Abstract
DNA polymerase θ protects against genomic instability via an alternative end-joining repair pathway for DNA double-strand breaks. Breast, lung and oral cancers over-express polymerase θ, and reduction of its activity in mammalian cells increases sensitivity to double-strand break inducing agents, including ionizing radiation. Reported here are crystal structures of the C-terminal polymerase domain from human polymerase θ, illustrating two potential modes of dimerization. One structure depicts insertion of ddATP opposite an abasic site analog during translesion DNA synthesis. The second structure describes a cognate ddGTP complex. Polymerase θ employs a specialized thumb subdomain to establish unique upstream contacts to the primer DNA strand, including an interaction to the 3’-terminal phosphate from one of five distinctive insertion loops. These observations demonstrate how polymerase θ grasps the primer to bypass DNA lesions, or extend poorly annealed DNA termini to mediate end-joining.
Introduction
Human polymerase θ (pol θ) is a multi-domain protein of 2590 amino acids, with homologs throughout multicellular organisms1–3. The human POLQ gene encodes the pol θ protein, which conserves superfamily 2 helicase (SF2) and family A DNA polymerase domains at the N- and C-termini, respectively, linked by a large central region4. In vertebrates, pol θ and polymerase ν (pol ν)5 are the only family A polymerases present in the cell nucleus. The mitochondrial polymerase γ (pol γ) is distantly related to pol θ, and 5’-deoxyribose phosphate (5’-dRP) lyase activity has been attributed to both pol θ and pol γ6,7. Pol θ, however, does not synthesize DNA with the accuracy or processivity of the mitochondrial replicase8,9. Several conserved insertion loops, absent from bacterial homologs, intervene within the family A polymerase fold of pol θ. Deletion of residues 2264–2315, which include much of the penultimate insertion loop, was shown to abrogate pol θ’s ability to extend unannealed single-stranded oligonucleotides10, and bypass AP sites or thymine glycol11.
Early biochemical characterization of pol θ hinted at a role in translesion DNA synthesis (TLS), given pol θ’s lesion bypass activity12,13, and the enzyme indeed appears to be involved in TLS of oxidative DNA damage in vivo14. Assays utilizing chicken DT40 cells deduced overlapping roles for pol θ and polymerase β (pol β) in base excision repair (BER)15. Pol θ can also substitute for pol β during BER in C. elegans16. It has been suggested that pol θ is involved in somatic hypermutation of immunoglobulin genes, but any role appears minimal17. Pol θ does, however, participate in a subset of immunoglobulin gene class-switch recombination events in mouse B cells, dependent on an alternative (Ku-independent) DNA end-joining pathway18. Pol θ contributes to alternative end-joining of double-strand breaks (DSBs) in D. melanogaster19 and mammalian cells18. In C. elegans, pol θ limits extensive deletions at DNA replication fork barriers, but generates small indels, templated by DNA adjacent to the excision site20,21. Pol θ also produces templated indels in Drosophila22,23 and mice18. Furthermore, pol θ interacts with the origin recognition complex in human cells during G1, and may affect DNA replication timing24.
POLQ codes for the only nuclear DNA polymerase over-expressed in breast cancer, and higher expression correlates with unsuccessful treatment25,26. Elevated POLQ expression also occurs in oral squamous cell carcinomas27. Higher POLQ expression associates with disease free survival of lung cancers, however28. Knockout or knockdown of POLQ in mouse18,29 and human30 cells increases sensitivity to double-strand break inducing agents, including ionizing radiation. Complementation of this sensitivity phenotype requires the polymerase activity of pol θ, but not the ATPase activity of the helicase domain18. The presence of distinctive insertion loops in pol θ has encouraged speculation that these loops bestow unique properties to the enzyme. However, the molecular mechanisms that confer on pol θ an ability to prime DNA synthesis from non-optimal base pairing in the context of alternative end-joining have remained elusive until now. We set out to determine the crystal structures of two active constructs, comprising the entire C-terminal polymerase domain and its associated inactivated exonuclease-like subdomain, which are reported here. The structural work, in conjunction with a biochemical analysis of point variants, illuminates specific interactions from the specialized thumb subdomain of pol θ to the primer terminus that are necessary for TLS, and contribute to elongation of single-stranded DNA oligonucleotides.
Results
Human pol θ inserts ddATP opposite an abasic site
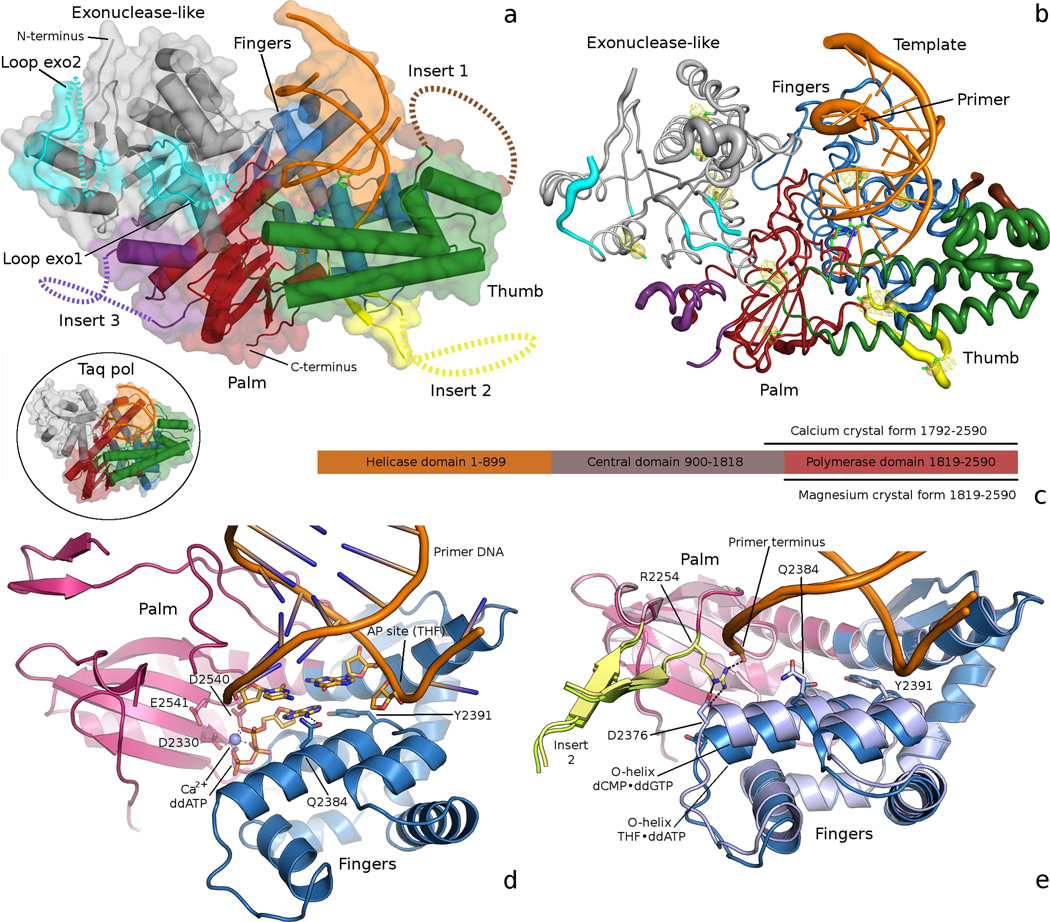
To identify structural components providing for the unique enzymatic activities of pol θ, we determined the crystal structures of two DNA polymerase domain constructs. Molecular replacement31, using a ternary complex of Taq DNA polymerase (1QSY)32 as the search model, sufficed to place the four similar molecules in the crystal asymmetric unit (ASU) of the THF–ddATP complex, which captured human pol θ inserting ddATP opposite tetrahydrofuran (THF), a stable abasic (AP) site analog. Refinement33 at 3.9 Å to an RFree of 30.2 % (RWork= 24.1 %; Table 1) revealed an overall fold reminiscent of bacterial homologs—with exonuclease, thumb and fingers subdomains oriented about a right hand palm subdomain (Supplementary Table 1)—but disrupted by five unique insertion loops (Fig. 1a–c and Supplementary Table 2). The THF–ddATP complex showed clearly the nascent base pair in the polymerase active site: The strictly conserved catalytic aspartate and glutamate residues (D2330, D2540, and E2541) of the palm subdomain coordinate a divalent Ca2+ ion, associated with the triphosphate tail of the ddATP nucleotide (Fig. 1d and Supplementary Fig. 1). As a known inhibitor of DNA polymerases, Ca2+ was essential for trapping the closed complex, because the primer strand retains a 3’-hydroxyl moiety for nucleophilic attack (Supplementary Fig. 2). The highly conserved lysine (K2383) and arginine (R2379) residues of the fingers subdomain O-helix contact non-bridging oxygens of the α and γ phosphates, respectively, as seen in other closed ternary complexes of family A polymerases (Supplementary Fig. 2)34. The conserved O-helix residue Y2391 was fully displaced from its template-occluding position in open or “ajar” structures35 (Fig. 1d and Supplementary Fig. 1), demonstrating that the pol θ structure represents the first fully closed model of any family A DNA polymerase inserting adenine opposite a non-templating DNA lesion. Previously, trapping ternary complexes destabilized by templating THF has required the purine analog 5-nitro-1-indolyl-2’-deoxyribose-5’-triphosphate (5-NITP), because of its enhanced capacity for base stacking36,37. The 5-NITP and ddATP double rings show substantial overlap when the current pol θ model is superimposed onto these previous structures38.
Table 1. Data collection and refinement statistics.
Data collection and refinement statistics (molecular replacement)
| Pol θ Ca2+ THF•ddATP |
Pol θ Mg2+ dCMP•ddGTP |
Pol θ SeMet Mg2+ (dCMP•ddGTP) |
|
|---|---|---|---|
| Data collection | |||
| Space group | P212121 | P21221 | P21221 |
| Cell dimensions | |||
| a, b, c (Å) | 126.9, 137.0, 248.0 | 100.7, 135.8, 161.9 | 100.4, 134.8, 166.5 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 50–3.9(4.0–3.9)* | 40–3.9(4.0–3.9) | 40–4.6(4.8–4.6) |
| Rmerge (%) | 20.5(100) | 19.6(72.2) | 15.5(31.5) |
| RFriedel† (%) | 8.4(79.2) | 9.0 (39.4) | 16.9(27.4) |
| I/σI | 16.7(2.1) | 12.5(1.9) | 8.7(4.3) |
| Completeness (%) | 99.7(99.1) | 85.6(64.0) | 78.1(53.1) |
| Redundancy | 16.6(14.0) | 22.6(13.4) | 4.8(3.7) |
| Refinement | |||
| Resolution (Å) | 30–3.91(3.96–3.91) | 30–3.90(3.96–3.90) | |
| No. reflections | 74685(2403) | 31372(1987) | |
| Rwork/Rfree (%) | 24.1(39.4)/30.2(43.9) | 25.8(35.0)/31.6(45.4) | |
| No. atoms | 22448 | 10902 | |
| Protein | 19900 | 9950 | |
| DNA | 2428 | 890 | |
| Ligand/ion | 120 | 62 | |
| B-factors (Å2) | |||
| Protein | 190 | 144 | |
| DNA | 215 | 166 | |
| Ligand/ion | 217 | 100 | |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.015 | 0.014 | |
| Bond angles (°) | 1.362 | 1.169 |
Values in parentheses are for highest-resolution shells. Pol θ Ca2+ and Mg2+ datasets are the product of merging 3 and 4 crystals, respectively. The pol θ Semet Mg2+ data set was collected on a single crystal.
RFriedel = Σ(|I+ − I−|)/Σ<I>
Fig. 1.
(a) The overall THF–ddATP pol θ structure is shown in comparison to Taq polymerase (circle insert; PDBID 1QSY32), identifying the 5 insertion loops. Loops exo1 and exo2 (cyan) extend from the β-sheet of the exonuclease-like subdomain. Insert 1 (brown) occupies the tip of the thumb subdomain. Insert 2 (yellow) joins the thumb subdomain, adjacent to the active site. Insert 3 (purple) of the palm subdomain lies in close proximately to the exonuclease-like subdomain. The dotted lines represent regions of the loops that were not seen in the electron density map and thus not built in the crystallographic model (b) A putty representation of pol θ, in the same orientation as panel a, displays the tube radius of the backbone trace proportionally to the refined atomic displacement parameters. Peaks from an NCS averaged anomalous difference electron density map (yellow mesh, contoured at 4 σ) pinpoint the location of methionines. (c) The full-length human pol θ domain architecture schematic describes the crystallization constructs, which encompass the entire C-terminal polymerase domain (residues 1819 to 2590) and vestigial exonuclease-like domain (residues 1819 to 2090). (d) A close-up view of the pol θ active site shows ddATP opposite THF in the closed conformation. Contacts (black dashes) are mediated from the O-helix residue Q2384 to the incoming nucleobase. (e) The THF–ddATP (dark pink, dark blue, and yellow-green) and dCMP–ddGTP (lighter hues) models appear superimposed, based on palm subdomain residues. Subtle rearrangements with cognate dCMP–ddGTP in the active site reposition the C-terminal end of the O-helix, forming a putative salt bridge from R2254 of insert 2 to D2376 of the fingers. All molecular illustrations were made with PyMOL (The PyMOL Molecular Graphics System, Version 1.5.0.4 Schrödinger, LLC).
Pol θ and pol ν both contain unique signature residues at mutable O-helix sites39, adjacent to the essential arginine and lysine side chains that contact the triphosphate tail and make pyrophosphate a better leaving group40. A glutamine residue occupies position 2384 of pol θ (Q2384), where pol ν conserves a lysine (K679) and bacterial polymerases generally harbor an alanine or threonine. Q2384 appeared in the vicinity of the Hoogsteen face of the incoming nucleotide, within hydrogen bonding distance to the exocyclic N6 of the ddATP molecule (Fig. 1d). Mutation of the analogous residue to alanine in pol ν (K679A) has reduced both bypass of Tg, and pol ν’s propensity for dTMP–dGTP mismatch formation39. An additional contact to the incoming nucleotide was established in pol θ between the β-phosphate and O-helix residue Y2387 (Supplementary Fig. 1), which is also conserved in DNA polymerases from T-odd bacteriophages and pol ν34,39.
The fingers subdomain closes on a cognate base pair
We pursued a related pol θ structure in the presence of Mg2+, the likely physiological divalent metal ion, and with a cognate dCMP–ddGTP base pair in the active site. This crystallization construct encompassed an additional 27 amino acid residue truncation from the N-terminus (Fig. 1c), and the DNA sequence was blunted to remove a 3’ template overhang. We also modified the 5’ sequence of the template to complement consecutive incoming ddGTP molecules, in order to promote enzymatic chain termination of the primer strand before trapping the closed ternary complex. These crystals diffracted to similar resolution as the Ca2+ crystals, although the diffraction data was deemed inferior, due to an off-origin native Patterson map peak and corresponding non-crystallographic symmetrical (NCS) translation, relating the two protein molecules of the ASU. Coupled with anisotropic diffraction, skewed intensity distribution statistics led to systematic data loss and reduced data completeness, even for highly redundant data sets compiled with extensive cross-crystal data merging (Table 1). A molecular replacement solution was readily obtained, however, based on the Ca2+ model, and refined to an RFree of 31.6 % (RWork= 25.8 %; Table 1). Subsequently, this approach yielded crystals (Table 1) of a selenomethionyl variant in the presence of Mg2+, from which a 4.6 Å anomalous difference Fourier map pinpointing the incorporated Se atoms was calculated. Superimposing these anomalous peaks onto the Ca2+ model added confidence to the overall trace of the model, and in assignment of side chains in poorly conserved regions, by anchoring the pol θ sequence (Fig. 1c). The Mg2+ complex appeared globally similar to the complex obtained with Ca2+. The former depicted subtle rearrangements in the fingers subdomain, adjusting the relative position of helix O to the primer–template and palm subdomain, as the O-helix appears to have moved closer to the cognate dCMP–ddGTP than the THF–ddATP pair (Fig. 1e).
Unique insertion loops decorate pol θ
The locations of the three loop elements previously described in the palm and thumb subdomains12 are now firmly established by the current pol θ structure, in addition to two previously unreported insertion elements present in the N-terminal vestigial exonuclease-like subdomain (Fig. 1a and Supplementary Data Table 2). Intrinsic flexibility prevented modeling any of the pol θ insertion loops in their entirety (Fig. 1b). Nevertheless, the molecular replacement solutions yielded electron density for the base of each loop, which served to orient them about the global polymerase fold and provided a partial description of their structures.
The tip of the thumb subdomain in pol θ resembles that of other homologous bacterial and phage polymerase structures, with two long helical segments separated by several flexible loops and smaller helices. Often in crystal structures of family A DNA polymerases, the tip of the thumb develops relatively high B-factors, due to the flexibility of these loops32,34, and pol θ provided no exception (Fig. 1b). Refinement produced electron density only for the first residues on either side of insert 1 (residues P2144-F2177), although the proximity of insert 1 to the DNA minor groove demonstrates potential for contacts to the DNA9. The location of insert 1 resembles that of both the thioredoxin-binding motif12 in T7 DNA polymerase34, and the binding site of the pol γ processivity factor8, which is consistent with the ascribed role of insert 1 in the processivity of pol θ11.
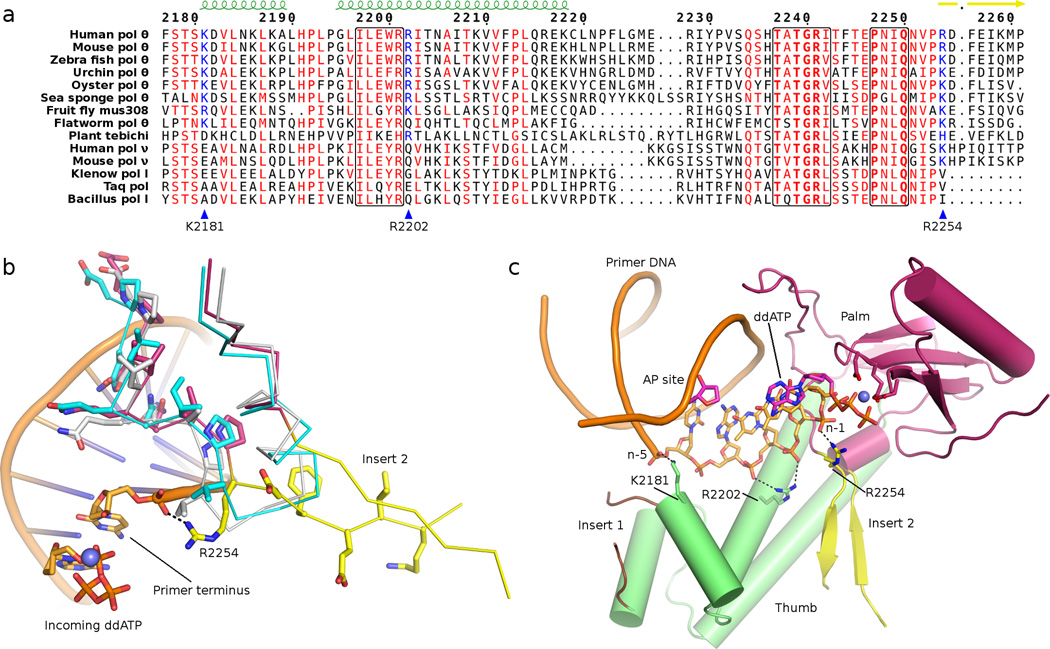
Insert 2 (residues R2254-S2313) departs the palm subdomain near the primer terminus binding site to join the thumb subdomain, where electron density for two β-strands was observed exiting the active site. These strands constitute the most highly conserved fragments of insertion loop 2 in pol θ and mus308-like proteins, from which a highly retained basic residue (R2254 in human pol θ; Fig. 2a) contacts the 3’-terminal phosphate of the primer DNA (Fig. 2b and Supplementary Fig. 2). A second contact of interest involving R2254 appeared only in the Mg2+ crystal form, where the O-helix has closed farther towards the active site. These subtle shifts reposition the acidic C-terminal end of the O-helix, from which a well conserved aspartate residue (D2376) engages in a salt bridge with R2254 of insertion loop 2 (Fig. 1e), pinning the guanidinium moiety between the terminal phosphate and O-helix. Unlike insert 3, which resides on the opposite site of the protein from the primer–template DNA, insert 2 is poised to align a poorly matched primer terminus for nucleophilic attack. Both inserts 2 and 3 have been shown necessary for TLS by pol θ, with insert 2 required for single-stranded oligonucleotide extension10,11.
Fig. 2.
(a) Structure-based sequence alignment illustrates the conservation of K2181, R2202, and R2254 (blue triangles) in multicellular organisms. Pol θ, mus308, and pol ν conserve an arginine or lysine at position 2254, whereas bacterial polymerases retain a hydrophobic side chain. Only pol θ-like enzymes (top nine proteins) conserve basic residues at positions 2181 and 2202. (b) Superposition of pol θ (warm pink), Taq (1QSY32, cyan), and Klenow (1KLN42, light grey) illustrates how insertion loop 2 (yellow) departs from the canonical family A polymerase fold. R2254 in human pol θ contacts the primer 3’-terminal (n-1) phosphate. (c) The palm (warm pink) and specialized thumb subdomain (green) of pol θ are illustrated to display unique contacts (black dashes) to the primer DNA (orange sticks). In addition to R2254 at the n-1 phosphate, R2202 contacts the n-2 and n-3 phosphates of the primer DNA. K2181 establishes a putative salt bridge to the n-5 phosphate, just after insertion loop 1 (brown).
The N-terminal exonuclease domains of family A DNA polymerases are generally divergent in sequence, despite conservation of the overall fold. A subset of these enzymes harbor 3’–5’ exonuclease activity in this domain, such as E. coli pol I, whereas this activity has been inactivated in other polymerases, such as Taq DNA polymerase. The crystallization construct used in this study includes approximately 300 residues upstream of the polymerase core, which is homologous to the E. coli pol I 3’–5’ exonuclease domain. The catalytic residues necessary for metal ion binding and primer degradation (D355, E357, D424 in E. coli pol I) are absent from the N-terminal subdomain of pol θ, and thus exonuclease activity has not been observed.
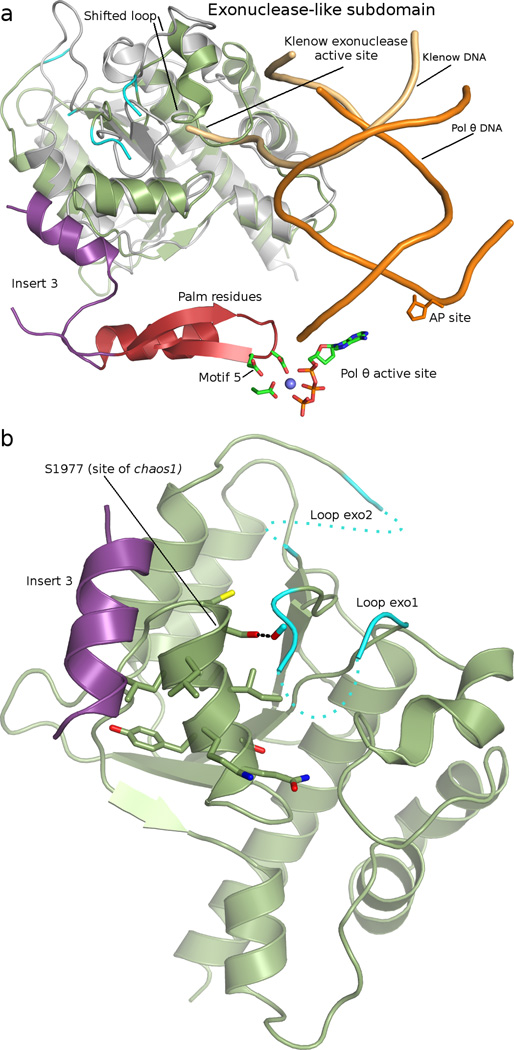
Density modification with cross-crystal NCS averaging provided detail to confidently place many side chains in regions of well-ordered secondary structure41, and this approach proved indispensable in modeling the N-terminal subdomain at 3.9 Å resolution, given the characteristic sequence divergence from available models. The current study identified two unreported insertions in the N-terminal subdomain of pol θ, in addition to the three inserts of the core polymerase subdomains. The second insert of the N-terminal subdomain (loop exo2; Q1918-D1936) extends an established surface loop, which is only 10 amino acids in bacterial pol I (386–396 in E. coli pol I). Residues K1858-L1899 form the first insertion (loop exo1), of which only 8 amino acids could be modeled. Loops exo1 and exo2 appear in close proximity, where together they extend from the conserved β-sheet of the N-terminal subdomain (Fig. 1a and Fig. 3a,b). In full-length pol θ, these insertion elements might provide contacts to the helicase domain or central region, considering their proximity to the extreme N-terminus of the polymerase domain crystallization construct.
Fig. 3.
(a) The N-terminal inactivated exonuclease-like subdomain of pol θ (smudge green) appears superimposed onto the 3’–5’ exonuclease domain of Klenow fragment (grey, 1KLN42). The Klenow structure shows the path of DNA into the exonuclease active site (tan), which is blocked by a shifted loop in the pol θ model. Insert 3 (purple) is resolved in close proximity to the exonuclease-like subdomain, and near loops exo1 and exo2 (cyan). Insert 3 is sequentially close to polymerase motif 5 of the palm (red), thus providing a structural linkage between the exonuclease-like subdomain and polymerase active site. (b) A close-up view of loops exo1 and exo2 (cyan) is provided, with the location of S1977 indicated. S1977P is homologous to the chaos1 mouse allele of pol θ. The current model indicates potential for hydrogen bond (black dashes) from S1977 to a backbone carbonyl of loop exo1.
Insert 3 (S2503-F2534) terminates with two consecutive glycine residues that lead into a 6-residue β-strand of the palm subdomain, directly prior to polymerase motif 5 (HDELLY) (Fig. 3a). Motif 5 contains two of the strictly conserved carboxylate residues, D2540 and E2541. A helical segment of insert 3 appears at the interface between the N-terminal and the palm subdomains of pol θ, lying at the base of the cleft present in the N-terminal subdomain. In E. coli pol I Klenow fragment, this cleft provides a path for DNA to migrate into the exonuclease proofreading active site42. In pol θ, a small loop (G2022-E2039) in the exonuclease-like domain is shifted that would block DNA from taking a trajectory identical to that previously observed in Klenow fragment (Fig. 3a).
The homologous chaos1 variant (S1977P) catalyzes TLS
A genetic screen for genomic stability factors in mice implicated pol θ in DSB repair43,44. Mouse cells carrying the chaos1 allele of POLQ have elevated spontaneous levels of micronuclei, and increased micronuclei following treatment with ionizing radiation or mitomycin C18,29,44. The chaos1 allele was subsequently verified to encode a serine to proline missense mutation (S1977P in human pol θ) in the N-terminal exonuclease-like subdomain. Partial synthetic lethality results when the chaos1 allele is introduced into an ATM knockout background, allowing only 10 % of mice to survive past the neonatal period44.
The human pol θ model illustrates that S1977 caps the C-terminal end of a hydrophobic helix in the vestigial exonuclease-like subdomain. In the current model, S1997 could provide a hydrogen bond to the backbone carbonyl of D1897 of loop exo1 (Fig. 3b). We generated the S1977P mutant to characterize the biochemical characteristics of the homologous human chaos1 variant. Assays aimed to evaluate TLS (Fig. 4) and single-stranded primer extension (data not shown) failed to reveal a dramatic biochemical phenotype, however. These findings support the suggestion that cellular levels of the pol θ protein are depleted in the chaos1 mice due to poor in vivo expression or stability18.
Fig. 4.
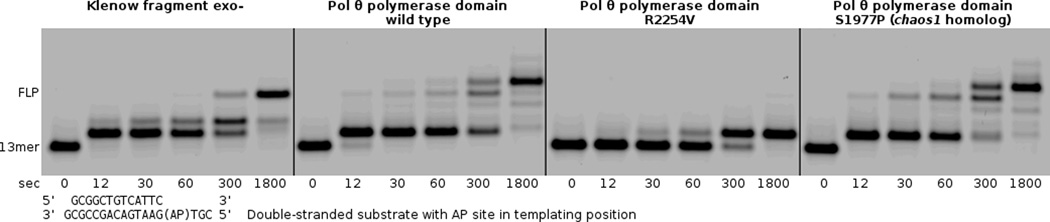
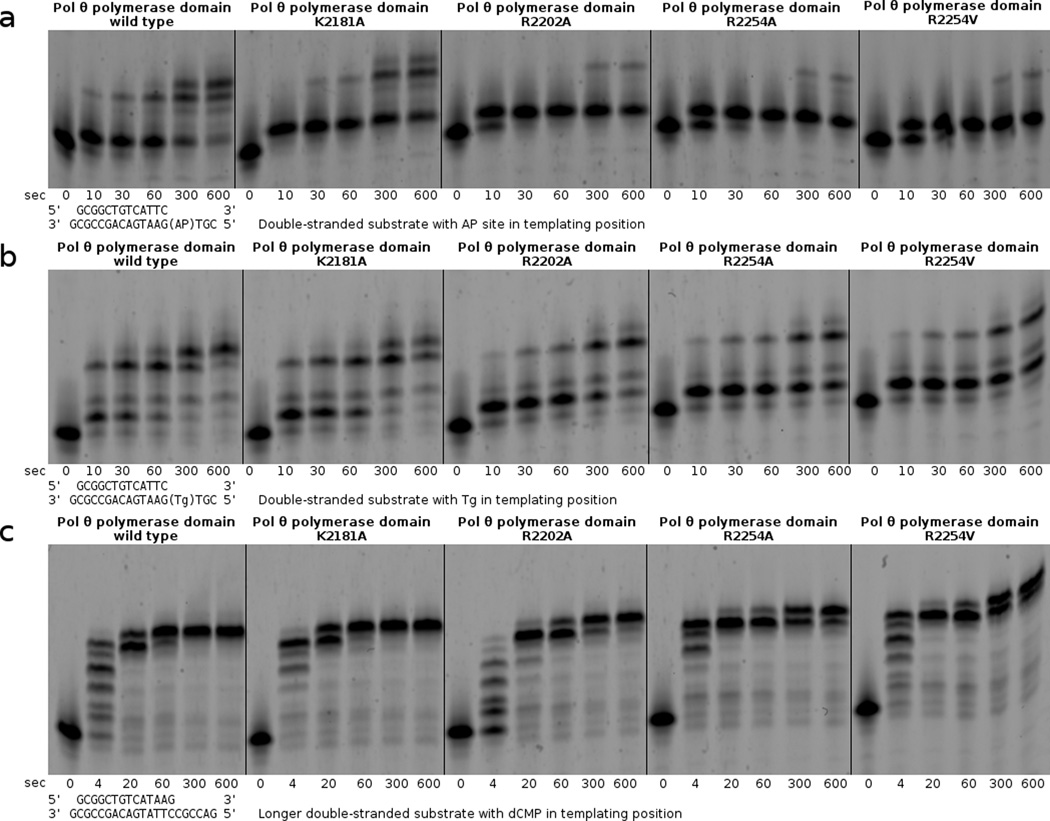
The primer extension assay demonstrates that pol θ R2254V cannot perform the bypass step of TLS, even when all nucleotides are provided (500 µM each). Pol θ and the human chaos1 allele homolog (pol θ S1977P) bypass the AP site to a full-length product, with terminal transferase activity extending the primer by one additional nucleotide. Klenow fragment exo- is able to bypass the AP site under these reaction conditions (125 nM enzyme + 250 nM primer–template), without terminal transferase activity.
Pol θ R2254V fails to bypass AP sites
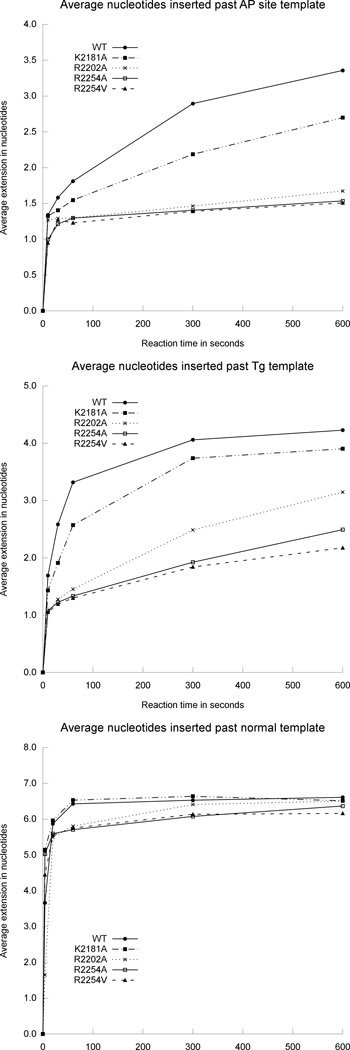
We designed the R2254V variant to evaluate how the conserved basic residue of insertion loop 2 contributes to pol θ’s activity on single-stranded DNA oligonucleotides, and bypass of AP sites or Tg lesions. Family A DNA polymerases from bacteria conserve a hydrophobic amino acid (valine or isoleucine) at the equivalent position of R2254 (Fig. 2a), and the R2254V variant therefore mimics these bacterial enzymes. Pol θ R2254V, although active on double-stranded DNA, failed to bypass AP sites or Tg (Figs. 4–6), and was marginally hindered during extension of unannealed single-stranded DNA oligonucleotides, especially when provided with only pyrimidine nucleotides (Fig. 7a,b). The salt bridge from R2254 to the primer 3’-terminal phosphate appears essential in compensating for interactions missing from the templating strand, due to DNA lesions or distorted base pairing.
Fig. 6.
Bands from the primer extension assays shown in Fig. 5 were quantified in order to plot the average extension by pol θ thumb subdomain variants opposite an AP site, Tg, and an undamaged template.
Fig. 7.
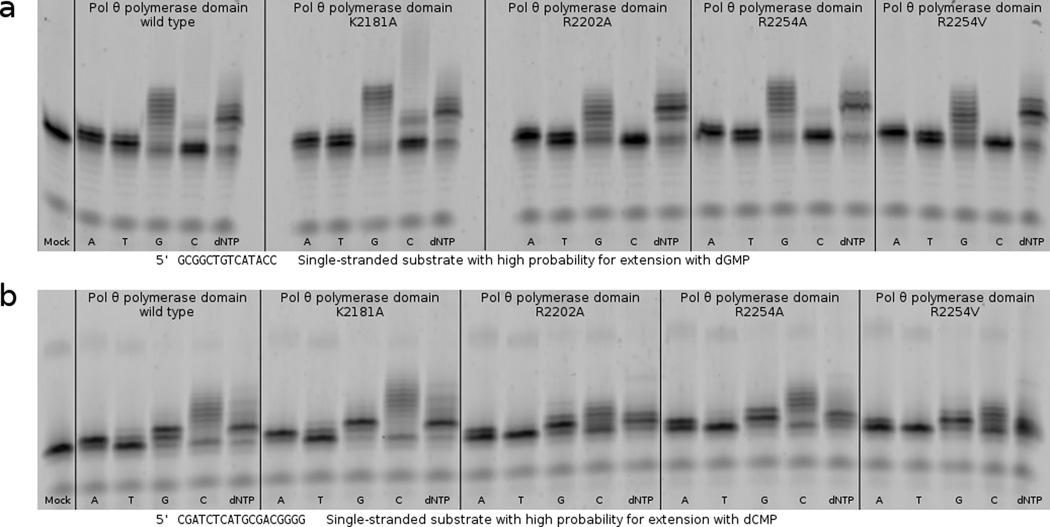
(a) Primer extension assays utilizing a single-stranded substrate show that pol θ variants K2181A, R2202A, R2254A and R2254V are all able to catalyze extension of a single-stranded oligonucleotide after 10 min in the indicated sequence context. (b) A primer extension assay in an alternate sequence context shows that variants of R2202 and R2254 produce shorter products when forced to utilize pyrimidines. In this sequence context, K2202A, R2254A, and R2254V appear unable to utilize dTTP and show reduced incorporation of dCTP compared to the parental enzyme.
The pol θ structure revealed other unique upstream contacts to the primer DNA strand mediated by the specialized pol θ thumb subdomain (Fig. 2a). R2202 inserts its guanidinium moiety between the n-2 and n-3 primer strand phosphates, and is therefore poised to make two contacts to the DNA backbone. An additional contact is possible from K2181 of the thumb subdomain to the primer n-5 phosphate (Fig. 2c). Including the contacts from R2254 to the n-1 phosphate, the pol θ thumb subdomain appears specialized to provide salt bridges to the primer strand phosphate backbone in excess of those conserved by all other family A polymerases, including pol ν (Fig. 2a). Like pol ν and bacterial pol I, pol θ retains arginine residues at positions 2201 and 2315 (631 and 690 in E. coli pol I), which provide absolutely conserved interactions to the n-4 and n-1 phosphate, respectively.
Generating alanine substitutions at K2181, R2202, or R2254 in pol θ, for the purpose of evaluating the contribution of the specialized thumb subdomain during bypass of an AP site or Tg, revealed that the greatest TLS defect associates with a loss of contacts closest to the extreme primer terminus (Fig. 5). Although the extension step of TLS opposite an AP site challenged all variants, the mutations made at R2254 (to alanine or valine) afforded the most dramatically deficient TLS phenotypes, followed closely by R2202A. K2181A reduced bypass of the AP site compared to the wild type enzyme, although not to the same extent as other variants (Fig. 6). When provided with a substrate placing Tg in templating position, the same pattern emerged, albeit R2202A possessed a lesser defect, closer to that of K2181A. Even though pol ν harbors a lysine (K584) residue in insertion 2, homologous to R2254 in pol θ, R2202 and K2182 appear in neither pol ν nor bacterial homologs (Fig. 2a). These differences could explain why only pol θ grasps the primer tightly enough to bypass AP sites and extend certain minimally annealed primer–templates45. Pol ν is less adept at bypassing blocking lesions, and has only been shown to bypass 5S-Tg effectively13. Likewise, pol ν does not extend single-stranded oligonucleotides in vitro, a reaction readily catalyzed by pol θ10. Considering that pol θ retained substantial ability to extend single-stranded substrates in the context of individual mutations at R2254, R2202 or K2181 in vitro (Fig. 7a), the transiently templated nature of this reaction is emphasized18, and pol θ must therefore employ a different set of amino acid side chains to manipulate the single-stranded substrate as its own template for primer extension.
Fig. 5.
(a) Primer extension assays compare bypass of a templating AP site by pol θ thumb subdomain variants K2181A, R2202A, R2254A, and R2254V. (b) The variant polymerases are also challenged during bypass of a Tg lesion in otherwise identical primer extension assays. (c) Nearly full activity for all variant polymerases is observed in primer extension assays with an undamaged template.
Pol θ crystals reveal two modes of dimerization
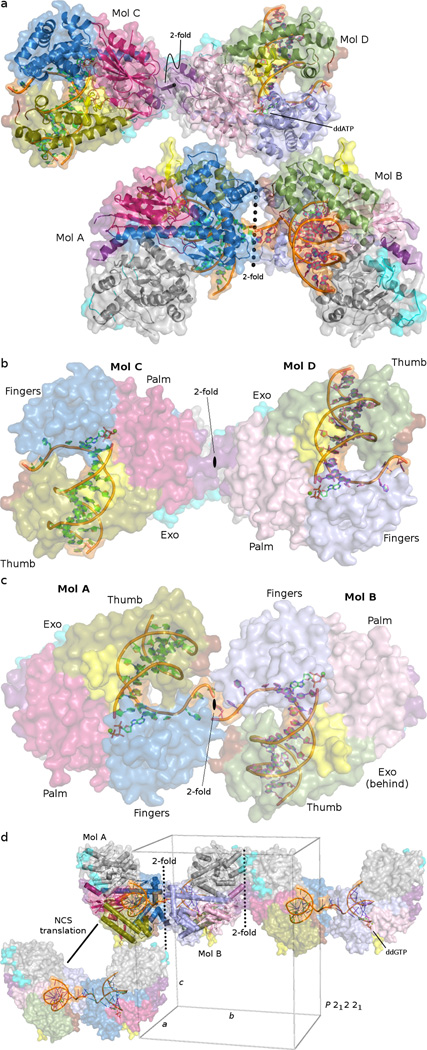
The pol θ ASU in the Ca2+ crystal form contains 4 protein-DNA complexes, assembled as a dimer of 2-fold dimers (Fig. 8a). Two dissimilar types of contacts mediated packing of the DNA ends distal from the active site: The 3’ ends of each template DNA strand provide a dGMP overhang, two of which stack against conserved tryptophan residues (W1907) of adjacent protein molecules. The remaining template 3’ ends appear in close proximity, forming inter-DNA contacts outside the molecular footprint of pol θ.
Fig. 8.
The NCS 2-fold axes in pol θ crystals are visualized and compared. (a) The ASU of the pol θ THF–ddATP model contains four protein-DNA complexes. Assembling the ASU requires two skew NCS 2-fold axes (dotted lines). (b) The NCS 2-fold relating chains C and D passes near insert 3 of the palm (pink hues) subdomain, adjacent to the N-terminal (grey shades) subdomain (c) The 5’-template DNA propagated NCS 2-fold relates chains A and B, passing near the fingers (blue hues) and thumb (green hues) subdomains. (d) Crystal packing in the dCMP–ddGTP crystal form is depicted, showing that the two molecules per ASU pack differently from the four molecules of the THF–ddATP crystal form (described in panel a). The identical NCS 2-fold axes are observed, but parallel to the 21 screw along cell edge c. The dimer at the right of the panel is generated by a crystallographic translation of the unit cell (green rectangular prism) along the b cell axis. The dimer at the bottom left is generated by the 21 screw along cell axis c (vertical axis), which in the context of the 5’-template DNA propagated NCS 2-fold axis gives rise to the NCS translation.
Three perpendicular 2-fold screw axes generate the space group P212121. However, two alternate pure 2-fold axes assemble the ASU of the Ca2+ crystal form, which exist as NCS operators. The first NCS 2-fold axis passes near insert 3 and the N-terminal subdomain (Fig. 8b). The second NCS 2-fold axis passes adjacent to the 5’-template DNA, upstream of the polymerase active site, and near the putative location of insertion loop 1 (Fig. 8c). The ASU in the current Ca2+ model therefore presents two potentially biologically relevant assemblies, where the 5’-template DNA 2-fold symmetry axis relates protein chains A and B, and the 2-fold symmetry axis of insert 3 relates molecules C and D. Interestingly, the pseudosymmetry observed in the Mg2+ crystal form was a consequence of similar configurations within the ASU, where the identical NCS 2-fold dimeric relationships about the 5’-template DNA and insert 3 appeared once again. In this case, both NCS 2-fold axes appeared nearly parallel to the crystallographic 21 screw axis along c, thus generating the NCS translation (Fig. 8d).
Discussion
An alternative DNA end-joining pathway in eukaryotes absolutely requires the POLQ gene product, where the C-terminal polymerase activity is applied specifically in a role for which other DNA polymerases or helicases are unable to compensate18–20. The current study informs a mechanism by which the human pol θ polymerase domain maintains a tight hold on the primer DNA strand, by establishing interactions from positively charged residues K2181, R2202 and R2254 to the phosphate backbone. The crystal structures also establish a precedence for the pol θ protein to dimerize. Although a multimer was not observed in solution by gel filtration chromatography, resolving identical NCS 2-fold axes in two different crystal forms indicates multimerization of the pol θ polymerase domain is possible at sufficient concentration. Analysis of these potential dimer interfaces in PISA46 attributes 12,400 and 13,100 Å2 of buried surface area for the 5’ DNA propagated and insert 3 NCS 2-fold interfaces (Fig. 8b and 8c), respectively, neglecting the flexible segments omitted from the models. In the context of alternative end-joining, pol θ multimerization might be essential for bringing two DNA ends together, or sequestering free DNA ends from classical nonhomologous end-joining factors, such as Ku47.
Higher expression of POLQ correlates with decreased survival in breast cancer patients48,49, and knockdown of the gene product in several malignant cell lines induces radiosensitivity30. Given that POLQ is non-essential in healthy cells44, the potential to pharmacologically target unique features of the pol θ protein, such as the specialized thumb subdomain revealed by this study, is an appealing approach for adjuvant radiation cancer therapy26. Future experiments will expand our understanding of the mechanisms by which pol θ protects cancer cells from radiation or chemotherapeutics. The extent to which transient pol θ dimers might bridge DSBs, for example, must also be considered as POLQ is evaluated as a potential target for next generation cancer drugs.
Online Methods
Crystallization of recombinant human pol θ
The pol θ polymerase crystallization construct (residues 1792–2590) was expressed from the pSUMO3 vector in Rosetta2(DE3)pLysS cells (Novagen) by autoinduction, and purified to homogeneity utilizing a nickel-NTA resin (Thermo Scientific), a HiTrap Heparin column (GE Healthcare Life Sciences), and a Superdex 200 Increase GL gel filtration column (GE Healthcare Life Sciences)11. The selenomethionyl pol θ protein was also autoinduced, but with the Overnight Express System 2 kit (EMD Millipore) using 125 mg/L of L-selenomethionine (Acros Organics). The protein was concentrated to 5 mg/mL in a buffer of ammonium acetate (150 mM), KCl (150 mM), Tris-HCl buffer pH 8.0 (40 mM), TCEP (2.5 mM), and glycerol (1% v/v). Reacting pol θ (2.5 mg/mL) with a 13–18mer oligonucleotide (50 µM) containing a THF abasic site analog (Midland), with annealed primer=gcggctgtcattc and template =cgt[THF]gaatgacagccgcg, in the presence of ddATP (1 mM), sucrose monolaurate (300 µM) and spermine tetrahydrochloride (20 mM) prepared the sample for hanging drop vapor diffusion over a 1 mL reservoir containing PEG 2000mme (9.5% v/v), MPD (2% v/v), CaCl2 (50 mM), KCl (200 mM), and Tris-HCl buffer pH 8.5 (100 mM), by mixing 0.6 µL of the reservoir solution with an equal part reaction solution. Narrow, long crystals of approximate final dimensions 400 × 60 × 60 µm3 grew over the next 3 days. Cryoprotection was achieved by adding 1 µL of a solution containing PEG 2000mme (25% v/v), MPD (25% v/v), CaCl2 (20 mM), spermine tetrahydrochloride (10 mM) and KCl (260 mM) and nucleotide (1 mM) directly to the drop, prior to flash cooling into liquid nitrogen.
The alternative Mg2+ crystal form was discovered in an attempt to obtain better diffracting pol θ crystals. An additional 27 residues were cleaved from the N-terminus of the crystallization construct, leaving residues 1819–2590. The DNA sequence was also altered to allow for the formation of a Watson-Crick base pair in the active site and eliminating the 3’ template dGMP overhang. Two consecutive dCMP were introduced in the template to allow for enzymatic chain termination of the primer strand by incubation with ddGMP, yielding the primer=gcggctgtcatt and template=cgtccaatgacagccgc. Crystals were obtained from similar conditions as before, except that the reaction mixture was supplemented with 1 mM MgCl2, and ddGTP replaced the ddATP. The PEG 2000mme in the reservoir was increased to 12%, and the MPD to 10%. Although these crystals grew to similar length, they were wider (80 µm) but thinner (20 µm). Increasing the precipitant slightly to 12.5% allowed for crystallization similarly of the selenomethionyl variant of the Δ1818 construct in complex with the same primer–template, although these crystals were approximately half the size of the native Mg2+ crystals. Both native and selenomethionyl forms were cryoprotected by the prior procedure, except the cryoprotection solution was supplemented with MgCl2 instead of CaCl2.
Data collection, structure solution, and refinement
Pol θ crystals of the Ca2+ form diffracted at the APS synchrotron (beamline 23-ID-D) past 3.9 Å at 12 kEV, and complete data sets were collected from these radiation-sensitive primitive orthorhombic (P212121) crystals at 80 K by merging approximately 20 image segments, as the 20 µm beam was translated along the length of the crystal. Due to anisotropy of diffraction, multiple crystals were merged in order to obtain a complete data set with substantial redundancy, to be used in structure solution and refinement. An identical strategy was utilized to collect native and derivative datasets at Se-peak (12.67 kEV) for the Mg2+ crystal forms.
Structure solution of the Ca2+ crystal form was accomplished by molecular replacement, utilizing the Taq DNA polymerase ternary complex 1QSY32 to search for the 4 protein/DNA complexes per ASU. Long segments of secondary structure were rigid body fit into a reduced bias “prime and switch” map53 with NCS averaging. Rebuilding in Coot54 and refinement in Phenix55 allowed most of the DNA to be built around the active site, although uncertainty remained concerning insertion elements, and in assigning sequence to the trace of the N-terminal vestigial exonuclease-like subdomain.
The Mg2+ crystal form diffracted to similar resolution as the Ca2+ form, but was plagued by a strong off-origin native Patterson peak consistent with an NCS translation of (0.1, −0.5, 0.5) fractional units, which was verified by molecular replacement in the space group P21221, utilizing the Ca2+ structure as a search model. The identical NCS 2-fold dimeric configurations were resolved, as observed of crystals grown in Ca2+, although in this case a single dimer constituted the ASU. The NCS 2-folds occurred parallel to the crystallographic 21 screw operator along c, which gave rise to pseudosymmetry and potential twinning (Fig. 5d). Refinement against potential twin laws in all related monoclinic cells failed to improve refinement statistics, however, and twinning was ruled out, despite the presence of suspect intensity distribution. Moreover, the electron density maps were of lesser quality. Nevertheless, phases obtained by refining the rigid-body solution of the Mg2+ crystal form in Phenix33 proved useful in generating cross-crystal density-modified NCS-averaged maps, which provided high quality electron density for the placing side chains of the final model, and helped resolve ambiguities in the tracing of the inserts and N-terminal exonuclease-like subdomain. Phases calculated from the transformed model also allowed visualization of anomalous peaks at 4.6 Å, due to selenomethionine substitution in the SeMet Mg2+ crystal form, and thereby provided invaluable verification of the backbone trace in the N-terminal domain and insertion element 2.
Concluding refinement steps were conducted in Phenix55, fitting protein domains and nucleic acid chains as rigid bodies. Torsion NCS, secondary structure, and Ramachandran restraints were enforced during individual xyz refinement, prior to TLS and group_ADP refinement, at which point the Rwork reached 28.3 % (Rfree= 31.2 %) for the Ca2+ model. Finally, the individual_ADP strategy was applied for 2 cycles of refinement to yield the completed model, for which statistics are reported. Ramachandran analysis places 98.6% of residues in the Ca2+ model in favored or allowed regions of the plot, leaving 1.4% outliers. The final Mg2+ model was obtained by trimming several residues from the base of insertion loop 1, and omitting 3 base-pairs of the duplex DNA extruding from the molecule in the 5’ primer direction, before similarly running refinement in Phenix. The completed Mg2+ model identifies 99.3% of residues in the favored or allowed region of the Ramachandran plot, with 0.7% outliers.
Variant production and biochemical analysis
Variant pol θ constructs were generated by site-directed mutagenesis with the QuikChange XL kit (Stratagene). Biochemical reactions were conducted by pre-incubating the primer–template (250 nM) with pol θ variants (125 nM) in Tris-HCl buffer pH 8.0 (20 mM), KCl (25 mM), and β-mercaptoethanol (1 mM), prior to adding deoxynucleotides (500 µM each or individually) and MgCl2 (10 mM). Aliquots were quenched at indicated time points by mixing in equal parts of a quench solution made of formamide (95 %), EDTA (20 mM), and trace bromophenol blue. Products were separated on a SequaGel UreaGel (National Diagnostics) denaturing sequencing gel (12 %), and visualized by excitation of a 5’ tetrachlorofluorescein tag on the primer strand at 532 nm setting on the PharosFX (BioRad) imaging device. Bands were quantified with the QuantityOne (BioRad) software and plotted in GNUplot via the function , where ρi is the normalized density of the band corresponding to the ith extension product, to yield the average extension of the primer strand in fractional nucleotides.
Supplementary Material
Acknowledgements
These studies were funded by US National Institutes of Health grants R01 CA052040 (S.D.) and CA097175 (R.D.W.) and grant RP130297 from the Cancer Prevention and Research Institute of Texas (R.D.W.) and the Grady F. Saunders Ph.D. Distinguished Research Professorship (R.D.W.). The GM/CA beamline at APS has been funded in whole or in part with Federal funds from the US National Cancer Institute (ACB-12002) and the US National Institute of General Medical Sciences (AGM-12006). This research used resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility operated for the US DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
Footnotes
Note added in proof:
During final revision of this manuscript, several papers linking pol θ function to alternative end-joining of double-strand breaks were published online50–52. Kent et al52 suggest that a dimeric form of pol θ could function in repair, with dimerization mediated by insertion loop 2. Our crystal structures indicate multimerization of the pol theta polymerase domain (Figure 8), but do not readily implicate insertion loop 2 in dimerization of the this domain.
Accession Codes
Coordinates and structure factor amplitudes for the ternary complexes of human pol θ obtained with Ca2+ (THF–ddATP) and Mg2+ (dCMP–ddGTP) were deposited to the protein data bank (www.pdb.org) with accession codes 4XOP and 4XOQ, respectively.
Author Contributions
K.E.Z. performed crystallization, data collection and structure refinement. A.M.A. expressed and purified native and SeMet pol θ protein. P.A. obtained preliminary diffraction images at Diamond Light Source beamline I04. K.E.Z. expressed and purified mutant proteins, and performed biochemical experiments. S.D. and R.D.W. oversaw the project. K.E.Z. wrote the manuscript with S.D. and R.D.W. All authors discussed the results and commented on the manuscript.
References
- 1.Harris PV, et al. Molecular cloning of Drosophila mus308, a gene involved in DNA cross-link repair with homology to prokaryotic DNA polymerase I genes. Mol Cell Biol. 1996;16:5764–5771. doi: 10.1128/mcb.16.10.5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Inagaki S, et al. Arabidopsis TEBICHI, with helicase and DNA polymerase domains, is required for regulated cell division and differentiation in meristems. Plant Cell. 2006;18:879–892. doi: 10.1105/tpc.105.036798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yousefzadeh MJ, Wood RD. DNA polymerase POLQ and cellular defense against DNA damage. DNA Repair (Amst) 2013;12:1–9. doi: 10.1016/j.dnarep.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seki M, Marini F, Wood RD. POLQ (Pol theta), a DNA polymerase and DNA-dependent ATPase in human cells. Nucleic Acids Res. 2003;31:6117–6126. doi: 10.1093/nar/gkg814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marini F, Kim N, Schuffert A, Wood RD. POLN, a nuclear PolA family DNA polymerase homologous to the DNA cross-link sensitivity protein Mus308. J Biol Chem. 2003;278:32014–32019. doi: 10.1074/jbc.M305646200. [DOI] [PubMed] [Google Scholar]
- 6.Longley MJ, Prasad R, Srivastava DK, Wilson SH, Copeland WC. Identification of 5'-deoxyribose phosphate lyase activity in human DNA polymerase gamma and its role in mitochondrial base excision repair in vitro. Proc Natl Acad Sci U S A. 1998;95:12244–12248. doi: 10.1073/pnas.95.21.12244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prasad R, et al. Human DNA polymerase theta possesses 5'-dRP lyase activity and functions in single-nucleotide base excision repair in vitro. Nucleic Acids Res. 2009;37:1868–1877. doi: 10.1093/nar/gkp035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee YS, Kennedy WD, Yin YW. Structural insight into processive human mitochondrial DNA synthesis and disease-related polymerase mutations. Cell. 2009;139:312–324. doi: 10.1016/j.cell.2009.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arana ME, Seki M, Wood RD, Rogozin IB, Kunkel TA. Low-fidelity DNA synthesis by human DNA polymerase theta. Nucleic Acids Res. 2008;36:3847–3856. doi: 10.1093/nar/gkn310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hogg M, Sauer-Eriksson AE, Johansson E. Promiscuous DNA synthesis by human DNA polymerase theta. Nucleic Acids Res. 2012;40:2611–2622. doi: 10.1093/nar/gkr1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hogg M, Seki M, Wood RD, Doublié S, Wallace SS. Lesion bypass activity of DNA polymerase theta (POLQ) is an intrinsic property of the pol domain and depends on unique sequence inserts. J Mol Biol. 2011;405:642–652. doi: 10.1016/j.jmb.2010.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seki M, et al. High-efficiency bypass of DNA damage by human DNA polymerase Q. EMBO J. 2004;23:4484–4494. doi: 10.1038/sj.emboj.7600424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takata K, Shimizu T, Iwai S, Wood RD. Human DNA polymerase N (POLN) is a low fidelity enzyme capable of error-free bypass of 5S-thymine glycol. J Biol Chem. 2006;281:23445–23455. doi: 10.1074/jbc.M604317200. [DOI] [PubMed] [Google Scholar]
- 14.Yoon JH, Roy Choudhury J, Park J, Prakash S, Prakash L. A role for DNA polymerase theta in promoting replication through oxidative DNA lesion, thymine glycol, in human cells. J Biol Chem. 2014;289:13177–13185. doi: 10.1074/jbc.M114.556977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoshimura M, et al. Vertebrate POLQ and POLbeta cooperate in base excision repair of oxidative DNA damage. Mol Cell. 2006;24:115–125. doi: 10.1016/j.molcel.2006.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Asagoshi K, et al. Single-nucleotide base excision repair DNA polymerase activity in C. elegans in the absence of DNA polymerase beta. Nucleic Acids Res. 2012;40:670–681. doi: 10.1093/nar/gkr727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martomo SA, Saribasak H, Yokoi M, Hanaoka F, Gearhart PJ. Reevaluation of the role of DNA polymerase theta in somatic hypermutation of immunoglobulin genes. DNA Repair (Amst) 2008;7:1603–1608. doi: 10.1016/j.dnarep.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yousefzadeh MJ, et al. Mechanism of Suppression of Chromosomal Instability by DNA Polymerase POLQ. PLoS Genet. 2014;10:e1004654. doi: 10.1371/journal.pgen.1004654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan SH, Yu AM, McVey M. Dual roles for DNA polymerase theta in alternative end-joining repair of double-strand breaks in Drosophila. PLoS Genet. 2010;6:e1001005. doi: 10.1371/journal.pgen.1001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koole W, et al. A Polymerase Theta-dependent repair pathway suppresses extensive genomic instability at endogenous G4 DNA sites. Nat Commun. 2014;5:3216. doi: 10.1038/ncomms4216. [DOI] [PubMed] [Google Scholar]
- 21.Roerink SF, van Schendel R, Tijsterman M. Polymerase theta-mediated end joining of replication-associated DNA breaks in C. elegans. Genome Res. 2014;24:954–962. doi: 10.1101/gr.170431.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu AM, McVey M. Synthesis-dependent microhomology-mediated end joining accounts for multiple types of repair junctions. Nucleic Acids Res. 2010;38:5706–5717. doi: 10.1093/nar/gkq379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.White TB, Lambowitz AM. The retrohoming of linear group II intron RNAs in Drosophila melanogaster occurs by both DNA ligase 4-dependent and -independent mechanisms. PLoS Genet. 2012;8:e1002534. doi: 10.1371/journal.pgen.1002534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernandez-Vidal A, et al. A role for DNA polymerase theta in the timing of DNA replication. Nat Commun. 2014;5:4285. doi: 10.1038/ncomms5285. [DOI] [PubMed] [Google Scholar]
- 25.Lemee F, et al. DNA polymerase theta up-regulation is associated with poor survival in breast cancer, perturbs DNA replication, and promotes genetic instability. Proc Natl Acad Sci U S A. 2010;107:13390–13395. doi: 10.1073/pnas.0910759107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Higgins GS, et al. Overexpression of POLQ confers a poor prognosis in early breast cancer patients. Oncotarget. 2010;1:175–184. doi: 10.18632/oncotarget.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lessa RC, et al. Identification of upregulated genes in oral squamous cell carcinomas. Head Neck. 2013;35:1475–1481. doi: 10.1002/hed.23169. [DOI] [PubMed] [Google Scholar]
- 28.Allera-Moreau C, et al. DNA replication stress response involving PLK1, CDC6, POLQ, RAD51 and CLASPIN upregulation prognoses the outcome of early/mid-stage non-small cell lung cancer patients. Oncogenesis. 2012;1:e30. doi: 10.1038/oncsis.2012.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goff JP, et al. Lack of DNA polymerase theta (POLQ) radiosensitizes bone marrow stromal cells in vitro and increases reticulocyte micronuclei after total-body irradiation. Radiat Res. 2009;172:165–174. doi: 10.1667/RR1598.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Higgins GS, et al. A small interfering RNA screen of genes involved in DNA repair identifies tumor-specific radiosensitization by POLQ knockdown. Cancer Res. 2010;70:2984–2993. doi: 10.1158/0008-5472.CAN-09-4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y, Mitaxov V, Waksman G. Structure-based design of Taq DNA polymerases with improved properties of dideoxynucleotide incorporation. Proc Natl Acad Sci U S A. 1999;96:9491–9496. doi: 10.1073/pnas.96.17.9491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adams PD, et al. The Phenix software for automated determination of macromolecular structures. Methods. 2011;55:94–106. doi: 10.1016/j.ymeth.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doublié S, Tabor S, Long AM, Richardson CC, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 35.Wu EY, Beese LS. The structure of a high fidelity DNA polymerase bound to a mismatched nucleotide reveals an "ajar" intermediate conformation in the nucleotide selection mechanism. J Biol Chem. 2011;286:19758–19767. doi: 10.1074/jbc.M110.191130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zahn KE, Belrhali H, Wallace SS, Doublié S. Caught bending the A-rule: crystal structures of translesion DNA synthesis with a non-natural nucleotide. Biochemistry. 2007;46:10551–10561. doi: 10.1021/bi7008807. [DOI] [PubMed] [Google Scholar]
- 37.Motea EA, Lee I, Berdis AJ. Quantifying the energetic contributions of desolvation and pi-electron density during translesion DNA synthesis. Nucleic Acids Res. 2011;39:1623–1637. doi: 10.1093/nar/gkq925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Obeid S, Welte W, Diederichs K, Marx A. Amino acid templating mechanisms in selection of nucleotides opposite abasic sites by a family a DNA polymerase. J Biol Chem. 2012;287:14099–14108. doi: 10.1074/jbc.M111.334904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takata K, Arana ME, Seki M, Kunkel TA, Wood RD. Evolutionary conservation of residues in vertebrate DNA polymerase N conferring low fidelity and bypass activity. Nucleic Acids Res. 2010;38:3233–3244. doi: 10.1093/nar/gkq048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Castro C, et al. Nucleic acid polymerases use a general acid for nucleotidyl transfer. Nat Struct Mol Biol. 2009;16:212–218. doi: 10.1038/nsmb.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Terwilliger TC. Statistical density modification with non-crystallographic symmetry. Acta Crystallogr D Biol Crystallogr. 2002;58:2082–2086. doi: 10.1107/S0907444902016360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beese LS, Derbyshire V, Steitz TA. Structure of DNA polymerase I Klenow fragment bound to duplex DNA. Science. 1993;260:352–355. doi: 10.1126/science.8469987. [DOI] [PubMed] [Google Scholar]
- 43.Shima N, et al. Phenotype-based identification of mouse chromosome instability mutants. Genetics. 2003;163:1031–1040. doi: 10.1093/genetics/163.3.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shima N, Munroe RJ, Schimenti JC. The mouse genomic instability mutation chaos1 is an allele of Polq that exhibits genetic interaction with Atm. Mol Cell Biol. 2004;24:10381–10389. doi: 10.1128/MCB.24.23.10381-10389.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seki M, Wood RD. DNA polymerase theta (POLQ) can extend from mismatches and from bases opposite a (6-4) photoproduct. DNA Repair (Amst) 2008;7:119–127. doi: 10.1016/j.dnarep.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 47.Ribes-Zamora A, Indiviglio SM, Mihalek I, Williams CL, Bertuch AA. TRF2 interaction with Ku heterotetramerization interface gives insight into c-NHEJ prevention at human telomeres. Cell Rep. 2013;5:194–206. doi: 10.1016/j.celrep.2013.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang S, Zhang H, Guo L, Zhao Y, Chen F. Reconstructing the coding and non-coding RNA regulatory networks of miRNAs and mRNAs in breast cancer. Gene. 2014;548:6–13. doi: 10.1016/j.gene.2014.06.010. [DOI] [PubMed] [Google Scholar]
- 49.Santarpia L, et al. DNA repair gene patterns as prognostic and predictive factors in molecular breast cancer subtypes. Oncologist. 2013;18:1063–1073. doi: 10.1634/theoncologist.2013-0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ceccaldi R, et al. Homologous-recombination-deficient tumours are dependent on Poltheta-mediated repair. Nature. 2015 doi: 10.1038/nature14184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mateos-Gomez PA, et al. Mammalian polymerase theta promotes alternative NHEJ and suppresses recombination. Nature. 2015 doi: 10.1038/nature14157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kent T, Chandramouly G, McDevitt SM, Ozdemir AY, Pomerantz RT. Mechanism of microhomology-mediated end-joining promoted by human DNA polymerase theta. Nat Struct Mol Biol. 2015 doi: 10.1038/nsmb.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
References for online methods
- 53.Terwilliger T. SOLVE and RESOLVE: automated structure solution, density modification and model building. J Synchrotron Radiat. 2004;11:49–52. doi: 10.1107/s0909049503023938. [DOI] [PubMed] [Google Scholar]
- 54.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Headd JJ, et al. Use of knowledge-based restraints in phenix.refine to improve macromolecular refinement at low resolution. Acta Crystallogr D Biol Crystallogr. 2012;68:381–390. doi: 10.1107/S0907444911047834. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.