Abstract
The ends of coiled-coil tropomyosin molecules are joined together by nine to ten residue-long head-to-tail “overlapping domains”. These short four-chained interconnections ensure formation of continuous tropomyosin cables that wrap around actin filaments. Molecular Dynamics simulations indicate that the curvature and bending flexibility at the overlap is 10 to 20% greater than over the rest of the molecule, which might affect head-to-tail filament assembly on F-actin. Since the penultimate residue of striated muscle tropomyosin, Ser283, is a natural target of phosphorylating enzymes, we have assessed here if phosphorylation adjusts the mechanical properties of the tropomyosin overlap domain. MD simulations show that phosphorylation straightens the overlap to match the curvature of the remainder of tropomyosin while stiffening it to equal or exceed the rigidity of canonical coiled-coil regions. Corresponding EM data on phosphomimetic tropomyosin S283D corroborate these findings. The phosphorylation-induced change in mechanical properties of tropomyosin likely results from electrostatic interactions between C-terminal phosphoSer283 and N-terminal Lys12 in the four-chain overlap bundle, while promoting stronger interactions among surrounding residues and thus facilitating tropomyosin cable assembly. The stiffening effect of D283-tropomyosin noted correlates with previously observed enhanced actin-tropomyosin activation of myosin S1-ATPase, suggesting a role for the tropomyosin phosphorylation in potentiating muscle contraction.
Keywords: Actin, tropomyosin, phosphorylation, cardiomyopathy, muscle regulation, Molecular Dynamics
Introduction
Post-translational phosphorylation and modification of cardiac muscle proteins are hallmarks of cardiovascular responses and adjustments to physiological challenges [1,2]. Sarcomeric proteins targeted for phosphorylation include cardiac myosin light chains (MLC-2), myosin binding protein C (MyBP-C), troponin I (TnI), troponin T (TnT), tropomyosin and titin [1–12]. However, the interplay between the effects of post-translational modification remains unclear. In fact, unlike the phosphorylation of most proteins, tropomyosin phosphorylation, studied here, is not subject to minute-to-minute change [7], suggesting that the modulation is a comparatively long-term functional adaptation. Tropomyosin phosphorylation in striated muscle appears to be restricted to serine 283, the penultimate residue in the 284 amino acid long tropomyosin sequence [7,8]. S283 phosphorylation is found to be highest in fetal hearts (~70 % phosphorylated) but declines slowly post-partum and with increasing age (to ~20 to 30%), suggesting a developmental role possibly related to myofilament assembly [13]. In some cases, a variable increase in phosphorylation is observed in diseased adult hearts [14], correlated with possible protein expression regressing to that of the fetus.
The regulation of cardiac muscle actin-myosin interaction and hence contractile force is governed by the thin filament regulatory proteins tropomyosin and troponin and their interactions with actin, myosin and Ca2+ [15,16]. 40 nm long tropomyosin coiled-coils associate head-to-tail on thin filaments to form a continuous cable that follows the helical path defined by actin subunit interactions. Each molecule of tropomyosin spans seven actin monomers and binds to one troponin complex containing troponin subunits TnT, TnI and TnC. Under the control of Ca2+-binding to troponin and myosin binding to actin, tropomyosin translocates azimuthally across the actin filament, to transiently block or open myosin-binding sites on actin, thereby regulating myosin-head cross-bridge cycling on actin and consequently contractility [17–19].
Single tropomyosin molecules bind to actin filaments with exceedingly low affinity. Tropomyosin only binds to actin filaments with appreciable affinity after head-to-tail polymerized molecules populate actin filaments and begin to form a cable [20]. Thus, effective binding results from the collective interactions of tropomyosin molecules in the cable and the successive actin subunits along thin filaments [18]. Since the tropomyosin molecule is semi-rigid, relatively long stretches of the cable, greater than the 40 nm length of single tropomyosin molecules, can move azimuthally as a unit over actin, implying that the head-to-tail connection also is semi-rigid [21]. Thus once assembled, the actin-tropomyosin co-filament cooperatively activates myosin ATPase and cross-bridge cycling.
Tropomyosin is a canonical coiled-coil along most of its length [21–23]. However, the two-chained coiled-coil architecture is disrupted over C-terminal 20 to 25 residues during tropomyosin polymerization [24–26]. Here tropomyosin splays apart to accommodate a more compact N-terminal end of an adjacent molecule [24–27]. Thus, the resulting four-coiled bundle/overlap domain is the “glue” responsible for tropomyosin polymerization into a cable. Phosphorylation of serine 283, located at the center of the head-to-tail tropomyosin junctional complex, may therefore influence tropomyosin cable assembly and stability. In the current investigation, we examined this possibility by studying the structural and functional effects of striated muscle α-tropomyosin (i.e. Tpm1.1st (a.b.b.a) [28]) phosphorylation in order to determine whether or not phosphoserine-283 affects the mechanical and chemical properties of the tropomyosin overlap domain. MD simulation and EM data acquired indicate that Ser283 phosphorylation straightens the tropomyosin overlap domain to match the curvature of the remainder of tropomyosin and to fit better to the actin-filament helix. Moreover, the phosphorylation stiffens the overlapping domain to equal or exceed the rigidity of canonical coiled-coil regions of tropomyosin. The conformational adjustment may potentiate tropomyosin cable translation across the actin filament and account, at least in part, for the phosphorylation-induced enhanced acto-S1 activation by actin-tropomyosin [29,30].
Materials and Methods
Reference models for MD simulations
The initial reference models used for MD of the striated muscle α-tropomyosin (Tpm1.1st (a.b.b.a)) head-to-tail overlap domain segment were based on a corresponding NMR structure (PDB ID code 2g9j:conformer model-1 ) [26]. In the work reported here and in reference 31, we replaced the peripheral non-tropomyosin residues used to stabilize overlap domain peptides examined in the NMR study with native coiled-coil sequences to build an 80-residue-long fragment of the tropomyosin polymer consisting of 40 residue contributions from the C- and N-termini connected by the 10-residue long head-to-tail nexus. We used native acetylated N-terminal methionine residues [cf. 31–33] instead of the amino acid mimetics shown in the PDB structure [26]. The initial reference model containing phosphorylated serine 283 was then built from the average MD structure of the domain [31] and the serine hydroxyl side-chain replaced with phosphate using the CHARMM SP2 patch [34]. Thus, the initial reference models built in the current study and used for MD simulations here are exactly the same as those in our previous work [31], except for the substitution of phosphoserine for serine at residue 283 of tropomyosin. Control studies examining the effect of phosphoserine 283 on isolated 40-residue long “non-polymerized” C-terminal fragments of tropomyosin were initiated from reference models cut from full length tropomyosin [21].
Molecular Dynamics and Analysis
The reference models were energy minimized [21,31,35] and Molecular Dynamics simulations performed in explicit solvent including 150 mM NaCl at 310° K, using NAMD version 2.9 [36] and the CHARMM27 force field [34,37,38] as previously described [21,31,35]. Analysis was carried out after discarding the first 4 to 5 ns of MD, ensuring that the variance of the measurements had stabilized [21,31,35]. MD trajectories were averaged as previously after discarding the first 4 to 5 ns in each simulation; averages were composed of snapshots sampled every picosecond during the trajectory [21,31,35]. The curvature of the tropomyosin overlap domain was quantified by determining the angle between the respective central axes of the C- and N-terminal coiled-coils fragments as diagramed in Figure 2 of reference [31]. Persistence length was calculated by applying the tangent correlation method on snapshots taken from the MD trajectory as described previously [21] and in detail in [22]. The program Chimera [39] was used to display the MD averages.
Protein Preparation
Standard methods were used to purify actin and to prepare F-actin and myosin subfragment 1 (S1) [40,41].
Preparation of recombinant wild-type and mutant tropomyosin
The cDNA of α-tropomyosin (Tpm1.1st (a.b.b.a)) was obtained using mouse heart total RNA as a template and following standard protocols of the Fermentas First Strand cDNA Syntheses kit (Thermo Scientific, Pittsburgh, PA). The cloning of the cDNA was further carried out using a TA PCR cloning kit (Invitrogen-Life Technologies, Grand Island, NY). Nine extra nucleotides (ATGGCTAGC) translating into Met-Ala-Ser were added at N-termini of the cDNA to mimic acetylation required for head-to-tail tropomyosin assembly [42]. Site directed mutagenesis was performed using the QuikChange II Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) to replace serine 283 in tropomyosin with either aspartic acid to mimic the phosphorylated state (S283D) or alanine as a control (S283A). A pET-24 (Novagen, Madison, Wisc.) vector containing the T7 promoter, lac operator and a kanamycin resistant gene was used for the expression of wild-type and mutant proteins in E. coli (BL21). The DNA sequences of the expression constructs were verified by DNA sequencing and confirmed the presence of Ala-Ser at the tropomyosin N-terminus. The expressed tropomyosin was extracted from bacterial cells by sonication in 25 mM Tris buffer (pH 8), 25 mM NaCl, 2 mM EDTA, and 0.1% Triton X-100. The protein was then purified by three cycles of precipitation at pH 4.6 and resuspension in 1M KCl buffer (pH 7), followed by ammonium sulfate fractionation. The protein precipitated between 65% and 70% (NH4)2SO4 saturation was collected and dialyzed against 2 mM β-mercaptoethanol and then lyophilized. The purity of the protein was assessed by SDS PAGE.
Electron Microscopy and Persistence Length Determination
Samples of control (wild-type), S283D and S283A tropomyosin constructs were rotary shadowed and electron microscopy carried out as previously [21,43]. Two samples of control and three samples of both S283D and S283A expressed proteins were examined. Proteins were dissolved in a solution of 5 mM Tris (pH 7.0), 5 mM KCl, 2 mM MgCl2, 3 mM DTT, containing 40% glycerol and sprayed onto mica at a concentration of 0.3 μM. Platinum was then evaporated onto the rotating samples at a 6° angle and carbon then deposited using a Cressington 380R Coating System (Cressington Scientific Instruments, Walford, UK). Electron microscopy was carried out on the shadowed molecules using a Philips CM120 electron microscope (FEI, Hillsboro, OR) and images digitized on a 2Kx2K F224 CCD camera (TVIPS, Gauting, Germany). Images of ~80 nm long tropomyosin dimers were selected and skeletonized for persistence length determination of 20 to 30 nm long segments [21,43] following manual assignment of points every 4 to 5 nm along the center of the molecules longitudinal axis [21,43]. The persistence length (PL) was calculated by the tangent correlation method using customized algorithms [21,43], where θ, deviation angles along tropomyosin from an idealized straight rod, was related to segment length (s) using the equation <cos(θ)(s))> = e−s/2PL, with the factor of 2 accounting for the two-dimensionality of the images.
Results and Discussion
Phosphorylation-induced changes in C-terminal residues of unpolymerized tropomyosin
The effect of Ser283 phosphorylation on isolated α-tropomyosin (Tpm1.1st (a.b.b.a)) was followed by MD in explicit water. Here 40-residue long C-terminal fragments disconnected from their N-terminal partners were studied. Comparison of averaged MD trajectories of phosphoserine-containing and control tropomyosin fragments shows that both structures retain coiled-coil conformation along their length, except for the last few C-terminal residues which disorder during MD (Fig. 1a). While these results confirm previous observations on the control fragment [27], they also provide new information about the phosphorylation. The coiled-coil sections of the phosphorylated and unphosphorylated structures superpose with identical inter-helix chain-chain dimensions. In contrast, the C-terminal ends of the two are distinct. Residues 279 to 284 of the phosphorylated structure unravel during MD (a consistent result found in four parallel MD runs), whereas only residues 281 to 284 are unfolded in the control (Fig. 1a). It is plausible that extra phosphorylation-induced unfolding more effectively favors greater head-to-tail junction formation in vitro.
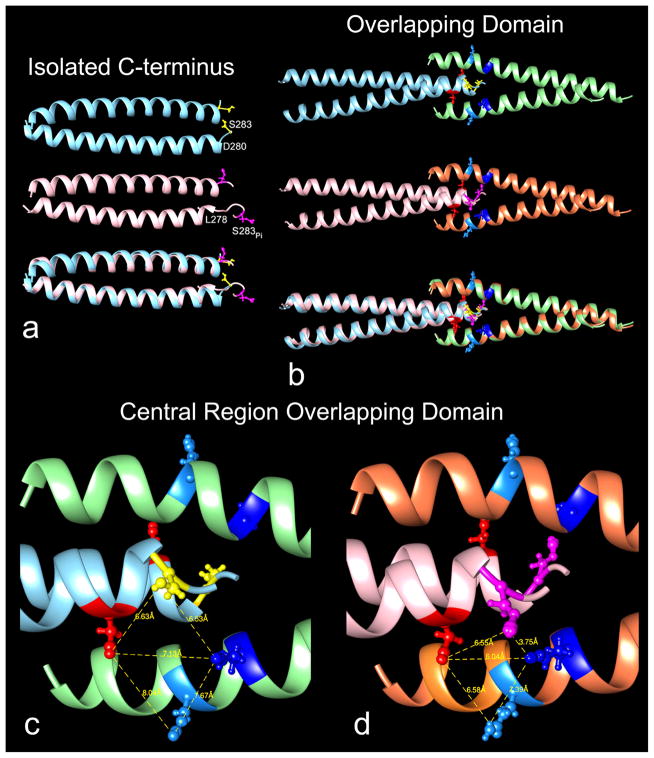
Fig. 1.
The effect of phosphorylation on tropomyosin structure. Ribbon representation of (a) isolated C-terminal domains, (b–d) N-C-terminal overlapping domains, all averaged from MD trajectories; (the coiled-coil rotation differs in (a) and (b,c,d) to best display the serine and phosphoserine 283 residues). (a) upper and middle rows: unphosphorylated (cyan) and phosphorylated (pink) C-terminal domains, bottom row, superposition of the two. Note that the last C-terminal residues are disordered (particularly at the ends one of the two component coiled-coil helices) beginning at residue 281 in unphosphorylated structure and at 279 in the phosphorylated one. The last respective helical residues (viz. 280 and 278) on one chain are labeled as are serine 283 (yellow) and phosphoserine 283 (magenta). (b) upper and middle rows: unphosphorylated and phosphorylated overlapping domains, bottom row superposition of the two (unphosphorylated C-terminal helices (cyan), N-terminal helices (green); phosphorylated C-terminal helices (pink), N-terminal helices (brown)). Average side-chain positions of serine 283 (yellow) and phosphoserine 283 (magenta); lysine 12 (dark blue), glutamine 9 (light blue), glutamate 280 (red) also indicated. Note that the phosphorylated overlapping domain is slightly straighter than unphosphorylated domain (see Table 1), and thus the superposition is not perfect. (c,d) Enlargement of the central region of overlapping domains shown in (b): unphosphorylated (left) and phosphorylated (right). Note the close interaction of phosphoserine 283 (magenta) and lysine 12 (dark blue) in the phosphorylated structure; and that its junctional region becomes more compact with corresponding shorter distances between lysine 12 and glutamate 280 and glutamine 9 and glutamate 280 (distances measured between the set of respective side chain pairs viewed face-on). Graphics and alignment of helices done with Chimera [39].
Phosphorylation-induced changes in tropomyosin overlap domain conformation
Molecular Dynamics simulations were previously carried out on 80-residue long head-to-tail overlapping segments of α-tropomyosin (40 residue contributions from both N- and from C-terminal sequences) responsible for tropomyosin polymerization on actin [31]. The starting structure in these simulations was based on NMR-models of the tropomyosin overlap domain [26]. New 46 ns simulations have now been performed in explicit solvent on the same structure but with the Ser283 hydroxyl side chain replaced by a phosphate group (see Methods section). Both control and phosphorylated structures stay intact throughout respective MD simulations, and here no unfolding or separation of the coiled-coils or the helices in the 4-helix overlap nexus is observed. In each case, the serine side chains point away from the hydrophobic core of the nexus and remain well separated throughout simulation.
The averaged backbone structures taken from MD trajectories of phosphorylated and unphosphorylated tropomyosin are virtually the same as each other and the two are essentially superposable (Fig. 1b). However, closer inspection of the region surrounding Ser283 indicates that the local geometry of side chain contacts between N- and C-terminal segments is rearranged following phosphorylation (Fig. 1b–d). The phosphorylated Ser283 side chains now closely approach residue Lys12 on the neighboring N-terminal chain at an average distance of ~3.7 Å to form a salt bridge, an effect which is seen almost instantaneously during simulation (see Supplementary Item Fig. S1). The tightening of the overlap domain caused by the lysine - phosphoserine interaction brings Lys12 closer to Asp280, and in turn Asp280 draws in Gln9, making the entire region more compact, and possibly more stable. Hence our results are consistent with earlier measurements of phosphorylation-induced increases in tropomyosin viscosity, considered to reflect enhanced head-to-tail tropomyosin interaction [29,30]. However, our MD additionally shows that the side-chains of tropomyosin residues Lys7, Asp20 and Glu23 thought to normally face and contact actin on thin filaments [24] retain their original external orientations during simulation. Thus potential interactions with actin are not compromised by the phosphorylation-induced changes, suggesting that the equilibrium binding site of tropomyosin on actin is not altered by phosphorylation.
Tropomyosin overlap domain curvature and flexibility
MD studies
The curvature of the averaged tropomyosin overlap domain MD structure decreases following phosphorylation, and then matches the bending angle found along the rest of the molecule (Table 1) [cf. 31,32]. In addition, the variance in curvature, measuring flexibility, is lower for the phosphorylated structure, indicating that phosphorylation stiffens the overlap segment.
Table 1.
Molecular Dynamics determination of tropomyosin overlap domain curvature and flexibility.
| Sample Region of Tropomyosin Sampled | Bending Angle ± Std. Deviation (°) | Apparent Persistence Length (nm) | Intrinsic Persistence Length (nm) | Dynamic Persistence Length (nm) |
|---|---|---|---|---|
| Unphosphorylated Tm overlap domain | 9.3 ± 4.8° | 91 nm | 124 nm | 342 nm |
| Phosphorylated Tm overlap domain | 8.4 ± 4.1° | 117 nm | 156 nm | 469 nm |
| Full-length Tm control (from ref. [21]) | 8.6 ± 4.3° | 106 nm | 141 nm | 423 nm |
Persistence length determinations gauging the curvature and bending stiffness of “rod-like” tropomyosin were also carried out. Standard persistence length analysis quantifies fluctuations of straight rods from their average vertical orientation. Because tropomyosin is a curved rather than a straight molecule, bending deviations from a hypothetical straight reference orientation yields a so-called “apparent” persistence length value (PLa) [21,22]. The apparent persistence length value obtained reflects both the intrinsic curvature of tropomyosin and the bending fluctuations about its continuously curved, average shape. However, the apparent persistence length of an MD structure can be decomposed into two terms: an intrinsic or static persistence length (PLi), which reflects tropomyosin’s average curvature, and a dynamic persistence length (PLd), which measures its deviations from the average structure and hence the protein’s flexibility [21,22]. Persistence length evaluation of respective MD trajectories confirms that phosphorylation both straightens and stiffens the overlap domain (Table 1).
Electron Microscopy
In the above in silico work, direct assessment of the effect of phosphorylation on the mechanics of tropomyosin was done computationally by substituting phosphoserine for serine in the reference model used to initiate MD. Corresponding in vitro work on fully phosphorylated tissue isolated tropomyosin is not as straightforward, since the requisite phosphorylating enzyme has not been characterized. Therefore, we and others [30] have instead expressed and then studied “phosphomimetic” tropomyosin analogs. In fact, our own observations and those of others indicate that S283D and S283E phosphomimetics behave like the phosphorylated protein to increase tropomyosin viscosity. Standard addition of Ala-Ser residues at N-termini of E. coli expressed tropomyosin mimics acetylation required for head-to-tail tropomyosin association [42].
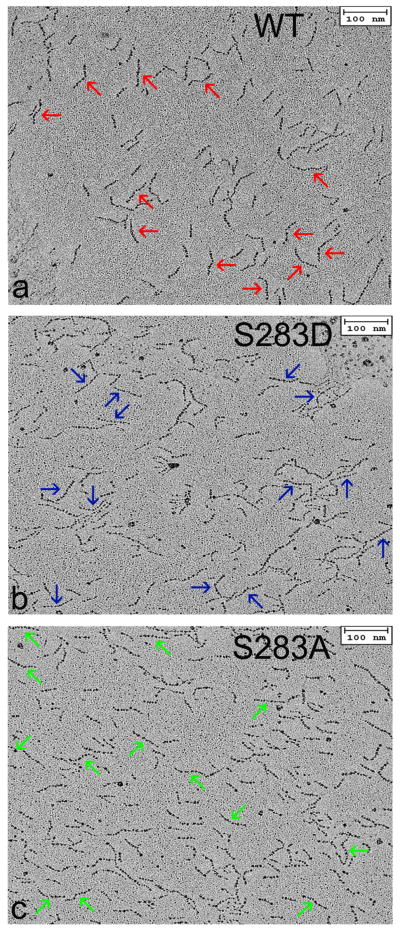
To evaluate the contribution of charged serine 283 on the bending stiffness of tropomyosin experimentally, samples of full-length wild-type and phosphomimetic S283D tropomyosin were rotary shadowed and directly visualized by EM. The EM images reveal the outlines of the gently curved elongated molecules (Fig. 2). The samples contain a mixture of single 40 nm long coiled-coils, 80 nm long end-to-end linked molecules (referred to here as dimers) and oligomeric polymers of varied length, which are all easily distinguished from each other.
Fig. 2.
Electron microscopy of isolated tropomyosin molecules. (a, b, c) Rotary shadowed wild-type, S283D and S283A tropomyosin molecules. Micrographs show a mixture of tropomyosin monomers, dimers and oligomers. Arrows indicate examples of dimers used to compare potential contributions of the tropomyosin head-to-tail overlap domains to the stiffness of the tropomyosin cable. For this purpose, thirty or more dimers were skeletonized and persistence lengths determined (see Table 2). Scale bars – 100 nm.
End-to-end linked tropomyosin dimers have the simplest unit dimensions needed to estimate local contour variance at the overlap domain and along the rest of the tropomyosin polymer [43], since the center of the dimer contains the head-to-tail domain and the periphery the canonical coiled-coil. Hence, the dimers, which represent about 15 percent of the total population of tropomyosin particles observed, offer a convenient means of assessing and comparing overlap contours of phosphomimetic and control tropomyosin. While quantitation of these contours provides persistence length information, unlike the MD work done in silico, EM images lack three-dimensionality but rather are two-dimensional projections of three-dimensional objects. The two-dimensionality, relatively low-resolution and the static nature of the EM data limit analysis to “apparent” persistence measurements.
We previously quantified EM images of wild-type tropomyosin dimers to determine apparent persistence lengths along the structures [43]. Our results showed that the central segment, containing the overlapping domain, had the lowest persistence length, as our current analysis and our MD work also indicate (Table 2). We now also repeated the procedure on full-length S283D tropomyosin to mimic the effect of tropomyosin phosphorylation. In this case, the persistence length of the central region of the dimer increases to match the value for the rest of the molecule. Thus, the in vitro experimental work confirms the MD study that serine 283 phosphorylation stiffens and/or straightens the tropomyosin overlap domain.
Table 2.
Apparent persistence length determination of rotary shadowed tropomyosin. Note that engineered tropomyosin constructs reported here contained unacetylated Ala-Ser N-termini and that phosphomimetic glutamate was used to mimic phosphoserine residue 283 with Ala283 as a control. These substitutions may account for the small quantitative differences in PLa values from those reported in Table 1 obtained by MD on unphosphorylated and phosphorylated models. Note, however, that the wild-type apparent persistence length values for the overlapping and non-overlapping regions of expressed tropomyosin (63, 105 nm) are virtually the same as those previously calculated for tissue purified tropomyosin (63, 108 nm), and that the values for overlapping and non-overlapping regions of S283D tropomyosin are almost identical to those of the non-overlapping region of the wild-type (105, 107, 105 nm)[cf. 20,40]. A diminution of persistence length at the overlapping region of “control” S282A tropomyosin is evident but the effect appears to be dampened relative to that of the wild-type.
The reliability of the data was assessed by comparing the persistence lengths of “half-data sets”. When image data were split in this way, comparable persistence length values for each related half-data set pair were obtained, suggesting that the measurement errors are small. Moreover, persistence length values obtained for tropomyosin monomers present in our samples of tissue purified [21,43] and corresponding values for the 3 tropomyosin constructs examined here were all the same, again indicating the reliability of the data (data not shown).
| Sample | Apparent Persistence Length (nm) |
|---|---|
| WT tropomyosin overlap region | 63 nm |
| WT tropomyosin non-overlap region | 105 nm |
| S283D tropomyosin overlap region | 105 nm |
| S283D tropomyosin non-overlap region | 107 nm |
| S283A tropomyosin overlap region | 84 nm |
| S283A tropomyosin non-overlap region | 99 nm |
Conclusions
Our data indicate that Ser283 phosphorylation straightens and stiffens the tropomyosin overlap domain, which then better matches the helical curvature and mechanical parameters of the rest of the tropomyosin molecule. In this way, phosphorylation enables tropomyosin molecules to fit more readily onto preformed or growing actin filaments and consequently polymerize head-to-tail. Hence, phosphorylation will result in tropomyosin producing a more mechanically uniform cable along the actin filament helix. In turn, a stiffer, seamless tropomyosin cable is likely to enhance thin filament activation, as is evident from the potentiation of acto-S1 ATPase by phosphorylated tropomyosin [29], and confirmed by our corresponding comparison of the S283D and S283A tropomyosin stimulation of acto-S1 ATPase (see Supplementary Item, Fig. S2). It follows that regulatory movement of tropomyosin on thin filaments in response to Ca2+ binding to troponin and myosin binding to actin on troponin-tropomyosin regulated filaments will become more sensitive, without compromising the extent of thin filament relaxation [cf. references 29, 44]. Phosphorylation-induced enhanced thin filament responsiveness to narrower cellular Ca2+ concentrations ranges will be advantageous to fetal muscle, where the control of intracellular Ca2+-levels may not be fully developed [45–47], and provide compensatory mechanisms in myopathic muscle [48], where Ca2+ metabolism may be compromised.
Without a high resolution model of troponin-tropomyosin, we cannot determine with any certainty the higher order structural impact of phosphorylation, for example, on interactions of closely linked tropomyosin and TnT. Despite well-known indications that the C-terminal TnT domains localize near to the phosphorylation site on the tropomyosin overlapping domain [15,23,49], no residue-residue specific structure has been solved for the binary complex. In fact, our own data suggest that tropomyosin phosphorylation may actually weaken TnT – tropomyosin interaction (Table 3, see Supplementary Item, Fig. S3). Acquiring a fuller appreciation of the global role played by tropomyosin phosphorylation in modulating thin filament behavior requires detailed maps of the interactions of all thin filament components.
Table 3.
Binding of TnT to actin-tropomyosin.
| Sample tested | Kd Actin-Tropomyosin to TnT1 (μM) |
|---|---|
| Actin-WT tropomyosin | 0.201 ± 0.030 |
| Actin-S283D tropomyosin | 0.284 ± 0.043 |
| Actin-S283A tropomyosin | 0.128 ± 0.018 |
A cosedimentation assay [50] was used to estimate the binding of various actin-tropomyosin complexes to TnT1. TnT1, the tropomyosin binding domain of TnT, was expressed in E. coli [51]. (TnT1, which is soluble in aqueous solution, was used since full-length TnT is insoluble when not part of the troponin complex). Curves (Supplementary Item Fig. S3) plotting the binding of TnT1 to actin-tropomyosin samples were used to determine the apparent Kd values (± std. dev.) cited, which are the average of three independent measurements. Note that engineered tropomyosin constructs used here contained Ala-Ser N-termini.
Supplementary Material
Highlights.
Molecular Dynamics simulations were carried out on phosphorylated tropomyosins.
MD shows that phosphorylated Ser283 binds to Lys12 on adjacent tropomyosins.
Phosphorylation straightens and strengthens tropomyosin end-to-end overlap domains.
The structural contour of the overlap domain then matches the rest of the molecule.
Phosphorylation is likely to facilitate both thin filament assembly and activation.
Acknowledgments
These studies were supported by NIH grants R37HL036153 (to W.L.), R01HL077280 (to J.R.M.), R01HL011197 (to M.R.), R01HD048895 (to M.R.; M.J. Bamshad, P.I.) and by Wellcome Trust grant 085309 and British Heart Foundation grant 30200 (to M.A.G). The Massachusetts Green High Performance Computing Center provided computational resources.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Barany M, Barany K. Protein phosphorylation in cardiac and vascular smooth muscle. Am J Physiol. 1981;241:H117–H128. doi: 10.1152/ajpheart.1981.241.2.H117. [DOI] [PubMed] [Google Scholar]
- 2.Solaro RJ, Kobayashi T. Protein phosphorylation and signal transduction in cardiac thin filaments. J Biol Chem. 2011;286:9935–9940. doi: 10.1074/jbc.R110.197731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lim MS, Walsh MP. Phosphorylation of skeletal and cardiac muscle C-proteins by the catalytic subunit of cAMP-dependent protein kinase. Biochem Cell Biol. 1986;64:622–630. doi: 10.1139/o86-086. [DOI] [PubMed] [Google Scholar]
- 4.Pfuhl M, Gautel M. Structure, interactions and function of the N-terminus of cardiac myosin binding protein C (MyBP-C): who does what, with what, and to whom? J Muscle Res Cell Motil. 2012;33:83–94. doi: 10.1007/s10974-012-9291-z. [DOI] [PubMed] [Google Scholar]
- 5.Solaro RJ, Henze M, Kobayashi T. Integration of troponin I phosphorylation with cardiac regulatory networks. Circ Res. 2013;112:355–366. doi: 10.1161/CIRCRESAHA.112.268672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Streng AS, de Boer D, van der Velden J, van Dieijen-Visser MP, Wodzig WK. Posttranslational modifications of cardiac troponin T: an overview. J Mol Cell Cardiol. 2013;63:47–56. doi: 10.1016/j.yjmcc.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 7.Heeley DH. Phosphorylation of tropomyosin in striated muscle. J Muscle Res Cell Motil. 2013;34:233–237. doi: 10.1007/s10974-013-9351-z. [DOI] [PubMed] [Google Scholar]
- 8.Peng Y, Yu D, Gregorich Z, Chen X, Beyer AM, Gutterman DD, Ge Y. In-depth proteomic analysis of human tropomyosin by top-down mass spectrometry. J Muscle Res Cell Motil. 2013;34:199–210. doi: 10.1007/s10974-013-9352-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marston SB, Copeland O. Tropomyosin isoform expression and phosphorylation in the human heart in health and disease. J Muscle Res Cell Motil. 2013;34:189–197. doi: 10.1007/s10974-013-9347-8. [DOI] [PubMed] [Google Scholar]
- 10.Wang CL, Coluccio LM. New insights into the regulation of the actin cytoskeleton by tropomyosin. Int Rev Cell Mol Biol. 2010;281:91–128. doi: 10.1016/S1937-6448(10)81003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Linke WA, Hamdani N. Gigantic business: titin properties and function through thick and thin. Circ Res. 2014;114:1052–1068. doi: 10.1161/CIRCRESAHA.114.301286. [DOI] [PubMed] [Google Scholar]
- 12.LeWinter MM, Granzier HL. Cardiac titin and heart disease. J Cardiovasc Pharmacol. 2014;63:207–212. doi: 10.1097/FJC.0000000000000007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heeley DH, Moir AJG, Perry SV. Phosphorylation of tropomyosin during development in mammalian striated muscle. FEBS Lett. 1982;146:115–118. doi: 10.1016/0014-5793(82)80716-3. [DOI] [PubMed] [Google Scholar]
- 14.Schulz EM, Wilder T, Chowdhury SA, Sheikh HN, Wolska BM, Solaro RJ, Wieczorek DF. Decreased tropomyosin dephosphorylation results in compensated cardiac hypertrophy. J Biol Chem. 2012;287:44478–44489. doi: 10.1074/jbc.M112.402040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tobacman LS. Thin filament-mediated regulation of cardiac contraction. Annu Rev Physiol. 1996;58:447–481. doi: 10.1146/annurev.ph.58.030196.002311. [DOI] [PubMed] [Google Scholar]
- 16.Gordon AM, Homsher E, Regnier M. Regulation of contraction in striated muscle. Physiol Rev. 2000;80:853–924. doi: 10.1152/physrev.2000.80.2.853. [DOI] [PubMed] [Google Scholar]
- 17.Poole KJ, Lorenz M, Evans G, Rosenbaum G, Pirani A, Tobacman LS, Lehman W, Holmes KC. A comparison of muscle thin filament models obtained from electron microscopy reconstructions and low-angle X-ray fibre diagrams from non-overlap muscle. J Struct Biol. 2006;155:273–284. doi: 10.1016/j.jsb.2006.02.020. [DOI] [PubMed] [Google Scholar]
- 18.Holmes KC, Lehman W. Gestalt-binding of tropomyosin to actin filaments. J Muscle Res Cell Motil. 2008;29:213–219. doi: 10.1007/s10974-008-9157-6. [DOI] [PubMed] [Google Scholar]
- 19.Lehman W, Craig R. Tropomyosin and the steric mechanism of muscle regulation. Adv Exp Med Biol. 2008;644:95–109. doi: 10.1007/978-0-387-85766-4_8. [DOI] [PubMed] [Google Scholar]
- 20.Wegner A. The interaction of α,α- and α,β-tropomyosin with actin filaments. FEBS Lettr. 1980;119:245–248. doi: 10.1016/0014-5793(80)80263-8. [DOI] [PubMed] [Google Scholar]
- 21.Li XE, Holmes KC, Lehman W, Jung H, Fischer S. The shape and flexibility of tropomyosin coiled coils: implications for actin filament assembly and regulation. J Mol Biol. 2010;395:327–339. doi: 10.1016/j.jmb.2009.10.060. [DOI] [PubMed] [Google Scholar]
- 22.Li XE, Lehman W, Fischer S. The relationship between curvature, flexibility and persistence length in the tropomyosin coiled-coil. J Struct Biol. 2010;170:313–318. doi: 10.1016/j.jsb.2010.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown JH, Cohen C. Regulation of muscle contraction by tropomyosin and troponin: how structure illuminates function. Adv Protein Chem. 2005;71:121–159. doi: 10.1016/S0065-3233(04)71004-9. [DOI] [PubMed] [Google Scholar]
- 24.Orzechowski M, Li XE, Fischer S, Lehman W. An atomic model of the tropomyosin cable on F-actin. Biophys J. 2014;107:694–699. doi: 10.1016/j.bpj.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, Mui S, Brown JH, Strand J, Reshetnikova L, Tobacman LS, Cohen C. The crystal structure of the C-terminal fragment of striated-muscle alpha-tropomyosin reveals a key troponin T recognition site. Proc Natl Acad Sci U S A. 2002;99:7378–7383. doi: 10.1073/pnas.102179999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greenfield NJ, Huang YJ, Swapna GV, Bhattacharya A, Rapp B, Singh A, Montelione GT, Hitchcock-DeGregori SE. Solution NMR structure of the junction between tropomyosin molecules: implications for actin binding and regulation. J Mol Biol. 2006;364:80–96. doi: 10.1016/j.jmb.2006.08.033. [DOI] [PubMed] [Google Scholar]
- 27.Greenfield NJ, Swapna GV, Huang Y, Palm T, Graboski S, Montelione GT, Hitchcock-DeGregori SE. The structure of the carboxyl terminus of striated alpha-tropomyosin in solution reveals an unusual parallel arrangement of interacting alpha-helices. Biochemistry. 2003;42:614–619. doi: 10.1021/bi026989e. [DOI] [PubMed] [Google Scholar]
- 28.Geeves MA, Hitchcock-DeGregori SE, Gunning PW. A systematic nomenclature for mammalian tropomyosin isoforms. J Muscle Res Cell Motil. 2014 doi: 10.1007/s10974-014-9389-6. in press http://DOI:10.1007/s10974-014-9389-6. [DOI] [PMC free article] [PubMed]
- 29.Heeley DH, Watson MH, Mak AS, Dubord P, Smillie LB. Effect of phosphorylation on the interaction and functional properties of rabbit striated muscle αα-tropomyosin. J Biol Chem. 1989;264:2424–2430. [PubMed] [Google Scholar]
- 30.Sano K, Maéda K, Oda T, Maéda Y. The effect of single residue substitutions of serine-283 on the strength of head-to-tail interaction and actin binding properties of rabbit skeletal muscle alpha-tropomyosin. J Biochem. 2000;127:1095–1102. doi: 10.1093/oxfordjournals.jbchem.a022703. [DOI] [PubMed] [Google Scholar]
- 31.Li XE, Orzechowski M, Lehman W, Fischer S. Structure and flexibility of the tropomyosin overlap junction. Biochem Biophys Res Commun. 2014;446:304–308. doi: 10.1016/j.bbrc.2014.02.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hitchcock-DeGregori SE. Tropomyosin: function follows structure. Adv Exp Med Biol. 2008;644:60–72. doi: 10.1007/978-0-387-85766-4_5. [DOI] [PubMed] [Google Scholar]
- 33.Lehman W, Li XE, Orzechowski M, Fischer S. The structural dynamics of α-tropomyosin on F-actin shape the overlap complex between adjacent tropomyosin molecules. Arch Biochem Biophys. 2014;552–553:68–73. doi: 10.1016/j.abb.2013.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brooks BR, Brooks CL, MacKerell AD, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M. CHARMM: The biomolecular simulation program. J Comput Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li XE, Tobacman LS, Mun JY, Craig R, Fischer S, Lehman W. Tropomyosin position on F-actin revealed by EM reconstruction and computational chemistry. Biophys J. 2011;100:1005–1013. doi: 10.1016/j.bpj.2010.12.3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J Comp Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 38.Mackerell AD, Feig M, Brooks CL. Extending the treatment of backbone energetics in protein force fields: Limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J Comput Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 39.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 40.Spudich JA, Watt S. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J Biol Chem. 1971;246:246, 4866–4871. [PubMed] [Google Scholar]
- 41.Margossian SS, Lowey S. Preparation of myosin and its subfragments from rabbit skeletal muscle. Meth Enzymol. 1982;85B:55–71. doi: 10.1016/0076-6879(82)85009-x. [DOI] [PubMed] [Google Scholar]
- 42.Monteiro PB, Lataro RC, Ferr JA, Reinach FC. Functional alpha-tropomyosin produced in Escherichia coli. A dipeptide extension can substitute the amino-terminal acetyl group. 1994;269:10461–10466. [PubMed] [Google Scholar]
- 43.Sousa D, Cammarato A, Jang K, Graceffa P, Tobacman LS, Li XE, Lehman W. Electron microscopy and persistence length analysis of semi-rigid smooth muscle tropomyosin strands. Biophys J. 2010;99:1–7. doi: 10.1016/j.bpj.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rao VS, Marongelli EN, Guilford WH. Phosphorylation of tropomyosin extends cooperative binding beyond a single regulatory unit. Cell Motil Cytoskeleton. 2009;66:10–23. doi: 10.1002/cm.20321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Katz AM. Physiology of the Heart. 3. Lippincott/Williams & Wilkins; Philadelphia: 2001. [Google Scholar]
- 46.Chen F, Ding S, Lee BS, Wetzel GT. Sarcoplasmic reticulum Ca2+ ATPase and cell contraction in developing rabbit heart. J Mol Cell Cardiol. 2000;32:745–755. doi: 10.1006/jmcc.2000.1116. [DOI] [PubMed] [Google Scholar]
- 47.Kawamura Y, Ishiwata T, Takizawa M, Ishida H, Asano Y, Nonoyama S. Fetal and neonatal development of Ca2+ transients and functional sarcoplasmic reticulum in beating mouse hearts. Circ J. 2010;74:1442–1145. doi: 10.1253/circj.cj-09-0793. [DOI] [PubMed] [Google Scholar]
- 48.Schulz EM, Wilder T, Chowdhury SA, Sheikh HN, Wolska BM, Solaro RJ, Wieczorek DF. Decreasing tropomyosin phosphorylation rescues tropomyosin-induced familial hypertrophic cardiomyopathy. J Biol Chem. 2013;288:28925–28935. doi: 10.1074/jbc.M113.466466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jin JP, Chong SM. Location of the two tropomyosin-binding sites of troponin-T. Arch Biochem Biophys. 2010;500:144–150. doi: 10.1016/j.abb.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Coulton AT, Lehrer SS, Geeves SSMA. Functional homodimers and heterodimers of recombinant smooth muscle tropomyosin. Biochemistry. 2006;45:12853–12858. doi: 10.1021/bi0613224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Coulton AT, Koka K, Lehrer SS, Geeves MA. Role of the head-to-tail overlap region in smooth and skeletal muscle beta-tropomyosin. Biochemistry. 2008;47:388–397. doi: 10.1021/bi701144g. [DOI] [PubMed] [Google Scholar]
- 52.Glyn H, Sleep J. Dependence of adenosine triphosphatase activity of rabbit psoas muscle fibres and myofibrils on substrate concentration. J Physiol. 1985;365:259–276. doi: 10.1113/jphysiol.1985.sp015770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lehrer SS, Morris EP. Dual effects of tropomyosin and troponin-tropomyosin on actomyosin subfragment 1 ATPase. J Biol Chem. 1982;257:8073–8080. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.