfbr2 in CNC cells (Tgfbr2fl/fl;Wnt1-Cre) exhibit craniofacial deformities such as calvarial defects, a small maxilla and mandible, and cleft palate (Ito et al., 2003; Iwata et al., 2010). To investigate bone formation in Tgfbr2fl/fl;Wnt1-Cre mice, we scanned E14.5, E16.5, and E18.5 Tgfbr2fl/fl;Wnt1-Cre and littermate control mice using microCT with 20 µm resolution and reconstructed the images in 3D (Fig. 1–Fig. 2). These embryonic stages were selected based on the timing of the initiation, expansion and maturation of craniofacial bones. Because only a small amount of bone formation was observed at E14.5, we did not include this time point in our data analysis. We performed most of our data analysis at E18.5 because it was the latest embryonic stage at which we could harvest the samples. We observed variations in development at post-natal day 0 (P0). Moreover, all Tgfbr2fl/fl;Wnt1-Cre mice died at P0. Therefore, we did not include P0 in this study. Overall, we found that the CNC–derived bones (premaxilla, maxilla, palatine bone, frontal bone, and mandible) were deformed and smaller than their counterparts in Tgfbr2fl/fl;Wnt1-Cre mice.
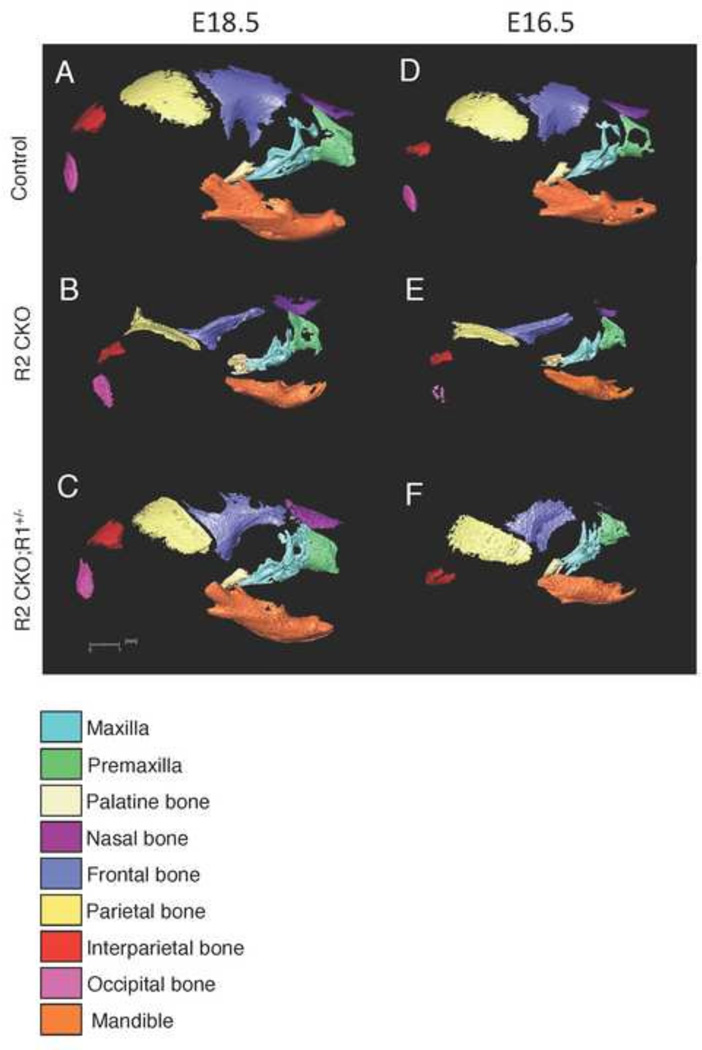
Figure 1. 3D microCT images of control, Tgfbr2fl/fl;Wnt1-Cre, and Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ mice at E16.5 and E18.5.
A–C: 3D reconstruction from microCT images of E18.5 control (Control; A), Tgfbr2fl/fl;Wnt1-Cre (R2 CKO; B), and Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ (R2 CKO;R1+/−; C) mice. D–F: 3D reconstruction from microCT images of E16.5 control (Control; D), Tgfbr2fl/fl;Wnt1-Cre (R2 CKO; E), and Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ (R2 CKO;R1+/−; F) mice. Colors identify the maxilla (turquoise), premaxilla (green), palatine bone (beige), nasal bone (purple), frontal bone (blue), parietal bone (yellow), interparietal bone (red), occipital bone (pink), and mandible (orange). Scale bar: 1 millimeter.
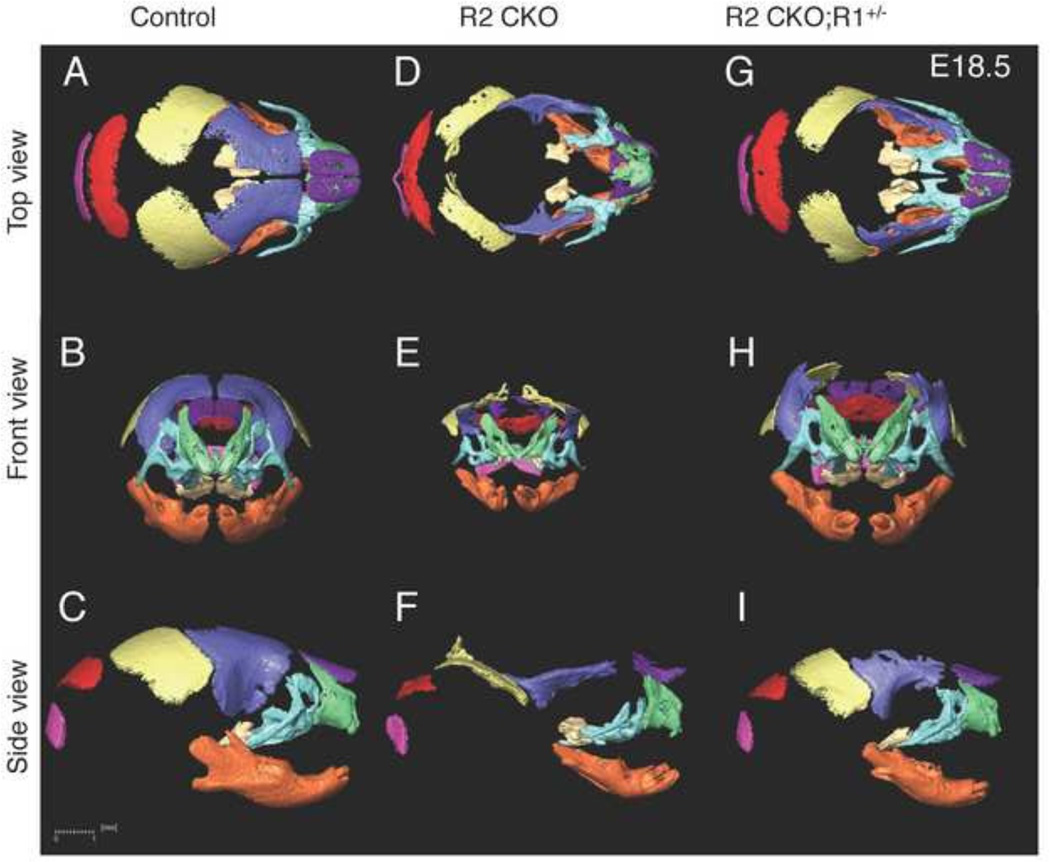
Figure 2. 3D microCT images of control, Tgfbr2fl/fl;Wnt1-Cre, and Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ mice.
A–I: 3D reconstruction from microCT images of E18.5 control (Control; A–C), Tgfbr2fl/fl;Wnt1-Cre (R2 CKO; D–F), and Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ (R2 CKO;R1+/−; GI) mice. (A, D, G) Top view; (B, E, H): front view; (C, F, I) top view. Scale bar: 1 millimeter.
We first defined anatomical landmarks on the premaxilla, maxilla, palatine bone, frontal bone and mandible in 3D microCT images of wild type control mice (Fig. 3–Fig. 7). We have recently reported that a haploinsufficiency of Tgfbr1/Alk5 (Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+) rescues cleft palate and largely restores other craniofacial deformities in Tgfbr2 mutant mice. A haploinsufficiency of Tgfbr1 disrupts ectopic non-canonical TGFβ signaling in Tgfbr2 mutant mice whereas canonical TGFβ signaling remains compromised (Iwata et al., 2012). Therefore, comparison of the size and shape of craniofacial bones across these mouse models allows us to identify the areas that are regulated by canonical and/or non-canonical TGFβ signaling during craniofacial development. To analyze the size and shape of the craniofacial bones, specifically the premaxilla, maxilla, palatine bone, frontal bone, and mandible, we highlighted and compared each bone from control, Tgfbr2fl/fl;Wnt1-Cre, and Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ mice (Fig. 2) and established new anatomical landmarks as well as measurement values for control samples.
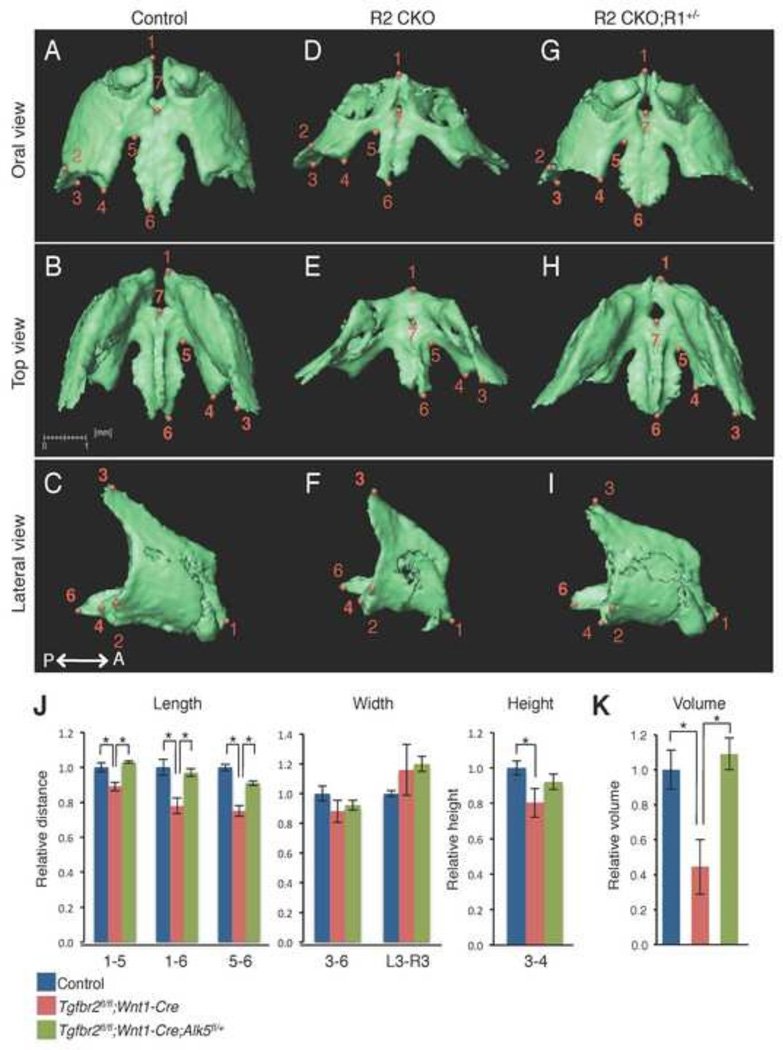
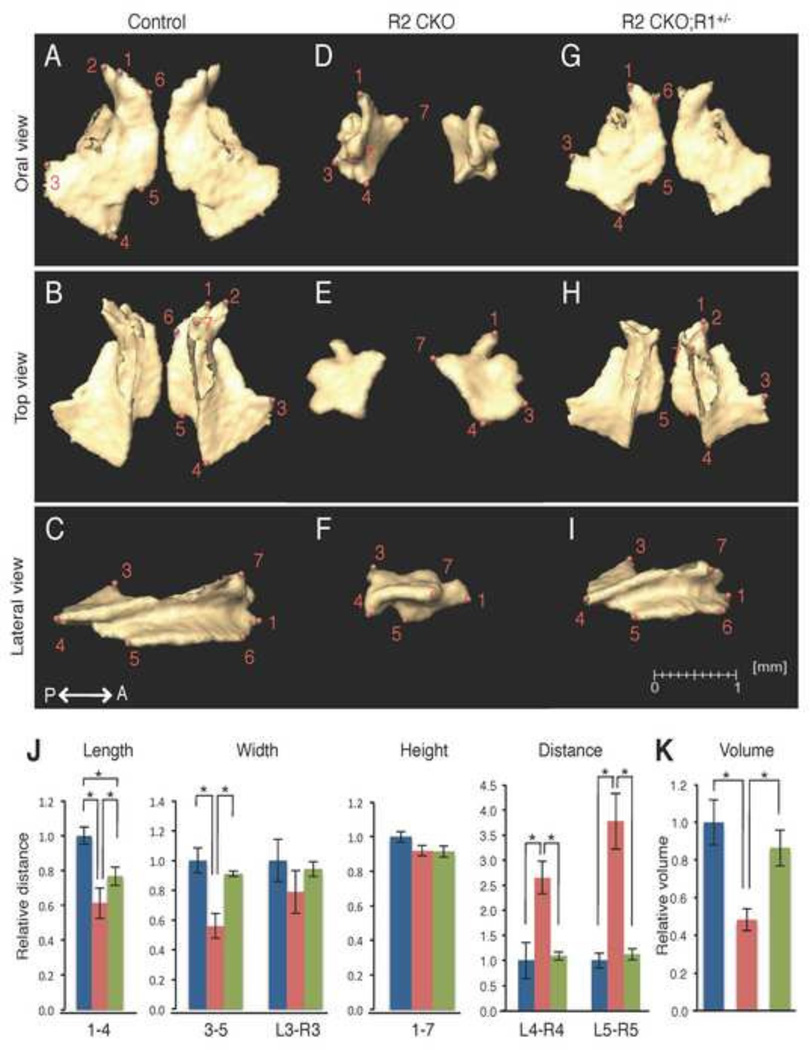
Figure 3. Comparison of the size and volume of the premaxilla at E18.5.
A–I: Isolated premaxilla from control (A–C), Tgfbr2fl/fl;Wnt1-Cre (R2 CKO; D–F), and Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ mice (R2 CKO;R1+/−;G–I) mice. P←→A: Posterior to Anterior J: Quantification of the size (length, width, and height) of the premaxilla from control (blue bars), Tgfbr2fl/fl;Wnt1-Cre (red bars), and Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ (green bars) mice. *p<0.01. K: Quantification of the volume of the premaxilla from control (blue bar), Tgfbr2fl/fl;Wnt1-Cre (red bar), and Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ mice (green bar) mice. *p<0.01. Definitions of landmarks: 1. Most anterior superior point of the premaxilla; 2. Most lateral point of the premaxillary-maxillary suture; 3. Tip of the frontal process; 4. Most medial point of the premaxillary-maxillary suture; 5. Most anterior point of the anterior palatine foramen; 6. Most posterior point of the premaxilla; 7. Most posterior point of the incisive foramen. Scale bar: 1 millimeter.
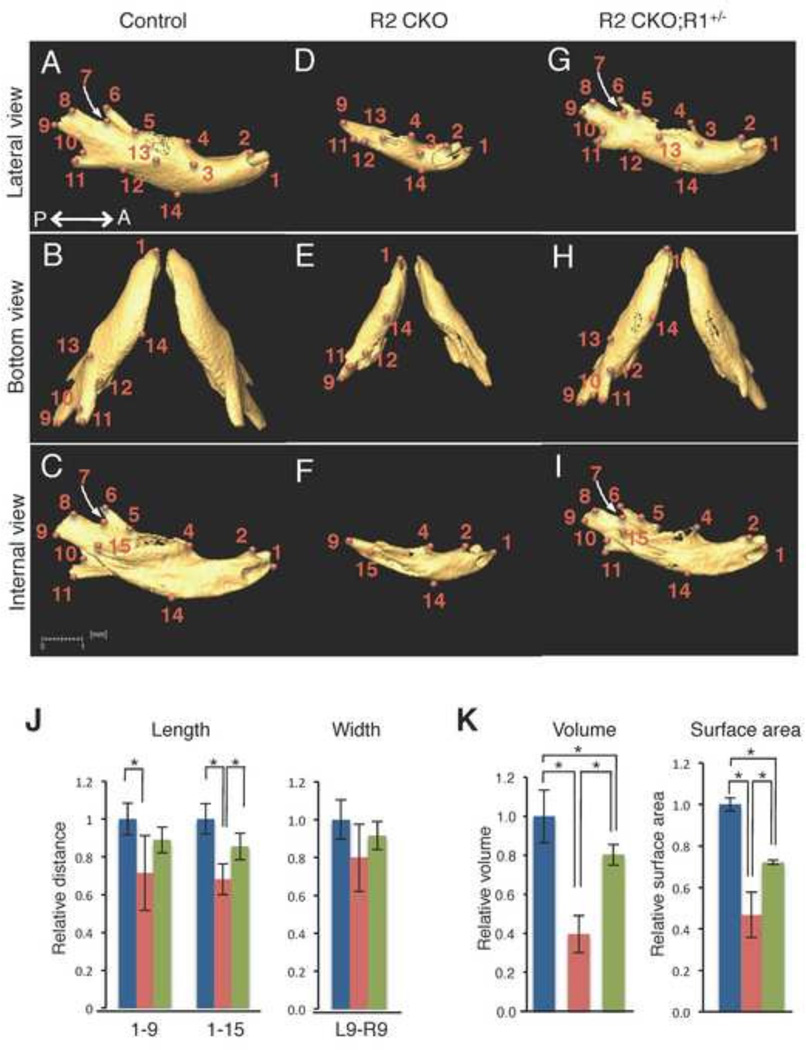
Figure 7. Comparison of the size and volume of the mandible at E18.5.
A–I: Isolated mandible from control (A–C), Tgfbr2fl/fl;Wnt1-Cre (R2 CKO; D–F), and Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ (R2 CKO;R1+/−; G–I) mice. P←→A: Posterior to Anterior J: Quantification of the size (length and width) of the mandible from control (blue bars), Tgfbr2fl/fl;Wnt1-Cre (red bars), and Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ (green bars) mice. *p<0.01. K: Quantification of the volume and surface area of the mandible from control (blue bars), Tgfbr2fl/fl;Wnt1-Cre (red bars), and Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ (green bars) mice. *p<0.01. Definitions of landmarks: 1. Most anterior point of the mandible; 2. Anterior-superior point of the mandible; 3. Mental foramen; 4. Molar alveolus of dentary; 5. Anterior junction of the mandibular ramus and body; 6. Superior tip of the coronary process; 7. Most inferior point of the mandibular notch; 8. Anterior point of the condylar process; 9. Posterior point of the condylar process; 10. Superior point of the angular process; 11. Secondary cartilage of the angular process; 12. Inferior junction of the mandibular ramus and body; 13. Midpoint of the external oblique ridge; 14. Inferior point of the mandibular body; 15. Mandibular foramen. Scale bar: 1 millimeter.
Comparison of Size and Shape of Craniofacial Bones
Premaxilla
We isolated the premaxilla from control, Tgfbr2fl/fl;Wnt1-Cre, and Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ mice and labeled the bones with defined anatomical landmarks (Fig. 3A–I). Next, we compared the sizes of the bones across groups using the landmarks as reference points (Fig 3J). With respect to the premaxilla, we found that the length and height were reduced in Tgfbr2fl/fl;Wnt1-Cre mice compared to controls. The shape of the premaxilla was also affected in Tgfbr2fl/fl;Wnt1-Cre mice, resulting from the smaller size of the body of the premaxilla (area of points 1–6) in Tgfbr2fl/fl;Wnt1-Cre mice compared to controls. The anterior portion of the premaxilla was fused (Fig. 3D, point 1) in Tgfbr2fl/fl;Wnt1-Cre mice and was not restored in Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ mice (Fig. 3G, point 1). The volume and the surface area were approximately 60% smaller in Tgfbr2fl/fl;Wnt1-Cre mice than in controls, whereas both volume and surface area were completely restored in Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ mice (Fig. 3K and Table 1).
Table 1.
Comparison of the volume of craniofacial bones of control, Tgfbr2f/f;Wnt1-Cre (R2 CKO), and Tgfbr2f/f;Wnt1-Cre;Alk5f/+ (R2 CKO;R1+/−) mice at E18.5.
| total mass volume | surface area | |||||
|---|---|---|---|---|---|---|
| control | R2 CKO | R2 CKO; R1+− |
control | R2 CKO | R2 CKO; R1+− |
|
| premaxilla | 100% | 46%* | 109% | 100% | 48%* | 103% |
| maxilla | 100% | 41%* | 85% | 100% | 38%* | 80% |
| palatine bone | 100% | 48%* | 87% | 100% | 42%* | 76% |
| frontal bone | 100% | 39%* | 69%* | 100% | 30%* | 54%* |
| mandible | 100% | 40%* | 80%* | 100% | 47%* | 72%* |
p< 0.01.
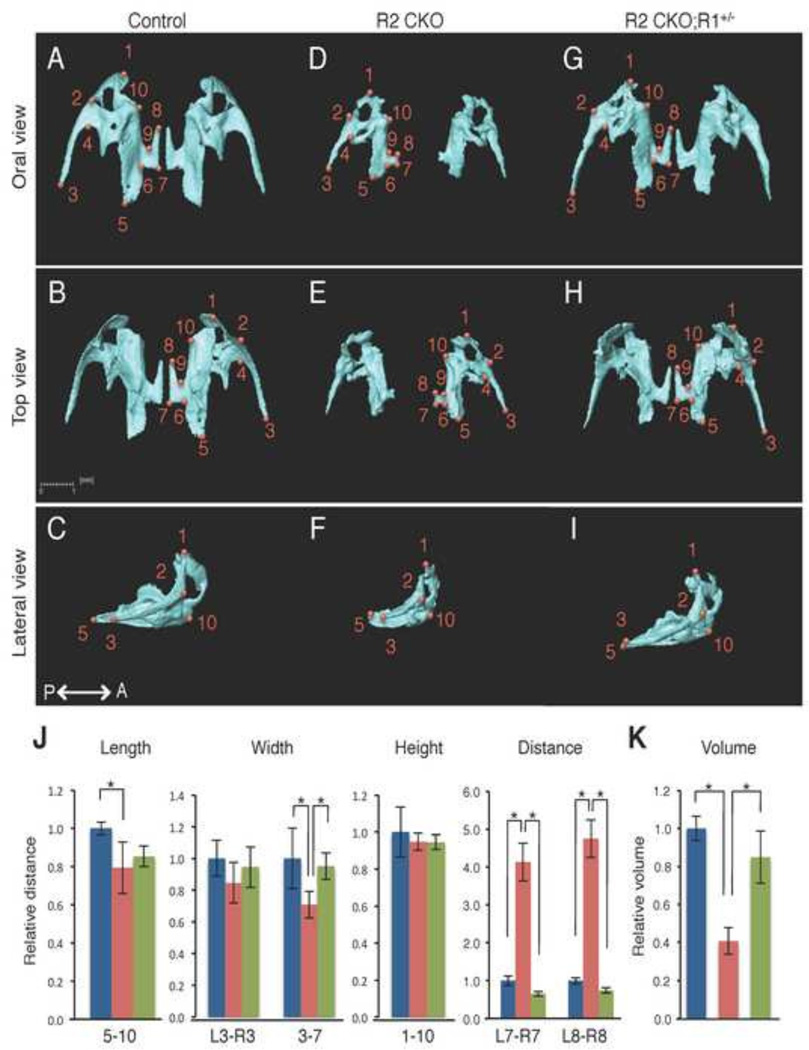
Maxilla
Similarly, we isolated the maxilla from control, Tgfbr2fl/fl;Wnt1-Cre, and Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ mice and defined the anatomical landmarks (Fig. 4A–I). Next, we compared their sizes using the landmarks (Fig 4J). The maxilla was severely affected in Tgfbr2fl/fl;Wnt1-Cre mice. The body of the maxilla (length 5–10 and width 3–7) was approximately 20% smaller than controls. The palatine process of the maxilla (area of points 6–9) was most severely deformed in Tgfbr2fl/fl;Wnt1-Cre mice. A gap between the palatine processes of the maxilla (distance L7-R7 and L8-R8), corresponding to cleft palate in Tgfbr2fl/fl;Wnt1-Cre mice, was completely restored in Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ mice. The shape of the maxilla was restored in Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ mice, whereas the volume and the surface area were reduced by approximately 40% in Tgfbr2fl/fl;Wnt1-Cre mice and only partially normalized (to 85% of the volume and 80% of the surface area of the controls) in Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ mice (Fig. 4K and Table 1).
Figure 4. Comparison of the size and volume of the maxilla at E18.5.
A–I: Isolated maxilla from control (A–C), Tgfbr2fl/fl;Wnt1-Cre (R2 CKO; D–F), Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ mice (R2 CKO;R1+/−; G–I) mice. P←→A: Posterior to Anterior J: Quantification of the size (length, width, height, and distance) of the maxilla from control (blue bars), Tgfbr2fl/fl;Wnt1-Cre (red bars), and Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ (green bars) mice. *p<0.01. K: Quantification of the volume of the maxilla from control (blue bar), Tgfbr2fl/fl;Wnt1-Cre (red bar), and Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ mice (green bar) mice. *p<0.01. Definitions of landmarks: 1. Anterior point of the maxilla; 2. Lateral inferior intersection of the frontal and zygomatic process; 3. Tip of the zygomatic process of maxilla; 4. Anterio-medial point to the zygomatic process; 5. Posterior point of the maxilla; 6. Posterior-lateral point of the palatine process of the maxilla; 7. Posterior-medial point of the palatine process of the maxilla; 8. Most anterior-medial point of the palatine process of the maxilla; 9. Anterior-lateral point of the palatine process of the maxilla; 10. Medial point of the premaxillary-maxillary suture. Scale bar: 1 millimeter.
Palatine bone
Next, we isolated the palatine bone from control, Tgfbr2fl/fl;Wnt1-Cre, and Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ mice and located the anatomical landmarks (Fig. 5A–I). We found that the orbital and perpendicular processes of the palatine bone (points 2 and 7, respectively) were compromised in Tgfbr2fl/fl;Wnt1-Cre mice. The horizontal plate (points 1, 5, and 6) was abnormal in Tgfbr2fl/fl;Wnt1-Cre mice (Figs. 5D and 5F). Next, we compared the sizes using the landmarks (Fig. 5J). The palatine bone was smaller than that of controls by approximately 40% in length (length 1–4), 40% in width (width 3–5), and 10% in height (height 1–7) in Tgfbr2fl/fl;Wnt1-Cre mice, and the shape was severely affected. A gap in the median palatine suture (distance L4-R4 and L5-R5) was normalized in Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ mice. The volume and the surface area were approximately 52% and 58% smaller, respectively, in Tgfbr2fl/fl;Wnt1-Cre mice than in controls and partially restored (to 87% and 76% of the control values, respectively) in Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ mice (Fig. 5K and Table 1).
Figure 5. Comparison of the size and volume of the palatine bone at E18.5.
A–I: Isolated palatine bone from control (A–C), Tgfbr2fl/fl;Wnt1-Cre (R2 CKO; D–F), Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ mice (R2 CKO;R1+/−; G–I) mice. P←→A: Posterior to Anterior J: Quantification of the size (length, width, height, and distance) of the palatine bone from control (blue bars), Tgfbr2fl/fl;Wnt1-Cre (red bars), and Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ (green bars) mice. *p<0.01. K: Quantification of the volume of the palatine bone from control (blue bar), Tgfbr2fl/fl;Wnt1-Cre (red bar), and Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ (green bar) mice. *p<0.01. Definitions of landmarks: 1. Most anterio-lateral point of the palatine plate; 2. Tip of the orbital process; 3. Lateral point of the palatine bone; 4. Posterior point of the palatine bone; 5. Posterior-medial point of the horizontal plate of the palatine bone; 6. Anterior-medial point of the horizontal plate of the palatine bone; 7. Anterior superior point of the perpendicular plate. Scale bar: 1 millimeter.
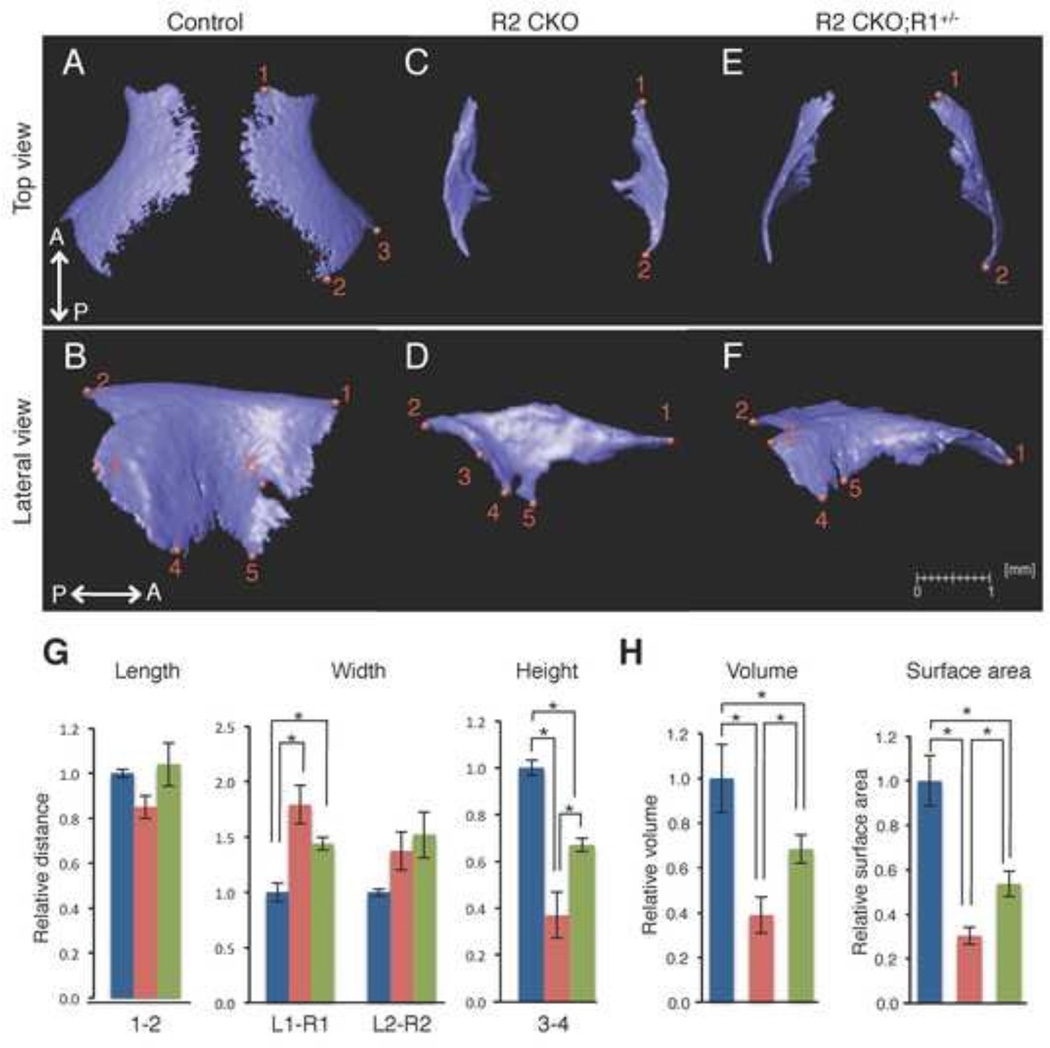
Frontal bone
We isolated the frontal bone from control, Tgfbr2fl/fl;Wnt1-Cre, and Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ mice and located the anatomical landmarks (Fig. 6A–F). We found that the orbital area (point 3–6) of the frontal bone was compromised in Tgfbr2fl/fl;Wnt1-Cre mice (Fig. 6D). Next, we compared the sizes using the landmarks (Fig 6G). The frontal bone formation was severely affected in the vertical dimension in Tgfbr2fl/fl;Wnt1-Cre mice (60% reduction in the distance between points 3–4). The shape of the frontal bone was also affected in Tgfbr2fl/fl;Wnt1-Cre mice, resulting from the altered distance of L1-R1. In Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ mice, the height of the frontal bone was not restored (height 3–4; 60% of the controls), and the orbital area was still compromised (missing point 6 in Fig. 6F), suggesting that frontal bone development is dependent on canonical TGFβ signaling. The length was slightly affected in Tgfbr2fl/fl;Wnt1-Cre mice. The volume and the surface area were reduced by approximately 60% and 70%, respectively, in Tgfbr2fl/fl;Wnt1-Cre mice. These were not restored (remained approximately 70% and 50% of the controls, respectively) in Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ mice (Fig. 6H and Table 1).
Figure 6. Comparison of the size and volume of the frontal bone at E18.5.
A–F: Isolated frontal bone from control (A and B), Tgfbr2fl/fl;Wnt1-Cre (R2 CKO; C and D), and Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ (R2 CKO;R1+/−; E and F) mice. P←→A: Posterior to Anterior G: Quantification of the size (length, width, and height) of the frontal bone from control (blue bars), Tgfbr2fl/fl;Wnt1-Cre (red bars), and Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ (green bars) mice. *p<0.01. H: Quantification of the volume and surface area of the frontal bone from control (blue bars), Tgfbr2fl/fl;Wnt1-Cre (red bars), and Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ mice (green bars). *p<0.01. Definitions of landmarks: 1. Most anterior-superior point of the frontal bone; 2. Most posterior-superior point of the frontal bone; 3. Most posterior-lateral intersection of the frontal bone and parietal bone; 4. Most posterior-inferior point of the frontal bone; 5. Most anterior-inferior point of the frontal bone; 6. Most posterior point of the orbitocranial canal. Scale bar: 1 millimeter.
Mandible
We isolated the mandible from control, Tgfbr2fl/fl;Wnt1-Cre, and Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ mice and identified anatomical landmarks (Fig. 7A–I). We found that the length of the mandible was smaller in Tgfbr2fl/fl;Wnt1-Cre mice than in control mice (Fig. 7J). The reduction in the length was partially restored in Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ mice (to approximately 90% of the controls). We found that the proximal region of the mandible (the condylar, coronoid, and angular processes, corresponding to the area of points 5–12) was largely affected in Tgfbr2fl/fl;Wnt1-Cre mice, indicating that TGFβ signaling mainly contributes to the development of the proximal region of the mandible. Furthermore, the defects in this area were almost restored in Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ mice indicating that the activation of non-canonical TGFβ signaling is responsible for causing defects in the proximal region of the mandible. The volume and the surface area were reduced by approximately 60% and 50%, respectively, in Tgfbr2fl/fl;Wnt1-Cre mice and partially restored (to 80% and 72% of the controls) in Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ mice (Fig. 7K and Table 1).
In this study, we investigated the relative involvement of canonical and non-canonical TGFβ signaling during craniofacial bone formation. We found that canonical and non-canonical TGFβ signaling pathways regulate the growth of craniofacial bones to different degrees in different bones (Tables 1 and 2). For example, in Tgfbr2fl/fl;Wnt1-Cre mice, the palatine bone was smaller than the controls in length and width, whereas the frontal bone was affected in width and height (Fig. 3J, 4J, and 6G). A haploinsufficiency of Tgfbr1 restored bone formation to different degrees; however, the length of the palatine bone and the height of the frontal bone were not restored in Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ mice. In addition, the total mass, volume and surface area of each craniofacial bone were compared across models (Table 1). We found that all of the craniofacial bones were smaller by 52% to 61% in volume in Tgfbr2fl/fl;Wnt1-Cre mice than in controls. Most of the craniofacial bones, except the frontal bone, were restored to 80–90% of the corresponding control volume in Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ mice.
Table 2.
Measurements of selected landmarks of craniofacial bones of control, Tgfbr2f/f; Wnt1-Cre (R2 CKO), and Tgfbr2f/f;Wnt1-Cre;Alk5f/+ (R2 CKO;R1+/−) mice at E18.5.
| Point | Control (mm) |
R2 CKO (mm) |
R2 CKO; R1+/− (mm) |
Tgfβ signaling involved |
|
|---|---|---|---|---|---|
| Premaxilla | 1–5 | 0.94±0.05 | 0.84±0.05* | 0.98±0.02 | non-canonical |
| 1–6 | 1.84±0.09 | 1.44±0.09* | 1.79±0.05 | non-canonical | |
| 5–6 | 0.96±0.04 | 0.72±0.06* | 0.87±0.03 | non-canonical | |
| 3–6 | 1.93±0.10 | 1.70±0.15 | 1 78±0.08 | non-canonical | |
| L3-R3 | 2.14±0.04 | 2.48±0.34 | 257±0.11 | non-canonical | |
| 3–4 | 1.74±0.08 | 1.40±0.16* | 1.61±0.09 | non-canonical | |
| Maxilla | 5–10 | 2.63±0.03 | 2.10±0.27* | 2.25±0.11 | non-canonical |
| L3-R3 | 5.43±0.11 | 4.60±0.23 | 5.13±0.26 | non-canonical | |
| 3–7 | 2.63±0.27 | 1.87±0.38* | 2.51±0.17 | non-canonical | |
| 1–10 | 1.84±0.10 | 1.52±0.01 | 1.52±0.06 | non-canonical | |
| Palatine | 1–4 | 1.84±0.10 | 1.13±0.17* | 1.42±0.10* | non-canonical |
| 3–5 | 1.22±0.04 | 0.68±0.17* | 1.11±0.04 | non-canonical | |
| L3-R3 | 2.54±0.10 | 2.00±0.29 | 2.40±0.01 | non-canonical | |
| 1–7 | 0.43±0.06 | 0.40±0.02 | 0.39±0.05 | non-canonical | |
| Frontal | 1–2 | 36.2±0.03 | 3.08±0.10 | 3.75±0.22 | canonical |
| L1-R1 | 1.68±0.18 | 3.01±0.35* | 2.42±0.13* | canonical | |
| L2-R2 | 2.70±0.16 | 3.71 ±0.35 | 4.10±0.47 | canonical | |
| 3–4 | 1.52±0.06 | 0.56±0.20* | 1.01±0.06* | non-canonical | |
| Mandible | 1–9 | 5.62±0.17 | 4.01 ±0.39* | 5.00±0.15 | non-canonical |
| 1–15 | 4.52±0.16 | 3.08±0.16* | 3.87±0.16 | non-canonical | |
| 9–15 | 1.01±0.06 | 0.98±0.11 | 1.10±0.06 | non-canonical | |
| L9-R9 | 5.52±.0.21 | 4.41 ±0.36 | 5.05±0.15 | non-canonical |
p< 0.01.
Discussion
The development of 3D microCT analysis has revolutionized our ability to visualize and analyze normal and abnormal anatomical structures. Craniofacial bones are unique in that most of them are irregularly shaped and connect with each other through sutures (with the exception of the temporomandibular joint). Because microCT allows craniofacial bones to be viewed and measured both individually and collectively, researchers can analyze how a genetic mutation affects an individual craniofacial bone and how this defect in turn affects the rest of the skull. In this study, we took a comprehensive approach by defining detailed new anatomical landmarks on selected craniofacial bones, which then allowed our analyses to reveal how TGFβ may exert its signaling specificity in regulating craniofacial development. Recently, with the support of the FaceBase Consortium, National Institute of Dental and Craniofacial Research (NIDCR) (Hochheiser et al., 2011), we have developed resources where users can perform interactive 3D analysis of mouse skulls in control and various mutant animals (https://www.facebase.org/ or http://face.usc.edu/). This tool allows users to perform virtual dissections and carry out specific measurements based on defined anatomical landmarks of each craniofacial bone in control and mutant samples, just as we presented in this study. This type of robust and quantitative analysis fills a void in craniofacial morphometric studies and will further illuminate the regulatory mechanisms that control craniofacial morphogenesis.
Previous studies have relied on whole mount skeletal staining analyses to evaluate normal and abnormal mouse skull development. Studies are now using different mouse models along with 3D microCT to analyze different stages of craniofacial bone development, for example, in Apert Syndrome mice (Perrine et al., 2014; Percival et al., 2014). However, the underlying mechanisms of craniofacial bone development remain to be elucidated. Because shape and size are critical factors in the classification of craniofacial birth defects, 3D microCT analyses are essential for the study of structural malformations in mice if they are to be used as models of human disorders (Rajion et al., 2006; Chan et al., 2007). The computational atlases generated by such analyses have many applications (Sharir et al., 2011; Wong et al., 2012). TGFβ signaling plays a critical role in craniofacial development as described in our previous studies. In this study, we defined new anatomical landmarks, performed comprehensive morphometric analysis, and analyzed 3D microCT images with the goal of more precise quantitative evaluation of the skull defects in TGFβ mutant mouse models.
Consistent with our previous findings based on skeletal staining, the reduction in the size of craniofacial bones in Tgfbr2fl/fl;Wnt1-Cre mice was partially restored in Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ mice. The size of the palatine bone and the formation of the palatine processes of the premaxilla and maxilla were almost completely restored in Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ mice, consistent with the rescued cleft palate. In contrast, the orbital area of the frontal bone was not restored in Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ mice. Thus, our data indicate that both canonical and non-canonical TGFβ signaling cascades are involved in regulating craniofacial bone formation. Significantly, differential TGFβ signaling mechanisms control the formation of different craniofacial bones.
The size and volume of the body, ramus, and coronoid process of the mandible, which are formed through intramembranous ossification, were smaller in Tgfbr2fl/fl;Wnt1-Cre mice than in controls, and partially restored in Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ mice. Similarly, the condylar and the angular processes of the mandible, which are formed through endochondral ossification, were diminished in Tgfbr2fl/fl;Wnt1-Cre mice and these areas were restored in Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ mice. Given that the condylar process is partially comprised of secondary cartilage and its formation is different than that of primary cartilage, our results suggest that TGFβ signaling plays a role in intramembranous as well as primary and secondary endochondral ossification.
We also observed the volume of the parietal bone was affected in Tgfbr2fl/fl;Wnt1-Cre mice (control: 0.843mm3±0.116; R2 CKO: 0.369mm3±0.079; p=0.0031) and was partially restored in Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ (control: 0.843mm3±0.116; R2 CKO;R1+/−: 0.639mm3; p=0.0395) mice even though the parietal bone is of mesodermal origin. Yoshida et al., (2008) demonstrated that the coronal suture (connecting the frontal and parietal bones) was the neural crest mesodermal boundary. Evidence from previous reports demonstrated that cranial sutures are major sites for craniofacial bone growth (Heuzé et al., 2014; Opperman, 2000). Therefore, we hypothesize that the changes to the parietal bone in Tgfbr2fl/fl;Wnt1-Cre embryos may be associated with the cranial sutures. Similarly, the volume of the interparietal bone was affected in Tgfbr2fl/fl;Wnt1-Cre mice (control: 0.330mm3±0.059; R2 CKO: 0.186mm3±0.059; p=0.0196) and was partially restored in Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ mice (control: 0.330mm3±0.059; R2 CKO;R1+/−: 0.288mm3±0.029; p=0.1736). We did not observe a significant change in volume in the occipital bone (control: 0.297mm3±0.069; R2 CKO: 0.236mm3±0.052; p=0.1432). Previous studies have shown that the developmental processes of craniofacial bones with different origin are interrelated (Perrine et al., 2014; Percival et al., 2014; Yoshida et al., 2008). Therefore, the effects on the parietal bone, interparietal bone and other non-CNC-derived craniofacial bones may be secondary. Further studies are needed to test these hypotheses.
Although 3D imaging data was initially applied to assist with clinical management of patients with craniofacial deformities, we envision that quantitative analysis of individual craniofacial bones will be informative for case analysis. Furthermore, this type of individual craniofacial bone analysis will provide more information regarding the defects associated with mutant animal models (Rajion et al., 2006), resulting in a better understanding of the mechanisms underlying craniofacial malformations. The ability to isolate each bone of the craniofacial complex makes it easier to visualize and evaluate phenotypic details. In this study, we establish new anatomical landmarks that can be easily identified in mouse 3D microCT images. We highlight the advantages of 3D microCT for evaluating the morphology of the mouse skull and show how morphogenetic analysis of prenatal craniofacial bones helps us elucidate the role of TGFβ signaling in regulating their development. Therefore, 3D microCT analysis can be added to the tool kit for studying craniofacial development. As we move into additional functional studies of craniofacial structures, these 3D data and other biochemical data will aid our study to gain a better understanding of the regulatory mechanism and integrated function of craniofacial bones.
Highlights.
Anatomical landmarks, size and volume established for craniofacial bones.
Differential regulatory mechanisms for specific sub-domains of craniofacial bones.
Loss of TGFβ signaling leads to compromised craniofacial bone formation.
This study serves as an important anatomical resource for developmental biologists.
ACKNOWLEDGMENTS
We are grateful to Dr. Bridget Samuels and Dr. Julie Mayo for critical reading of the manuscript, to Dr. Harold Moses for Tgfbr2fl/fl mice, and to Dr. Vesa M. Kaartinen for Alk5fl/fl mice. We thank the Molecular Imaging Center at USC for technical assistance. This study was funded by grants from the National Institute of Dental and Craniofacial Research, NIH (DE024421 and DE012711) to Yang Chai.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- Brewer S, Feng W, Huang J, Sullivan S, Williams T. Wnt1-Cre-mediated deletion of AP-2alpha causes multiple neural crest-related defects. Dev Biol. 2004;267:135–152. doi: 10.1016/j.ydbio.2003.10.039. [DOI] [PubMed] [Google Scholar]
- Chai Y, Ito Y, Han J. TGF-beta signaling and its functional significance in regulating the fate of cranial neural crest cells. Crit Rev Oral Biol Med. 2003;14:78–88. doi: 10.1177/154411130301400202. [DOI] [PubMed] [Google Scholar]
- Chai Y, Maxson RE., Jr Recent advances in craniofacial morphogenesis. Dev Dyn. 2006;235:2353–2375. doi: 10.1002/dvdy.20833. [DOI] [PubMed] [Google Scholar]
- Chan HJ, Woods M, Stella D. Three-dimensional computed craniofacial tomography (3D-CT): potential uses and limitations. Aust Orthod J. 2007;23:55–64. [PubMed] [Google Scholar]
- Ford-Hutchinson AF, Cooper DM, Hallgrimsson B, Jirik FR. Imaging skeletal pathology in mutant mice by microcomputed tomography. J Rheumatol. 2003;30:2659–2665. [PubMed] [Google Scholar]
- Heuzé Y, Singh N, Basilico C, Jabs EW, Homes G, Richtsmeier JT. Morphological comparison of the craniofacial phenotypes of mouse models expressing the Apert FGFR2 S252W mutation in neural crest- or mesoderm-derived tissues. Bone. 2014;63:101–109. doi: 10.1016/j.bone.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochheiser H, Aronow BJ, Artinger K, Beaty TH, Brinkley JF, Chai Y, Clouthier D, Cunningham ML, Dixon M, Donahue LR, Fraser SE, Iwata J, Marazita ML, Murray JC, Murray S, Postlethwait J, Potter S, Shapiro L, Spritz R, Visel A, Weinberg SM, Trainor PA. The Facebase Consortium: A comprehensive program to facilitate craniofacial research. Dev Biol. 2011;355:175–182. doi: 10.1016/j.ydbio.2011.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Yeo JY, Chytil A, Han J, Bringas P, Jr, Nakajima A, Shuler CF, Moses HL, Chai Y. Conditional inactivation of Tgfbr2 in cranial neural crest causes cleft palate and calvaria defects. Development. 2003;130:5269–5280. doi: 10.1242/dev.00708. [DOI] [PubMed] [Google Scholar]
- Iwata J, Suzuki A, Pelikan RC, Ho TV, Sanchez-Lara PA, Chai Y. Modulation of lipid metabolic defects rescues cleft palate in Tgfbr2 mutant mice. Hum Mol Genet. 2014;23(1):182–193. doi: 10.1093/hmg/ddt410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata J, Hacia JG, Suzuki A, Sanchez-Lara PA, Urata M, Chai Y. Modulation of noncanonical TGF-beta signaling prevents cleft palate in Tgfbr2 mutant mice. J Clin Invest. 2012;122:873–885. doi: 10.1172/JCI61498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata J, Hosokawa R, Sanchez-Lara PA, Urata M, Slavkin H, Chai Y. Transforming growth factor-beta regulates basal transcriptional regulatory machinery to control cell proliferation and differentiation in cranial neural crest-derived osteoprogenitor cells. J Biol Chem. 2010;285:4975–4982. doi: 10.1074/jbc.M109.035105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata J, Parada C, Chai Y. The mechanism of TGF-beta signaling during palate development. Oral Dis. 2011;17:733–744. doi: 10.1111/j.1601-0825.2011.01806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Iseki S, Maxson RE, Sucov HM, Morriss-Kay GM. Tissue origins and interactions in the mammalian skull vault. Dev Biol. 2002;241:106–116. doi: 10.1006/dbio.2001.0487. [DOI] [PubMed] [Google Scholar]
- Loeys BL, Chen J, Neptune ER, Judge DP, Podowski M, Holm T, Meyers J, Leitch CC, Katsanis N, Sharifi N, Xu FL, Myers LA, Spevak PJ, Cameron DE, De Backer J, Hellemans J, Chen Y, Davis EC, Webb CL, Kress W, Coucke P, Rifkin DB, De Paepe AM, Dietz HC. A syndrome of altered cardiovascular, craniofacial, neurocognitive and skeletal development caused by mutations in TGFBR1 or TGFBR2. Nat Genet. 2005;37:275–281. doi: 10.1038/ng1511. [DOI] [PubMed] [Google Scholar]
- Mizuguchi T, Collod-Beroud G, Akiyama T, Abifadel M, Harada N, Morisaki T, Allard D, Varret M, Claustres M, Morisaki H, Ihara M, Kinoshita A, Yoshiura K, Junien C, Kajii T, Jondeau G, Ohta T, Kishino T, Furukawa Y, Nakamura Y, Niikawa N, Boileau C, Matsumoto N. Heterozygous TGFBR2 mutations in Marfan syndrome. Nat Genet. 2004;36:855–860. doi: 10.1038/ng1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motch Perrine SM, Cole TM, 3rd, Martinez-Abadias N, Aldridge K, Jabs EW, Richtsmeier JT. Craniofacial divergence by distinct prenatal growth patterns in Fgfr2 mutant mice. BMC Dev Biol. 2014;14:8. doi: 10.1186/1471-213X-14-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opperman LA. Cranial sutures as intramembranous bone growth sites. Dev Dyn. 2000;219:472–485. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1073>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Percival CJ, Huang Y, Jabs EW, Li R, Richtsmeir JT. Embryonic craniofacial bone volume and bone mineral density in Fgfr2(+/P253R) and nonmutant mice. Dev Dyn. 2014;243(4) doi: 10.1002/dvdy.24095. 541+451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajion ZA, Townsend GC, Netherway DJ, Anderson PJ, Yusof A, Hughes T, Shuaib IL, Halim AS, Samsudin AR, David DJ. A three-dimensional computed tomographic analysis of the cervical spine in unoperated infants with cleft lip and palate. Cleft Palate Craniofac J. 2006;43:513–518. doi: 10.1597/05-023. [DOI] [PubMed] [Google Scholar]
- Ross S, Hill CS. How the Smads regulate transcription. Int J Biochem Cell Biol. 2008;40:383–408. doi: 10.1016/j.biocel.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Sharir A, Ramniceanu G, Brumfeld V. High resolution 3D imaging of ex-vivo biological samples by micro CT. J Vis Exp. 2011 doi: 10.3791/2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Nagy A, Larsson J, Dudas M, Sucov HM, Kaartinen V. Defective ALK5 signaling in the neural crest leads to increased postmigratory neural crest cell apoptosis and severe outflow tract defects. BMC Dev Biol. 2006;6:51–64. doi: 10.1186/1471-213X-6-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong MD, Dorr AE, Walls JR, Lerch JP, Henkelman RM. A novel 3D mouse embryo atlas based on micro-CT. Development. 2012;139:3248–3256. doi: 10.1242/dev.082016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Vivabutsiri P, Morriss-Kay G, Saga Y, Iseki S. Cell lineage in mammalian craniofacial mesenchyme. Mech Dev. 2008;125:797–808. doi: 10.1016/j.mod.2008.06.007. [DOI] [PubMed] [Google Scholar]