Abstract
Cyclic nucleotide phosphodiesterases (PDEs) have been identified as important enzyme targets for drug development in both humans and in Trypanosoma brucei, the causative agent of human African trypanosomiasis (HAT). With this in mind, we recently reported the profiling of a range of human PDE inhibitors, showing that human PDE4 (hPDE4) inhibitors tend to display the best potency against the trypanosomal phosphodiesterase TbrPDEB1. Among these was GSK-256066, a potent inhibitor of hPDE4 and a weak inhibitor of TbrPDEB1. In this report, we describe the results of a structure-activity relationship study of this chemotype, leading to the discovery of analogs with improved potency against TbrPDEB1 and micromolar inhibition of T. brucei cellular growth. We rationalize the potency trends via molecular docking of the new inhibitors into a recently reported apo structure of TbrPDEB1. The studies in this article will inform future efforts in repurposing human PDE inhibitors as anti-trypanosomal agents.
Keywords: GSK-256066, Phosphodiesterase inhibitors, Trypanosoma brucei, TbrPDEB1, TbrPDEB2
Introduction
Human African trypanosomiasis (HAT) is a neglected tropical disease that infects approximately 10,000 patients in sub-Saharan Africa [1]. Caused by the protozoan parasite Trypanosoma brucei, the advanced stage of HAT consists of an infection of the central nervous system, a condition that is invariably fatal if untreated. Current treatments for the CNS stage of HAT are suboptimal, including eflornithine (alone, or now in combination with nifurtimox) [2], which is only active against the T. brucei gambiense subspecies, and melarsoprol, which is a toxic organoarsenical agent whose toxicity is such that approximately 5% of patients will die from its side effects [3].
Since HAT affects some of the poorest patients in the world, there is little financial incentive to pursue costly de novo drug discovery programs. With this in mind, we have applied a target repurposing approach to therapeutics discovery, wherein essential parasite targets and pathways are matched with druggable human homologs that have existing chemical matter that target them [4]. These compounds can provide new leads for antiparasitic drug discovery without undertaking a costly high-throughput screening campaign. We have pursued this approach with a variety of kinase [5, 6] and phosphodiesterase (PDE) inhibitors [7-10].
PDE inhibitors have been developed for a variety of indications, including treatment of erectile dysfunction and pulmonary hypertension (PDE5), and chronic obstructive pulmonary disease (PDE4). The success of these efforts is evident in the approval of various selective PDE inhibitors for clinical use [11-14]. T. brucei expresses five PDEs, including the homologs TbrPDEB1 and B2, which have been together demonstrated to be essential by RNAi, such that both enzymes must be inhibited in order to affect parasite survival.[15] These two enzymes are highly homologous (88.5%) [15], and we have shown previously [7] (and within this work) that inhibitors tested against both enzymes most frequently display similar potency against both. Furthermore, the essentiality data and sequence similarity between human and trypanosomal PDEs led us to believe that target repurposing could be a fruitful approach for new inhibitor discovery [7].
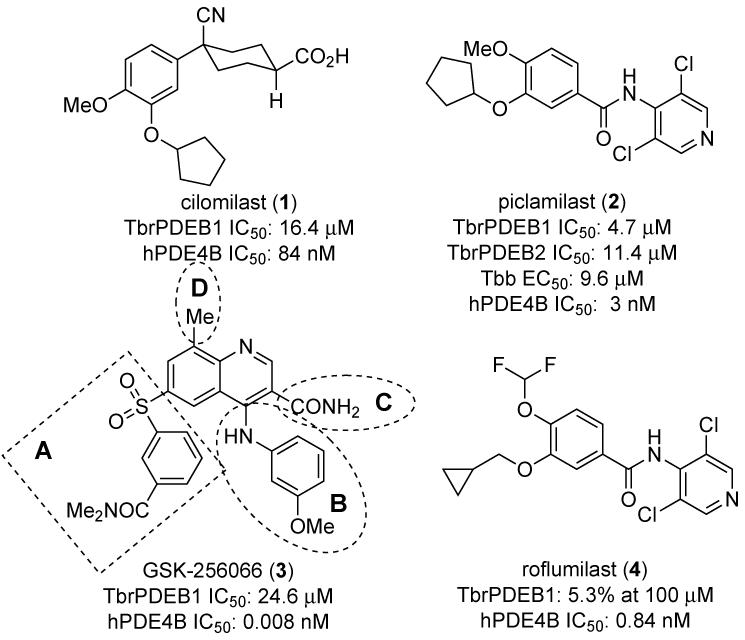
We previously reported the assessment of a range of established human PDE chemotypes against TbrPDEB1 and B2, and reported that these enzymes are susceptible to a number of chemotypes, primarily derivatives of established human PDE4 (hPDE4) inhibitors (Figure 1). Besides piclamilast (1) and cilomilast (2), we also identified GSK-256066 (3)[16], an investigational compound for chronic obstructive pulmonary disease (COPD), as a weak inhibitor of TbrPDEB1 [7].
Figure 1.
Previously benchmarked human PDE4 inhibitors [7, 16, 31, 32].
Besides improving potency at the trypanosomal target, another significant issue for any target repurposing program is to identify divergent structure-activity relationships (SAR) between the host and pathogen enzymes. Such selectivity is important in order to reduce potentially troublesome side-effect profiles, such as emesis, as observed with most hPDE4 inhibitors, which has been a significant challenge to date [7, 17].
Material and Methods
TbrPDEB1 Biochemical assay
Biochemical assays were performed as previously described [7] and are described in detail in the Supporting Information.
Human PDE4B biochemical assay
This assay was performed at Takeda Pharmaceuticals using methods previously reported [18].
Trypanosome cell culture assays
Bloodstream forms of Trypanosoma brucei brucei strain 427 were grown at 37 °C in a 5% CO2 atmosphere in HMI-11 medium supplemented with 10% fetal bovine serum (FBS, Sigma). Cells in the mid-logarithmic stage of growth were diluted to a density of 104 cells/ml and were incubated with a range of concentrations of inhibitor in DMSO or DMSO alone. The final concentration of DMSO was 1%. Cell densities were determined after 48 h using Alamar blue (Invitrogen) per the manufacturer’s instructions. All values are the mean of three or more independent experiments.
Chemical Synthesis
Unless otherwise noted, reagents were obtained from Sigma-Aldrich, Inc. (St. Louis, MO), Fisher Scientific, Frontier Scientific Services, Inc. (Newark, DE), Matrix Scientific (Columbia, SC) and used as received. Boronic acids/esters and aniline reagents were purchased, except for the boronates listed in the Supporting Information. Reaction solvents were purified by passage through alumina columns on a purification system manufactured by Innovative Technology (Newburyport, MA). Microwave reactions were performed using a Biotage Initiatior-8 instrument. NMR spectra were obtained with Varian NMR systems, operating at 400 or 500 MHz for 1H acquisitions as noted. LCMS analysis was performed using a Waters Alliance reverse-phase HPLC, with single-wavelength UV-visible detector and LCT Premier time-of-flight mass spectrometer (electrospray ionization). All newly synthesized compounds were that were submitted for biological testing were deemed >95% pure by LCMS analysis (UV and ESI-MS detection) prior to submission for biological testing. Preparative LCMS was performed on a Waters FractionLynx system with a Waters MicroMass ZQ mass spectrometer (electrospray ionization) and a single-wavelength UV-visible detector, using acetonitrile/H2O gradients with 0.1% formic acid. Fractions were collected on the basis of triggering using UV and mass detection. Yields reported for products obtained by preparative HPLC represent the amount of pure material isolated; impure fractions were not repurified.
ethyl 6-iodo-8-methyl-4-oxo-1,4-dihydroquinoline-3-carboxylate (6)
To 4-iodo-2-methylaniline (5a) (5.35 g, 22.96 mmol) was added diethyl 2-(ethoxymethylene)malonate (5.10 mL, 25.3 mmol) and the reaction mixture was heated at 100 °C for 1 hour. The heat was removed and the white solid that formed was collected, washed with cyclohexane (70 mL) and ethanol 30 mL (2×), and dried in vacuo at 40 °C overnight to give diethyl 2-(((4-iodo-2-methylphenyl)amino)methylene)malonate (Yield: 98%). 1H NMR (500 MHz, DMSO-d6) δ 10.83 (d, J = 13.6 Hz, 1H), 8.42 (d, J = 13.1 Hz, 1H), 7.65 (s, 1H), 7.56 - 7.61 (m, 1H), 7.25 (d, J = 8.3 Hz, 1H), 4.20 (q, J = 7.3 Hz, 2H), 4.11 (q, J = 7.3 Hz, 2H), 2.25 (s, 3H), 1.24 (td, J = 6.9, 10.5 Hz, 6H). LCMS found 404.16 [M+H]+. Then to the diethyl 2-((4-iodo-2-methylphenylamino)methylene)malonate (9.0 g, 22.3 mmol) was added diphenyl ether (35.5 mL, 223 mmol). The reaction was run for 45 min at 250 °C. The mixture was cooled and isohexane was added (30 mL). The solid formed (light yellow solid) was collected by filtration and washed further with isohexane (30 mL). The solid was dried under high vacuum to give the desired product as a light yellow solid (Yield: 100%). 1H NMR (400 MHz, DMSO-d6) δ 11.74 (br. s., 1H), 8.37 (s, 1H), 8.28 (s, 1H), 7.89 (s, 1H), 4.21 (q, J = 7.3 Hz, 2H), 1.26 (t, J = 7.3 Hz, 3H). LCMS found 357.95 [M+H]+.
4-chloro-6-iodo-8-methylquinoline-3-carboxamide (7)
NaOH (1.96 g, 49.1 mmol) was dissolved in water (40mL) and ethanol (20 mL). The resultant solution was added to 6 (7.97 g, 22.32 mmol) and the mixture was heated and refluxed for 1 hour with stirring. Then concentrated HCl was added until a white precipitate formed. The reaction was stirred overnight at room temperature. After stirring overnight the precipitate was filtered, washed with water and dried in vacuo to give 6-iodo-8-methyl-4-oxo-1,4-dihydroquinoline-3-carboxylic acid as a white solid. (LCMS found 329.89 [M+H]+). To 6-iodo-8-methyl-4-oxo-1,4-dihydroquinoline-3-carboxylic acid (7.34 g, 22.30 mmol) was added thionyl chloride (24.42 mL, 335 mmol), then 3 drops DMF were added. The mixture was refluxed for 2 hours. The excess thionyl chloride was evaporated in vacuo and the residue was azeotroped with toluene (5 mL, 1×). The crude product 4-chloro-6-iodo-8-methylquinoline-3-carbonyl chloride was used in the next step without further characterization. To stirred ammonium hydroxide (34.7 mL, 892 mmol) was added portion wise 4-chloro-6-iodo-8-methylquinoline-3-carbonyl chloride (8.16 g, 22.3 mmol) and the mixture was stirred at room temperature overnight. The solid formed was filtered, washed with water and dried under vacuum at 60 °C to give 7 as a white solid (Yield: 80%). 1H NMR (400 MHz, DMSO-d6) δ 8.89 (s, 1H), 8.46 (s, 1H), 8.20 (br. s., 1H), 8.10 (s, 1H), 7.99 (br. s., 1H), 2.70 (s, 3H). LCMS found 346.87 [M+H]+.
6-iodo-4-(3-methoxyphenylamino)-8-methylquinoline-3-carboxamide (8)
Compound 7 (174 mg, 0.499 mmol) was dissolved in acetonitrile and 3-methoxyaniline (0.059 mL, 0.52 mmol) was added. The mixture was heated and refluxed overnight. The precipitate formed was filtered, washed with acetonitrile and the solid (light yellow solid) obtained was dried under high vacuum to give the title compound (Yield: 90%). 1H NMR (400 MHz, DMSO-d6) δ 8.78 (s, 1H), 8.39 (br. s., 1H), 8.31 (br. s., 1H), 8.10 (br. s., 1H), 7.74 (br. s., 1H), 7.28 (t, J = 8.0 Hz, 1H), 6.73 - 6.86 (m, 3H), 3.72 (s, 3H), 2.65 (s, 3H). LCMS found 434.97 [M+H]+.
methyl 3-(3-carbamoyl-4-(3-methoxyphenylamino)-8-methylquinolin-6-ylthio)benzoate (9a)
To 8 (20.0 mg, 0.046 mmol) was added Pd2(dba)3 (8.4 mg, 9.23 μmol), then 2,2′-oxybis(2,1-phenylene)bis(diphenylphosphine) (9.9 mg, 0.018 mmol). The methyl 3-mercaptobenzoate (11.6 mg, 0.069 mmol) was dissolved in toluene (1.5 mL) and then added to the solids. Lastly, KOtBu (0.092 mL, 0.092 mmol) was added. The mixture was placed in the MW and heated for 40 minutes at 170 °C. The crude product was filtered through Celite after adding methanol and further washed with methanol. The solvent was concentrated. The crude product was chromatographed using 50-100% EtOAC in hexane to give the desired product 9a as an orange solid (Yield: 75%).1H NMR (400 MHz, CD3OD) δ 8.91 (s, 1H), 7.91 (d, J = 7.3 Hz, 1H), 7.85 (s, 1H), 7.50 (s, 1H), 7.42 - 7.46 (m, 2H), 7.33 - 7.40 (m, 1H), 6.99 (t, J = 8.0 Hz, 1H), 6.49 - 6.53 (m, 1H), 6.30 - 6.36 (m, 2H), 3.91 (s, 3H), 3.66 (s, 3H), 2.69 (s, 3H). LCMS found 474.01, [M+H]+.
4-((3-methoxyphenyl)amino)-8-methyl-6-(phenylthio)quinoline-3-carboxamide (9b)
To compound 8 (40.0 mg, 0.092 mmol) was added Pd2(dba)3 (16.9 mg, 0.018 mmol) and 2,2′-oxybis(2,1-phenylene)bis(diphenylphosphine) (19.9 mg, 0.037 mmol). The benzenethiol (0.014 mL, 0.138 mmol) was dissolved in toluene and added to the solids. Lastly, KOtBu (0.115 mL, 0.185 mmol) was added. The mixture was heated in the MW for 40 minutes at 170 °C. To the crude product was added methanol and the solution was filtered through Celite. Then, the solvent was concentrated. The crude product was chromatographed 0-100% EtOAc in hexane to give the desired product as a yellow solid. (Yield: 53%). 1H NMR (400 MHz, CD3OD) δ 8.89 (s, 1H), 7.44 (s, 2H), 7.18 - 7.29 (m, 5H), 7.08 (t, J = 8.0 Hz, 1H), 6.58 - 6.64 (m, 1H), 6.34 - 6.40 (m, 2H), 3.71 (s, 3H), 2.67 (s, 3H). LCMS found 416.01, [M+H]+.
methyl 3-(3-carbamoyl-4-(3-methoxyphenylamino)-8-methylquinolin-6-ylsulfonyl)benzoate (10a)
To 9a (103 mg, 0.218 mmol) was added oxone (401 mg, 0.653 mmol) in DMF (5 mL). The reaction was stirred at room temperature for 5 hours. Then, the reaction mixture was poured into water (40 mL) and extracted with DCM (5×), and the combined organic layers were dried (Na2SO4) and concentrated. The crude product was chromatographed using 0-10 % MeOH in DCM to give the desired product as an orange solid (Yield: 85%). 1H NMR (500 MHz, CDCl3) δ 10.86 (s, 1H), 8.93 (s, 1H), 8.44 (s, 1H), 8.23 - 8.26 (m, 1H), 8.21 (d, J = 7.8 Hz, 1H), 7.91 (s, 1H), 7.77 (d, J = 7.8 Hz, 1H), 7.54 (t, J = 7.8 Hz, 1H), 7.15 (t, J = 8.0 Hz, 1H), 6.75 (dd, J = 1.95, 8.3 Hz, 1H), 6.57 (d, J = 7.8 Hz, 1H), 6.54 (s, 1H), 5.90-6.10 (br. s.,2H), 3.97 (s, 3H), 3.65 (s, 3H), 2.75 (s, 3H). LCMS found 506.01, [M+H]+.
4-((3-methoxyphenyl)amino)-8-methyl-6-(phenylsulfonyl)quinoline-3-carboxamide (10b)
To compound 9b (13.4 mg, 0.032 mmol) was added oxone (59.5 mg, 0.097 mmol) and DMF (3 mL). The reaction was stirred for 4 h at rt. Then, the reaction mixture was poured into water (20 mL) and extracted with DCM (5×), the combined organics were washed with brine (1×) and dried under sodium sulfate. The crude product was chromatographed using 50-70% EtOAc in hexane to give the desired product as a light yellow solid. (Yield: 62%).1H NMR (400 MHz, CD3OD) δ 9.03 (s, 1H), 8.38 (s, 1H), 7.93 (s, 1H), 7.72 (d, J = 7.3 Hz, 2H), 7.64 (t, J = 7.3 Hz, 1H), 7.55 (t, J = 7.7 Hz, 2H), 7.24 (t, J = 8.0 Hz, 1H), 6.82 (dd, J = 1.8, 8.4 Hz, 1H), 6.63 (d, J = 7.3 Hz, 1H), 6.57 (s, 1H), 3.64 (s, 3H), 2.74 (s, 3H). LCMS found 448.01, [M+H]+.
3-(3-carbamoyl-4-(3-methoxyphenylamino)-8-methylquinolin-6-ylsulfonyl)benzoic acid (11)
A solution of 10a (60.0 mg, 0.119 mmol) in ethanol (2mL) was treated with NaOH (0.890 mL, 1.780 mmol) and the resulting solution was stirred at 45 °C overnight. The solvent was evaporated. The residue was dissolved in water and acidified with 1 M HCl to pH 4. The resulting precipitate was filtered, washed with water and dried in vacuo. The crude product was purified via preparative HPLC to give the desired product as a yellow solid (Yield: 31%). 1H NMR (400 MHz, CD3OD) δ 9.01 (s, 1H), 8.38 (s, 1H), 8.34 (s, 1H), 8.24 (d, J = 7.3 Hz, 1H), 7.99 (s, 1H), 7.86 (d, J = 8.0 Hz, 1H), 7.65 (t, J = 7.7 Hz, 1H), 7.18 (t, J = 8.0 Hz, 1H), 6.74 - 6.79 (m, 1H), 6.53 - 6.62 (m, 2H), 3.62 (s, 3H), 2.75 (s, 3H). LCMS found 492.01, [M+H]+.
6-(3-carbamoylphenylsulfonyl)-4-(3-methoxyphenylamino)-8-methylquinoline-3-carboxamide (12a)
A solution of 10a (18.0 mg, 0.036 mmol) in dioxane was added to a solution of ammonium hydroxide (0.761 mL, 19.55 mmol). The mixture was stirred at room temperature overnight. Then the solvent was concentrated, and the residue was partitioned between sat. NH4Cl and ethyl acetate. The aqueous layer was extracted with EtOAc (3×) and the combined organics were washed with brine and dried over Na2SO4. After filtration and evaporation, the crude product was chromatographed using 0-10% MeOH to give the desired product as a yellow solid (Yield: 18%). 1H NMR (400 MHz, CD3OD) δ 9.02 (s, 1H), 8.36 (s, 1H), 8.31 (s, 1H), 8.10 (s, 1H), 7.99 (s, 1H), 7.81 (d, J = 7.3 Hz, 1H), 7.65 (t, J = 8.0 Hz, 1H), 7.17 (t, J = 8.4 Hz, 1H), 6.75 - 6.80 (m, 1H), 6.55 - 6.60 (m, 2H), 3.64 (s, 3H), 2.75 (s, 3H). LCMS found 491.01, [M+H]+.
4-(3-methoxyphenylamino)-8-methyl-6-(3-(methylcarbamoyl)phenylsulfonyl)quinoline-3-carboxamide (12b)
To 11 (8.6 mg, 0.017 mmol) in DMF was added HATU (7.3 mg, 0.019 mmol). After 5 minutes methylamine HCl (1.14 mg, 0.017 mmol) and DIEA (6.4 μl, 0.037 mmol) were added. The resulting solution was stirred at room temperature for 6 hours. The solvent was concentrated. The crude product was purified via preparative HPLC to give the desired product as a yellow solid (Yield: 37%). 1H NMR (400 MHz, DMSO-d6) δ 10.77 (s, 1H), 9.08 (s, 1H), 8.71 (d, J = 4.4 Hz, 1H), 8.32 - 8.35 (m, 1H), 8.29 (br. s., 1H), 8.26 (s, 1H), 8.08 (d, J = 8.0 Hz, 1H), 7.99 (s, 1H), 7.76 (d, J = 7.3 Hz, 2H), 7.68 (t, J = 7.7 Hz, 1H), 7.12 (t, J = 8.0 Hz, 1H), 6.69 (dd, J = 1.8, 8.4 Hz, 1H), 6.60 (s, 1H), 6.52 (d, J = 7.3 Hz, 1H), 3.62 (s, 3H), 2.81 (d, J = 4.4 Hz, 3H), 2.69 (s, 3H). LCMS found 505.01, [M+H]+.
methyl 3-(3-carbamoyl-4-(3-methoxyphenylamino)-8-methylquinolin-6-ylamino)benzoate (13)
To 8 (40.0 mg, 0.092 mmol) was added 2,2′-oxybis(2,1-phenylene)bis (diphenylphosphine) (19.9 mg, 0.037 mmol), then Pd(dppf)Cl2·DCM (13.5 mg, 0.018 mmol) and methyl 3-aminobenzoate (27.9 mg, 0.185 mmol). Lastly, KOtBu (0.185 mL, 0.185 mmol) was added. The reaction mixture was heated in the MW for 20 minutes at 160 °C. After the reaction mixture cooled to ambient temperature the crude was filtered through Celite washing with MeOH (3×). The solvent was concentrated under reduced pressure. Then, the crude product was purified via preparative HPLC to give 13 as a yellow solid (Yield: 12%). 1H NMR (400 MHz, CD3OD) δ 8.76 (s, 1H), 7.55 (s, 1H), 7.46 (d, J = 8.0 Hz, 1H), 7.30 (d, J = 2.9 Hz, 1H), 7.26 (s, 1H), 7.13 (t, J = 8.0 Hz, 1H), 7.08 (t, J = 8.0 Hz, 1H), 6.95 (dd, J = 1.5, 8.0 Hz, 1H), 6.62 (dd, J = 1.8, 8.4 Hz, 1H), 6.50 (t, J = 2.2 Hz, 1H), 6.43 - 6.47 (m, 1H), 3.88 (s, 3H), 3.70 (s, 3H), 2.70 (s, 3H). LCMS found 457.01, [M+H]+.
8-ethyl-6-iodo-4-((3-methoxyphenyl)amino)quinoline-3-carboxamide (16b)
To 15b [19, 20] (302 mg, 0.838 mmol) was added acetonitrile and 3-methoxyaniline (0.099 mL, 0.879 mmol). The mixture was heated at 80 °C overnight. Then the solvent was concentrated and the crude product was purified via chromatography (0-6% MeOH in DCM) to give 16b as a light yellow solid, (Yield: 78%). LCMS found 447.99 [M+H]+. The product was taken to the next step without further characterization.
General procedure A
Synthesis of intermediates 23 was achieved via published methods [16, 19, 20], and are summarized in the Supporting Information. For example, compound 7 was dissolved in acetonitrile and the desired amine (1.1 equiv) was added. The mixture was generally heated under reflux overnight or run in the microwave at 145°C for 25 minutes unless stated otherwise in the protocol. Then the precipitate formed was filtered, washed with acetonitrile and the solid obtained was dried under high vacuum to give 23. These analogs were confirmed by LCMS and/or NMR and used in the next step (General procedure B) without further characterization.
General procedure B
Suzuki coupling protocol. To iodo-substituted templates 8 and 23 - (1 equiv.) was added the desired boronic acid or ester (1.5 equiv.), Pd(dppf)Cl2·DCM (0.1 equiv.), dioxane and sodium carbonate (6 equiv.). The reaction mixture was heated in the MW at 130 °C, 145 °C or 160°C as specified in Scheme 2 or Scheme 3 for 20 min. After the reaction mixture cooled to ambient temperature the crude was filtered through Celite washing with MeOH/DCM (1:9). The filtrate was concentrated under reduced pressure. Unless otherwise noted the crude products were chromatographed or purified via preparative HPLC to give the desired products of the general templates 17, 19, 22e,f and 24, which are characterized in the Supporting Information.
Scheme 2.
a Preparation of analogs 17-19.
aReagents and Conditions. a) EtOCH=C(CO2Et)2, 100 °C, 1h (80-98%). b) Ph2O, 250 °C, 45 min (81-100%). c) NaOH, EtOH, reflux, 1h, then conc. HCl, overnight. d) SOCl2, DMF, 80 °C, 2h e) NH4OH, rt, overnight (61-92%). f) 3-methoxyaniline, MeCN, 80 °C, overnight (78-94%). g) Ar-B(OH)2, Na2CO3, Pd(dppf)Cl2, dioxane, 130 or 145 °C, MW, 20 minutes (20-70%). h) 1-Methyl-1H-indazole-6-boronic acid, Na2CO3, Pd(dppf)Cl2, dioxane, 160 °C, MW, 20 minutes (13-56%). i) LiOH, H2O:THF:MeOH, rt, 12 h.
Scheme 3.
a Preparation of analogs exploring Region B.
aReagents and Conditions. a) Various amines (R2), MeCN, 80 °C, 24 h (61-80%) or Various amines (R2), MeCN, 145 °C, 25 min (59-80%). b) 1-Methyl-1H-indazole-6-boronic acid, Na2CO3, Pd(dppf)Cl2, dioxane, 130 or 145 °C, MW, 20 minutes (9-22%). and c) (3-(methoxycarbonyl) phenyl)boronic acid, Na2CO3, Pd(dppf)Cl2, dioxane, 130-145 °C, MW, 20 minutes (7-12%). d) (3-(methoxycarbonyl) phenyl)boronic acid or (3-(oxazol-2-yl)phenyl)boronic acid, Na2CO3, Pd(dppf)Cl2, dioxane, 150 °C, MW, 10 minutes (20a (30%), 20b (14%), (21a (5%), 21b (2%)). e) Various amines (R2), MeCN, 80 °C, 48 h (10-40%).
General procedure C
To 20 (1 equiv.) was added acetonitrile and the required amine (5-6 equiv.) was added. The reaction was run at 80°C in a sealed vial for 48 h. The solvent was concentrated and the crude product was purified via preparative HPLC or chromatography to afford the products (22a-d), which are characterized in the Supporting Information.
5-(3-carbamoyl-4-((3-methoxyphenyl)amino)-8-methylquinolin-6-yl)isophthalic acid (18a)
To 17e (7.00 mg, 0.014 mmol) was added LiOH (2.014 mg, 0.084 mmol) and the solids were dissolved in a mixture of water:THF:MeOH (1:1:1). The solution was stirred at 90 °C in a sealed vial for 2 hours. Then the solvent was concentrated. The crude product was purified via PREP HPLC to give 5-(3-carbamoyl-4-((3-methoxyphenyl)amino)-8-methylquinolin-6-yl)isophthalic acid (18a), (white solid, Yield: 25%). 1H NMR (500 MHz, DMSO-d6) δ 13.25 (br. s., 1H), 10.47 (s, 1H), 8.99 (s, 1H), 8.39 (s, 1H), 8.21 (s, 1H), 8.15 (m, 2H), 7.96 (s, 2H), 7.67 (s, 1H), 7.15 (t, J = 8.0 Hz, 1H), 6.62-6.56 (m, 2H), 6.54 (d, J = 7.0 Hz, 1H), 3.68 (s, 3H), 2.77 (s, 3H). LCMS found 472.01, [M+H]+.
3-(3-carbamoyl-4-((3-methoxyphenyl)amino)-8-methylquinolin-6-yl)-5-(methoxycarbonyl)benzoic acid (18b)
To 17e (11.40 mg, 0.023 mmol) was added LiOH (0.273 mg, 0.011 mmol) and the solids were dissolved in a mixture of water:THF:MeOH (1:1:1). The reaction mixture was stirred at rt for 12h. Then another 0.5 equiv. of LiOH was added, and the temperature was raised to 50 °C. The reaction ran 6 more hours at 50 °C, and then the solvent was concentrated. The crude product containing the diester and the monoacid was chromatographed (0-10% MeOH/DCM) to give 3-(3-carbamoyl-4-((3-methoxyphenyl)amino)-8-methylquinolin-6-yl)-5-(methoxycarbonyl) benzoic acid (18b), (white solid, Yield: 12%). 1H NMR (500 MHz, DMSO-d6) δ 10.50 (s, 1H), 9.00 (s, 1H), 8.42 (s, 1H), 8.24 (d, J = 7.3 Hz, 2H), 8.04 (br. s., 1H), 7.95 (s, 2H), 7.69 (br. s., 1H), 7.17 (t, J = 8.0 Hz, 1H), 6.66 (dd, J = 1.9, 8.3 Hz, 1H), 6.62 (s, 1H), 6.55 (d, J = 7.8 Hz, 1H), 3.90 (s, 3H), 3.68 (s, 3H), 2.77 (s, 3H). LCMS found 486.01, [M+H]+.
methyl 3-(3-carbamoyl-4-chloro-8-methylquinolin-6-yl)benzoate (20a), dimethyl 3,3′-(3-carbamoyl-8-methylquinoline-4,6-diyl)dibenzoate (21a), and methyl 3-(3-carbamoyl-8-methylquinolin-6-yl)benzoate (21b)
To 7 (200 mg, 0.577 mmol) was added (3-(methoxycarbonyl)phenyl)boronic acid (104 mg, 0.577 mmol), Pd(dppf)Cl2·DCM (84 mg, 0.115 mmol), dioxane (4 mL) and sodium carbonate (1.73 mL, 3.46 mmol). The reaction mixture was heated in the MW at 150 °C for 10 minutes. After the reaction mixture cooled to ambient temperature the crude was filtered through Celite washing with MeOH/DCM (1:9). The filtrate was concentrated under reduced pressure. The crude mixture was chromatographed using hexanes/EtOAc to give as the major product 20 as a white solid (Yield: 29%) and a mixture of 21a and 21b. This mixture was separated using preparative HPLC to give the clean products 21a (white solid, Yield: 5%) and 21b (white solid, Yield: 2%). (20).1H NMR (500 MHz, DMSO-d6) δ 8.90 (s, 1H), 8.30 - 8.35 (m, 2H), 8.21 (s, 1H), 8.13 - 8.17 (m, 2H), 8.03 (d, J = 7.8 Hz, 1H), 7.99 (s, 1H), 7.70 (t, J = 7.8 Hz, 1H), 3.91 (s, 3H), 2.82 (s, 3H). LCMS found 354.01, [M+H]+. (21a). 1H NMR (500 MHz, DMSO-d6) δ 9.00 (s, 1H), 8.07 - 8.12 (m, 3H), 7.99 (s, 1H), 7.95 (d, J = 7.8 Hz, 1H), 7.83 - 7.89 (m, 2H), 7.69 - 7.75 (m, 2H), 7.60 (t, J = 7.8 Hz, 1H), 7.54 (s, 1H), 7.48 - 7.50 (m, 1H), 3.32 (s, 6H), 2.87 (s, 3H). LCMS found 455.11, [M+H]+. (21b). 1H NMR (500 MHz, CDCl3) δ 9.31 (s, 1H), 8.69 (d, J = 2.4 Hz, 1H), 8.40 (s, 1H), 8.09 (d, J = 7.8 Hz, 1H), 7.97 (d, J = 7.3 Hz, 2H), 7.91 (d, J = 7.8 Hz, 1H), 7.59 (t, J = 7.5 Hz, 1H), 3.98 (s, 3H), 2.91 (s, 3H). LCMS found 321.01, [M+H]+.
4-chloro-6-iodo-N,8-dimethylquinoline-3-carboxamide (25a)
To sodium hydride (60 % dispersion in mineral oil, 10.9 mg, 0.27 mmol) in DMF (2 mL) was added intermediate 7 (100 mg, 0.289 mmol) in dry DMF (3 mL). The reaction mixture was stirred for 35 minutes at rt before adding iodomethane (0.017 mL, 0.274 mmol), after which the reaction mixture was stirred at rt overnight. Water and EtOAc was added, and the aqueous layer was extracted with EtOAc (3×). The combined organics were washed with brine and finally dried under sodium sulfate. The solvent was concentrated and the product was confirmed by LCMS, LCMS found 360.01, [M+H]+. The product was taken to the next step without further characterization.
4-chloro-6-iodo-N,N,8-trimethylquinoline-3-carboxamide (25b)
To sodium hydride (60 % dispersion in mineral oil, 23 mg, 0.57 mmol) in DMF (2 mL) was added intermediate 7 (100 mg, 0.289 mmol) in dry DMF (3 mL). The reaction mixture was stirred for 35 minutes at rt before adding iodomethane (0.020 mL, 0.317 mmol), after which the reaction mixture was stirred at rt overnight. Then, to the reaction mixture was added water and EtOAc. The aqueous layer was extracted with EtOAc (3×). The combined organics were washed with brine and finally dried under sodium sulfate. The solvent was concentrated and the product was confirmed by LCMS, LCMS found 374.01, [M+H]+. The product was taken to the next step without further characterization.
6-iodo-4-((3-methoxyphenyl)amino)-N,8-dimethylquinoline-3-carboxamide (26a)
To 25a (104 mg, 0.288 mmol) was added acetonitrile and 3-methoxyaniline (0.034 ml, 0.303 mmol). The mixture was heated under reflux overnight. The solution was filtered, and the solids were washed with acetonitrile. The filtrate was concentrated. The product 26a (Yield: 41%) was confirmed by LCMS (found 448.04 [M+H]+) and was taken to the next step without further characterization.
6-iodo-4-((3-methoxyphenyl)amino)-N,N,8-trimethylquinoline-3-carboxamide (26b)
To 25b (56.0 mg, 0.149 mmol) was added acetonitrile and 3-methoxyaniline (0.018 ml, 0.157 mmol). The mixture was heated under reflux overnight. The solution was filtered, and the solids were washed with acetonitrile. The filtrate was concentrated. The product (Yield: 61%) was confirmed by LCMS (462.01, [M+H]+), and was taken to the next step without further characterization.
4-((3-methoxyphenyl)amino)-N,8-dimethyl-6-(1-methyl-1H-indazol-6-yl)quinoline-3-carboxamide (27a)
(synthesized using General procedure B from 26a, yellow solid, Yield: 8%). 1H NMR (500 MHz, DMSO-d6) δ 10.05 (s, 1H), 8.87 (s, 1H), 8.59 (d, J = 4.4 Hz, 1H), 8.14 (s, 1H), 8.07 (s, 1H), 8.06 (s, 1H), 7.80 (d, J = 8.3 Hz, 1H), 7.69 (s, 1H), 7.34 - 7.37 (m, 1H), 7.21 (t, J = 8.0 Hz, 1H), 6.64 - 6.68 (m, 2H), 6.58 (d, J = 8.3 Hz, 1H), 4.07 (s, 3H), 3.70 (s, 3H), 2.80 (s, 3H), 2.66 (d, J = 4.4 Hz, 3H). LCMS found 452.01, [M+H]+.
4-((3-methoxyphenyl)amino)-N,N,8-trimethyl-6-(1-methyl-1H-indazol-6-yl)quinoline-3-carboxamide, formic acid salt (27b)
(synthesized using General procedure B from 26b, yellow solid, Yield: 17%). 1H NMR (500 MHz, DMSO-d6) δ 9.11 (s, 1H), 8.55 (s, 1H), 8.51 (s, 1H), 8.13 (s, 2H), 8.07 (s, 1H), 8.04 (s, 1H), 7.86 (d, J = 8.3 Hz, 1H), 7.66 (d, J = 8.3 Hz, 1H), 7.17 (t, J = 8.5 Hz, 1H), 6.63 (d, J = 8.3 Hz, 1H), 6.55 - 6.59 (m, 2H), 4.12 (s, 3H), 3.71 (s, 3H), 2.79 (s, 3H), 2.75 (s, 3H), 2.38 (s, 3H). LCMS found 466.01, [M+H]+.
4-(cyclopentylamino)-8-methyl-6-(3-(oxazol-2-yl)phenyl)quinoline-3-carboxamide (28)
To 20b (13.00 mg, 0.036 mmol) was added acetonitrile and lastly cyclopentanamine (0.021 mL, 0.214 mmol). The reaction was run at 80°C in a sealed vial for 48 h. The solvent was concentrated and the crude product was chromatographed using 0-7% MeOH in DCM to give the desired product as a yellow solid (Yield: 34%).1H NMR (500 MHz, CDCl3) δ 9.50 (d, J = 7.3 Hz, 1H), 8.75 (s, 1H), 8.39 (d, J = 4.8 Hz, 2H), 8.07 (d, J = 7.8 Hz, 1H), 7.87 (s, 1H), 7.76 (s, 1H), 7.75 (d, J = 7.8 Hz, 1H), 7.59 (t, J = 7.6 Hz, 1H), 7.29 (s, 1H), 5.90 (br. s., 2H), 4.55 - 4.64 (m, 1H), 2.80 (s, 3H), 2.11 - 2.20 (m, 2H), 1.78 - 1.90 (m, 4H), 1.68 - 1.75 (m, 2H). LCMS found 413.01, [M+H]+.
Molecular Modeling
Docking for all compounds was performed using POSIT v 1.0.2 (OpenEye Scientific Software: Santa Fe, NM). The coordinates of the apo crystal structure of TbrPDEB1 were downloaded from the Protein Data Bank (PDB), PDB code: 4I15 [21]. The protein was prepared for docking and mild ligand–protein clashes were allowed in order to account for the average coordinate error expected in PDB structures. The “combine-receptors” option was used in order to include the binding mode observed in the human phosphodiesterase 4B2B in complex with a quinoline Inhibitor during docking in TbrPDEB1 (PDB code: 3FRG) [19]. Mild clashes were also used in pose prediction. All the other options in POSIT were kept as default. The docked poses were energy minimized using SZYBKI (v 1.7.0 OpenEye Scientific Software, Santa Fe, NM) allowing partial relaxation of the protein residues in the direct proximity to the ligand. The docking program FRED v 3.0.0 (OpenEye Scientific Software: Santa Fe, NM) was also used to investigate alternative binding modes (such as for compound 28).
Water molecule predictions were performed using SZMAP v 1.1.0 (OpenEye Scientific Software: Santa Fe, NM) on three different protein-ligand complex structures: (i) the crystal structure of the human phosphodiesterase 4B2B[16], (ii) the TbrPDEB1-19a complex, and (iii) the TbrPDEB1-24g complex. The conformations of the ligands of the two latter protein-ligand complexes were obtained with molecular docking. The protein-ligand complexes were used as input to SZMAP, which was then used to predict the solvation of the active binding site in complex with the ligands using the semi-continuum solvation theory, in combination with a single explicit water probe. The results were visualized with VIDA (OpenEye Scientific Software, Santa Fe, NM).
Results
In the interest of exploring the SAR around 3, we divided the compound into 4 regions, highlighted in Figure 1. First, we synthesized a set of analogs to determine the preferred aryl substituent and connection to the quinoline core for Region A, following Scheme 1. Synthesis commenced with the condensation of 5a with diethyl ethoxymethylenemalonate, followed by cyclization to afford 6. The ester was hydrolyzed to the acid, and the resulting product was chlorinated and quenched with aqueous ammonia affording the primary carboxamide 7. Reaction of 7 with m-anisidine under reflux provided 8 in high yield. This template was reacted with commercially available aryl thiols using palladium catalysis to provide the sulfides, 9, which could be oxidized to the appropriate sulfones 10 using oxone.
Scheme 1.
a Preparation of analog 13, exploring Region A.
aReagents and conditions. a) EtOCH=C(CO2Et)2, 100 °C, 1h (98%). b) Ph2O, 250 °C, 45 min (100%). c) NaOH, EtOH, reflux, 1h, then conc. HCl, overnight. d) SOCl2, DMF, 80 °C, 2h e) NH4OH, rt, overnight (80%). f) 3-methoxyaniline, MeCN, 80 °C, overnight (90%). g) 3-substituted (R1) benzenethiols, Pd2(dba)3, DPEphos, KOtBu, toluene, 170 °C, MW, 30 minutes (70-85%). h) Oxone, DMF, rt, 12-24h (70-90%). i) NaOH, EtOH, 45°C, overnight (31%). j) 11, Me2NH or MeNH2·HCl, HATU, DIEA, DMF, rt (for 3 (21%) and 12b (37%). k) for 10a: NH4OH, dioxane, rt, overnight (12a,18%) l) methyl 3-aminobenzoate, Pd(dppf)Cl2, (oxybis(2,1-phenylene))bis (diphenylphosphine), KOtBu, dioxane, 160 °C, MW, 20 minutes (12%).
Amide substituents were introduced via hydrolysis of the ester functionality of 10a under basic conditions to give 11, and then standard amide coupling with the appropriate amine provided 3 and 12b. Treatment of 10a with ammonium hydroxide gave analog 12a. To explore a nitrogen linker to Region A, the reaction of the commercial methyl 3-aminobenzoate with 8 using a modified palladium-catalyzed coupling procedure provided 13 [22, 23].
Following synthesis, the analogs were tested at a single concentration (10 μM) against TbrPDEB1, and those compounds with >65 % inhibition, and those that represented key SAR points, were tested in a dose-response assay. We noted that removal of N-alkyl groups from the benzamide of 3 results in little change in percentage inhibition, the key data in selecting compounds for IC50 determination (3, 12a-b, Table 1). Replacement of the carboxamide with an ester (10a) or carboxylic acid (11) improved activity by approximately 8-fold over 3. The sulfone linker between the quinoline and sidechain was better than either a sulfide (9a) or amine (13).
Table 1.
a Biochemical potency data for analogs of 3.

|
|||||
|---|---|---|---|---|---|
| Compound | R1 | X | TbrPDEB1 (% inh) |
TbrPDEB1 (IC50 μM) |
TbrPDEB2 (IC50 μM) |
| 3 | CONMe2 | SO2 | 51 ± 3.9 | 24 ±1.7 | 29.8 ± 7.5 |
| 9a | CO2Me | S | 12 ±6.1 | - | - |
| 9b | H | S | 10.0b | ||
| 10a | CO2Me | SO2 | 70 ± 7.9 | 3.5 ± 0.3 | 15.1 ± 3.9 |
| 10b | H | SO2 | 21 ± 16 | - | - |
| 11 | CO2H | SO2 | 82 ± 10 | 2.9 ± 0.8 | 6.8 ± 0.6 |
| 12a | CONH2 | SO2 | 64 ± 7 | - | - |
| 12b | CONHMe | SO2 | 66 ± 6.1 | 31 ± 5.8 | - |
| 13 | CO2Me | NH | 84 ± 7.7 | 16 ± 1.7 | - |
All values are the mean of three or more replicates ± SEM.
n=1
Having explored the sulfone, sulfide, and amine-linker of the A-region of the inhibitor, we designed analogs with no intervening atom between the core template and the A-region via a biaryl linkage. This was accomplished using the route shown in Scheme 2. The iodo template 16 was prepared using a route analogous to the preparation of 8, followed by reaction with various boronic acids using Suzuki coupling chemistry.
The potency of the biaryl analog 17a on TbrPDEB1 (Table 2) was within two-fold of the IC50 of the matched sulfone analog 10a (Table 1). The observation that the matched biaryl linker was essentially equivalent to the more synthetically intensive sulfone linker allowed us to access a larger set of analogs via one step synthesis from the iodo template (8 or 16) and boronates using Suzuki couplings. We explored the SAR for this region with the biaryl linkage using various aryl boronic acids or esters obtained from commercial sources, or via synthesis (Supporting Information) [24-26]. Besides close-in analogs of 17a, we explored small, heterocyclic replacements and bioisosteres for the methyl ester moiety (Table 2, 17o-17r). A small set of analogs bearing fused bicyclic heteroaromatic rings was also prepared (17t-v, 19a). Notably, the potency of the N-methyl indazole analog 19a was within two-fold of the potency of 17a. Additionally, the hydrolysis of the diester groups of 17e provided the mono- and di-acids 18a-b, which showed low (<50%) inhibition at 10 μM.
Table 2. a Analogs exploring the Region A of 3.

| Compound | R1 | TbrPDEB1 (% inh) |
TbrPDEB1 (IC50 μM) |
TbrPDEB2 (IC50 μM) |
|---|---|---|---|---|
| 17a |

|
94 ± 3.5 | 6.4 ± 1.7 | 6.2 ± 0.6 |
| 17b |

|
37 ± 14 | ||
| 17c |

|
6 ±1.4 | - | |
| 17d |

|
54 ± 8.8 | - | |
| 17e |

|
50 ± 6.1 | - | |
| 17f |

|
35 ± 10 | 23 ± 4.0 | |
| 17g |

|
65 ± 5.5 | ndb | |
| 17h |

|
9 ±7.1 | - | |
| 17i |

|
8.6 ± 5.2 | - | |
| 17j |

|
74 ± 13 | 9 ± 0.6 | |
| 17k |

|
10 ± 6.9 | - | |
| 17l |

|
43 ±16 | - | |
| 17m |

|
83± 7.5 | 6.7 ± 2.9 | 4.6± 0.1 |
| 17n |

|
13 ± 8 | - | |
| 17o |

|
83 ± 1.3 | 6 ± 1.7 | |
| 17p |

|
53 ± 24 | 5.9 ± 0.4 | |
| 17q |

|
27 ± 8.3 | - | |
| 17r |

|
31 ± 6.3 | - | |
| 17s |

|
68 ± 6.3 | 12 ± 2.7 | |
| 17t |

|
85 ± 6.5 | 5.9 ±1.0 | 13.5 ± 3.5 |
| 17u |

|
52 ± 8.3 | - | |
| 17v |

|
43 ± 3.3 | - | |
| 18a |

|
10 ± 5.3 | - | |
| 18b |

|
31 ±14 | - | |
| 19a |

|
84 ± 9.1 | 3.1 ± 0.5 | 8.0 ± 3.9 |
All values are the mean of three or more replicates ± SEM.
No IC50 obtained due to limited solubility at higher concentrations.
The 3-methyl ester substitution was preferred over the 2- or 4-position (Table 2, 17a-17c). We noted that several compounds bearing replacements for the methyl ester of 17a (17m, 17o, 17p, 17t), were approximately equipotent, though none were significantly better. Given their increase in complexity and size over 17a, these analogs were less desirable. The tetrazole analog 17m was more potent than the corresponding bioisosteric carboxylic acid (17d), which was not potent enough to advance to IC50 determination.
The potency of the best biaryl compound in Table 2 (19a) is similar to the best sulfone from Table 1 (11), leading us to focus upon these biaryl compounds given their ease of synthesis and purification compared to the sulfone analogs. With this in mind, we next turned our attention to preparation of Region B variants, keeping Region A constant with either of the two best Region A substituents: the methyl indazole fragment of 19a or the methylbenzoate fragment of 17a.
These analog syntheses were accomplished using one of the two routes shown in Scheme 3. Reaction of 7 with (3-(methoxycarbonyl)phenyl)boronic acid using Suzuki coupling afforded 20, the bis-arylated product 21 and the dehalogenated byproduct 21b. The final step required heating of 20 with the desired amine to give 22a-d, and 28. Additionally, reaction of 7 with various amines under reflux provided intermediates 23 (General procedure A and Supporting Information), followed by reaction of these intermediates via Suzuki coupling with 1-methyl-1H-indazol-6-ylboronic acid to give 24a-g or 3-(methoxycarbonyl)phenylboronic acid to afford 22e-f.
As shown in Table 3, compound activity is affected by the regiochemistry of the methoxy substituent on the R2 group: para substitution is not tolerated (24a), whereas the ortho methoxy analog 24b is only slightly less potent than 19a. We observe that the ethyl analog 24d, which is isosteric to 19a, is just as potent. This led us to question the essentiality of this oxygen atom, though some sort of substitution at this position seems important (compare 19a versus 24a or 24g).
Table 3. a Biochemical data for analogs exploring Region B.

| |||
|---|---|---|---|
| Compound | R2 | TbrPDEB1 (% inh) |
TbrPDEB1 (IC50 μM) |
| 17a |

|
94 ± 3.5 | 6.4 ± 1.7 |
| 21a |

|
41 ± 5.8 | - |
| 21b |

|
0 | - |
| 22a |

|
19 ± 6.6 | - |
| 22b |

|
19 ± 4.2 | - |
| 22c |

|
22 ± 20 | 28 ± 3.4 |
| 22d |

|
80 ± 4.8 | ndb |
| 22e |

|
59 ± 3.6 | - |
| 22f |

|
72 ± 14 | 26 ± 1.3 |
| 19a |

|
83.7 ± 9.1 | 3.8 ± 0.4 |
| 24a |

|
34 ± 0.3 | - |
| 24b |

|
60 ±16 | 7 ± 0.9 |
| 24c |

|
82 ±12 | 4.2 ± 0.6 |
| 24d |

|
67 ± 7.5 | 3.5 ± 0.5 |
| 24e |

|
85 ± 11 | 3.5 ± 0.6 |
| 24f |

|
69 ±16 | 3.7 ± 0.2 |
| 24g |

|
21 ± 6.5 | - |
| 28 |

|
76 ± 2.2 | 5.9 ± 0.67 |
All values are the mean of three or more replicates ± SEM.
No IC50 obtained due to limited solubility at higher concentrations.
Analogs of 17a are sensitive to the spacing between the B-region substituent and the rest of the molecule. Extending this spacing by an additional carbon (e.g. 22a) led to loss of activity. Removal of this substituent altogether resulted in a total loss of activity (21b). Finally, comparison of 17a and the N-methylated analog 22e reveals loss in activity.
Synthesis of analogs varying Regions C and D of 19a is shown in Schemes 2 and 4. Reaction of 7 with NaH and iodomethane in DMF provided intermediates 25a-b (Scheme 4). Heating of these intermediates with m-anisidine in acetonitrile afforded 26a-b, which were subjected to Suzuki reaction to provide the Region C variants 27a-b.
Scheme 4.
a Preparation of analogs 27 exploring Region C.
aReagents and Conditions. a) NaH, DMF, 30 min, rt, then CH3I, rt, overnight (52-100%). b) 3-methoxyaniline, MeCN, 80 °C, overnight (40-60%). c) 1-Methyl-1H-indazole-6-boronic acid, Na2CO3, Pd(dppf)Cl2, dioxane, 160 °C, MW 20 minutes (8-17%).
The variations at Region D were introduced at the beginning of the synthesis when the appropriate aniline 5 was used (Scheme 2). The methylindazole fragment was introduced in the last step via Suzuki chemistry to provide analogs 19a-c.
As shown in Table 4, the primary amide in Region C is required; substitution with one or two methyl groups leads to dramatic loss of activity (19a vs. 27a and 27b). Removal of the Region D methyl group leads to a ~4-fold loss in activity (19a vs. 19b), while extension to an ethyl group (i.e. 19c) is approximately equipotent.
Table 4.
a Exploring Regions C and D of 19.

| Compound | R3 | R4 | TbrPDEB1 (% inh) |
TbrPDEB1 (IC50 μM) |
|---|---|---|---|---|
| 19a | Me | NH2 | 84 ± 9.1 | 3.1 ± 0.5 |
| 19b | H | NH2 | 84 ± 2.4 | 12 ± 1.3 |
| 19c | Et | NH2 | 79 ± 8.9 | 3.7 ± 0.5 |
| 27a | Me | NHCH3 | 34 ±11 | - |
| 27b | Me | NMe2 | 22 ± 3.8 | - |
All values are the mean of three or more replicates ± SEM.
The range of activity in this SAR study is relatively narrow, spanning potency differences of about 100-fold, though we were pleased that we were able to improve compound potency about eight-fold from 3. Inhibition of both TbrPDEB1 and B2 are required, though we have previously noted close correlation in TbrPDEB1 and B2 IC50 values [7]. Therefore, we periodically obtained TbrPDEB2 IC50 values for compounds, and tested against both PDEs prior to assessment in T. brucei cells. We note that, with the exception of one compound 10a, those compounds tested against TbrPDEB2 showed IC50 values within ~2-fold of TbrPDEB1. Testing against human PDE4B unfortunately confirmed that these compounds remain extraordinarily potent inhibitors (Table 5). Nonetheless, testing of some of the most potent TbrPDEB1/B2 inhibitors do show cellular growth inhibition approximating the biochemical potency. One notable exception is the lead compound 3; surprisingly, though this compound is a weak TbrPDEB1/B2 inhibitor, it inhibits cell growth with an EC50 of 7.8 uM. We presume this must be due to other mechanisms of inhibition operating in the cell.
Table 5. Biochemical and cellular characterization of the most potent TbrPDEB1 inhibitors.
| Compound | Reg Number | cLogP | TbrPDEB1 (IC50 μM)a |
TbrPDEB2 (IC50 μM)a |
hPDE4 (IC50 μM)a |
T brucei cell (EC50, μM)a |
|---|---|---|---|---|---|---|
| 3 | NEU-355 | 4.33 | 24 ± 1.7 | 29.8 ± 7.5 | 7.9×10−6 | 7.8 ± 2.5 |
| 10a | NEU-356 | 5.04 | 3.5 ± 0.3 | 15.1 ± 3.9 | - | 7.3 ± 0.97 |
| 17a | NEU-433 | 5.73 | 6.4 ± 1.7 | 6.2 ± 0.6 | 4.2×10−5 | 7.0± 3.9 |
| 17t | NEU-489 | 5.82 | 5.9 ± 1.0 | 13.5 ± 3.5 | - | 5.6 ± 1.9 |
| 19a | NEU-462 | 5.17 | 3.1 ± 0.5 | 8.0 ± 3.9 | 8.3×10−5 | 6.8 ± 0.82 |
| 19c | NEU-528 | 5.62 | 3.7 ± 0.5 | 5.6 ± 3.4 | - | 7.9 ± 2.7 |
| 24d | NEU-542 | 6.29 | 3.5 ± 0.5 | 12.1 ± 3.2 | - | 8.7± 1.8 |
All values are the mean of three or more replicates ± SEM.
We looked towards molecular modeling to help explain our observations, utilizing the recently published X-ray crystal structure of TbrPDEB1 to perform docking experiments of key analogs of 3. The binding mode of a similar quinoline derivative in complex with the human PDE4B2B was reported in a crystal structure published by Lunniss and co-workers [19]. This crystal structure reveals that the quinoline nitrogen of 3 interacts with the amide NH2 of Gln874, and that the ligand amide NH2 (Region C in Figure 1) interacts with carbonyl group of Asn825 residue (Figure 2A). Both residues (Gln874 and Asn825 in TbrPDEB1) are conserved between the human and parasite PDE and are therefore predicted to give similar interactions with this class of ligands. The amide carbonyl (Region C) is predicted to give an internal hydrogen bond with the NH group of the substituent occupying area B (Figure 1 and 2A).
Figure 2.
(A). Overlay of the crystal structure of the human phosphodiesterase 4B2B in complex with a quinoline Inhibitor (PDB code: 3FRG, cyan carbon atoms) with the docked conformation of 11 in TbrPDEB1 (green carbon atoms). TbrPDEB1 residue numbering is used, with the corresponding hPDE4B residues in brackets. (B). Overlay of the docking poses of (i) 17a (yellow carbon atoms), (ii) 11 (green carbon atoms) and (iii) 17m (brown carbon atoms) in TbrPDEB1. (C). Docked conformation of 28 in TbrPDEB1. This pose was generated using FRED. The side chain of Asn825 has been flipped in order to allow a hydrogen bond of the amidic nitrogen with the pyridine nitrogen of the ligand. In all these figures, Mg2+ and Zn2+ are shown as green and grey spheres respectively, and the interactions of the ligands with Gln 874, Asn 825 and Asn 717 are shown in blue dashed lines. The surface of the active site and the P-pocket can also be seen in B and C. Images generated using the PyMOL Molecular Graphics System, Version 1.5.0.4 Schrödinger, LLC.
The Region A substituents are predicted to point towards the N-terminus of an alpha-helix, which is slightly positively charged in consequence of the dipole moment [27]. The fact that electron-rich substituents occupying Region A (e.g. 11, Figure 2A) display generally better potency values could be therefore explained by their favorable interaction with this dipole. Within the helix, an important key amino acid change between the human PDE4B and parasite PDE is Ser454 to Asn717 (Figure 2A & B)[19]. The docking poses of compounds 11, 17a and 17m are shown in Figure 2B, highlighting the hydrogen bonds to the amide NH2 of Asn717. In addition, the linker in the substituent in region A appears to play an important role in placing the electron rich substituent in contact with the Asn717 residue in the N-terminal helix. Woodrow et al. previously described the detrimental effect of replacing the sulfone linker with a sulfide (such as in the case of 9a vs. 10a), likely due to the ease of the sulfone to pre-orient the molecule in the proper conformation compared to the more flexible sulfide[16]. We infer that the C-C linker-containing compounds must be able to pre-orient similarly to their sulfone congeners given the similar activity profile (e.g. 10a and 17a).
The substituents in Region B are predicted to occupy the space near the magnesium ion (Figure 2A). Analysis of the crystal structure of hPDE4B2B shows that the m-methoxy substituent is located in the proximity of magnesium (~4 Å), indirectly interacting with the metal via intervening water molecules (Figure S1A, Supporting Information). However, in the case of TbrPDEB1, we note the interesting observation that analogs with ethyl or methyl replacements of the m-methoxy group retain potency (compare 19a with 24d and 24e), though an unsubstituted phenyl ring (24g) is detrimental for potency (Table 3). Water prediction calculations (SZMAP v1.1.0, OpenEye Scientific Software) suggest that, in absence of a meta-position substitution in this phenyl group, there is a small, unoccupied cavity in the protein-ligand complex (Figure S1C). This unfavorable situation may explain the lack of activity of 24g.
We predict the cyclopentyl-substituted compounds 22d and 28 to orient in an alternative binding mode, accommodated by an alternative rotamer of Asn825 that is able to interact with the pyridine nitrogen of the ligand (Figure 2C) [28, 29]. We prepared these analogs, and while 28 is a 5.9 μM inhibitor of TbrPDEB1, 22d was not sufficiently soluble to test in the dose-response experiment, though we observed the compound to inhibit TbrPDEB1 80% at 5 μM.
Methylation of the amide occupying Region C is detrimental for potency (compare 19a, 27a, and 27b, Table 4). This is consistent with the hypothesized essential H-bonding interaction between this amide NH2 with Asn825 (Figures 2A and B). Finally, the R3 substituent (Region D) is predicted to occupy an area, adjacent to the “parasite-” or “P-pocket” (Figure 2B) [7, 21], defined by the side chain of Met861 and the alpha helix located at the back pocket of the active site. The presence of a methyl or ethyl group in Region D is predicted to increase potency by partially filling the P-pocket, and this potency improvement was confirmed experimentally as can be seen by comparing the potency values for compounds 19a, 19b, and 19c in Table 4.
Discussion and conclusions
We have applied a target repurposing approach to identify inhibitors of TbrPDEB1, an essential enzyme of the protozoan pathogen T. brucei. One of the advantages of this approach is the ability to launch optimization studies for new inhibitors with minimal up-front screening efforts. Indeed, our efforts to re-optimize chemotypes related to piclamilast [7, 10], cilomilast [30], and now GSK-256066 for improved potency against TbrPDEB1 were launched by a broad assessment of 20 existing human PDE inhibitors [7]. In this report the SAR of the GSK-256066 chemotype was elaborated in a rapid fashion and provided improved analogs such as 10a, 17a, 17t, 19a, 19c and 24d. This was possible by systematically studying the four regions of compound 3 (regions A, B, C and D; Figure 1) that we set out to explore. Our work revealed some general requirements needed for increased activity. First, for Region A the most favorable functionalities were the meta substituted (e.g. ester, carboxylic acid, small heterocycles) aryls (e.g. 10a, 11, 17m, 17o, 17p) or the methylindazole fragment (19a). This highlights that while Region A needs to be filled with lipophilic aromatic cores, this is not sufficient for activity. More important is a requirement for hydrogen acceptor groups that may interact with Asn717 (Figure 2B). Furthermore, for Region B we discovered that substituted aryls in the meta position with groups such as methyl, ethyl, methoxy or ethoxy (i.e. 24e, 24d, 17a and 24c) are preferred for increased activity, as there is a small unoccupied cavity in the protein-ligand complex that that we posit is now filled by these lipophilic groups (Figure S1C, Supporting Information). For Region C the amide functionally was shown to be essential as it interacts with Asn825, which is present in both hPDE4 and TbrPDEB1 (shown in Figure 2A). Lastly, for Region D we show that by occupying the P-pocket by a methyl (19a) or ethyl substituent (19c) we observe an increase in activity (Figure 2B). This region would benefit from additional exploration, though this is synthetically the most challenging part of the molecule to vary. In the end, our medicinal chemistry efforts provided compound 19a, which is 6.5-fold more active in the TbrPDEB1 biochemical assay, 10.5-fold less potent against hPDE4, and shows modest potency against T. brucei cells.
Unfortunately, the compounds we have identified still retain significant (sub-nanomolar) potency against human PDE4 (e.g. 19a IC50=0.083 nM). Though this represents a slight improvement in overall selectivity, more must be done to improve selectivity for a new HAT therapeutic. For us, and for others [17, 18], this appears to be a significant issue that has yet to be overcome, though with recent structural biology reports of TbrPDEB1 [21], we expect that understanding about the ligand-target interactions that drive selectivity will become more clear in the near future.
Importantly, in most cases, the structure-activity relationships we observe are consistent with the expected binding modality of this chemotype, based on previous structural biology reports of 3 bound to hPDE4B [16]. For example, the requirement for an unsubstituted 3-quinoline carboxamide moiety is retained. On the other hand, we have made a surprising observation regarding the importance of the m-methoxy substituent in Region B: in hPDE4, this functionality provides ~7 fold potency over an alkyl group at this position [16], presumably by interacting with the catalytic metal ion via intervening water molecules. On the other hand, in the parasite enzyme, replacement of this methoxy group with an isosteric ethyl group shows surprisingly good activity (compare 19a with 24d, which have equivalent TbrPDEB1 and cellular potency, Table 5).
Supplementary Material
Acknowledgements
This work was funded by the National Institutes of Health (R01AI082577). We are grateful to Dr Emanuele Amata for helpful discussions. A free academic license to OpenEye Scientific Software and ChemAxon for their suites of programs is gratefully acknowledged.
Abbreviations
- COPD
chronic obstructive pulmonary disease
- HAT
Human African trypanosomiasis
- hPDE
human phosphodiesterase
- NECT
nifurtimox/eflornithine combination therapy
- TbrPDEB1, TbrPDEB2
Trypanosoma brucei phosphodiesterase B1 or B2
Footnotes
Conflict of Interest. The authors declare that no conflict of interest exists.
Synthesis of select boronic acid/ester intermediates, additional molecular modeling images, and tabulation of the compounds with their Northeastern registry numbers and screening data is available in the Supporting Information. The screening data has been made freely available as a shared data set at www.collaborativedrugdiscovery.com.
References
- [1].Savioli L, Daumerle D. Sustaining the drive to overcome the global impact of neglected tropical diseases. World Health Organization; 2013. Available from: http://apps.who.int/iris/bitstream/10665/77950/1/9789241564540_eng.pdf. [Google Scholar]
- [2].Priotto G, Kasparian S, Mutombo W, Ngouama D, Ghorashian S, Arnold U, et al. Nifurtimox-eflornithine combination therapy for second-stage African Trypanosoma brucei gambiense trypanosomiasis: a multicentre, randomised, phase III, non-inferiority trial. The Lancet. 2009;374:56–64. doi: 10.1016/S0140-6736(09)61117-X. [DOI] [PubMed] [Google Scholar]
- [3].Nok AJ. Arsenicals (melarsoprol), pentamidine and suramin in the treatment of human African trypanosomiasis. Parasitol Res. 2003;90:71–79. doi: 10.1007/s00436-002-0799-9. [DOI] [PubMed] [Google Scholar]
- [4].Pollastri MP, Campbell RK. Target repurposing for neglected diseases. Future Med. Chem. 2011;3:1307–1315. doi: 10.4155/fmc.11.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ochiana SO, Pandarinath V, Wang Z, Kapoor R, Ondrechen MJ, Ruben L, et al. The human Aurora kinase inhibitor danusertib is a lead compound for anti-trypanosomal drug discovery via target repurposing. Eur. J. Med. Chem. 2013;62:777–784. doi: 10.1016/j.ejmech.2012.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Patel G, Karver CE, Behera R, Guyett PJ, Sullenberger C, Edwards P, et al. Kinase scaffold repurposing for neglected disease drug discovery: Discovery of an efficacious, lapatanib-derived lead compound for trypanosomiasis. J. Med. Chem. 2013;56:3820–3832. doi: 10.1021/jm400349k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Bland ND, Wang C, Tallman C, Gustafson AE, Wang Z, Ashton TD, et al. Pharmacological validation of Trypanosoma brucei phosphodiesterases B1 and B2 as druggable targets for African sleeping sickness. J. Med. Chem. 2011;54:8188–8194. doi: 10.1021/jm201148s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ochiana SO, Gustafson A, Bland ND, Wang C, Russo MJ, Campbell RK, et al. Synthesis and evaluation of human phosphodiesterases (PDE) 5 inhibitor analogs as trypanosomal PDE inhibitors. Part 2. Tadalafil analogs. Bioorg. Med. Chem. Lett. 2012;22:2582–2584. doi: 10.1016/j.bmcl.2012.01.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wang C, Ashton TD, Gustafson A, Bland ND, Ochiana SO, Campbell RK, et al. Synthesis and evaluation of human phosphodiesterases (PDE) 5 inhibitor analogs as trypanosomal PDE inhibitors. Part 1. Sildenafil analogs. Bioorg. Med. Chem. Lett. 2012;22:2579–2581. doi: 10.1016/j.bmcl.2012.01.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Woodring JL, Bland ND, Ochiana SO, Campbell RK, Pollastri MP. Synthesis and assessment of catechol diether compounds as inhibitors of trypanosomal phosphodiesterase B1 (TbrPDEB1) Bioorg. Med. Chem. Lett. 2013;23:5971–5974. doi: 10.1016/j.bmcl.2013.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Setter SM, Iltz JL, Fincham JE, Campbell RK, Baker DE. Phosphodiesterase 5 inhibitors for erectile dysfunction. Ann. Pharmacother. 2005;39:1286–1295. doi: 10.1345/aph.1E487. [DOI] [PubMed] [Google Scholar]
- [12].Greulich T, Koczulla AR, Vogelmeier C. Chronic obstructive pulmonary disease: New pharmacotherapeutic options. Internist. 2012;53:1364–1370. doi: 10.1007/s00108-012-3119-1. [DOI] [PubMed] [Google Scholar]
- [13].Maiga M, Agarwal N, Ammerman NC, Gupta R, Guo H, Maiga MC, et al. Successful shortening of tuberculosis treatment using adjuvant host-directed therapy with FDA-approved phosphodiesterase inhibitors in the mouse model. PLoS One. 2012;7:e30749. doi: 10.1371/journal.pone.0030749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Yano M, Kohno M, Ohkusa T, Mochizuki M, Yamada J, Hisaoka T, et al. Effect of milrinone on left ventricular relaxation and Ca(2+) uptake function of cardiac sarcoplasmic reticulum. Am. J. Physiol. Heart Circ. Physiol. 2000;279:H1898–1905. doi: 10.1152/ajpheart.2000.279.4.H1898. [DOI] [PubMed] [Google Scholar]
- [15].Oberholzer M, Marti G, Baresic M, Kunz S, Hemphill A, Seebeck T. The Trypanosoma brucei cAMP phosphodiesterases TbrPDEB1 and TbrPDEB2: flagellar enzymes that are essential for parasite virulence. FASEB J. 2007;21:720–731. doi: 10.1096/fj.06-6818com. [DOI] [PubMed] [Google Scholar]
- [16].Woodrow MD, Ballantine SP, Barker MD, Clarke BJ, Dawson J, Dean TW, et al. Quinolines as a novel structural class of potent and selective PDE4 inhibitors. Optimisation for inhaled administration. Bioorg. Med. Chem. Lett. 2009;19:5261–5265. doi: 10.1016/j.bmcl.2009.04.012. [DOI] [PubMed] [Google Scholar]
- [17].de Koning HP, Gould MK, Sterk GJ, Tenor H, Kunz S, Luginbuehl E, et al. Pharmacological validation of Trypanosoma brucei phosphodiesterases as novel drug targets. J. Infect. Dis. 2012;206:229–237. doi: 10.1093/infdis/jir857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Orrling KM, Jansen C, Vu XL, Balmer V, Bregy P, Shanmugham A, et al. Catechol pyrazolinones as trypanocidals: fragment-based design, synthesis, and pharmacological evaluation of nanomolar inhibitors of trypanosomal phosphodiesterase B1. J. Med. Chem. 2012;55:8745–8756. doi: 10.1021/jm301059b. [DOI] [PubMed] [Google Scholar]
- [19].Lunniss CJ, Cooper AW, Eldred CD, Kranz M, Lindvall M, Lucas FS, et al. Quinolines as a novel structural class of potent and selective PDE4 inhibitors: optimisation for oral administration. Bioorg. Med. Chem. Lett. 2009;19:1380–1385. doi: 10.1016/j.bmcl.2009.01.045. [DOI] [PubMed] [Google Scholar]
- [20].Baldwin IR, Barker MD, Dean AW, Eldred CD, Evans B, Gough SL, et al. Preparation of quinoline derivatives as phosphodiesterase inhibitors WO2004103998A1. 2004 [Google Scholar]
- [21].Jansen C, Wang H, Kooistra AJ, de Graaf C, Orrling KM, Tenor H, et al. Discovery of novel Trypanosoma brucei phosphodiesterase B1 inhibitors by virtual screening against the unliganded TbrPDEB1 crystal structure. J. Med. Chem. 2013;56:2087–2096. doi: 10.1021/jm3017877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Surry DS, Buchwald SL. Biaryl phosphane ligands in palladium-catalyzed amination. Angew. Chem. 2008;47:6338–6361. doi: 10.1002/anie.200800497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Schopfer U, Schlapbach A. A general palladium-catalyzed synthesis of aromatic and heteroaromatic thioethers. Tetrahedron. 2001;57:3069–3073. [Google Scholar]
- [24].Buckman B, Nicholas JB, Beigelman L, Serebryany V, Stoycheva AD, Thrailkill T, et al. Preparation of macrocyclic peptides, especially proline-containing peptides, as inhibitors of hepatitis C virus replication for treating hepatitis C infection and liver fibrosis WO2011038293A1. 2011 [Google Scholar]
- [25].Wu W-L, Burnett DA, Stamford AW, Cumming JN, Bennett CE, Gilbert EJ, et al. Preparation of 5-substituted iminothiazines and their mono- and dioxides as BACE inhibitors WO2012139425A1. 2012 [Google Scholar]
- [26].Young J, Czako B, Altman M, Guerin D, Martinez M, Rivkin A, et al. Pyridazinones as tyrosine kinase inhibitors and their preparation and use in the treatment of cancer WO2011084402A1. 2011 [Google Scholar]
- [27].Wada A. The alpha-helix as an electric macro-dipole. Adv. Biophys. 1976:1–63. [PubMed] [Google Scholar]
- [28].Higman VA, Boyd J, Smith LJ, Redfield C. Asparagine and glutamine side-chain conformation in solution and crystal: a comparison for hen egg-white lysozyme using residual dipolar couplings. J. Biomol. NMR. 2004;30:327–346. doi: 10.1007/s10858-004-3218-y. [DOI] [PubMed] [Google Scholar]
- [29].Word JM, Lovell SC, Richardson JS, Richardson DC. Asparagine and glutamine: using hydrogen atom contacts in the choice of side-chain amide orientation. J. Mol. Biol. 1999;285:1735–1747. doi: 10.1006/jmbi.1998.2401. [DOI] [PubMed] [Google Scholar]
- [30].Amata E, Bland ND, Hoyt CT, Settimo L, Campbell RK, Pollastri MP. Repurposing human phosphodiesterase 4 inhibitors for neglected tropical diseases. 1 Design, synthesis and evaluation of cilomilast analogues as Trypanosoma brucei phosphodiesterase B1 (TbrPDEB1) inhibitors. Bioorg. Med. Chem. Lett. 2014 doi: 10.1016/j.bmcl.2014.07.063. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Duplantier AJ, Bachert EL, Cheng JB, Cohan VL, Jenkinson TH, Kraus KG, et al. SAR of a Series of 5,6-Dihydro-(9H)-pyrazolo[3,4-c]-1,2,4-triazolo[4,3-α]pyridines as Potent Inhibitors of Human Eosinophil Phosphodiesterase. J. Med. Chem. 2007;50:344–349. doi: 10.1021/jm060904g. [DOI] [PubMed] [Google Scholar]
- [32].Ehrman TM, Barlow DJ, Hylands PJ. In silico search for multi-target anti-inflammatories in Chinese herbs and formulas. Bioorg. Med. Chem. 2010;18:2204–2218. doi: 10.1016/j.bmc.2010.01.070. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.