Abstract
Humoral immunity, including antibody switching and somatic hypermutation, is critically regulated by CD4+ T cells. T follicular helper (Tfh) cells have been recently shown to be a distinct T cell subset important in germinal center reations. The transcriptional regulation of Tfh cell development and function has not been well understood. Here, we report that the CCAAT enhancer binding protein alpha (C/EBPα), a basic region/leucine zipper (bzip) transcription factor, is highly expressed in Tfh cells. Cebpa-deficient CD4+ T cells exhibit enhanced IFN-γ expression in vitro and in vivo. T cell-specific Cebpa knockout (KO) mice, though not defective in Tfh cell generation, produce significantly increased levels of IgG2a/b and IgG3 following immunization with a protein antigen. Moreover, C/EBPα binds to the Ifng gene and inhibits T-bet-driven Ifng transcription in a DNA binding-dependent manner. Our study thus demonstrates that C/EBPα restricts IFN-γ expression in T cells to allow proper class switching by B cells.
Introduction
Antibody production is a major defense mechanism by the immune system. To generate effective antibodies against various pathogens, B cells need to receive cognate help from CD4+ T cells, especially in germinal center (GC), in which somatic hyper-mutation (SHM) and class switch recombination (CSR) take place (1). CSR, by generating different isotypes of immunoglobulin (Ig) that vary in binding to Fc receptors, half lives and activation of the complement system as well as tissue localization (2), is necessary for optimal humoral immunity. Both Th1 and Th2 cells have been shown to regulate class-switching: IL-4 is able to promote B cell proliferation and class switching, especially to IgE and IgG1, whereas IFN-γ regulates IgG2 and IgG3 antibody production. T follicular helper (Tfh) cells, which produce substantial amounts of IL-21 and IL-4, promote the production of isotype-switched, high-affinity antibodies in the germinal center (3–7).
Helper T (Th) cell differentiation is programmed by lineage-specific master transcription factors (8). T-bet, encoded by Tbx21, is an essential factor to induce IFN-γ and Th1 differentiation. GATA3 is a master regulator for Th2 differentiation. ROR-γ and ROR-α are critical transcription factors for inducing IL-17 and Th17 differentiation. B cell lymphoma 6 (Bcl6) has been shown to be a critical transcription factor that regulates the generation of Tfh cells (9–11). Bcl6 does not appear to regulate early Tfh cell generation (7), and our new data indicate that Ascl2 initiates CXCR5 expression in T cells and their migration to the B cell follicles (12).
In T-dependent (TD) responses, B cells make cognate interaction with activated CD4+ T cells at the T-B border and differentiate along either extrafollicular or follicular pathways. It is important to note that this initial T-B interaction initiates the process of isotype switching toward IgG1 and IgG2a in B cells (13). Although extrafollicular antibody produced by short-lived plasma cells is relatively low affinity, this early antibody is can be important for fast protection against virus infection (14–16). IgG2a-producting B cells are enriched in the extrafollicular region and relatively low in frequencies in GCs of normal mice, where IgG1+ cells prevail, possibly because IFN-γ expression is downregulated in Tfh cells (14, 17). However, IgG2a production is dramatically increased in a lupus model (Roquinsan/san) in an IFN-γ dependent manner (18). In several other mouse lupus models including MRL/lpr mice, among the IgG isotypes elevated in the mice, IgG2a is most dramatically increased in a T cell-dependent manner and closely linked to lupus pathogenesis (19, 20). Heightened IgG2a production may thus have implications in autoimmunity. However, mechanisms whereby antigen-specific CD4+ T cells regulate class switching toward IgG2 in GCs and extrafollicular regions have not fully been understood.
The CCAAT enhancer binding protein alpha (C/EBPα) is the founding member of a family of basic region/leucine zipper (bzip) transcription factor. C/EBP family proteins share highly homologous C-terminal dimerization (leucine zipper) domains and DNA binding [basic region (BR)] motifs but differ in their N-terminal transactivation domains and bind DNA either as homo- or heterodimers (21). C/EBPα has been reported to play critical roles in lineage-specific gene regulation in several cell types during hepatic, adipogenic, granulocytic, skin, lung and placenta development (22). However, roles of C/EBPα in T cell differentiation remain unknown
Here, we report that C/EBPα is highly expressed in Tfh cells compared to other Th subsets. C/EBPα suppressed Th1 and Th17 differentiations in vitro. The deficiency of Cebpa in T cells resulted in enhanced IFN-γ expression in vivo and increased antigen-specific IgG2a/b and IgG3 production. Furthermore, C/EBPα binds to the Ifng gene in Tfh cells and suppresses T-bet-mediated Ifng gene transcription. Taken together, C/EBPα expressed in T cells plays a crucial role in negative regulation of IgG2 and IgG3 antibody responses in vivo by controlling IFN-γ production. This study provides a new mechanism whereby appropriate T cell function is regulated in humoral immunity.
Materials and Methods
Mice
Cebpa f/f (33) and Cd4-cre Tg mice (34) were provided by The Jackson Laboratory (Bar Harbor, Main) and by Dr. Wilson. T cell-specific Cebpa conditional KO mice were produced by breeding Cebpa f/f mice with Cd4cre Tg mice. Screening of Cebpa conditional KO mice was carried out, as previously described (33, 34). Mice 6–10 weeks of age were used in experiments following protocols approved by Institutional Animal Care and Use Committee, MD Anderson Cancer Center.
Helper T cell differentiation and stimulation of activated T cells
CD44lo CD62Lhi CD25− naïve CD4+ T cells from lymph nodes and spleens of mice were purified by FACS sorting. For Th differentiation, naïve CD4 T cells were stimulated with plate-bound anti-CD3 (0.5 μg/ml; 2C11; BioXcell) plus anti-CD28 (0.5 μg/ml; 37.51, BioXcell) in the presence of neutralizing antibodies [10 μg/ml anti-IL-4 (11B11, BioXcell), 10 μg/ml anti-IFN-γ (XMG 1.2, BioXcell) and anti-TGF-β (1D11, BioXcell)] or with polarizing cytokines for Th0;10 μg/ml anti-IL-4, 10 ng/ml IL-12 (210-12, Peprotech) and 50 U/ml human IL-2 for Th1; 10 μg/ml anti-IFN-γ, 10 ng/ml IL-4 and 50 U/ml human IL-2 for Th2; 20 ng/ml IL-6 (216-16; Peprotech), 5 ng/ml TGF-β, anti-IFN-γ and anti-IL-4 for Th17; 50U/ml human IL-2, 5 ng/ml TGF-β, anti- IFN-γ and anti- IL-4 for iTreg; 20 ng/ml IL-6, anti- IFN-γ, anti- IL-4 and anti-TGF-β for Tfh-like cells. For stimulation with peptide-loaded APC, FACS-sorted naïve CD4+ T cells were cultured with irradicated splenocytes in the presence of 10 μg/ml OTII peptide (chicken OVA peptide 323–339). After 4 d of culture, cells were washed and re-stimulated with plate-bound anti-CD3 (0.5 μg/ml) for 4 h, and cells were then collected for RNA extraction. For cytokine measurement by ELISA, culture supernatants were collected at 24 h. For intracellular cytokine analysis, cells were restimulated with 500 ng/ml of ionomycin and 50 ng/ml of PMA in the presence of Golgi Stop (BD Pharmingen) for 5 h. Cells were then permeabilized with Cytofix/Cytoperm Kit (BD Pharmingen) or Foxp3 2staining buffer set (e-bioscience) and analyzed for the expression of intracellular cytokines with anti-IFN-γ (XMG1.2), IL-4 (11B11) and IL-17A (TC11-18H10) Abs [BD (Flanklin Lakes, NJ)]. Intracellular Bcl6 and Foxp3 were detected with anti-Bcl6 (K112-471.3.93) and Foxp3 (FJK-16s) Abs. The reagents for ELISA, anti-IFN-γ (R4-6A2 and XMG1.2 biotin), anti-IL-2 (JES6-1A12 and JES6-5H4 biotin) anti-IL-4 (BVD4-1D11 and BVD6-24G2 biotin) and anti-IL-17 (TC11-18H10 and TC11-8H4.1 biotin) were purchased from BD.
Immunization
Mice of 6–10 wks old were immunized either with KLH (0.5 mg/ml) or NP27-KLH emulsified in CFA (0.5 mg/ml) at the base of the tail (100 μl each mouse) or OVA (1 mg/ml) emulsified in Alum in peritoneal cavity. The germinal center B cells were stained with anti-GL7, anti-Fas and anti-B220 Abs (BD). Tfh cell and extrofollicular T cell generation were determined by staining with anti-CD4, anti-BTLA, anti-Bcl6, anti-CXCR5 and anti-PD-1 Abs (BD). KLH or NP-specific IgM and IgG Abs in sera from immunized mice were measured with ELISA. Serum samples were added in a 3-fold serial dilution onto plates pre-coated with OVA, KLH or NP-BSA (NP4-BSA for high affinity and NP26-BSA for global affinity). Antigen-specific antibodies were detected with horseradish peroxidase (HRP)-conjugated goat anti-mouse IgM or rat anti-mouse IgG Abs (Southern Biotechnology Associates). The relative affinity of anti-NP Abs was estimated by calculation of the ratio of anti-NP4 /anti-NP26 Abs. For analysis of cytokine expression, draining LN (dLN) cells and splenocytes from immunized mice were stimulated as triplicates with 0, 5, 20 and 100 μg/ml KLH or 0, 4, 20 and 100 μg/ml OVA. After 4 days of re-stimulation, measurement of IFN-γ and IL-17 was carried out by ELISA. For intracellular cytokine staining, dLN cells and splenocyte from immunized mice were restimulated with 0 and 100 μg/ml KLH or 0 and 250 μg/ml OVA for 24 hr. PMA and Ionomycin or KLH-loaded APC were used for re-stimulation of ex vivo PD-1 hi Tfh (CD4+, PD-1hi and CXCR5+), Tef (CD4+, PD-1lo and CXCR5+) and non-Tfh (CD4+, CD44hi and CXCR5−) cells to detect intracellular IFN-γ. In the last 5 hrs, Golgi-stop (BD) was added, and IFN-γ producing cells were detected in CD4+ population, described above. To measure mRNA expression or for western blotting, CD4+ T cell subpopulation were purified by FACS sorting from dLNs, on the basis of their specific markers. For mRNA measurement, cells were re-stimulated with anti-CD3 Ab for 4hrs. Anit-C/EBPα Ab (C-18) was purchased from Santa Cruz Biotechnology for western blotting.
Retroviral transduction
pGFP-RV-Cebpa were generated by cloning Cebpa cDNA into pGFP-RV or pMIGRII retroviral vector. HA tag was inserted in N terminus of Cebpa cDNA by PCR. Mutant Cebpa which lacking basic region (BR:287-247a.a.) was cloned by PCR and constructed in pGFP RV (pGFP-RV-ΔBR Cebpa). The plasmid pGFP-RV and pMIGRII contain an internal ribosomal entry site (IRES)-regulated green fluorescent protein and human CD2 genes, respectively. pGFP-RV-Bcl6 was previously described (10). Naive CD4+CD25−CD62LhiCD44lo T cells from OT-II Tg mice were sorted by flow cytometry and activated with OT-II peptide (10ug/ml) and syngeneic APC (irradiated splenocytes) in the presence of polarizing cytokines. 24 hours after activation, cells were spin-infected with retrovirus expressing C/EBPα or control empty vector. 4 days after activation, GFP+ or hCD2+ cells were sorted by flow cytometry and gene expression was assessed by quantitative real-time PCR. For cytokine measurement, ELISA and intracellular cytokine staining were performed as described above. For proliferation assay, Vybrant dye cycle violet stain (Molecular Probes) was used as an indicator for cell division.
Quantitative RT-PCR (qRT-PCR)
Total RNA extracted using Trizol reagent (Invitrogen) was used to generate cDNA using oligo (dT), random hexamers, and MMLV reverse transcriptase (Invitrogen). For quantitation of cytokine, cDNA samples were amplified in IQ SYBR Green Supermix (Bio-Rad Laboratories). The data were normalized to an Actb reference. The primer pairs for analysis were previously described (17). Primer pair for Cebpa (endo+exo) : forward, 5′-TGCTGGAGTTGACCAGTGAC-3′; reverse, 5′-CCTTGACCAAGGAGCTCTCA-3′, Cebpa (endo) : forward, 5′-ACAATCGATCCATCCCAGAG-3′; reverse, 5′-AGCATAGACGTGCACACTGC-3′
Reporter assay
293T were transfected with pGFP- RV-T-bet (35) together with luciferase construct containing the human Ifng promoter (30). We generated luciferase mutant constructs (Δ −56 to −48 and Δ −51 to −48) by PCR. Total amount of plasmid DNA remained constant in each sample. These cells were further transfected with various amount of pGFP-RV-Cebpa or pGFP-RV-ΔBR Cebpa. Firefly and renilla luciferase activity was measured with a Dual-Luciferase Reporter system (Promega). Data was normalized by the activity of Renilla luciferase.
Chromatin immunoprecipitation assay
ChIP assays were done as described (36) (37) with anti-H3K9/14Ac (Upstate), anti-H3K4me Ab (Upstate), anti-H3K9me3 Ab, anti-H3K27me3 (Upstate), anti-RNA polymerase II (SantaCruz) anti-C/EBPα Ab (sc-61) or control IgG. The immuno-precipitated DNA was analyzed by real-time PCR. Primers were as follows: CNS-55 forward, 5′-TGTCTCGGTGACACATCCTT-3′, and reverse, 5′-GGGAGGCAGGAGGAACTTTA-3′; CNS-34 forward, 5′-AAAAGAGTCCAAGATATGAAAGCAA-3′, and reverse, 5′-GGCTTTGGGAATTCTACCTTG-3′; CNS-22 forward, 5′-ATGACAAAATGCAGGGCTTC-3′, and reverse, 5′-CCCACACTAGATGATATATGATTTTCC-3′; CNS1 (CNS-5) forward, 5′-CACTTCTGTGCAACCCTTGA-3′, and reverse, 5′-AAGCACTCACTGGGTCATTG-3′; Ifn-γ Promoter forward, 5′-CCCCACCTATCTGTCACCAT-3′, and reverse, 5′-CACCTCTCTGGCTTCCAGTT-3′; CNS2 (CNS+17) forward, 5′-AACTGGAAAATGGCAGGCTA-3′, and reverse, 5′-CCCGAGATAAATTCCATCCA-3′; H19 ICR forward, 5′-GCATGGTCCTCAAATTCTGCA-3′, and reverse, 5′-GCATCTGAACGCCCCAATTA-3′; Actb exon1 forward, 5′-CACGATGGAGGGGAATACAG-3′, and reverse, 5′-TCTTGATAGTTCGCCATGGAT-3′
Immunohistochemical analysis
Inguinal lymph nodes were isolated from either WT or Cebpa KO mice 7 days after immunization with KLH in CFA and embedded in OCT and frozen with isopentane. Staining of PNA, CD4, IgG2a and IgG1 was performed as described previously (17). Anti- IgG2a and IgG1 Abs were purchased from BD. Cultured cells were put on slide glasses by cytospin (Thermo) and fixed and permeabilized. Anti-HA Ab (6E2) (Cell Signaling) was used for staining.
Statistical analysis
All data represented with mean±SD, and statistical analysis was performed with the Student’s t test. Differences were recognized significant with p value less than 0.05. *p<0.05 and **p<0.01
Results
C/EBPα is highly expressed in Tfh cells
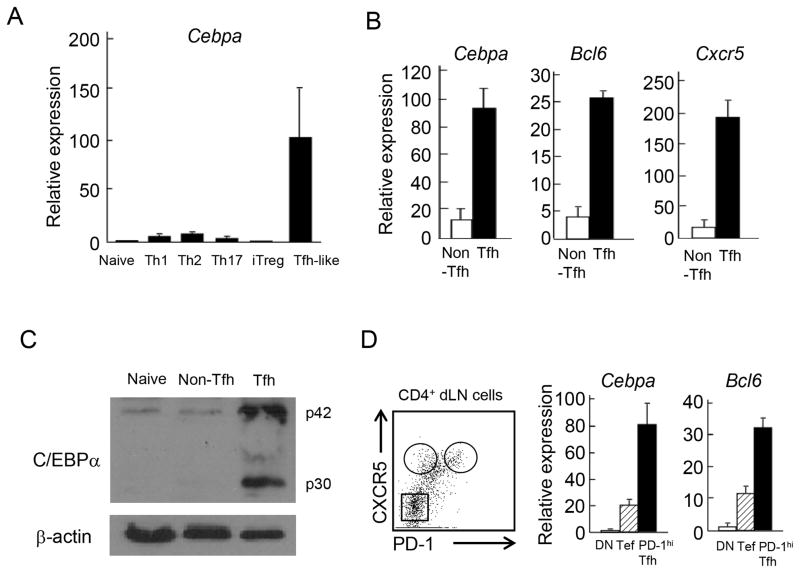
In the gene expression microarray analysis, Cebpa mRNA expression was higher in CXCR5+ T follicular helper (Tfh) than Th1, Th2 and Th17 cells (17). We thus confirmed Cebpa mRNA expression in Th1, Th2, Th17, inducible regulatory T cells (iTreg) and Tfh-like cells by real-time RT-PCR analysis. We cultured naive CD4+ T cells isolated from OT-II transgenic (Tg) mice under Th1, Th2, Th17 and iTreg as well as Tfh-like promoting culture conditions with OVA peptide-loading irradiated antigen-presenting cells (APCs). The expression of Cebpa mRNA was highly induced in Tfh-like cells but not in other helper T (Th) cell lineages upon re-stimulation on day 5 (Figure 1A). IL-6 or IL-21 alone was not sufficient in upregulating Cebpa mRNA expression in purified T cells (data not shown), suggesting that a collective signal from APC and cytokines is required for its regulation.
Figure 1. C/EBPα is highly expressed in Tfh cells.
(A) CD4+ T cells isolated from OT-II TCR Tg mice were stimulated with OVA peptide loaded APCs under Th1, Th2, Th17, iTreg, and T follicular helper (Tfh) cell-like skewing conditions for 5 days. The cultured cells were re-stimulated with anti-CD3 Ab. Then Cebpa mRNA expression was measured by quantitative PCR analysis. Data were normalized to bactin. The expression in naïve CD4+ T cells was determined as one. (B) B6 mice (n=3) were immunized with KLH CFA. After 7 days, CXCR5+ Tfh and CXCR5− non-Tfh cells were isolated from CD4+CD44hi dLNs by FACS sorting. The mRNA expression of Cebpa, Bcl6 and Cxcr5 in stimulated Tfh and non-Tfh cells with anti-CD3 Ab was measured by quantitative PCR. Data are a representative of three independent experiments. (C) C/EBPα was, by western blotting, detected in nave (CD44lo CD4+), non-Tfh, and Tfh cells isolated from immunized B6 mice. β-actin was used for loading control. Data are a representative of two independent experiments. (D) The mRNA expression of Cebpa and Bcl6 in PD-1hi Tfh (PD-1hi CXCR5+ CD4+), extrafollicular T (Tef) (PD-1lo CXCR5+ CD4+) and double negative (DN) (PD-1− CXCR5− CD4+) cells was measured by quantitative PCR. Data are a representative of two independent experiments.
We further assessed the expression of C/EBPα in vivo. C57BL/6 (B6) mice were immunized with keyhole limpet hemocyanin (KLH) in complete Freund’s adjuvant (CFA). After 7 days, CXCR5+CD44hi Tfh and CXCR5−CD44hi non-Tfh CD4+ cells were isolated from draining Lymph nodes (dLN) and analyzed. Tfh cells expressed higher levels of mRNA for Cxcr5, Bcl6 and Cebpa than non-Tfh cells (Figure 1B). In addition to the mRNA analysis, the expression of C/EBPα in naïve T, non-Tfh and Tfh cells was further confirmed at the protein level (Figure 1C). Extrafollicular T (Tef) cells, which regulate extrafollicular antibody response, have been identified as PD-1lo CXCR5+ CD4+ T cells outside of follicles (14). Tef cells but not PD-1 and CXCR5 double negative (DN) cells expressed mRNA for Cebpa and Bcl6, although their expression levels were less than those of PD-1hi Tfh cells (Figure 1D). These data indicates that C/EBPα is highly expressed both in Tef and PD-1hi Tfh cells in vivo.
C/EBPα negatively regulates IFN-γ expression as well as IgG2a/b and IgG3 antibody response in vivo
Since C/EBPα is highly expressed in Tfh cells (Figure 1), we speculated that C/EBPα may regulate the generation or function of Tfh cells. To examine this, we crossed Cebpa f/f mice with Cd4-Cre transgenic (Tg) mice. Staining for CD4 and CD8 in the thymus and CD62L and CD44 in peripheral CD4+ T cells did not show any abnormality of T cell subpopulations in the absence of Cebpa. The frequency of CD4+Foxp3+CD25+ regulatory T (Treg) cells was not changed, indicating no obvious defect in T cell development as a result of Cebpa deficiency (Figure S1A). When we evaluated Cebpa gene deletion at genomic and mRNA levels, Cebpa was preferentially disrupted in CD4+ T cells but not in B cells and DCs (Figure S1B). when we measured the expression of Cebpb and Cebpd in naive, Tef and PD-1hi Tfh cells isolated from Cebpa f/f (WT) and Cebpa f/f-Cd4-Cre Tg (KO) mice immunized with KLH in CFA, we did not find they were changed in the absence of Cebpa (Figure S1C), indicating that the functional defects caused by Cebpa deficiency in T cells may not be compensated by increased expression of C/EBPβ and C/EBPδ in vivo.
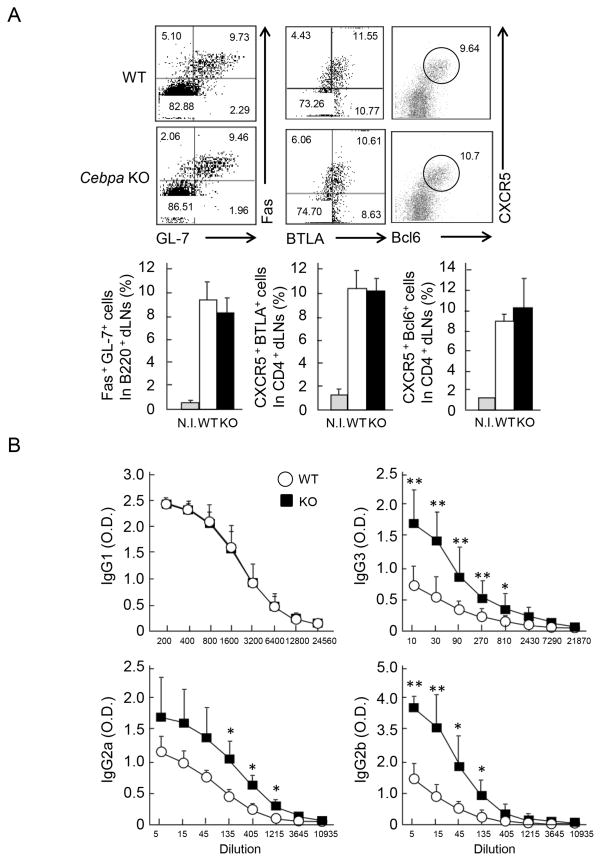
To examine the function of C/EBPα in T cells, WT and Cebpa KO mice were immunized with KLH plus CFA. After 7 days, germinal center (GC) B cells were detected by staining for GL7 and Fas as well as CXCR5 and Bcl6 in B220+ dLN cells. CXCR5 with BTLA or Bcl6 staining was used to identify Tfh cells in dLNs. GC B cell and Tfh cells developed normally in Cebpa-deficient mice (Figure 2A and Figure S1D). In addition, we analyzed the localization of GC B cells and Tfh cells either in WT and the KO mice by immunohistochemical staining with PNA (brown) and anti-CD4 (blue). There was no significant difference between the two groups of mice (Figure S1E), suggesting that the KO T cells were able to migrate properly and induce GC reactions. The expression of Tfh-related genes such as Bcl6, Cxcr5, Il21r, Il6ra, Il6st, Icos, Il21, Sap, Cd40lg and Il4 was further analyzed by real-time RT-PCR in Tfh cells isolated from immunized WT and Cebpa KO mice. While C/EBPα expression in Tfh cells isolated from KO mice was completely eliminated, the expression levels of the Tfh-related genes were found to be comparable in Cebpa-deficient and -sufficient Tfh cells (Figure S1F and data not shown), indicating that C/EBPα is not necessary for the development of Tfh cells and their migration and/or localization.
Figure 2. Enhanced IgG2a/b and IgG3 production in immunized Cebpa KO mice.
(A) Cebpa f/f (WT n=4; white) and Cebpa f/f-Cd4cre (Cebpa KO n=4; black) mice were immunized with KLH CFA. 7 days later, germinal center (GC) B cells were identified by staining of GL7/Fas in B220+ dLN cells as well as Tfh cells by CXCR5/BTLA or CXCR5/Bcl6 in CD4+dLN cells (upper). Frequency of Tfh and GC B cells was shown by combining results from four independent experiments (lower). (B) The sera were isolated from WT and Cebpa KO mice after 7 days of immunization and subjected to a 3-fold serial dilution. The concentration of KLH specific IgG1, IgG2a, IgG2b, and IgG3 were determined by ELISA. Data are a representative of three independent experiments with similar results.
We further analyzed KLH-specific antibody production and found IgG2a/b and IgG3 production in sera of immunized Cebpa KO mice to be significantly enhanced (2~3 fold at 135, 405 and 1215 dilution for IgG2a, at 5, 15, 45 and 135 dilution for IgG2b, and at 10, 30, 90, 270 and 810 for IgG3) compared to those of WT mice, while the production of other antibody isotypes such as IgM and IgG1 was similar between WT and Cebpa KO mice (Figure 2B and data not shown). Since Tfh cells controls affinity maturation of antibodies in the responding B cells (23), we examined whether C/EBPα expressed in T cells regulates this process. WT and Cebpa KO mice were immunized with 4-hydroxy-3-nitrophenyl (NP)-conjugated KLH in CFA and further boosted with NP-KLH in incomplete Freund’s adjuvant (IFA). Anti-NP IgM, IgG1, IgG2a and IgG3 titers were determined by NP-specific ELISA and ratio of high affinity versus global affinity Ab titers were compared between WT and the KO mice. Although the ratio of high to global affinity antibodies in each isotype was increased after boosting, no significant difference in the ratio between WT and KO mice was observed (Figure S2A), indicating C/EBPα is dispensable for affinity maturation and C/EBPα KO Tfh cells may provide similar help to B cells as WT Tfh cells do in terms of GC formation and affinity maturation. To test whether the enhanced IgG2 production found in Cebpa KO mice originated from GCs or outside of follicles, we conducted histological and flow cytometric analyses of IgG1 and IgG2a expression. On day 7 after immunization, the co-staining of IgG1 or IgG2a together with PNA and the detection of IgG1 and IgG2a in GL-7+ (GCs) or GL-7− (outside of GCs) cells in B220+ dLNs were performed. The C/EBPα deficiency led to the enhanced frequency of IgG2a-but not IgG1-forming cells both inside and outside of GCs (Figure S2B and Figure S2C), indicating that the enhanced IgG2a production occurs GCs and outside follicles.
Cebpa deficiency in T cells leads to enhanced IFN-γ production
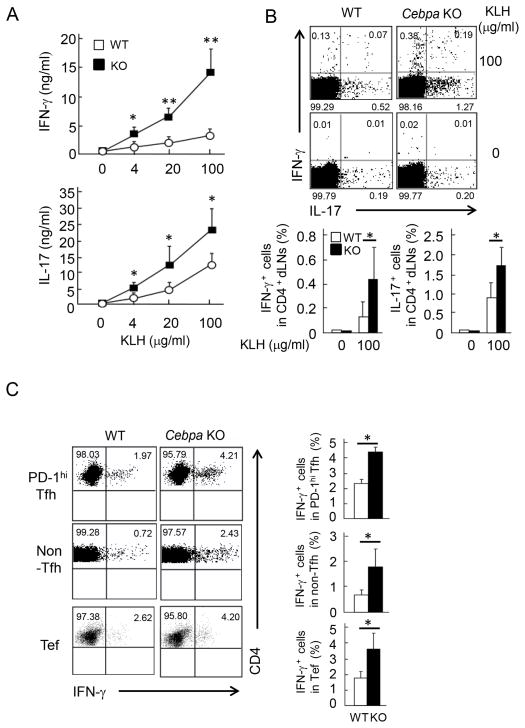
To understand the mechanism underlying the enhanced IgG2 and IgG3 production in KO mice, splenocytes from immunized WT and Cebpa KO mice were stimulated with various concentrations of KLH protein. IL-2 expression was measured by ELISA on day 1 and incorporation of [3H] thymidine, on day 3 of re-stimulation. Cebpa KO T cells did not show any difference of IL-2 production or proliferation, when compared to WT cells (Figure S3A). Because the class switching of IgG2 and IgG3 is regulated by IFN-γ (24–27), IFN-γ and IL-17 production by KLH-specific T cells in immunized mice were detected by ELISA and intracellular cytokine staining (ICS). In response to KLH re-stimulation, Cebpa KO CD4+ T cells in dLNs expressed increased levels of IFN-γ and IL-17 than WT cells by ELISA and ICS (Figure 3A and 3B). Taken together, C/EBPα appears to negatively regulate IFN-γ and IL-17 expression in antigen-specific T cell responses in vivo.
Figure 3. Enhanced IFN-γ and IL-17 production in T cells from immunized Cebpa KO mice.
(A) Total dLN cells were isolated from WT (n=4) and Cebpa KO mice (n=4) after immunization. Then, cells were stimulated with indicated concentrations of KLH. IFN-γ and IL-17 were measured by ELISA after 7 days or 5 days of treatment, respectively. (B) The cells isolated in (A) were stimulated with 0 or 100 μg/ml KLH. After 24 hrs, IFN-γ and IL-17 were detected in CD4+ cells by ICS (upper). Frequency of IFN-γ+ and IL-17+ cells was shown by combining three experiments (Lower). Data are the mean of three independent experiments **p<0.01 and *p<0.05 by the Student’s t-test. (C) PD-1hi Tfh, Tef and non-Tfh cells were isolated from WT (n=4) and Cebpa KO mice (n=4) after 7 day of immunization. IFN-γ expression was detected in Tfh, Tef and non-Tfh cells stimulated with PMA and Ionomycin or KLH-loaded APCs by ICS (left). Frequency of these IFN-γ-expressing T cell pupulations was shown by combining results from two independent experiments (lower).
We next asked which T cell populations contributed to the enhanced IFN-γ expression in Cebpa KO mice. Tfh, Tef and non-Tfh cells were isolated from WT or Cebpa KO mice after immunization and stimulated with phorbol myristate acetate (PMA) plus ionomycin or KLH-loaded APC. The frequency of IFN-γ+ Tfh cells, IFN-γ+ Tef, IFN-γ+ non-Tfh cells were 2 to 3 fold increased in the KO mice, compared to those in WT mice upon stimulation (Figure 3C). This data indicates that IFN-γ expression in effector CD4+ T cells localized both in outside and inside of follicles was enhanced in the absence of Cebpa and that the suppressive activity of C/EBPα in IFN-γ expression might not be restricted to Tfh cells.
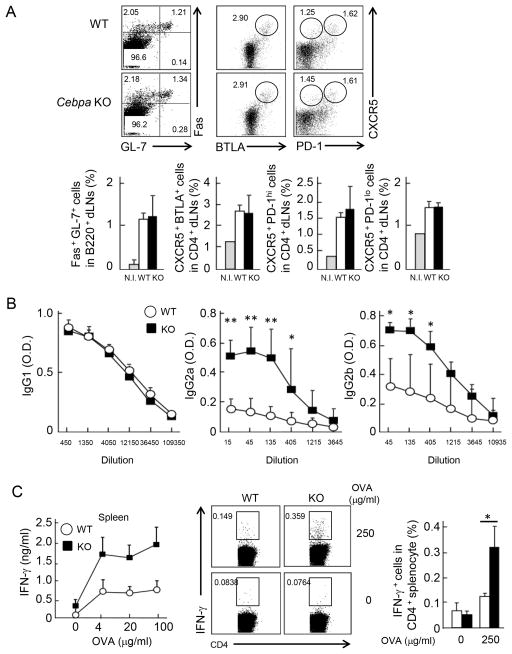
To further confirm the suppressive role of C/EBPα in IFN-γ expression in vivo, we immunized WT and Cebpa KO mice with ovalbumin (OVA) in aluminium sulfate (Alum) which induces Th2-biased immune response. Although the generation of GC B cells, Tfh cells and Tef cells was not affected (Figure 4A), OVA-specific IgG2a/b production were enhanced in the absence of C/EBPα in T cells (Figure 4B). Furthermore, upon re-stimulation with OVA, Cebpa KO CD4+ T cells in spleen expressed increased levels of IFN-γ than WT cells by ELISA and ICS (Figure 4C). These results further emphasize the importance of suppressive activity of C/EBPα in IFN-γ expression in T cells.
Figure 4. Enhanced IgG2a/b in Cebpa KO mice immunized with OVA in alum.
(A) WT (n=4; white) and Cebpa KO (n=5; black) mice were immunized with OVA alum (day 0 and day 7) and boosted with OVA on day 14. On day 21, GC B, Tef and PD-1+ Tfh cells were identified by FACS staining in spleen (upper). Frequency of these populations was shown (lower). (B) The sera were isolated from WT and Cebpa KO mice on day 21 after boosting and subjected to a 3-fold serial dilution. The concentration of OVA specific IgG1, IgG2a and IgG2b were determined by ELISA. Data are a representative of three independent experiments with similar results. (C) Total splenocyte was isolated from WT and Cebpa KO mice after immunization. Then, cells were stimulated with indicated concentrations of OVA. IFN-γ was measured after three days of re-stimulation by ELISA (left). The splenocytes were stimulated with 0 or 250 μg/ml OVA. After 24 hrs, IFN-γ was detected in CD4+ cells by ICS (middle). Frequency of IFN-γ+ cells was shown (right). Data are a representative of three independent experiments with similar results.
C/EBPα inhibits Th1 and Th17 cell differentiation
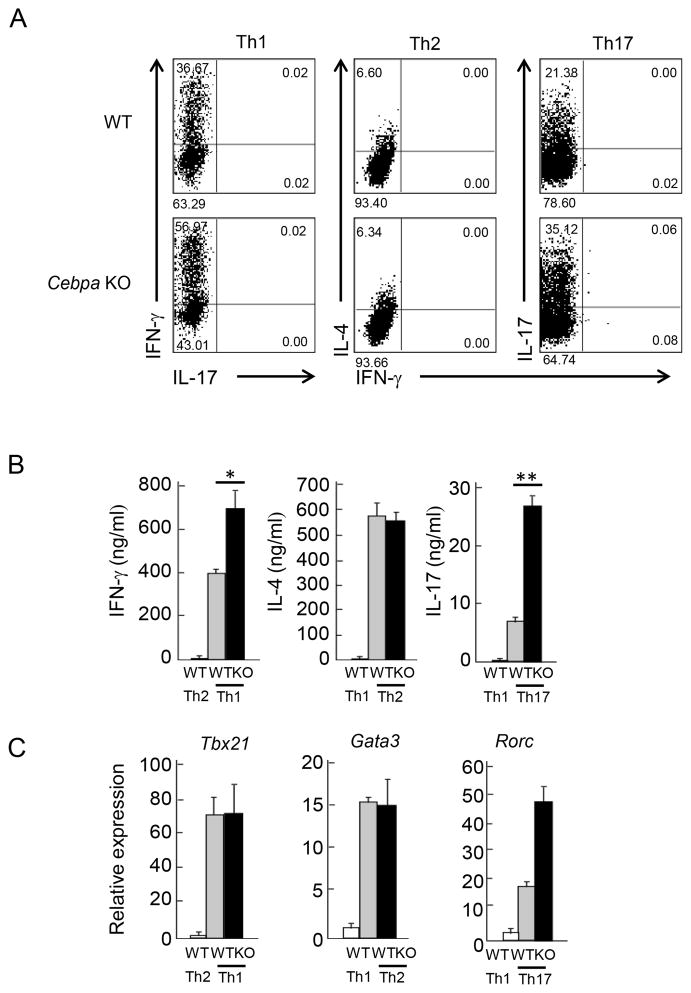
The results from the above in vivo experiment indicate that C/EBPα is not required for Tfh cell generation but may negatively regulate their IFN-γ expression. To further confirm the role of C/EBPα in Th differentiation, naïve CD4+ T cells were isolated from WT and the KO mice by FACS sorting and stimulated with anti-CD3 and CD28 Abs under Th1, Th2 and Th17 skewing condition. After 4 days, cells were re-stimulated and cytokine expression was detected by ELISA and ICS. Under Th1 and Th17 condition, Cebpa KO CD4+ T cells exhibited enhanced amounts of IFN-γ and IL-17, respectively. In contrast, loss of C/EBPα did not affect IL-4 production under Th2 condition (Figure 5A). Similar results were obtained by ELISA assay (Figure 5B). To further address whether C/EBPα inhibits Th cell differentiation, the expression of master transcription factors were measured by quantitative RT-PCR. Tbx21 and Gata3 mRNA expression in Th1 or Th2 cells was not changed by C/EBPα deficiency, respectively. However, Rorc expression was significantly enhanced in Cebpa KO cells at the mRNA level (Figure 5C). These observations suggest that C/EBPα plays an important role in inhibition of Th1 and Th17 differentiation.
Figure 5. Enhanced Th1 and Th17 differentiation in vitro in the absence of C/EBPα.
(A) Naive CD4+ T cells (CD25−CD44loCD62Lhi) isolated from WT (gray bar) and Cebpa KO (black bar) mice were stimulated by anti-CD3 and anti-CD28 mAbs under Th1, Th2 and Th17 polarizing culture. After 4 days, cultured cells were re-stimulated, then IFN-γ, IL-4 and IL-17 expression were detected by ICS, respectively. (B) IFN-γ, IL-4 and IL-17 in culture supernatant of re-stimulated Th1, Th2 and Th17 cells described in (A) were measured by ELISA. (C) Tbx21, Gata3 and Rorc mRNA were measured by quantitative RT-PCR in re-stimulated Th1, Th2 and Th17 cells cultured as described in (A), respectively. Data are the representative of three independent experiments.
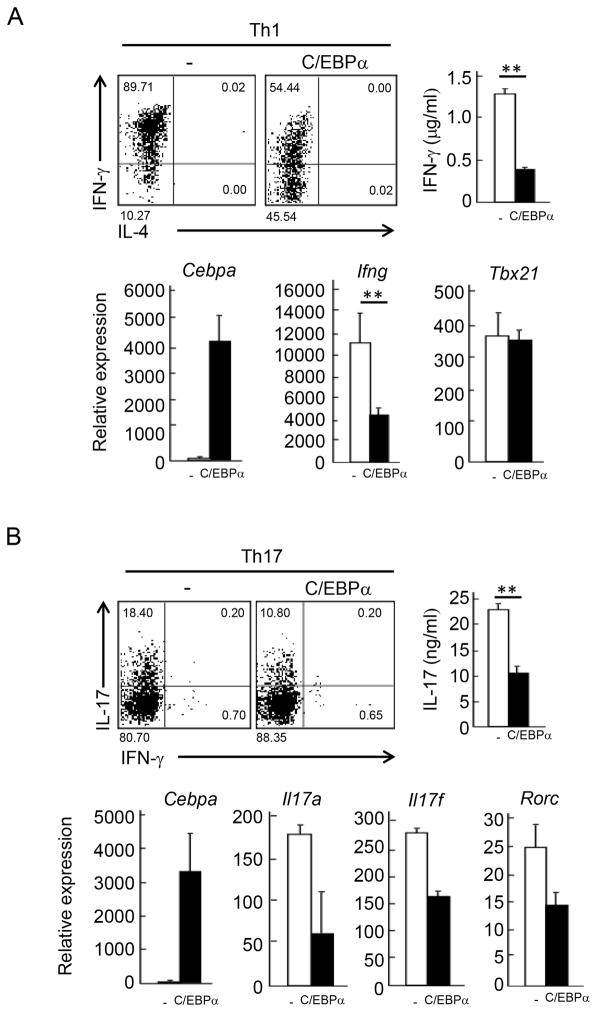
To better understand the function of C/EBPα, we overexpressed it retrovirally in naïve OT-II T cells stimulated with OVA peptide-loaded APCs under Th1 and Th17 skewing condition. After 4 days, cells were re-stimulated with anti-CD3 antibody or PMA /Ionomycin and cytokine production was measured by ELISA and ICS analysis. Compared with vector control, C/EBPα suppressed IFN-γ and IL-17 expression under Th1 and Th17 skewing conditions, respectively (Figure 6A and 6B). The C/EBPα-mediated IFN-γ suppression was observed before and after restimulation (Figure S3B). Then, quantitative RT-PCR analysis was performed to measure gene expression in the effector cells after re-stimulation with anti-CD3 Ab. In Th1 skewing condition, Ifng mRNA expression but not Tbx21 was reduced by 60 % with enforced expression of C/EBPα (Figure 6A lower). On the other hand, C/EBPα overexpression did not inhibit T cell proliferation (Figure S3C). Furthermore, the retroviral introduction of C/EBPα suppressed IFN-γ expression in WT and Cebpa KO CD4+ T cells cultured under Th1 condition to the same levels (Figure S3D). Overexpression of C/EBPα in Th17 condition also resulted in the suppression of Th17-related genes such as Il17a, Il17f and Rorc (Figure 6B lower). These results indicate that C/EBPα function to suppress both Th1 and Th17 cell differentiation.
Figure 6. Suppression of Th1 and Th17 differentiation by C/EBPα.
(A) Naive CD4+ T cells derived from OT-II TCR Tg mice were transduced with retroviruses carrying pGFP-RV(−) and pGFP-RV-Cebpa (C/EBPα) with primary stimulation by peptide-loaded APCs under Th1 skewing culture. After 4 day, transduced T cells were isolated by their expression of the indicator GFP. IFN-γ expression by re-stimulated Th1 cells was detected by ICS (upper left) and ELISA (upper right). mRNA expression of Cebpa, Ifng and Tbx21 was measured by quantitative PCR (lower). (B) Retroviral transduction of C/EBPα was performed under Th17 skewing condition. IL-17 expression by re-stimulated Th17 cells was detected by ICS (upper left) and ELISA (upper right). mRNA expression of Il17a, Il17f, and Rorc was measured by quantitative PCR (lower). Data are a representative of four independent experiments with similar results.
C/EBPα inhibits T-bet-mediated Ifng gene transcription
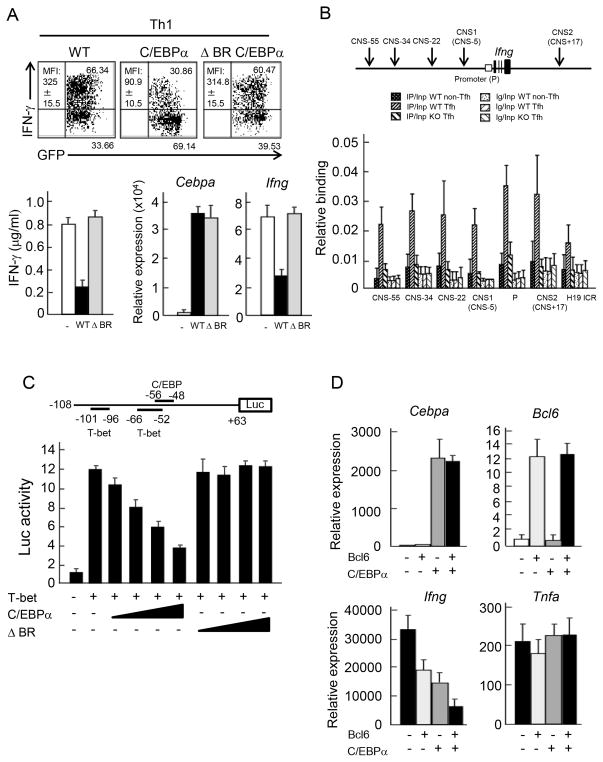
IFN-γ expression was significantly enhanced in Cebpa-deficient T cells, whereas T-bet expression was not affected and C/EBPα overexpression inhibits IFN-γ but not T-bet expression. Based on these results, we speculated that C/EBPα may antagonize T-bet function in activating Ifng transcription. To confirm the requirement of DNA binding by C/EBPα in suppression of IFN-γ, we deleted the DNA binding motif of C/EBPα and retrovirally introduced WT and mutant C/EBPα into CD4+ T cells cultured in Th1 skewing condition, the exogenous expression level of mutant C/EBPα was almost equal to that of WT. Overexpression of mutant C/EBPα failed to suppress IFN-γ expression at both protein and mRNA levels, while WT C/EBPα efficiently suppressed Th1 differentiation (Figure 7A). To verify C/EBPα-binding in Ifng locus in vivo, we performed chromatin-immunoprecipitation (ChIP) analysis with anti-C/EBPα Ab in Tfh cells isolated from immunized B6 mice by cell sorting. The possible regulatory regions of the Ifng gene have been previously identified as conserved non-coding sequence (CNS) (28) (29). We found that C/EBPα binding exist in each CNS such as Ifng promoter, CNS-55, CNS-34, CNS-22, CNS1 (CNS-5), and CNS2 (CNS+17) but not to the genomic region of H19 imprinting control region (H19 ICR), while the binding was not observed in WT non-Tfh cells and Cebpa KO Tfh cells (Figure 7B). This result confirms that C/EBPα directly bind to the Ifng gene locus in ex vivo Tfh cells. We next examined whether C/EBPα suppresses the transcription of the Ifng regulated by T-bet. We carried out a reporter assay using the Ifng promoter (30), which has been reported to contain two conserved putative binding sites for T-bet and one site for C/EBP, respectively (31) (Figure 7C upper). T-bet overexpression induced approximately 12-fold increased activation of Ifng promoter, compared to vector control. This activity was significantly suppressed by C/EBPα in a dose-dependent manner. Then, generated luciferase constructs lacking C/EBP binding site. However, we could not detect any Ifng promoter activity induced by T-bet. (Figure S4A). In line with a previous report (31), our data suggest that T-bet recognition site possibly overlaps with C/EBP site. On the other hand, the DNA binding mutant of C/EBPα failed to suppress Ifng promoter activity (Figure 7C lower). Of note, the failure of the suppression of Ifng promoter activity and IFN-γ expression by the delta BR mutant was not due to lack of nuclear localization (Figure S4B). Furthermore, overexpression of C/EBPα resulted in reduced accumulation of RNA polymerase on Ifng promoter without affecting histone modifications (Figure S4C). These data suggest that C/EBPα suppresses IFN-γ expression at the transcription level.
Figure 7. Inhibition of T-bet-mediated IFN-γ transcription by C/EBPα.
(A) Naive CD4+ T cells derived from OT-II TCR Tg mice were transduced with retroviruses carrying pGFP-RV(−), pGFP-RV- Cebpa (C/EBPα) and pGFP-RV-ΔBR Cebpa (ΔBR C/EBPα) with stimulation by peptide-loaded APCs under Th1 skewing culture. After 4 days, IFN-γ expression was analyzed in infected T cells by ICS (upper) and by ELISA (lower left). MFI of IFN-γ expression in infected cells was indicated. mRNA expression of Cebpa and Ifng was measured by quantitative PCR (lower right). Data is a representative of three independent experiments with similar results. (B) Upper panel indicates a schematic diagram of the Ifng locus. C/EBPα binding sites were accessed by ChIP analysis. PCR amplification was conducted with DNA prepared from chromatin precipitated with control Ig and anti-C/EBPα Abs in non-Tfh and Tfh cells isolated from immunized WT or Cebpa KO mice by FACS sorting. Primer set for H19 ICR locus was used as negative control. Data is a representative of two independent experiments with similar results. (C) A schematic diagram of the luciferase Ifng promoter reporter constructs (upper). +63 to −108 region from transcription start site in Ifng gene was used as a promoter. −56 to −48 region contains a C/EBP binding site. −66 to −52 and −101 to −96 regions shown contain T-box protein binding sites. Luciferase assay of 293T cell transduced with pGFP-RV(−), pGFP-RV-Cebpa (C/EBPα) or pGFP-RV-ΔBR Cebpa (ΔBR C/EBPα) together with pGFP-RV-T-bet was performed (lower). Data is a representative of three independent experiments with similar results. (D) Naive CD4+ T cells derived from OT-II TCR Tg mice were stimulated with peptide loaded APCs under Th1 skewing culture and further infected with combination of retroviruses carrying pGFP-RV(−), pGFP-RV-Bcl6 (Bcl6) pMIGRII-RV(−) and pMIGRII-RV-Cebpa (C/EBPα) as indicated. After 4 days, GFP+ hCD2+ cells were isolated and re-stimulated with anti-CD3 Ab. Then, mRNA expression of Cebpa, Bcl6, Ifng and Tnfa was measured by quantitative PCR. Data are a representative of two independent experiments.
Bcl6 has been reported to be a suppressor of IFN-γ expression (10). We finally examined whether Bcl6 and C/EBPα cooperatively suppress IFN-γ expression. Naïve OT-II CD4+ T cells were stimulated under Th1 skewing condition and infected with Bcl6 and /or C/EBPα. After 4 days of culture, doubly infected cells were sorted and re-stimulated. Single induction of either Bcl6 or C/EBPα significantly suppressed IFN-γ production. In the dual overexpression, C/EBPα and Bcl6 more potently suppressed IFN-γ expression, although TNF-α expression was not affected by the overexpression (Figure 7D). These data indicate that C/EBPα suppresses T-bet-mediated Ifng gene transcription, in collaboration with Bcl6.
Discussion
C/EBPα has been reported to regulate multiple biological events including differentiation of myeloid lineage, cell proliferation, metabolism and leukemogenesis. However, a role of C/EBPα in T cells remains unclear. Here, we reported that C/EBPα is highly expressed by Tfh cells. Although not required for Tfh cell differentiation, C/EBPα is a potent suppressor of Th1 and Th17 differentiation. Following KLH CFA immunization, we found an augmented IgG2a/b and IgG3 production in mice lacking C/EBPα in T cells. Consistent with the observation, antigen-specific IFN-γ and IL-17 production was enhanced in Cebpa KO T cells. Moreover, Tfh, Tef and non-Tfh cells expressed enhanced level of IFN-γ in the absence of C/EBPα. Furthermore, C/EBPα was able to bind to Ifng promoter and suppress its transcription mediated by T-bet. Therefore, our study altogether indicates that C/EBPα plays a pivotal role in regulation of humoral immunity by antagonizing IFN-γ expression in effector CD4+ T cells.
Since C/EBPα is highly expressed in Tfh cells, we examined whether deficiency of Cebpa in T cells resulted in abnormal GC reaction. However, both by KLH and OVA immunization, GC B cells and Tfh cells were normally generated in Cebpa KO mice. Furthermore, gene expressions related to Tfh cells such as Bcl6, Cxcr5, Il21, Il21r, Cd40l, Sap and Il4 in ex vivo Cebpa KO Tfh cells was not defective. This finding was supported by the observation that overexpression of C/EBPα could not induce expression of these genes (data not shown). Histological analysis also indicated normal migration of Cebpa-deficient T cells into GCs. One of the crucial roles of Tfh cells is facilitating affinity maturation of antibodies in B cells. We observed normal affinity maturation in the KO mice upon NP-KLH immunization. These data suggeste Cebpa-deficient T cells developed normally into Tfh cells and provided similar help to B cells as WT T cells do. This idea further supported by the observation that frequency of Bcl6+ GC B cells and its expression level in T cell specific Cebpa KO mice was comparable to those in WT mice after immunization.
On the other hand, IgG2a/b and IgG3 production were enhanced in inside and outside of follicles of immunized Cebpa KO mice. Consistent with this, increased IFN-γ expression was observed in PD-1hi Tfh, extrafollicular T (Tef) and non-Tfh cells isolated from Cebpa KO mice, although we did not observe the developmental defect of Tef cells in the KO mice after immunization (data not shown), indicating that C/EBPα may contribute to proper antibody production by suppressing IFN-γ in T cells both out and inside of follicles. These results also suggest that the suppressive effect of C/EBPα on IFN-γ expression in various CD4+ T cell subpopulations, despite higher expression of C/EBPα in Tfh cells.
C/EBPα was found to be a strong suppressor of Th1 and Th17 differentiation. In Th17 skewing condition, ectopic expression of C/EBPα suppressed the expression of Rorc as well as Il17 expression. Consistent with the observation, IL-17 and ROR-γ expression was enhanced in Cebpa KO T cells cultured under Th17 skewing condition. By contrast, in Th1 culture, IFN-γ expression but not T-bet was inhibited by enforced expression of C/EBPα. In addition, Cebpa KO T cells exhibited an enhanced activity to express IFN-γ but not T-bet. These findings suggest that C/EBPα is able to suppress Th17 differentiation by suppressing the expression of a key transcription factor. Meanwhile, whereas T-bet expression is not affected by C/EBPα, T-bet-mediated IFN-γ transcription is inhibited by C/EBPα. This idea is supported by our observations that C/EBPα bound to Ifng promoter as well as other regulatory CNS regions in Tfh cells generated in vivo and that ΔBR C/EBPα, which cannot bind to its target sequence, failed to repress IFN-γ expression as well as Ifng promoter activity induced by T-bet. Furthermore, C/EBPα suppressed DNA pol II recruitment on Ifng promoter, without affecting histone modifications. Taken together, C/EBPα suppresses IFN-γ expression at the transcriptional level. Cebpb expression was not affected in the absence of Cebpa, and Cebpd expression was modestly increased in Cebpa KO naïve, Th1, and Th2 cells but not in Th17, iTreg and Tfh-like cells (data not shown). Therefore, C/EBPβ or δ may not compensate for Cebpa deficiency. In our previous report, Bcl6 was demonstrated to be a crucial suppressor of IFN-γ production as well (10). Indeed, in our double overexpression experiment, C/EBPα and Bcl6 had additive effects in suppressing IFN-γ expression in Th1 cells. Thus, these observations indicate that C/EBPα plays a crucial role in antagonizing IFN-γ at the transcription level in a coordinated way with Bcl6 during Tfh differentiation. In addition, although Th1 cells and Th17 cells in vitro and non-Tfh cells in vivo express lower levels of C/EBPα than Tfh cells, it could still suppress IFN-γ and IL-17.
Our data show that Tfh cells express higher levels of C/EBPα than other helper T cell subsets both in vitro and in vivo, thus C/EBPα is thought to be a novel useful marker to identify Tfh cells among CD4+ T cell subsets. Interestingly, human Tfh cells also highly express C/EBPα (32). Further analysis of regulatory mechanisms governing C/EBPα during Tfh cell commitment may reveal novel regulation of this differentiation process.
Tfh cells are a newly identified T cell subset, whose regulation is not well understood. Bcl6 has been previously shown to be a key transcription factor not only governing Tfh cell development, but also antagonizing the alternate T cell differentiation processes. Here, our results have demonstrated C/EBPα, as the second transcription factor selectively expressed in Tfh cells, in regulation of a proper antibody response by suppressing IFN-γ expression. Our observations may aid a better understanding of humoral and pathogenesis of autoimmune disorders.
Supplementary Material
Acknowledgments
We thank BD for providing anti-Bcl6 antibody, Dr. Laurie Glimcher for T-bet retroviral vector, Dr. Chris Wilson for the Ifng promoter reporter and CD4-cre mice, the flow cytometry core facility at the M.D. Anderson Cancer Center and the entire Dong lab for their help and discussions.
This work is supported by research grants from the National Institutes of Health (C.D. and R.I.N.) and Ministry of Education, Culture, Sports, Science, and Technology of Japan (S.T. and T.K.). G.J.M. was a Schissler Foundation M.D. Anderson Cancer Center Fellow in cancer research. R.I.N. was a recipient of a Scientist Development Grant from the American Heart Association (AHA).
Footnotes
Disclosures
There is no patent interest for this work.
References
- 1.MacLennan IC. Germinal centers. Annu Rev Immunol. 1994;12:117–139. doi: 10.1146/annurev.iy.12.040194.001001. [DOI] [PubMed] [Google Scholar]
- 2.Stavnezer J, Guikema JE, Schrader CE. Mechanism and regulation of class switch recombination. Annu Rev Immunol. 2008;26:261–292. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breitfeld D, Ohl L, Kremmer E, Ellwart J, Sallusto F, Lipp M, Forster R. Follicular B helper T cells express CXC chemokine receptor 5, localize to B cell follicles, and support immunoglobulin production. J Exp Med. 2000;192:1545–1552. doi: 10.1084/jem.192.11.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bryant VL, Ma CS, Avery DT, Li Y, Good KL, Corcoran LM, de Waal Malefyt R, Tangye SG. Cytokine-mediated regulation of human B cell differentiation into Ig-secreting cells: predominant role of IL-21 produced by CXCR5+ T follicular helper cells. J Immunol. 2007;179:8180–8190. doi: 10.4049/jimmunol.179.12.8180. [DOI] [PubMed] [Google Scholar]
- 5.Kim CH, Rott LS, Clark-Lewis I, Campbell DJ, Wu L, Butcher EC. Subspecialization of CXCR5+ T cells: B helper activity is focused in a germinal center-localized subset of CXCR5+ T cells. J Exp Med. 2001;193:1373–1381. doi: 10.1084/jem.193.12.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schaerli P, Willimann K, Lang AB, Lipp M, Loetscher P, Moser B. CXC chemokine receptor 5 expression defines follicular homing T cells with B cell helper function. J Exp Med. 2000;192:1553–1562. doi: 10.1084/jem.192.11.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu X, Yan X, Zhong B, Nurieva RI, Wang A, Wang X, Martin-Orozco N, Wang Y, Chang SH, Esplugues E, Flavell RA, Tian Q, Dong C. Bcl6 expression specifies the T follicular helper cell program in vivo. J Exp Med. 2012;209:1841–1852. S1841–1824. doi: 10.1084/jem.20120219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dong C. TH17 cells in development: an updated view of their molecular identity and genetic programming. Nat Rev Immunol. 2008;8:337–348. doi: 10.1038/nri2295. [DOI] [PubMed] [Google Scholar]
- 9.Johnston RJ, Poholek AC, DiToro D, Yusuf I, Eto D, Barnett B, Dent AL, Craft J, Crotty S. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 2009;325:1006–1010. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nurieva RI, Chung Y, Martinez GJ, Yang XO, Tanaka S, Matskevitch TD, Wang YH, Dong C. Bcl6 mediates the development of T follicular helper cells. Science. 2009;325:1001–1005. doi: 10.1126/science.1176676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu D, Rao S, Tsai LM, Lee SK, He Y, Sutcliffe EL, Srivastava M, Linterman M, Zheng L, Simpson N, Ellyard JI, Parish IA, Ma CS, Li QJ, Parish CR, Mackay CR, Vinuesa CG. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 2009;31:457–468. doi: 10.1016/j.immuni.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 12.Liu X, Chen X, Zhong B, Wang A, Wang X, Chu F, Nurieva RI, Yan X, Chen P, van der Flier LG, Nakatsukasa H, Neelapu SS, Chen W, Clevers H, Tian Q, Qi H, Wei L, Dong C. Transcription factor achaete-scute homologue 2 initiates follicular T-helper-cell development. Nature. 2014;507:513–518. doi: 10.1038/nature12910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toellner KM, Luther SA, Sze DM, Choy RK, Taylor DR, MacLennan IC, Acha-Orbea H. T helper 1 (Th1) and Th2 characteristics start to develop during T cell priming and are associated with an immediate ability to induce immunoglobulin class switching. J Exp Med. 1998;187:1193–1204. doi: 10.1084/jem.187.8.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee SK, Rigby RJ, Zotos D, Tsai LM, Kawamoto S, Marshall JL, Ramiscal RR, Chan TD, Gatto D, Brink R, Yu D, Fagarasan S, Tarlinton DM, Cunningham AF, Vinuesa CG. B cell priming for extrafollicular antibody responses requires Bcl-6 expression by T cells. J Exp Med. 2011;208:1377–1388. doi: 10.1084/jem.20102065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luther SA, Maillard I, Luthi F, Scarpellino L, Diggelmann H, Acha-Orbea H. Early neutralizing antibody response against mouse mammary tumor virus: critical role of viral infection and superantigen-reactive T cells. J Immunol. 1997;159:2807–2814. [PubMed] [Google Scholar]
- 16.Odegard JM, Marks BR, DiPlacido LD, Poholek AC, Kono DH, Dong C, Flavell RA, Craft J. ICOS-dependent extrafollicular helper T cells elicit IgG production via IL-21 in systemic autoimmunity. J Exp Med. 2008;205:2873–2886. doi: 10.1084/jem.20080840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nurieva RI, Chung Y, Hwang D, Yang XO, Kang HS, Ma L, Wang YH, Watowich SS, Jetten AM, Tian Q, Dong C. Generation of T follicular helper cells is mediated by interleukin-21 but independent of T helper 1, 2, or 17 cell lineages. Immunity. 2008;29:138–149. doi: 10.1016/j.immuni.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee SK, Silva DG, Martin JL, Pratama A, Hu X, Chang PP, Walters G, Vinuesa CG. Interferon-gamma excess leads to pathogenic accumulation of follicular helper T cells and germinal centers. Immunity. 2012;37:880–892. doi: 10.1016/j.immuni.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 19.Peng SL, Madaio MP, Hughes DP, Crispe IN, Owen MJ, Wen L, Hayday AC, Craft J. Murine lupus in the absence of alpha beta T cells. J Immunol. 1996;156:4041–4049. [PubMed] [Google Scholar]
- 20.Jacobson BA, Rothstein TL, Marshak-Rothstein A. Unique site of IgG2a and rheumatoid factor production in MRL/lpr mice. Immunol Rev. 1997;156:103–110. doi: 10.1111/j.1600-065x.1997.tb00962.x. [DOI] [PubMed] [Google Scholar]
- 21.Khanna-Gupta A. Sumoylation and the function of CCAAT enhancer binding protein alpha (C/EBP alpha) Blood Cells Mol Dis. 2008;41:77–81. doi: 10.1016/j.bcmd.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fuchs O. Growth-inhibiting activity of transcription factor C/EBPalpha, its role in haematopoiesis and its tumour suppressor or oncogenic properties in leukaemias. Folia Biol (Praha) 2007;53:97–108. [PubMed] [Google Scholar]
- 23.Ye BH, Cattoretti G, Shen Q, Zhang J, Hawe N, de Waard R, Leung C, Nouri-Shirazi M, Orazi A, Chaganti RS, Rothman P, Stall AM, Pandolfi PP, Dalla-Favera R. The BCL-6 proto-oncogene controls germinal-centre formation and Th2-type inflammation. Nat Genet. 1997;16:161–170. doi: 10.1038/ng0697-161. [DOI] [PubMed] [Google Scholar]
- 24.Snapper CM, Paul WE. Interferon-gamma and B cell stimulatory factor-1 reciprocally regulate Ig isotype production. Science. 1987;236:944–947. doi: 10.1126/science.3107127. [DOI] [PubMed] [Google Scholar]
- 25.Abbas AK, Urioste S, Collins TL, Boom WH. Heterogeneity of helper/inducer T lymphocytes. IV. Stimulation of resting and activated B cells by Th1 and Th2 clones. J Immunol. 1990;144:2031–2037. [PubMed] [Google Scholar]
- 26.Hasbold J, Hong JS, Kehry MR, Hodgkin PD. Integrating signals from IFN-gamma and IL-4 by B cells: positive and negative effects on CD40 ligand-induced proliferation, survival, and division-linked isotype switching to IgG1, IgE, and IgG2a. J Immunol. 1999;163:4175–4181. [PubMed] [Google Scholar]
- 27.Huang S, Hendriks W, Althage A, Hemmi S, Bluethmann H, Kamijo R, Vilcek J, Zinkernagel RM, Aguet M. Immune response in mice that lack the interferon-gamma receptor. Science. 1993;259:1742–1745. doi: 10.1126/science.8456301. [DOI] [PubMed] [Google Scholar]
- 28.Hatton RD, Harrington LE, Luther RJ, Wakefield T, Janowski KM, Oliver JR, Lallone RL, Murphy KM, Weaver CT. A distal conserved sequence element controls Ifng gene expression by T cells and NK cells. Immunity. 2006;25:717–729. doi: 10.1016/j.immuni.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 29.Shnyreva M, Weaver WM, Blanchette M, Taylor SL, Tompa M, Fitzpatrick DR, Wilson CB. Evolutionarily conserved sequence elements that positively regulate IFN-gamma expression in T cells. Proc Natl Acad Sci U S A. 2004;101:12622–12627. doi: 10.1073/pnas.0400849101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Penix L, Weaver WM, Pang Y, Young HA, Wilson CB. Two essential regulatory elements in the human interferon gamma promoter confer activation specific expression in T cells. J Exp Med. 1993;178:1483–1496. doi: 10.1084/jem.178.5.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cho JY, Grigura V, Murphy TL, Murphy K. Identification of cooperative monomeric Brachyury sites conferring T-bet responsiveness to the proximal IFN-gamma promoter. Int Immunol. 2003;15:1149–1160. doi: 10.1093/intimm/dxg113. [DOI] [PubMed] [Google Scholar]
- 32.Rasheed AU, Rahn HP, Sallusto F, Lipp M, Muller G. Follicular B helper T cell activity is confined to CXCR5(hi)ICOS(hi) CD4 T cells and is independent of CD57 expression. Eur J Immunol. 2006;36:1892–1903. doi: 10.1002/eji.200636136. [DOI] [PubMed] [Google Scholar]
- 33.Zhang P, Iwasaki-Arai J, Iwasaki H, Fenyus ML, Dayaram T, Owens BM, Shigematsu H, Levantini E, Huettner CS, Lekstrom-Himes JA, Akashi K, Tenen DG. Enhancement of hematopoietic stem cell repopulating capacity and self-renewal in the absence of the transcription factor C/EBP alpha. Immunity. 2004;21:853–863. doi: 10.1016/j.immuni.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 34.Makar KW, Perez-Melgosa M, Shnyreva M, Weaver WM, Fitzpatrick DR, Wilson CB. Active recruitment of DNA methyltransferases regulates interleukin 4 in thymocytes and T cells. Nat Immunol. 2003;4:1183–1190. doi: 10.1038/ni1004. [DOI] [PubMed] [Google Scholar]
- 35.Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100:655–669. doi: 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- 36.Akimzhanov AM, Yang XO, Dong C. Chromatin remodeling of interleukin-17 (IL-17)-IL-17F cytokine gene locus during inflammatory helper T cell differentiation. J Biol Chem. 2007;282:5969–5972. doi: 10.1074/jbc.C600322200. [DOI] [PubMed] [Google Scholar]
- 37.Yang XO, Pappu BP, Nurieva R, Akimzhanov A, Kang HS, Chung Y, Ma L, Shah B, Panopoulos AD, Schluns KS, Watowich SS, Tian Q, Jetten AM, Dong C. T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR alpha and ROR gamma. Immunity. 2008;28:29–39. doi: 10.1016/j.immuni.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.