Abstract
In this study we used the yeast two-hybrid system to identify interactions between protein subunits of the virB type IV secretion system of Bartonella henselae. We report interactions between inner membrane and periplasmic proteins, the pilus polypeptide, and the core complex and a novel interaction between VirB3 and VirB5.
Gram-negative bacteria have adapted cell surface organelles for use in the delivery of macromolecules across kingdom boundaries by a cell contact-dependent mechanism. These organelles include the type III secretion systems, which have been extensively characterized in different bacterial systems (10, 20, 21) and are assembled from core components that are related to the flagellar machine (1, 22, 40). The type IV secretion system (TFSS) is ancestrally related to the core components of the conjugation machine (12, 13). The TFSS is able to transport diverse macromolecule substrates, including DNA and proteins (11, 18, 35, 55).
Historically, the TFSS of Agrobacterium tumefaciens, a phytopathogen, has been the reference TFSS (12). This system is assembled from products of the virB operon (virB1 to virB11), responsible for the delivery of the tumor-inducing DNA (T-DNA) to plant cells. Several human pathogens have also been shown to possess a TFSS, i.e., Brucella spp., Campylobacter jejuni, Coxiella burnetii, Legionella pneumophila, Rickettsia prowazekii, Helicobacter pylori, and Bordetella pertussis (2-4, 19, 34, 38, 39, 54). CagA (of H. pylori) and pertussis holotoxin (of B. pertussis) are known substrate effector proteins for these TFSSs.
More recently TFSS genes were found in several species of the genus Bartonella (46, 48). Bartonellae are fastidious, hemin-dependent, gram-negative bacteria. The genus Bartonella currently includes 19 species, seven of which are pathogenic for humans, with Bartonella henselae (the agent of cat scratch disease) and Bartonella quintana (urban trench fever) being the most prevalent (25, 33, 46, 48).
A common thread among Bartonella species is their hemotropic lifestyle in mammalian reservoir hosts and transmission by blood-sucking arthropods (17). The main diseases associated with B. henselae are cat scratch disease, which is most often seen in immunocompetent children, and bacillary angiomatosis, a vascular proliferative disease most commonly associated with long-standing human immunodeficiency virus infection or other significant immunosuppression (25). B. henselae has also been associated with bacillary peliosis, relapsing bacteremia, and endocarditis in humans.
The B. henselae TFSS was recently implicated in virulence (44, 46, 47). The TFSS is thought to be encoded by the vir operon, based on genetic homology with the well-characterized virB operon of A. tumefaciens. However, there are significant differences between the vir operon of B. henselae and that of A. tumefaciens. VirB1, a lytic transglycosylase, although not essential for the biogenesis of the TFSS, is absent from the vir operon of B. henselae.
To facilitate the transfer of effector proteins, the TFSS forms a stable conduit through the bacterial membranes to the host cell or external milieu. Protein-protein interactions are essential to the creation and stabilization of this TFSS structure. VirB2 is conserved among the TFSSs and is thought to be the main component of a hollow pilus structure involved in substrate transfer (28), believed to be energized by the VirB11 ATPase (9). VirB11, in association with VirB4, VirB6, VirB8, and VirB10, is assumed to form the base of the secretion apparatus, while VirB9 and VirB7 are thought to localize to the periplasm, connecting the base and the pilus structures (5, 15, 16, 24, 30, 50, 52).
In A. tumefaciens VirB3 is localized, in a VirB4 ATPase-dependent manner, to the outer bacterial membrane (29). It is suggested that VirB4 supplies energy for TFSS assembly or substrate transport (14); VirB4 itself is associated with the inner membrane, where it forms homodimers and interacts with VirB8 and VirB10 (52). VirB5 is an outer membrane protein involved in VirB2 pilus assembly (45). VirB6 is a highly hydrophobic subunit of the secretion system predicted to span the inner membrane multiple times, to stabilize VirB5, and to plays a role in VirB7 homo- and hetero- protein interactions (24, 30). VirB7, a periplasmic protein anchored to the outer membrane, interacts with VirB5 and, through intermolecular disulfide bonds, with VirB7 and VirB9 (30, 52). VirB9 (periplasmic) also interacts with VirB8 (inner membrane), VirB10 (inner membrane), and VirB11 (ATPase) (8, 16). VirB10 interacts with VirB8, as does VirB11. The ATPase activity of VirB11 is correlated with substrate transfer and gating molecules at the inner membrane.
The aim of this study was to determine interactions between the virB locus-encoded proteins VirB2 to VirB11 of B. henselae (Fig. 1). Week-old B. henselae colonies were harvested from Columbia agar plates, and genomic DNA was extracted by using a DNeasy kit from Qiagen. Accutaq LA DNA polymerase (Sigma) and standard thermal cycling protocols were used to amplify all of the virB locus genes. The plasmids and primer pairs used here are listed in Table 2 and 3, respectively.
FIG. 1.
Genetic organization of the TFSS of B. henselae.
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Reference or Source |
|---|---|---|
| pGBT9 | oriColE1 ori2μ TRP1 PADH::GAL4′ binding domain::MCS Apr | 6 |
| pGAD424 | oriColE1 ori2μ LEU1 PADH::GAL4′ activator domain::multiple cloning site Apr | 6 |
| pICC257 | pGBT9-virb2 | This study |
| pICC258 | pGBT9-virb3 | This study |
| pICC259 | pGBT9-virb4 | This study |
| pICC260 | pGBT9-virb5 | This study |
| pICC261 | pGBT9-virb6 | This study |
| pICC262 | pGBT9-virb7 | This study |
| pICC263 | pGBT9-virb8 | This study |
| pICC264 | pGBT9-virb9 | This study |
| pICC265 | pGBT9-virb10 | This study |
| pICC266 | pGBT9-virb11 | This study |
| pICC267 | pGAD424-virb2 | This study |
| pICC268 | pGAD424-virb3 | This study |
| pICC269 | pGAD424-virb4 | This study |
| pICC270 | pGAD424-virb5 | This study |
| pICC271 | pGAD424-virb6 | This study |
| pICC272 | pGAD424-virb7 | This study |
| pICC273 | pGAD424-virb8 | This study |
| pICC274 | pGAD424-virb9 | This study |
| pICC275 | pGAD424-virb10 | This study |
| pICC276 | pGAD424-virb11 | This study |
| pET28a | PT7 N-terminal His6 expression vector | Novagen |
| pMAL-c2X | Ptac malE fusion expression vector | New England Biolabs |
| pICC277 | pET28a-VirB3 | This study |
| pICC278 | pMal-c2X-VirB5 | This study |
TABLE 3.
Primer list
| Gene | Primer
|
|
|---|---|---|
| Forward | Reverse | |
| virB2 | GCG GAA TTC ATG ACA GAC ACT ATA TCC | GCG GTC GAC TTA TGA TGC ACT CG |
| virB3 | GCG GAA TTC ATG TTT GCC GGT GTT AC | GCG GTC GAC TCA CCT GTT TAG TTC C |
| virB4 | GCG GGA TCC AAA TGT CAA TGA TGA AAC GG | GCG GTC GAC TCA TTG ATT TTC TCT CC |
| virB5 | GCG GAA TTC ATG GCT GCC TAT ATT TC | GCG GTC GAC CTA AAG TCG GAC ATC |
| virB6 | GCG GAA TTC ATG TCC GAC TTT AG | GCG GTC GAC TTA AAA TCG ACC ACG |
| virB7 | GCG GGA TCC AAA TGG TGC ATT TTG TAA G | GCG GTC GAC TTA ATT TTC ACG CGC |
| virB8 | GCG GAA TTC ATG CAT GGC ATG CGA C | GCG GTC GAC TCA TTG TAT CAC CTC TGG |
| virB9 | GCG GAA TTC ATG ATG AGG ATT | GCG GTC GAC TCA TTC ATA ACC GTT TCC |
| virB10 | GCG GAA TTC ATG AAT GAT CCA ATG G | GCG GTC GAC TCA TTG CAA AAT TAC CGC |
| virB11 | GCG GAA TTC ATG AAC CAA AAC TTG C | GCG GTC GAC TTT AAT TCC CAC C |
Yeast two-hybrid system (YTHS) screening identifies multiple protein interactions.
The amplified genes virB2 to virB10 were cloned into EcoRI/SalI or BamHI/SalI sites of pGBT9 (“bait”) and pGAD424 (“prey”) yeast matchmaker vectors, respectively, producing fusions to the binding and activation domains of the yeast transcriptional activator GAL4. Both vectors carry an AmpR gene for selection in bacterial hosts; in addition, pGBT9 carries the yeast TRP1 marker gene and pGAD424 carries the yeast LEU2 marker gene. The constructs were introduced into Saccharomyces cerevisiae strain PJ69-4A (Table 1) by using the high-efficiency lithium acetate transformation procedure described by Geitz and Schiestl, and the plasmid-borne TRP1 and LEU2 genes were initially selected for (23). PJ69-4A contains three separate reporter genes (HIS3, ADE2, and lacZ), each under the independent control of three different GAL4 promoters (GAL1, GAL2, and GAL7, respectively), which provide a high level of sensitivity with respect to detecting weak interactions, coupled with a low background level of false positives.
TABLE 1.
Bacterial and yeast strains
| Strain | Relevant genotype | Reference |
|---|---|---|
| E. coli | ||
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15 Tn10(Tetr)] | Stratagene |
| BL21(DE3)/pLysS | F−ompT hsdSB (rB− mB−) gal dcm (DE3) pLysS (Cmr) | Novagen |
| S. cerevisiae PJ69-4A | MATatrpl-901 leu2-3112 ura3-52 his3-200 gal4Δ gal80Δ LYS2::GAL1-HIS3 GAL2-ADE2 met2::GAL7-lacZ | 27 |
The resulting transformants were replica plated onto 3-aminotriazole-containing medium to select for the HIS3 reporter and onto synthetic complete medium lacking Trp, Leu, and Ade to select for the ADE2 reporter. The function of the LacZ reporter was quantified in cell extracts by assaying for β-galactosidase activity with o-nitrophenyl-d-galactopyranoside as a substrate (37).
Of the cloned genes, virB8 was found to be self-activating in the bait plasmid (pGBT9) and therefore was excluded from the screen. We tested the remaining 90 possible combinations of the cloned genes for pairwise interactions. Protein interactions, indicated by growth on selective media, were further confirmed in the yeast by assessing relative expression from the lacZ reporter gene using semiquantitative β-galactosidase assays. Results were expressed as the increase (n-fold) in β-galactosidase activity compared to activity in strains harboring the bait or prey plasmids alone.
Seventeen pairs of interacting proteins were identified (Table 4). The yeast colonies were graded from strong to weak depending on whether yeast colonies were observed on selective media (positive YTH phenotype) at 2 to 3 days, at 4 to 7 days, or after 7 days (Table 4).
TABLE 4.
Yeast two-hybrid results
| Type of protein interaction | Bait | Prey | Growtha | β-Galactosidase units | Fold increase in β- galactosidase activityb |
|---|---|---|---|---|---|
| Periplasmic and outer membrane | VirB2 | VirB2 | +++ | 27.2 ± 3.1 | 102.2 |
| VirB5 | VirB2 | +++ | 10.4 ± 1.2 | 29 | |
| VirB3 | VirB5 | +++ | 110.7 ± 1.2 | 380.1 | |
| VirB7 | VirB5 | + | 3.8 ± 0.4 | 10.7 | |
| VirB7 | VirB7 | ++ | 26.8 ± 8.8 | 58.6 | |
| VirB7 | VirB9 | + | 3.4 ± 0.3 | 7.4 | |
| VirB9 | VirB7 | ++ | 37.4 ± 2.3 | 16.4 | |
| Inner membrane | VirB9 | VirB9 | ++ | 16.3 ± 3.6 | 28.6 |
| VirB9 | VirB10 | ++ | 29.4 ± 1.3 | 26.1 | |
| VirB9 | VirB8 | +++ | 14.3 ± 6.1 | 37.9 | |
| VirB9 | VirB11 | + | 8.2 ± 1.8 | 15 | |
| VirB10 | VirB8 | ++ | 17.8 ± 4.4 | 63.4 | |
| VirB10 | VirB9 | +++ | 30.8 ± 4.9 | 68.3 | |
| VirB11 | VirB11 | +++ | 41.1 ± 4.7 | 91.8 | |
| VirB10 | VirB4 | ++ | 19.6 ± 7.5 | 23.3 | |
| VirB4 | VirB10 | ++ | 43.4 ± 5.6 | 222.7 | |
| VirB4 | VirB4 | +++ | 75.5 ± 21.7 | 316.3 |
Yeast colones were observed at 2 to 3 days (+++), at 4 to 7 days (++), or after 7 days (+).
Increase over the β-galactosidase units of the strain expressing the bait alone together with those strains expressing the prey alone.
For clarity, we divided the interacting protein pairs into two groups, as follows.
(i) Periplasmic and outer membrane proteins.
VirB2, which in other TFSSs has been shown to be the major pilus component, expectedly exhibited homo- protein interactions (Table 4). As reported for other TFSSs, VirB2 (bait) of B. henselae also interacted with VirB5 (prey) (Table 4). Interestingly, VirB3 (bait), an outer membrane protein with unknown function, interacted strongly (based on the β-galactosidase activity) with VirB5 (prey) (Table 4). VirB5:VirB3 interaction in other TFSSs has not been reported. VirB5 also interacted with VirB7. The observed VirB7:VirB7 and VirB7:VirB9 interactions (Table 4) were also reported for A. tumefaciens.
(ii) Inner membrane proteins.
As a possible link across the periplasm we observed VirB9:VirB9 homo- protein interactions (Table 4). VirB9 (bait) also interacted with other core component proteins, namely, VirB8, VirB10, and VirB11. VirB10 (bait) interacted with VirB8, VirB9, and VirB4. VirB4 (bait) exhibited both homo- and hetero- protein interactions with VirB10. As expected, we observed VirB11:VirB11 homo- protein interaction. VirB4:VirB4 was the strongest protein interaction in this group, as determined by β-galactosidase activity (Table 4).
VirB3 binds VirB5 in column pulldown binding assays.
All the above interactions except VirB3:VirB5 were reported for other TFSSs. It was therefore important to confirm this novel interaction biochemically. In order to obtain purified recombinant polypeptides, a virB3 PCR product was cloned into pET28a for expression as a His-tagged protein, generating plasmid pICC277. A PCR product of virB5 was cloned into pMal (New England Biolabs) for expression as maltose binding protein (MBP) fusions, generating plasmid pICC278.
Overnight cultures of XL1-Blue(pICC278) or XL1-Blue(pMal) were diluted 1:100 in fresh L broth and grown with mild shaking at 37°C. Log-phase cultures (optical density at 600 nm = 0.6 to 0.8) were induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside), and the cultures were induced for a further 4 h. After centrifugation, bacterial pellets were resuspended in 15 ml of MBP column buffer (50 mM Tris, 0.2 M NaCl, 1 mM EDTA, 1 mM dithiothreitol), and cells were lysed by sonication. The cell lysates were clarified by centrifugation, 8 ml was loaded onto 2-ml MBP-amylose columns, and the columns were equilibrated by washing with MBP column buffer. For the production of a His-tagged protein, overnight cultures of BL21 were diluted 1:100 into 100 ml of L broth and grown with mild aeration to an optical density at 600 nm of 0.6. IPTG (1 mM) was added, and the cultures were incubated for a further 4 h. Bacteria were harvested by centrifugation, and the cell pellet was resuspended in 15 ml of MBP column buffer. After a single freeze-thaw step, cells were lysed through sonication, and the lysate was clarified by centrifugation. Consequently, the prepared amylose columns were overlaid with 8 ml of filtered crude lysate. After extensive washing (>20 packed column volumes) of the column with column buffer, the protein was eluted into 1-ml fractions by the addition of column buffer supplemented with 10 mM maltose. Fractions containing eluted protein were diluted with sodium dodecyl sulfate sample buffer and were subjected to separation on 12% polyacrylamide gels. The proteins were electrotransferred from the gels to nitrocellulose membranes and blocked for 1 h by the addition of phosphate-buffered saline-Tween 20 (0.05%) containing 5% (wt/vol) dried milk powder. Membranes were variously probed for 2 h to overnight with anti-MBP antibodies (New England Biolabs) or anti-His antibodies (Santa Cruz Biotechnology), all at a 1:1,000 dilution in PBS-Tween 20. After a standard washing step, blots were incubated with anti-rabbit immunoglobulin alkaline phosphatase-conjugated secondary antibody at a 1:5,000 dilution. Immunoblots were developed by the addition of one buffered tablet (Sigma) containing 0.15 mg of 5-bromo-4-chloro-3-indolyl phosphate and 0.30 mg of nitroblue tetrazolium liter−1 dissolved in 10 ml of distilled water.
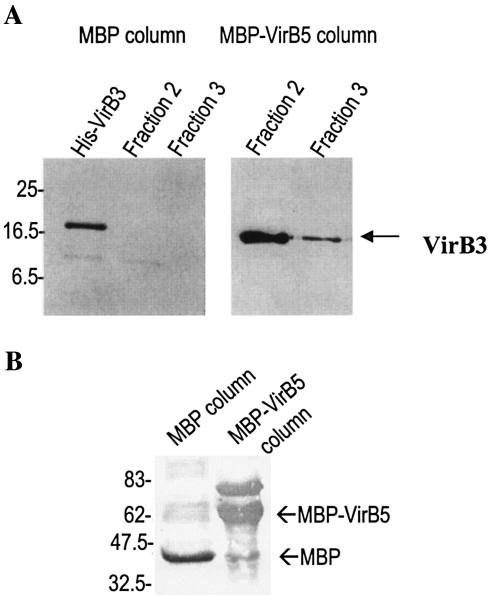
The binding assays revealed specific coelution of His-VirB3 with MBP-VirB5 (Fig. 2). No binding of His-VirB3 was detected to MBP only (Fig. 2A). These results confirm the novel VirB3:VirB5 protein interaction.
FIG. 2.
His-VirB3 copurifies with MBP-VirB5. MBP-VirB5 or MBP alone was immobilized on amylose columns and used to capture His-VirB3 from a crude lysate subsequently washed through the column. Fractions (1 ml) eluted from the column were analyzed by Western blotting for the presence of copurified His-VirB3 using anti-His antisera (A) or for MBP-VirB5 or MBP using anti-MBP antisera (B).
In this study we used the YTHS to determine pair protein interactions between structural components of the B. henselae secretion apparatus. This is the first study in which full-length protein subunits were successfully used to determine interactions between TFSS proteins of a human bacterial pathogen. We identified 17 protein interactions, which suggests a structural conduit between VirB11/VirB4 (two cytoplasm-facing ATPases) on one side of the cell wall and a pilus subunit on the other.
VirB4 was found to exhibit homo- protein interactions as well as forming heterodimers with VirB10. The steady-state levels of VirB4 have been suggested to be influenced by VirB7 (50). Accordingly, VirB4 could be affected through VirB7:VirB9, VirB9:VirB10, and VirB4:VirB10 protein interactions. Dang et al. and Jones et al. also suggested that VirB4 affects the membrane localization of VirB3 (14, 29). Since no VirB3:VirB4 protein interaction has been reported so far, membrane localization might be mediated via interactions with yet-unidentified proteins. Together with VirB11, VirB4 is implicated in supplying energy for assembly of the secretion apparatus and substrate transport (14). In this YTHS, VirB11 exhibited homo- protein interactions, which is in accordance with the finding for A. tumefaciens TFSS (40, 41). The VirB11-homologous protein in the H. pylori TFSS, HP0525, has shown to form a hexameric structure where each monomer consists of two domains formed by the N- and C-terminal halves of the protein. These domains form two rings which function as a chamber, open on one side and closed on the other. This hexametric structure has features reminiscent of eukaryotic p97 and NSF, which are involved in assembly and disassembly of the vesicle fusion apparatus (43, 53). A virB11 A. tumefaciens mutant accumulated wild-type levels of other VirB proteins but was impaired in pilus production and substrate export (42), indicative of a gating activity at the inner membrane.
VirB10 interacted with both VirB8 and VirB9, interactions that have also been described for A. tumefaciens (8, 31). TrwE, a VirB10-like protein, has been shown to be localized in the periplasm anchored to the inner membrane by its C terminus (36). The VirB10:VirB4 interaction could indicate that VirB4 is partly involved in the structure of the transport channel (52).
In our assay, VirB9 had a central position, since in addition to homo- protein interaction, it also interacted with VirB7, VirB8, VirB10, and VirB11. Our findings are in accordance with previous reports of A. tumefaciens VirB9 (5, 8, 16, 50, 52). Interaction between VirB9 and VirB7 was bidirectional, with the strongest interaction being between VirB9 prey and VirB7 bait. Until recently, VirB7 of B. henselae was thought to be a 15-kDa protein and together with a 17-kDa antigen was considered Bartonella specific and unrelated to the respective VirB5 and VirB7 counterparts encoded at the corresponding positions within the A. tumefaciens VirB T-DNA transfer locus. However, Schulein and Dehio recently renamed the genes virB5 and virB7, since a higher sequence similarity with their respective counterparts in the avh locus of A. tumefaciens was identified (48). Moreover, almost the same level of identity was observed between VirB5 and VirB7 of Sinorhizobium meliloti and their counterparts in Bartonella.
A novel finding in this screen was the interaction between VirB3 and VirB5, the strongest interaction of all 17 combinations, as measured by β-galactosidase assay. So far, VirB3 has been found to be localized mainly to outer membrane, although it was also detected, in small quantities, in the inner membrane (49). Accumulation of VirB3 in the outer membrane was dependent on VirB4 (29). This suggests that VirB3 could play an important role in correct transmission of energy for pilus assembly. Our findings strengthen this hypothesis and show that VirB3 is in close contact with the pilus via VirB5 and may play a role in stabilizing the pilus structure. However, as the sequence of the Bartonella VirB5 is distinct among its TFSS counterparts, it is impossible to extrapolate whether the VirB5:VirB3 protein interaction is specific to Bartonella or represents a general phenomenon. This would have to be determined experimentally.
VirB7 formed a homodimer and also interacted with VirB9, presumably via intermolecular disulfide bonds, as described for A. tumefaciens (50). Most recently, in an immunofluorescence microscopy experiment, it was shown that the filamentous structure of H. pylori TFSS apparatus is associated with proteins homologous to VirB7 or VirB9 (51). In a gel filtration assay, VirB7 and VirB2 of A. tumefaciens were found in the same fractions, suggesting an interaction between these two proteins (42). However, we could not detect VirB2:VirB7 protein interaction but observed interactions between VirB7 and VirB5, which might link the export channel to the pilus in B. henselae. VirB5 is the minor compartment of the pilus (45) and requires VirB6 for its stability (24).
We have shown that VirB2, the major component of the pilus (28, 32), could from homo- protein interactions. B. henselae VirB2 has homology with the conjugative pilin TraA on the Escherichia coli F plasmid (49). B. henselae expresses pilin-like structures when initially isolated from a clinical specimen (7). Direct correlation was found between pilus production and cell adhesion and invasion, suggesting that the pili are a major virulence factor.
We found no protein interactions between VirB6 and other VirB proteins. This finding was also reported by Ward et al. (52) and could be explained by the extreme hydrophobic nature of this polytopic inner membrane protein. However, more recently Jakubowski et al. (26) used immunoprecipitation and protein affinity tags to show that VirB6 is required for biogenesis of the A. tumefaciens T pilus and the secretion channel and interacts with both VirB7 and VirB9. Unfortunately, due to the fact that VirB8 as bait was self-activating in yeast, we had to omit it from the screen. Therefore, we could not rule out the possibility that we might have missed some VirB8 protein interactions. Indeed, previous reports demonstrated VirB8:VirB8, VirB8:VirB11, and VirB8:VirB4 interactions that were not detected in the present screen. However, VirB8 as prey interacted with both VirB9 and VirB10. In addition, using polypeptides rather than full-length proteins, Ward et al. (52) identified other interactions that were not observed in this study (i.e., VirB11:VirB4, VirB11:VirB8, and VirB11:VirB10). It is likely that the differences are due to incorrect folding in the yeast nucleus rather than being of biological significance.
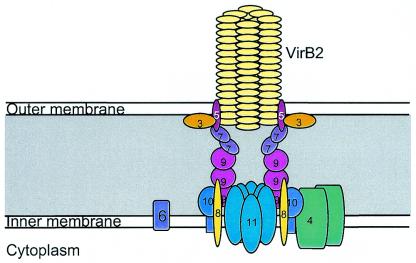
Based on the findings in this YTHS screen and those reported for others, we propose a new model of TFSS apparatus structure (Fig. 3), in which we assign a link between VirB3, VirB5, and the VirB2-associated putative pilus structure.
FIG. 3.
Model depicting the location of the VirB proteins in the structure of the B. henselae TFSS apparatus.
Acknowledgments
This study was supported by the Swedish Society for Medical Research and the Wenner-Gren Foundations.
REFERENCES
- 1.Aizawa, S. I. 2001. Bacterial flagella and type III secretion systems. FEMS Microbiol. Lett. 202:157-164. [DOI] [PubMed] [Google Scholar]
- 2.Andersson, S. G., A. Zomorodipour, J. O. Andersson, T. Sicheritz-Ponten, U. C. Alsmark, R. M. Podowski, A. K. Naslund, A. S. Eriksson, H. H. Winkler, and C. G. Kurland. 1998. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396:133-140. [DOI] [PubMed] [Google Scholar]
- 3.Bacon, D. J., R. A. Alm, D. H. Burr, L. Hu, D. J. Kopecko, C. P. Ewing, T. J. Trust, and P. Guerry. 2000. Involvement of a plasmid in virulence of Campylobacter jejuni 81-176. Infect. Immun. 68:4384-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baron, C., D. O'Callaghan, and E. Lanka. 2002. Bacterial secrets of secretion: EuroConference on the biology of type IV secretion processes. Mol. Microbiol. 43:1359-1365. [DOI] [PubMed] [Google Scholar]
- 5.Baron, C., Y. R. Thorstenson, and P. C. Zambryski. 1997. The lipoprotein VirB7 interacts with VirB9 in the membranes of Agrobacterium tumefaciens. J. Bacteriol. 179:1211-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartel, P., C. T. Chien, R. Sternglanz, and S. Fields. 1993. Elimination of false positives that arise in using the two-hybrid system. BioTechniques 14:920-924. [PubMed] [Google Scholar]
- 7.Batterman, H. J., J. A. Peek, J. S. Loutit, S. Falkow, and L. S. Tompkins. 1995. Bartonella henselae and Bartonella quintana adherence to and entry into cultured human epithelial cells. Infect. Immun. 63:4553-4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beaupre, C. E., J. Bohne, E. M. Dale, and A. N. Binns. 1997. Interactions between VirB9 and VirB10 membrane proteins involved in movement of DNA from Agrobacterium tumefaciens into plant cells. J. Bacteriol. 179:78-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berger, B. R., and P. J. Christie. 1994. Genetic complementation analysis of the Agrobacterium tumefaciens virB operon: virB2 through virB11 are essential virulence genes. J. Bacteriol. 176:3646-3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blocker, A., K. Komoriya, and S. Aizawa. 2003. Type III secretion systems and bacterial flagella: insights into their function from structural similarities. Proc. Natl. Acad. Sci. USA 100:3027-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burns, D. L. 2003. Type IV transporters of pathogenic bacteria. Curr. Opin. Microbiol. 6:29-34. [DOI] [PubMed] [Google Scholar]
- 12.Christie, P. J. 2001. Type IV secretion: intercellular transfer of macromolecules by systems ancestrally related to conjugation machines. Mol. Microbiol. 40:294-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christie, P. J., and J. P. Vogel. 2000. Bacterial type IV secretion: conjugation systems adapted to deliver effector molecules to host cells. Trends Microbiol. 8:354-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dang, T. A., X. R. Zhou, B. Graf, and P. J. Christie. 1999. Dimerization of the Agrobacterium tumefaciens VirB4 ATPase and the effect of ATP-binding cassette mutations on the assembly and function of the T-DNA transporter. Mol. Microbiol. 32:1239-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Das, A., L. B. Anderson, and Y. H. Xie. 1997. Delineation of the interaction domains of Agrobacterium tumefaciens VirB7 and VirB9 by use of the yeast two-hybrid assay. J. Bacteriol. 179:3404-3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Das, A., and Y. H. Xie. 2000. The Agrobacterium T-DNA transport pore proteins VirB8, VirB9, and VirB10 interact with one another. J. Bacteriol. 182:758-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dehio, C. 2001. Bartonella interactions with endothelial cells and erythrocytes. Trends Microbiol. 9:279-285. [DOI] [PubMed] [Google Scholar]
- 18.Deng, W., L. Chen, W. T. Peng, X. Liang, S. Sekiguchi, M. P. Gordon, L. Comai, and E. W. Nester. 1999. VirE1 is a specific molecular chaperone for the exported single-stranded-DNA-binding protein VirE2 in Agrobacterium. Mol. Microbiol. 31:1795-1807. [DOI] [PubMed] [Google Scholar]
- 19.Farizo, K. M., T. G. Cafarella, and D. L. Burns. 1996. Evidence for a ninth gene, ptlI, in the locus encoding the pertussis toxin secretion system of Bordetella pertussis and formation of a PtlI-PtlF complex. J. Biol. Chem. 271:31643-31649. [DOI] [PubMed] [Google Scholar]
- 20.Francis, M. S., H. Wolf-Watz, and A. Forsberg. 2002. Regulation of type III secretion systems. Curr. Opin. Microbiol. 5:166-172. [DOI] [PubMed] [Google Scholar]
- 21.Frankel, G., A. D. Phillips, I. Rosenshine, G. Dougan, J. B. Kaper, and S. Knutton. 1998. Enteropathogenic and enterohaemorrhagic Escherichia coli: more subversive elements. Mol. Microbiol. 30:911-921. [DOI] [PubMed] [Google Scholar]
- 22.Galan, J. E., and A. Collmer. 1999. Type III secretion machines: bacterial devices for protein delivery into host cells. Science 284:1322-1328. [DOI] [PubMed] [Google Scholar]
- 23.Gietz, R. D., and R. H. Schiestl. 1991. Applications of high efficiency lithium acetate transformation of intact yeast cells using single-stranded nucleic acids as carrier. Yeast 7:253-263. [DOI] [PubMed] [Google Scholar]
- 24.Hapfelmeier, S., N. Domke, P. C. Zambryski, and C. Baron. 2000. VirB6 is required for stabilization of VirB5 and VirB3 and formation of VirB7 homodimers in Agrobacterium tumefaciens. J. Bacteriol. 182:4505-4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacomo, V., P. J. Kelly, and D. Raoult. 2002. Natural history of Bartonella infections (an exception to Koch's postulate). Clin. Diagn. Lab. Immunol. 9:8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jakubowski, S. J., V. Krishnamoorthy, and P. J. Christie. 2003. Agrobacterium tumefaciens VirB6 protein participates in formation of VirB7 and VirB9 complexes required for type IV secretion. J. Bacteriol. 185:2867-2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones, A. L., E. M. Lai, K. Shirasu, and C. I. Kado. 1996. VirB2 is a processed pilin-like protein encoded by the Agrobacterium tumefaciens Ti plasmid. J. Bacteriol. 178:5706-5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones, A. L., K. Shirasu, and C. I. Kado. 1994. The product of the virB4 gene of Agrobacterium tumefaciens promotes accumulation of VirB3 protein. J. Bacteriol. 176:5255-5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krall, L., U. Wiedemann, G. Unsin, S. Weiss, N. Domke, and C. Baron. 2002. Detergent extraction identifies different VirB protein subassemblies of the type IV secretion machinery in the membranes of Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. USA 99:11405-11410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar, R. B., Y. H. Xie, and A. Das. 2000. Subcellular localization of the Agrobacterium tumefaciens T-DNA transport pore proteins: VirB8 is essential for the assembly of the transport pore. Mol. Microbiol. 36:608-617. [DOI] [PubMed] [Google Scholar]
- 32.Lai, E. M., and C. I. Kado. 1998. Processed VirB2 is the major subunit of the promiscuous pilus of Agrobacterium tumefaciens. J. Bacteriol. 180:2711-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.La Scola, B., Z. Zeaiter, A. Khamis, and D. Raoult. 2003. Gene-sequence-based criteria for species definition in bacteriology: the Bartonella paradigm. Trends Microbiol. 11:318-321. [DOI] [PubMed] [Google Scholar]
- 34.Liles, M. R., V. K. Viswanathan, and N. P. Cianciotto. 1998. Identification and temperature regulation of Legionella pneumophila genes involved in type IV pilus biogenesis and type II protein secretion. Infect. Immun. 66:1776-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Llosa, M., F. X. Gomis-Ruth, M. Coll, and F. F. de la Cruz. 2002. Bacterial conjugation: a two-step mechanism for DNA transport. Mol. Microbiol. 45:1-8. [DOI] [PubMed] [Google Scholar]
- 36.Llosa, M., S. Zunzunegui, and F. de la Cruz. 2003. Conjugative coupling proteins interact with cognate and heterologous VirB10-like proteins while exhibiting specificity for cognate relaxosomes. Proc. Natl. Acad. Sci. USA 100:10465-10470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 38.O'Callaghan, D., C. Cazevieille, A. Allardet-Servent, M. L. Boschiroli, G. Bourg, V. Foulongne, P. Frutos, Y. Kulakov, and M. Ramuz. 1999. A homologue of the Agrobacterium tumefaciens VirB and Bordetella pertussis Ptl type IV secretion systems is essential for intracellular survival of Brucella suis. Mol. Microbiol. 33:1210-1220. [DOI] [PubMed] [Google Scholar]
- 39.Odenbreit, S., J. Puls, B. Sedlmaier, E. Gerland, W. Fischer, and R. Haas. 2000. Translocation of Helicobacter pylori CagA into gastric epithelial cells by type IV secretion. Science 287:1497-1500. [DOI] [PubMed] [Google Scholar]
- 40.Plano, G. V., J. B. Day, and F. Ferracci. 2001. Type III export: new uses for an old pathway. Mol. Microbiol. 40:284-293. [DOI] [PubMed] [Google Scholar]
- 41.Rashkova, S., G. M. Spudich, and P. J. Christie. 1997. Characterization of membrane and protein interaction determinants of the Agrobacterium tumefaciens VirB11 ATPase. J. Bacteriol. 179:583-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sagulenko, E., V. Sagulenko, J. Chen, and P. J. Christie. 2001. Role of Agrobacterium VirB11 ATPase in T-pilus assembly and substrate selection. J. Bacteriol. 183:5813-5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Savvides, S. N., H. J. Yeo, M. R. Beck, F. Blaesing, R. Lurz, E. Lanka, R. Buhrdorf, W. Fischer, R. Haas, and G. Waksman. 2003. VirB11 ATPases are dynamic hexameric assemblies: new insights into bacterial type IV secretion. EMBO J. 22:1969-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmid, M. C., R. Schulein, M. Dehio, G. Denecker, I. Carena, and C. Dehio. 2004. The VirB type IV secretion system of Bartonella henselae mediates invasion, proinflammatory activation and antiapoptotic protection of endothelial cells. Mol. Microbiol. 52:81-92. [DOI] [PubMed] [Google Scholar]
- 45.Schmidt-Eisenlohr, H., N. Domke, C. Angerer, G. Wanner, P. C. Zambryski, and C. Baron. 1999. Vir proteins stabilize VirB5 and mediate its association with the T pilus of Agrobacterium tumefaciens. J. Bacteriol. 181:7485-7492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmiederer, M., and B. Anderson. 2000. Cloning, sequencing, and expression of three Bartonella henselae genes homologous to the Agrobacterium tumefaciens VirB region.DNA Cell Biol. 19:141-147. [DOI] [PubMed] [Google Scholar]
- 47.Schmiederer, M., R. Arcenas, R. Widen, N. Valkov, and B. Anderson. 2001. Intracellular induction of the Bartonella henselae virB operon by human endothelial cells. Infect. Immun. 69:6495-6502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schulein, R., and C. Dehio. 2002. The VirB/VirD4 type IV secretion system of Bartonella is essential for establishing intraerythrocytic infection. Mol. Microbiol. 46:1053-1067. [DOI] [PubMed] [Google Scholar]
- 49.Shirasu, K., and C. I. Kado. 1993. Membrane location of the Ti plasmid VirB proteins involved in the biosynthesis of a pilin-like conjugative structure on Agrobacterium tumefaciens. FEMS Microbiol. Lett. 111:287-294. [DOI] [PubMed] [Google Scholar]
- 50.Spudich, G. M., D. Fernandez, X. R. Zhou, and P. J. Christie. 1996. Intermolecular disulfide bonds stabilize VirB7 homodimers and VirB7/VirB9 heterodimers during biogenesis of the Agrobacterium tumefaciens T-complex transport apparatus. Proc. Natl. Acad. Sci. USA 93:7512-7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tanaka, J., T. Suzuki, H. Mimuro, and C. Sasakawa. 2003. Structural definition on the surface of Helicobacter pylori type IV secretion apparatus. Cell Microbiol. 5:395-404. [DOI] [PubMed] [Google Scholar]
- 52.Ward, D. V., O. Draper, J. R. Zupan, and P. C. Zambryski. 2002. Peptide linkage mapping of the Agrobacterium tumefaciens vir-encoded type IV secretion system reveals protein subassemblies. Proc. Natl. Acad. Sci. USA 99:11493-11500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yeo, H. J., S. N. Savvides, A. B. Herr, E. Lanka, and G. Waksman. 2000. Crystal structure of the hexameric traffic ATPase of the Helicobacter pylori type IV secretion system. Mol. Cell 6:1461-1472. [DOI] [PubMed] [Google Scholar]
- 54.Zamboni, D. S., S. McGrath, M. Rabinovitch, and C. R. Roy. 2003. Coxiella burnetii express type IV secretion system proteins that function similarly to components of the Legionella pneumophila Dot/Icm system. Mol. Microbiol. 49:965-976. [DOI] [PubMed] [Google Scholar]
- 55.Zupan, J. R., D. Ward, and P. Zambryski. 1998. Assembly of the VirB transport complex for DNA transfer from Agrobacterium tumefaciens to plant cells. Curr. Opin. Microbiol. 1:649-655. [DOI] [PubMed] [Google Scholar]