Abstract
A novel Gram-stain positive, non-motile, non-sporeforming coccus-shaped, obligately anaerobic bacterium was isolated from a fecal sample of an individual residing in a traditional Peruvian community. The organism was characterized using biochemical, chemotaxonomic and phylogenetic methods. Comparative 16S rRNA gene sequence analyses and phenotypic characteristics demonstrated that the organism was biochemically and phenotypically related, but distinct, from a group of organisms referred to as the Gram-stain positive anaerobic cocci (GPAC). The major cellular fatty acids of the novel isolate were determined to be C16:0 (18.3%), C18:1ε9c (39.8%), C18:2ε6,9c/C18:0 ANTE (13.2%). Fermentation end products from PYG are acetate and formate. Cell-wall peptidoglycan was found to be A4∋ (L-Lys-L-Ala-L-Glu) and the G + C content was determined to be 38.4 mol%. Based on the phenotypic, chemotaxonomic, and phylogenetic results, Ezakiella peruensis gen. nov., sp. nov., is now proposed. The type strain is M6.X2T (DSM 27367T = NBRC 109957 T = CCUG 64571T).
Keywords: Ezakiella peruensis gen. nov., sp. nov., 16S rRNA-phylogeny, taxonomy
1. Introduction
To date, the majority of studies on the human microbiome have focused on western populations, implementing the use of culture-independent methods[1-3]. These molecular inventories have provided tremendous insights into the microbial diversity and the richness of taxa present, suggesting that in the human gut, roughly 80% of the phylotypes represent uncultured bacteria[1]. In order to truly appreciate if there is a “core microbiome,” individuals from a variety of geographic regions with diverse diets must also be included in these investigations [4,5]. Initial studies are now revealing that the microbiome of indigenous communities may be significantly different than those derived from “westernized” communities [6,7]. There is now a renewed interest in culture-dependent approaches to recover organisms and characterize their physiological and metabolic properties in order to better understand the ecology of these microbial communities and the role they play in both health and disease processes [8-10].
The Gram-stain positive anaerobic cocci (GPAC) are part of the commensal flora of humans and animals and are also associated with a variety of human infections [11,12]. This group of organisms has undergone extensive taxonomic changes with many former members of the Peptostreptococcaceae being transferred to a number of novel genera that encompass Peptoniphilus, Anaerococcus, Finegoldia, Gallicola, and Parvimonas [12,13]. In addition, the genera Anaerosphaera, Helcococcus and Murdochiella have also been described and are phylogenetically related and phenotypically similar to the aforementioned genera [14-16]. Until recently the exact relationship of this group of organisms with other close close members of Firmicutes was somewhat uncertain, and was reflected in their placement in the Family XI Incertae Sedis (order Clostridiales, class Clostridia, phylum Firmicutes) in the current edition of Bergey's Manual of Systematic Bacteriology [17,18]. However, Johnson et al., (2014)[19] recently described the family Peptoniphilaceae to accommodate the genera Peptoniphilus, Anaerococcus, Anaerosphaera, Finegoldia, Gallicola, Helcococcus, Murdochiella, and Parvimonas.
A study focused on a Peruvian community to examine microbial diversity of geographically remote, traditional native communities, resulted in the isolation and characterized of a Gram-positive staining, obligately anaerobic, coccus-shaped organism recovered from a fecal sample. The organism displayed phenotypic traits consistent with the GPAC but phylogenetic analysis demonstrated that it represented a novel lineage within this group. Based on the results of a polyphasic taxonomic study, we describe and propose a novel genus and species for which the name Ezakiella peruensis gen. nov. sp. nov. is proposed. The type strain is M6.X2T (DSM 27367T = NBRC 109957T = CCUG 64571T).
2. Materials and methods
2.1. Cultures and cultivation
Strain M6.X2T (DSM 27367T = NBRC 109957T = CCUG 64571T) was isolated from a freshly voided fecal sample obtained from a member of the Afro-Peruvian community of Cruz Verde located in Ica, Peru. The sample was collected, processed anaerobically, and transported back to the laboratory on ice for further processing. Multiple enrichments using an array of substrates were constructed and inoculated with 1 ml of fecal slurry. Strain M6.X2T was isolated from an enrichment of Medium 2 [20] supplemented with xylan (per 100 mL distilled water): Casitone (1.0g), yeast extract (0.25g), minerals solution (A), minerals solution (B), clarified, sterile rumen fluid (20 mL), resazurin (0.0001g), sodium lactate (70% w/v) (1.0g), xylan (0.2g), cysteine HCl (0.05g), sodium bicarbonate (0.4g), distilled water (to 100 mL). Minerals solution (A) contains (per 1000 mL); K2HP04 (3.0g). Minerals solution (B) contains (per 100 mL); KH2P04 (3.0g), (NH4)2S04 (6.0g), NaCl (6.0g), MgS04.7H20 (0.6g), CaCl2 (0.6g). The enrichment was incubated for 7 days at 37°C with a gas mix of 5% Hydrogen, 10% Carbon Dioxide, and 85% Nitrogen. After incubation, a sample of the enrichment was inoculated onto Medium 2 agar and isolates were then sub-cultured onto BD Bacto™ Brain Heart Infusion (Sparks, MD, USA) agar plates supplemented with 5% defibrinated sheep’s blood until pure colonies were obtained.
2.2. Phenotypic and biochemical characterization
Unless indicated otherwise, all analyses were performed with cells grown on BD Bacto™ Brain Heart Infusion (Sparks, MD, USA) agar plates supplemented with 5% defibrinated sheep’s blood or in anoxic modified peptone-yeast extract (PY) broth medium (DSMZ Medium #104 with glucose excluded) at 37°C (pH 7.2). Cells were examined with an Olympus CX41 microscope using phase contrast at 1000X magnification. For biochemical characterization, API Rapid ID 32A and API 50 CH test systems (API bioMérieux, Marcy l’Etoile, France) were used following the manufacturer’s instructions except for the following modifications; both test systems were incubated anaerobically and cupules for the API 50 CH test strips were overlaid with mineral oil. Additional physiological characteristics were determined at varying temperature growth ranges (4-60 °C, in increments of ~5 °C) and pH values (5.0-9.5, in increments of 0.5 pH units). Salt tolerance was examined using different concentrations of NaCl (0-0.5 % (w/v), and 1-9% (w/v), in increments of 1%).
Optimum growth conditions were determined by monitoring the optical density using a spectrophotometer at 600 nm (Spectronic 20D, Milton Roy, DE). Growth resulting in an increase of OD600nm of >0.1 was considered to indicate growth. All tests were performed in duplicate.
Metabolic end products were determined from cultures grown under anaerobic conditions in PYG broth. Sample analyses were carried out in duplicate on an Aminex HPX-87H organic acid analysis column (Bio-Rad), using ion-exclusion HPLC with 0.015 HCL running buffer at a flow rate of 0.9 mL/min. Retention times and peak areas of fermentation products were compared to standards of acetate, butyrate, lactate, succinate, formate, and propionate.
2.3. DNA isolation and 16S rRNA gene sequencing and phylogenetic analysis
For phylogenetic analysis, DNA of strain M6.X2T was extracted using the UltraClean® Microbial DNA Isolation Kit (MoBio Laboratories, Inc.) following manufacturer’s instructions. 16S rRNA gene fragments were generated by PCR using universal primers pA (positions 8 to 28, Escherichia coli numbering) and pH* (1542 to 1522)[23]. The amplicon was purified using Exo-SapIt (USB Corporation) and the sequence determined using the Big Dye terminator cycle sequencing kit (ver. 3.1), with an automatic DNA sequencer (model 3100 Avant, Applied Biosystems). The closest known relatives of the new isolate based on the 16S rRNA gene sequence were determined by performing database searches using the program EzTaxon-e server (http://eztaxon-e.ezbiocloud.net/;[24]. These sequences and those of other related strains were aligned with the sequence derived from M6.X2T using the program ClustalW. Phylogenetic reconstructions were performed in MEGA (version 4) [25] using the neighbour-joining method [26], applying evolutionary genetic distances that had been calculated by the Kimura two-parameter model [27].
2.4. GenBank accession numbers
The 16S rRNA gene sequence for strain M6.X2T was deposited with the EBI Sequence Database under the following accession number KJ469554.
2.5. Chemotaxonomic methods
Biomass for fatty acid analysis was collected from a plate of BHI agar amended with 5% sheep’s blood after a 6 day incubation at 37°C. Analysis was performed at the Center for Microbial Identification and Taxonomy (University of Oklahoma, Norman, Oklahoma). Fatty acid methyl esters were extracted using the Sherlock Microbial Identification System (MIDI) version 6.1 as described previously [21,22]. Analysis was performed using an Agilent Technologies 6890N gas chromatograph equipped with a phenyl methyl silicone fused silica capillary column (HP-2 25m × 0.2 mm × 0.33 μm film thickness) and a flame ionization detector with hydrogen used as the carrier gas. The temperature program was initiated at 170°C and increased at 5 °C min−1 reaching a final temperature of 270°C. Fatty acids were identified and expressed in the form of percentages using the QBA1 peak naming database. Peptidoglycan analysis was performed using the method of Hamada et al., [28] at the Biological Resource Center, National Institute of Technology and Evaluation (NBRC), Japan. The mol% G+C was determined according to the method of Mesbah et al. [29] and was carried out by the Leibniz-Institut DSMZ, Germany).
3. Results and discussion
3.1. Phenotypic and biochemical characterization
M6.X2T is a non-motile Grain-stain positive, diplococci that is strictly anaerobic, non-spore forming and non-hemolytic. Growth on BHI blood agar plates after 6 days at 37°C colonies are small (<1mm in diameter), clear, circular, and convex. Indole is produced but catalase and urease are not while nitrate is not reduced. Using the API Rapid 32A test system, positive reactions were observed for alkaline phosphatase, arginine arylamidase, leucyl glycine arylamidase, phenylalanine arylamidase, leucine arylamidase, pyroglutamic acid arylamidase, tyrosine arylamidase, alanine arylamidase, glycine arylamidase, histidine arylamidase, and glutamyl glutamic acid. Negative reactions were obtained for arginine dihydrolase, proline arylamidase, D-galactosidase, β-galactosidase, β-galactosidase-6 phosphate, D-glucosidase, β-glucosidase, D-arabinosidase, β-glucuronidase, N-acetyl-β-glucosaminidase, glutamic acid decarboxylase, D-fucosidase, mannose, raffinose, serine arylamidase and urease. Using the API 50 CH test system, negative reactions were observed for glycerol, erythritol, D-arabinose, L-arabinose, D-ribose, D-xylose, L-xylose, D-adonitol, methyl-ßd-xylopyranoside, D-galactose, D-glucose, D-fructose, D-mannose, L-sorbose, L-rhamnose, dulcitol, inositol, D-mannitol, D-sorbitol, methyl-αd-mannopyranoside, methyl-αd-glucopyranoside, N-acetylglucosamine, amygdalin, arbutin, esculin, salicin, D-cellobiose, D-maltose, D-lactose, D-melibiose, D-sucrose, D-trehalose, inulin, D-melezitose, D-raffinose, starch, glycogen, xylitol, gentiobiose, D-turanose, D-lyxose, D-tagatose, D-fucose, L-fucose, D-arabitol, L-arabitol, potassium gluconate, potassium 2-ketogluconate, and potassium 5-ketogluconate. Metabolic end products from PYG were determined to be Acetate and Formate. Temperature range for growth is 30–37 °C with an optimum temperature of 37 °C. The pH range for growth is pH 7.0–8.5 with an optimum pH of pH 7.75. Growth occurs at NaCl concentrations of 0.5% (w/v) only.
3.2 Phylogenetic analysis
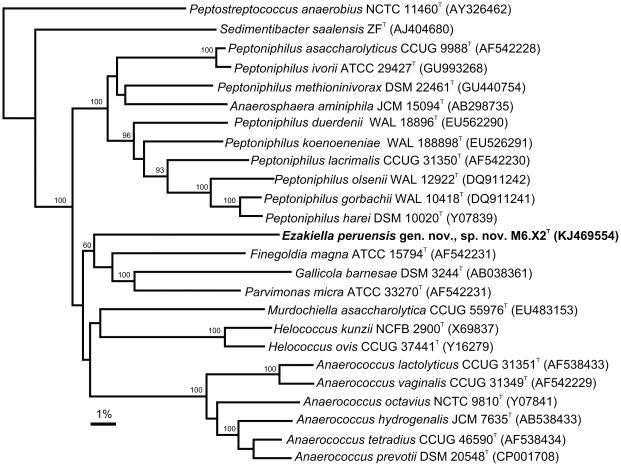
Phylogenetic analysis demonstrated that the organism was a member of the Firmicutes sharing a loose relationship with members of the GPAC (Fig. 1). These genera, Peptoniphilus, Anaerococcus, Anaerosphaera, Finegoldia, Gallicola, Helcococcus, Murdochiella and Parvimonas have recently been assigned to a newly described family Peptoniphilaceae [19]. The pairwise comparisons showed that strain M6.X2T formed a loose cluster with the genera Finegoldia (86.7% sequence similarity), Gallicola (83.5% sequence similarity) and Parvimonas (84.4% sequence similarity). All the major groupings in the neighbor-joining tree were confirmed using the maximum parsimony program (data not shown). In addition, database searches recovered only two closely related sequences (>99% sequence similarity) corresponding to uncultured organisms, GQ016861 and GQ016867. These sequences formed a tight cluster with strain M6.X2T but did not change the topology of the tree with respect to the other close relatives included in the analysis (data not shown). Although not recovered from fecal material they were however isolated from human skin[30].
Fig. 1.
Phylogenetic tree of 16S rRNA gene sequences indicating the position of Ezakiella peruensis gen. nov. sp. nov within members of related genera within the phylum Firmicutes. The tree was constructed using the neighbour-joining algorithm from MEGA (version 4) with Peptostreptococcus anaerobius as the outgroup. Bootstrap values (%) were obtained with 1000 replicates and are displayed on their relative branches.
3.3. Chemotaxonomic analysis
The fatty acid data of strain M6.X2T represented the following: major fatty acids were C16:0 (18.3%), C18:1ε9c (39.8%) and C18:2ε6,9C/C18:0 ANTE (13.2%). The minor fatty acids were C10:0 (7.3%), C13:0 ANTEISO (3.1%), C14:0 (3.0%), C17:1 ε8c (1.6%), C18:0/17:0 CYCLO (5.2%), C18:1 ε7c (4.2%), C15:0 (1.6%), C16:1 ε7c/C16:1 ε6c (1.4%), C17:0 (5.2%). The novel organism and its nearest relatives all produce C16:0, C18:1ε9c and C18:2ε6,9C/C18:0 ANTE as the major products. The peptidoglycan analysis revealed the presence of a A4α type with an interpeptide bridge comprising L-Lys-L-Ala-L-Glu. The DNA G+ C content of strain M6.X2T was determined to be 38.4 mol%.
The unidentified organism from human feces was found to possess biochemical and chemotaxonomic traits consistent with organisms belonging to the GPAC but could clearly be distinguished from its nearest phylogenetic relatives using characteristics shown in Tables 1 and 2. Although there is much discussion on the validity of describing novel genera and species on a single strain, this continues to be a common practice. We feel it is important to name this novel organism in order to allow members of the scientific community to identify additional strains or species of this novel genus. With the increasing number of cultured strains recovered from individuals from indigenous communities, a clearer picture of the ecology of these environments will emerge. Although originally recovered from an enrichments containing xylan, it was later determined that the isolate was not utilizing this substrate instead using components contained within the peptone and yeast extract. For instance, the novel strain M6.X2T appears to prefer proteinaceous materials such as amino acids or peptides that can be derived from either the diet or the host itself.
Table 1.
Fatty acid profiles of strain M6.X2T and close relatives.
| Fatty Acid a | M6.X2T |
Anaerococcus
prevotii CCUG 41932T |
Anaerosphaera
aminiphila JCM 15094T |
Finegoldia
magna CCUG 17636T |
Murdochiella
asaccharolytica CCUG 55976T |
Helcococcus
kunzii CCUG 32213T |
Parvimonas
micra CCUG 46357T |
Peptoniphilus
asaccharolyticus CCUG 9988T |
|---|---|---|---|---|---|---|---|---|
| C10:0 | 7.3 | 9.4 | ||||||
| C12:0 | 4.8 | 12.0 | ||||||
| anteiso-C13:0 | 3.1 | |||||||
| C14:0 | 3.0 | 2.0 | 2.0 | 5.9 | 11.0 | 2.5 | 1.6 | 5.4 |
| C15:0 | 1.6 | |||||||
| iso-C15:0 | 2.6 | |||||||
| C16:0 | 18.3 | 17.1 | 9.4 | 17.6 | 34.0 | 30.0 | 13.4 | 14.4 |
| C16:0 alde | 6.8 | 6.4 | ||||||
| C16:0 DMA | 6.5 | |||||||
| C16:1 ω7c/ C16:1ω6c |
1.4 | |||||||
| C16:1 ω7c | 2.0 | 2.8 | 2.0 | 3.9 | ||||
| C16:1 ω9c | 7.4 | |||||||
| C17:1ω8c | 1.6 | 14.3 | ||||||
| C17:1ω9c | 7.1 | |||||||
| iso-C17:1ω5c | 3.9 | 3.0 | ||||||
| anteiso-C17:0 | 1.7 | 4.5 | 1.6 | |||||
| C17:0 DMA | ||||||||
| iso-C17:1/C16:0
DMA |
18.2 | |||||||
| Cyclo C17:0/
C18:0 |
5.2 | |||||||
| C18:0 | 11.5 | 16.0 | 6.8 | 9.4 | ||||
| C18:0 ald | 4.4 | |||||||
| C18:0 DMA | 2.0 | |||||||
| C18:1 ω7c | 4.2 | 6.9 | ||||||
| C18:1 ω7 DMA | 12.2 | |||||||
| C18:1 ω9c | 39.8 | 19.3 | 2.0 | 3.6 | 54.0 | 19.3 | 15.5 | 20.2 |
| C18:2 ω6,9c/
C18:0 ANTE |
13.2 | 27.0 | 5.6 | 29.4 | 58.3 | 22.0 | ||
| C18:1 ω9c DMA |
6.4 | 11.1 | 6.6 | |||||
| iso-C19:1 | 2.0 | 1.5 | ||||||
| Unknown C18:177 |
6.9 | 13.1 | 5.1 | |||||
| C20:4ω6,9,12,15c | 1.4 |
Predominant products are shown in bold, values below 1% are not shown. Profiles for Gallicola have not been performed.
Table 2.
Morphological, biochemical and chemotaxonomic properties that are useful in the differentiation of strain M6.X2T and the type species of its close relatives.
| Characteristic | Ezakiella | Anaerococcus | Anaerosphaera | Finegoldia | Gallicola | Helcococcus | Murdochiella | Parvimonas | Peptoniphilus |
|---|---|---|---|---|---|---|---|---|---|
| Spore formation or thermotolerant cell |
− | − | + | − | − | − | − | − | − |
| Fermentation End Product (PYG) |
A, F | A, B L, | A, B | A | A, B | L, A | L | A | A, B |
| Metabolism | Obligate anaerobe |
Obligate anaerobe |
Obligate anaerobe |
Obligate anaerobe |
Obligate anaerobe |
Facultative anaerobe |
Obligate anaerobe | Obligate anaerobe |
Obligate anaerobe |
| Production of Indole |
+ | − | − | − | w | − | + | − | + |
|
Fermentation of: Mannose Raffinose |
− − |
− w |
− − |
− − |
− − |
+ + |
w w |
− − |
− − |
| Activity of: Alkaline phosphatase Arginine arylamidase Histine arylamidase Leucine arylamidase Proline arylamidase Phenylalanine arylamidase Pyroglutamic acid arylamidase |
+ + + + − + + |
− + + w − w + |
nd nd nd nd nd nd nd |
+ + w + − − + |
− − − − − − − |
− − − − − + − |
− + + + + + − |
+ + + + + + + |
− + + − − − − |
| API rapid 32A profile |
0000657707 | 5444017305 | nd | 2000456605 | nd | 0517004020 | 2006272705 | 2000477707 | 2000210001 |
| Cell-wall murein | Lys | Lys, D-Glu | Lys | Lys, D-Asp | D-Asp | D-Asp | ND | Lys | Orn, D-Glu |
| DNA G + C content (mol%) |
38.4 | 30-35 | 32.5 | 32-34 | 27-34 | ND | ND | 27-28 | 30-34 |
| Source | Human feces | Human intestine, clinical material, vaginal discharges, ovarian, peritoneal and other bodily abscesses, skin, and nasal passage |
Methanogenic cattle manure reactor |
Vagina, oral cavity and other bodily abscesses |
Chicken feces | Skin, human clinical material, sheep |
Human clinical material |
Oral cavity and human abscesses |
Human intestine and clinical material, vaginal discharges, ovarian, peritoneal and other bodily abscesses, cattle mastitis and swine manure |
Data: Ezakiella peruensis (M6.X2T), this study; Anaerococcus [31]; Anaerosphaera[14]; Finegoldia[32]; Gallicola[31]; Helcococcus[15]; Murdochiella[16]; Parvimonas[33]; Peptoniphilus[31]. Additional data obtained from de Vos et al., (2009) and the CCUG web site. (www.ccug.se)
+, positive; -, negative; nd, no data; A, acetate; B, Butyrate, F, Formate, L, Lactate; m-Dpm, meso-diaminopimelic acid
Based on the biochemical, chemotaxonomic and phylogenetic data presented, we consider the coccus-shaped organism from human feces represents a novel genus within the GPAC for which the name Ezakiella peruensis gen. nov., sp. nov. is proposed.
4. Description of Ezakiella gen. nov.
Ezakiella (E.za.ki.el’la N.L. fem. dim. Ezakiella named after the Japanese microbiologist Takayuki Ezaki who has contributed immensely to the taxonomy of the anaerobic Gram-stain positive cocci group of bacteria).
Cells are Gram-stain positive, non-motile, cocci that are strictly anaerobic. Catalase and urease negative. The predominant fatty acids are C16:0, C18:1 ε9c, C18:2 ε6,9C/18:0 ante. Cell-wall peptidoglycan is A4∋ (L-Lys-L-Ala-L-Glu). The DNA G+ C content of the type strain of the type species is 38.4 mol%. The genus Ezakiella is a member of the phylum Firmicutes within the family Peptoniphilaceae. The type species is Ezakiella peruensis.
Description of Ezakiella peruensis sp. nov.
Ezakiella peruensis (pe.ru.en'sis. N.L. fem. adj. peruensis pe.ru.en'sis N. L. gen. n. pertaining to the country of Peru from where the organism was first isolated). Displays the following properties in addition to those given in the genus description. After 6 days of growth on BHI Blood agar plates colonies are small (<1mm in diameter), clear, circular, and convex. Catalase and urease negative. Nitrate is not reduced and indole is produced. The API Rapid ID 32An test system, positive reactions are obtained for alkaline phosphatase, arginine arylamidase, leucyl glycine arylamidase, phenylalanine arylamidase, leucine arylamidase, pyroglutamic acid arylamidase, tyrosine arylamidase, alanine arylamidase, glycine arylamidase, histidine arylamidase, and glutamyl glutamic acid. Negative reactions are obtained for arginine dihydrolase, proline arylamidase, D-galactosidase, β-galactosidase, β-galactosidase-6 phosphate, D-glucosidase, β-glucosidase, D-arabinosidase, β-glucuronidase, N-acetyl-β-glucosaminidase, glutamic acid decarboxylase, D-fucosidase, mannose, raffinose, serine arylamidase and urease. Using the API 50 CH test system under anaerobic conditions no positive reactions are observed. Metabolic end products from PYG are acetate and formate.
The type strain M6.X2T (DSM 27367T = NBRC 109957T = CCUG 64571T) was isolated from a fecal sample of an individual from a traditional Peruvian community in the region Ica.
Highlights.
This work demonstrates the importance of cultivation studies to augment the large number of molecular-based studies on the human gut microbiome. The use of these methods will help further our understanding of this important ecosystem. Any “yet-to-becultivated” organisms remain to be identified and characterized.
Acknowledgements
This study was supported by the U.S. National Institutes of Health; RO1 GM089886. We would like to thank Universidad Cientifica del Sur, the personnel at the medical post in Tambo de Mora and the field team of the project Metagenomica e Inclusion Social for sample collection. We also thank Ken-ichiro Suzuki at Biological Resource Center, National Institute of Technology and Evaluation (NBRC) for facilitating the analysis of the peptidoglycan.
Abbreviations
- BD
Becton, Dickinson and Company
- CCUG
Culture Collection-University of Goteborg
- GPAC
Gram-Positive Anaerobic Cocci
- HMP
Human Microbiome Project
- NBRC
NITE Biological Resource Center
- PY
peptone-yeast extract
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The 16S rRNA sequences of strain M6.X2T (DSM 27367 T = NBRC 109957 T = CCUG 64571T) have been deposited in GenBank under accession number KJ469554.
References
- [1].Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett C, Knight R, Gordon JI. The human microbiome project: exploring the microbial part of ourselves in a changing world. Nature. 2007;449:804–10. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Gordon JI. Honor thy gut symbionts redux. Science. 2012;336:1251–3. doi: 10.1126/science.1224686. [DOI] [PubMed] [Google Scholar]
- [3].Zhou Y, Mihindukulasuriya KA, Gao H, La Rosa PS, Wylie KM, Martin JC, et al. Exploration of bacterial community classes in major human habitats. Genome Biol. 2014;15:R66. doi: 10.1186/gb-2014-15-5-r66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Lewis CM, Jr, Obregón-Tito A, Tito RY, Foster MW, Spicer PG. The Human Microbiome Project: lessons from human genomics. Trends in Microbiology. 2012;20:1–4. doi: 10.1016/j.tim.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hattori M, Taylor TD. The Human Intestinal Microbiome: A New Frontier of Human Biology. DNA Res. 2009;16:1–12. doi: 10.1093/dnares/dsn033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Contreras M, Costello EK, Hidalgo G, Magris M, Knight R, Dominguez-Bello MG. The bacterial microbiota in the oral mucosa of rural Amerindians. Microbiology. 2010;156:3282–7. doi: 10.1099/mic.0.043174-0. [DOI] [PubMed] [Google Scholar]
- [7].Blaser MJ, Dominguez-Bello MG, Contreras M. Distinct cutaneous bacterial assemblages in a sampling of South American Amerindians and US residents. Isme J. 2012 doi: 10.1038/ismej.2012.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lagier JC, Armougom F, Million M, Hugon P, Pagnier I, Robert C, et al. Microbial culturomics: paradigm shift in the human gut microbiome study. Clinical Microbiology and Infection. 2012;18:1185–93. doi: 10.1111/1469-0691.12023. [DOI] [PubMed] [Google Scholar]
- [9].Kaeberlein T, Lewis K, Epstein SS. Isolating “uncultivable” microorganisms in pure culture in a simulated natural environment. Science. 2002;296:1127–9. doi: 10.1126/science.1070633. [DOI] [PubMed] [Google Scholar]
- [10].Wylie KM, Truty RM, Sharpton TJ, Mihindukulasuriya KA, Zhou Y, Gao H, et al. Novel bacterial taxa in the human microbiome. PLoS ONE. 2012;7:e35294. doi: 10.1371/journal.pone.0035294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Murdoch DA. Gram-positive anaerobic cocci. Clinical Microbiology Reviews. 1998;11:81–120. doi: 10.1128/cmr.11.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ezaki T, Li N, Kawamura Y. The Anaerobic Gram-Positive Cocci. In: Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E, editors. The Anaerobic Gram-Positive Cocci. Vol. 4. Springer US; New York, NY: 2006. pp. 795–808. [Google Scholar]
- [13].Ezaki T, Kawamura Y, Li N, Li ZY, Zhao L, Shu S. Proposal of the genera Anaerococcus gen. nov., Peptoniphilus gen. nov. and Gallicola gen. nov. for members of the genus Peptostreptococcus. Int J Syst Evol Microbiol. 2001;51:1521–8. doi: 10.1099/00207713-51-4-1521. [DOI] [PubMed] [Google Scholar]
- [14].Ueki A, Abe K, Suzuki D, Kaku N, Watanabe K, Ueki K. Anaerosphaera aminiphila gen. nov., sp. nov., a glutamate-degrading, Gram-positive anaerobic coccus isolated from a methanogenic reactor treating cattle waste. Int J Syst Evol Microbiol. 2009;59:3161–7. doi: 10.1099/ijs.0.011858-0. [DOI] [PubMed] [Google Scholar]
- [15].Collins MD, Facklam RR, Rodrigues UM, Ruoff KL. Phylogenetic Analysis of Some Aerococcus-Like Organisms from Clinical Sources: Description of Helcococcus kunzii gen. nov., sp. nov. Int J Syst Evol Microbiol. 1993;43:425–9. doi: 10.1099/00207713-43-3-425. [DOI] [PubMed] [Google Scholar]
- [16].Ulger-Toprak N, Liu C, Summanen PH, Finegold SM. Murdochiella asaccharolytica gen. nov., sp. nov., a Gram-stain-positive, anaerobic coccus isolated from human wound specimens. Int J Syst Evol Microbiol. 2010;60:1013–6. doi: 10.1099/ijs.0.015909-0. [DOI] [PubMed] [Google Scholar]
- [17].Ludwig W, Schleifer K-H, Whitman WB. Revised road map to the phylum Firmicutes. In: de Vos P, Garrity GM, Jones D, Kreig NR, Rainey FA, Schleifer K-H, et al., editors. Bergeys Manual of Systematic Bacteriology. Vol. 3. The Firmicutes, Springer; 2009. pp. 1–13. [Google Scholar]
- [18].de Vos P, Garrity G, Jones D, Krieg NR, Rainey FA, Schleifer K-H, et al. Family XI. Incertae Sedis. In: de Vos P, Garrity GM, Jones D, Kreig NR, Rainey FA, Schleifer KH, et al., editors. Bergyey's Manual of Systematic Bacteriology. Vol. 3. The Firmicutes, Systematic Bacteriology; 2009. pp. 1130–9. [Google Scholar]
- [19].Johnson CN, Whitehead T, Cotta MA, Rhoades RE, Lawson P. Peptoniphilus stercorisuis sp. nov. from a swine manure storage tank and description of Peptoniphilaceae fam. nov. Int J Syst Evol Microbiol. 2014;64:3538–3545. doi: 10.1099/ijs.0.058941-0. [DOI] [PubMed] [Google Scholar]
- [20].Hobson PN. Rumen Bacteria. Methods in Microbiology. 1969;3:133–49. [Google Scholar]
- [21].Sasser M. Identification of Bacteria by Gas Chromatography of Cellular Fatty Acids:Technical Note #101. n.d.
- [22].Kämpfer P, Kroppenstedt RM. Numerical analysis of fatty acid patterns of coryneform bacteria and related taxa. 2011;42:989–1005. [Google Scholar]
- [23].Hutson RA, Thompson DE, Lawson PA, Schocken-Itturino RP, Böttger EC, Collins MD. Genetic interrelationships of proteolytic Clostridium botulinum types A, B, and F and other members of the Clostridium botulinum complex as revealed by small-subunit rRNA gene sequences. Antonie Van Leeuwenhoek. 1993;64:273–83. doi: 10.1007/BF00873087. [DOI] [PubMed] [Google Scholar]
- [24].Kim O-S, Cho Y-J, Lee K, Yoon S-H, Kim M, Na H, et al. Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. International Journal of Systematic Bacteriology. 2012;62:716–21. doi: 10.1099/ijs.0.038075-0. [DOI] [PubMed] [Google Scholar]
- [25].Tamura K, Peterson DA, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Molecular Biology and Evolution. 2011;28:2731–9. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].The neighbor-joining method: a new method for reconstructing phylogenetic trees. 1987;4:406–25. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- [27].Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–20. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- [28].Hamada M, Yamamura H, Komukai C, Tamura T, Suzuki K-I, Hayakawa M. Luteimicrobium album sp. nov., a novel actinobacterium isolated from a lichen collected in Japan, and emended description of the genus Luteimicrobium. The Journal of Antibiotics. 2012;65:427–31. doi: 10.1038/ja.2012.45. [DOI] [PubMed] [Google Scholar]
- [29].Mesbah M, Premachandran U, Whitman WB. Precise Measurement of the G+C Content of Deoxyribonucleic Acid by High-Performance Liquid Chromatography. Int J Syst Evol Microbiol. 1989;39:159–67. [Google Scholar]
- [30].Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, et al. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324:1190–2. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ezaki T, Kawamura Y, Li N, Li ZY, Zhao L, Shu S. Proposal of the genera Anaerococcus gen. nov., Peptoniphilus gen. nov. and Gallicola gen. nov. for members of the genus Peptostreptococcus. Int J Syst Evol Microbiol. 2001;51:1521–8. doi: 10.1099/00207713-51-4-1521. [DOI] [PubMed] [Google Scholar]
- [32].Murdoch DA, Shah HN. Reclassification of Peptostreptococcus magnus (Prevot 1933) Holdeman and Moore 1972 as Finegoldia magna comb. nov. and Peptostreptococcus micros (Prevot 1933) Smith 1957 as Micromonas micros comb. nov. 1999;5:555–9. [Google Scholar]
- [33].Tindall BJ, Euzeby JP. Proposal of Parvimonas gen. nov. and Quatrionicoccus gen. nov. as replacements for the illegitimate, prokaryotic, generic names Micromonas Murdoch and Shah 2000 and Quadricoccus Maszenan 2002, respectively. Int J Syst Evol Microbiol. 2006;56:2711–3. doi: 10.1099/ijs.0.64338-0. [DOI] [PubMed] [Google Scholar]