Abstract
AIM: To evaluate interferon-λ3 (IFNL3) polymorphisms in response-guided pegylated interferon-α plus ribavirin (Peg-IFNα/RBV) therapy for genotype 2 (G2) chronic hepatitis C.
METHODS: Between January 2006 and June 2012, a total of 180 patients with chronic infections of G2 hepatitis C virus (HCV) were treated with response-guided Peg-IFNα/RBV therapy. The treatment duration was 24 wk for patients who achieved rapid virologic response (RVR), and 36 or 48 wk for patients who did not. Then, the impact of the IFNL3 single nucleotide polymorphism genotype (TT/non-TT at rs8099917) on treatment outcomes was evaluated in the 180 patients, and between patients infected with either HCV sub-genotype 2a or 2b.
RESULTS: Of the 180 patients evaluated, 111 achieved RVR, while the remaining 69 patients did not. In RVR patients, the sustained virologic response (SVR) rate was 96.4%, and the IFNL3 genotype did not influence the SVR rate (96.6% vs 95.8% in IFNL3 genotype TT vs non-TT). However, in non-RVR patients, the SVR rate decreased to 72.5% (P < 0.0001), and this rate was significantly different between the IFNL3 genotype TT and non-TT groups (80.0% vs 42.9%, P = 0.0146). Multivariate regression analysis in non-RVR patients identified the IFNL3 genotype TT as the only baseline-significant factor associated with SVR (OR = 5.39, 95%CI: 1.29-22.62; P = 0.0189). In analysis according to HCV sub-genotype, no significant difference in the SVR rate was found between HCV sub-genotypes 2a and 2b.
CONCLUSION: In response-guided Peg-IFNα/RBV combination therapy for chronically HCV G2-infected patients, the impact of the IFNL3 genotype on SVR was limited to non-RVR patients.
Keywords: Hepatitis C virus genotype 2, Interferon-λ3 single nucleotide polymorphism, Pegylated-interferon plus ribavirin response-guided therapy, Rapid virologic response, Sustained virologic response
Core tip: Interferon-λ3 (IFNL3) single nucleotide polymorphisms (SNPs), such as rs8099917 and rs12979860, affect the virologic responses of chronically hepatitis C virus genotype 1-infected patients to response-guided pegylated interferon-α plus ribavirin therapy. However, the significance of these SNPs in therapy for hepatitis C virus genotype 2 (G2)-infected patients is unclear. We show that rs8099917 significantly influences sustained virologic response (SVR) achievement only in patients who do not attain rapid virologic response. Therefore, IFNL3 SNP genotyping is valuable for predicting SVR only in non-rapid virologic response patients, irrespective of the G2 subtype, even when therapy is extended up to 48 wk.
INTRODUCTION
Hepatitis C virus (HCV) genotype 2 (G2) is the second-most frequent HCV genotype and accounts for approximately 30% of chronic HCV infections in Japan[1,2]. However, the prevalence of HCV G2 is decreasing due to successful treatment with standard 24-wk regimens of pegylated interferon-α plus ribavirin (Peg-IFNα/RBV) combination therapy; with > 80% of patients achieving sustained virologic response (SVR)[3,4]. However, a fraction of patients who do not achieve a rapid virologic response (RVR) may remain uncured, even when therapy is extended for 36 or 48 wk[5,6].
The impact of single nucleotide polymorphisms (SNPs) near the interferon-λ3 (IFNL3)/interleukin-28B gene on Peg-IFNα/RBV combination therapy for HCV genotype 1[7-9] has been firmly established. However, it remains controversial whether the IFNL3 genotype is useful in predicting virologic responses to peg-IFNα/RBV therapy in HCV G2 patients[10-13].
Previously, we demonstrated the value of response-guided therapy for HCV G2 patients who were treated for 24 wk with Peg-IFNα/RBV combination therapy if they achieved RVR, and for 36 or 48 wk if they did not achieve RVR[6]. In the present study, we assessed the impact of IFNL3 SNP (rs8099917) genotypes on virologic responses and outcomes of HCV G2 (subtype of G2a or G2b) patients who received response-guided Peg-IFNα/RBV combination therapy.
MATERIALS AND METHODS
Patients
Between January 2006 and June 2012, 180 chronically HCV G2-infected patients were treated with response-guided Peg-IFNα/RBV combination therapy at the Jikei University Katsushika Medical Center, the Jikei University Kashiwa Hospital, and the Shinmatsudo Central General Hospital. The treatment duration was 24 wk for patients who achieved RVR (RVR group) and 36 or 48 wk for patients who did not (non-RVR group). Patients received weekly subcutaneous injections of Peg-IFNα-2b (PegIntron; MSD K.K.; Tokyo, Japan) at a dose of 1.5 μg/kg, plus RBV (Rebetol; MSD K.K.) at a dose of 600-1000 mg/d according to body weight (< 60 kg: 600 mg/d; 60-80 kg: 800 mg/d; and > 80 kg: 1000 mg/d). Doses of Peg-IFNα-2b and/or RBV were appropriately adjusted if side effects were observed.
All the patients studied satisfied the following inclusion criteria: (1) serum HCV RNA levels ≥ 10000 copies/mL (Amplicor HCV Monitor Test, version 2.0; Roche Diagnostics, Basel, Switzerland; quantification limit: 50 IU/mL) or ≥ 5 log10 IU/mL (COBAS AmpliPrep/COBAS TaqMan HCV Test; Roche Diagnostics; quantification limit: 1.2 log10 IU/mL); (2) white blood cell counts ≥ 2000/mm3; (3) neutrophil counts ≥ 1500/mm3; (4) hemoglobin levels ≥ 11 g/dL; (5) platelet counts ≥ 60000/mm3; and (6) serotype 2 or genotype 2a or 2b (G2a or G2b) determined by serologic and conventional PCR-based methods, as reported previously[14,15]. Patients were excluded from this study if they were positive for hepatitis B surface antigen or anti-human immunodeficiency virus antibody, consumed > 20 g of alcohol/d, had psychiatric disorders or hepatocellular carcinoma, or were diagnosed with other liver diseases. Patients with established liver cirrhosis that was easily diagnosed by image inspection or for whom laboratory tests did not indicate the need for liver biopsy (e.g., low platelet count or prolonged prothrombin time) were not included in the present study. One hundred and sixty-five patients (91.7%) were treatment-naïve and the remaining 15 had previously been treated with 24-wk Peg-IFNα/RBV combination therapy.
This study complied with the standards of the Declaration of Helsinki (revised edition 2008) and current ethical guidelines, and was approved by the human ethics review committees of each institution. Written informed consent was obtained from all patients.
Histology, HCV sub-genotyping, and detection of HCV RNA
Liver biopsies and HCV G2 sub-genotyping were performed in 152/180 and 159/180 patients, respectively. Histologic grades of liver fibrosis were classified as F1-F4, according to the METAVIR scoring system[16]. HCV G2 sub-genotyping was performed by the conventional PCR-based method[14]. HCV serotypes were determined by enzyme-linked immune assay[15]. The presence or absence of serum HCV RNA was evaluated after 4 wk of therapy, at the end of therapy, and at 24 wk after the completion of therapy. Serum HCV RNA levels were evaluated with the qualitative Amplicor HCV Monitor Test between January 2006 and November 2007, and the COBAS AmpliPrep/COBAS TaqMan HCV test was used thereafter. To evaluate potential discrepancies due to the use of different tests, 21 samples that were originally analyzed using the Amplicor HCV Monitor Test were re-tested with the COBAS AmpliPrep/COBAS TaqMan HCV test, using serum stocks stored at -30 °C. Patients in whom serum HCV RNA levels were undetectable with the COBAS AmpliPrep/COBAS TaqMan HCV test at 4 wk after initiating therapy were designated as RVR patients, while the remaining patients were designated as non-RVR patients. The end point in this study was SVR (undetectable serum HCV RNA at 24 wk post-treatment).
Analysis of SNPs near IFNL3
Genomic DNA was extracted and isolated from whole blood using a MagNA Pure LC Instrument and the DNA Isolation Kit (Roche Diagnostics). Alleles of the rs8099917 SNP near IFNL3 were determined using TaqMan SNP genotyping assays (Applied Biosystems of Thermo Fisher Scientific, Waltham, MA, United States), as described previously[9]. The rs8099917 genotypes were classified into TT (major homozygous genotype) and non-TT genotypes (heterozygous genotype TG or minor homozygous genotype GG). The rs8099917 genotype of all patients was determined at the Research Center for Medical Science at the Jikei University School of Medicine.
Statistical analysis
The Mann-Whitney U-test was used to analyze differences in continuous variables. Fisher’s exact tests were used to analyze differences in categorical data. All tests of significance were two-tailed. P < 0.05 and < 0.1 were considered statistically significant and marginal, respectively. To determine which factors were associated with SVR, variables that were significant or marginal in univariate analyses were analyzed by multiple logistic regression analysis. All statistical analyses were performed using Statistica for Windows version 6 (StatSoft; Tulsa, OK, United States).
RESULTS
Treatment responses
Of the 180 patients evaluated, 111 (61.7%) achieved RVR and received a 24-wk treatment course (RVR group). The remaining 69/180 (38.3%) patients failed to achieve RVR and the treatment duration was extended to 36 or 48 wk (non-RVR group; Figure 1). HCV G2a was more frequently detected in the RVR group than in the non-RVR group (P = 0.0005; Table 1). With respect to the HCV sub-genotype, 69/98 (70.4%) G2a patients had RVR, whereas only 23/57 (40.4%) G2b patients had RVR. The baseline level of HCV RNA was significantly lower in the RVR group than in the non-RVR group (P < 0.0001). Serum albumin levels were significantly higher in the RVR group than in the non-RVR group (P = 0.0029). Multivariate analysis identified the baseline levels of HCV RNA and serum albumin as significant factors associated with RVR (OR = 4.40, 95%CI: 2.25-8.63, P < 0.0001; and OR = 0.13, 95%CI: 0.04-0.42, P = 0.0006; respectively). However, no difference was observed in the distribution of IFNL3 SNP genotypes between the RVR and non-RVR groups (Figure 1 and Table 1). The percentages of TT genotype patients were 78.4% (87/111) vs 79.7% (55/69) in the RVR and non-RVR groups, respectively.
Figure 1.
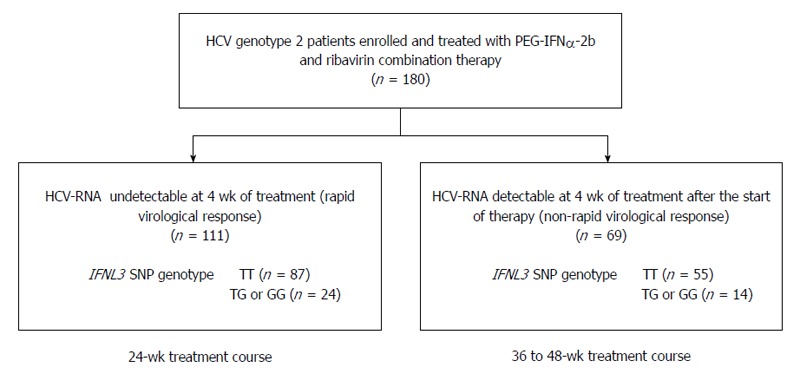
Study flow chart. PEG-IFNα: Pegylated interferon-α; HCV: Hepatitis C virus; SNP: Single nucleotide polymorphism.
Table 1.
Analysis of factors affecting rapid virologic response to pegylated interferon-α plus ribavirin combination therapy in patients infected with hepatitis C virus genotype 2
| RVR | non-RVR |
RVR vs non-RVR (1: non-RVR/2: RVR) |
||||
| Univariate analysis |
Multivariate analysis |
|||||
| P value | OR | 95%CI | P value | |||
| Baseline factors | ||||||
| Demographic data | ||||||
| Number of patients | 111 | 69 | ||||
| Gender (1:male/2:female) | 54/57 | 31/38 | 0.8705 | |||
| Age (yr) | 60 (18-76) | 60 (18-80) | 0.9623 | |||
| Body weight (kg) | 59.4 (35.6-90.9) | 56.0 (37.0-104) | 0.5154 | |||
| Body mass index (kg/m2) | 23.4 (17.1-31.2) | 22.8 (15.2-34.8) | 0.9645 | |||
| Histological fibrosis of liver (F0/1/2/3/4/ND )1 | 5/45/18/13/13/17 | 2/24/9/6/17/11 | ||||
| (F0-3/F4) | 81/13 | 41/17 | 0.0202 | |||
| Prior interferon and ribavirin treatment response | ||||||
| Naïve/Relapse/Non response | 101/10/0 | 64/3/2 | 0.8577 | |||
| Laboratory data | ||||||
| Genetic variation at rs8099917 (TT/TG or GG) | 87/24 | 55/14 | 0.9800 | |||
| HCV-RNA (log10IU/mL) | 5.5 (5.0-7.0) | 6.4 (5.0-7.3) | < 0.0001 | 4.40 | 2.24-8.63 (per 1.0 log10IU/mL) | < 0.0001 |
| HCV Genotype (2a/2b/2a + 2b/ND) | 69/23/2/17 | 29/34/2/4 | 0.0005 | |||
| White blood cells (/μL) | 4800 (2400-10300) | 4700 (2000-10200) | 0.5170 | |||
| Hemoglobin (g/dL) | 14.1(9.5-18.2) | 13.8 (9.3-16.9) | 0.1899 | |||
| Platelets (× 104/μL) | 17.1 (4.8-34.1) | 16.6 (5.5-34.1) | 0.7993 | |||
| Aspartate aminotransferase (IU/L) | 48 (16-317) | 48 (13-455) | 0.6753 | |||
| Alanine aminotransferase (IU/L) | 57 (10-361) | 56 (9-356) | 0.9699 | |||
| Gamma-glutamyl-transpeptitase (IU/L) | 43 (7-719) | 48 (9-331) | 0.7526 | |||
| Albumin (g/dL) | 4.1 (3.2-5.0) | 4.0 (2.9-4.8) | 0.0029 | 0.13 | 0.04-0.42 (per 1.0 g/dL) | 0.0006 |
| Fasting total cholesterol (mg/dL) | 170 (118-265) | 173 (106-270) | 0.8155 | |||
| Fasting low density lipoprotein-cholesterol (mg/dL) | 99 (45-158) | 103 (40-181) | 0.4186 | |||
| Fasting plasma glucose (mg/dL) | 99 (76-221) | 100 (76-320) | 0.2002 | |||
| Homeostasis model assessment-Insulin Resistance | 1.59 (0.56-10.83) | 1.38 (0.79-6.88) | 0.4722 | |||
| Alpha-fetoprotein (ng/mL) | 4.7 (1.6-193.6) | 5.0 (1.0-130) | 0.4101 | |||
Classified by METAVIR score. Data are expressed as number of patients or median (range); ND: Not done; HCV: Hepatitis C virus; RVR: Rapid virologic response.
SVR in RVR patients
Of the 111 RVR patients, 107 (96.4%) achieved SVR with the 24-wk treatment. Regarding the IFNL3 SNP genotype, 84/87 (96.6%) TT patients and 23/24 (95.8%) non-TT patients achieved SVR (Figure 2A). As for HCV G2 subtype, 66/69 (95.7%) patients with G2a and 23/23 (100%) patients with G2b achieved SVR. There were also no significant differences in other variables between patients with SVR and non-SVR. Although only four patients failed to achieve SVR, no characteristics distinguishing them from SVR patients were identified. All four of the non-SVR patients (1 male and 3 female; age: 34-65 years) were treatment-naïve and completed treatment as scheduled. They had a mild degree of liver fibrosis and baseline HCV RNA levels of 5.0 log IU/mL to 6.5 log IU/mL.
Figure 2.
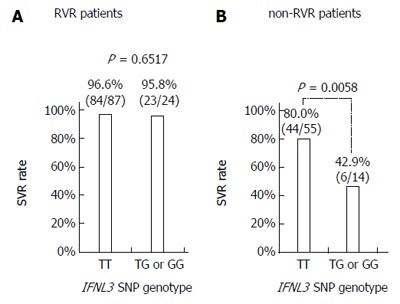
Sustained virologic response rates according to interferon-λ3 single nucleotide polymorphism genotype. A: Rapid virologic response (RVR); and B: Non-RVR patient groups. SVR: Sustained virologic response; SNP: Single nucleotide polymorphism; IFNL3: Interferon-λ3.
SVR in non-RVR patients
Of the 69 non-RVR patients, 50 (72.5%) achieved SVR with the extended treatment to 36 or 48 wk. The SVR rate in the non-RVR group was significantly lower than in the RVR group (72.5% vs 96.4%, P < 0.0001). Thirty-eight patients (55.1%) received a 48-wk treatment course and 31/69 (44.9%) received a 36-wk treatment course. SVR rates were higher in the 36-week treatment patient group than in the 48-wk group (80.6% vs 65.8%), but the difference was not significant. Regarding the IFNL3 SNP genotype, 44/55 (80.0%) patients with the TT genotype and 6/14 (42.9%) patients with the non-TT genotype achieved SVR (P = 0.0058; Figure 2B). Among patients with the IFNL3 TT genotype, the SVR rates were significantly different between RVR and non-RVR patient groups (96.6% vs 80.0%; P = 0.0033; Figure 2A and B). Similarly, among patients with non-TT genotypes, the SVR rates were significantly different between RVR and non-RVR patient groups (95.8% vs 42.9%, P = 0.0009). In HCV G2 sub-genotype patients, 21/29 (72.4%) patients with G2a and 24/34 (70.6%) patients with G2b achieved SVR.
Factors contributing to SVR in non-RVR patients
In the non-RVR patient group, the IFNL3 TT genotype was the only baseline factor that significantly related to SVR in univariate analysis (P = 0.0146). Among the other baseline factors, aspartate aminotransferase was marginal (P = 0.0751). The histologic stage of fibrosis and HCV G2 sub-genotypes were not significant factors for SVR. In multiple logistic regression analysis, only the IFNL3 TT genotype was identified as an independent factor that was significantly associated with SVR (OR = 5.87, 95%CI: 1.62-21.22; P = 0.0058; Table 2).
Table 2.
Analysis of factors affecting sustained virologic response in non-rapid virologic response hepatitis C virus genotype 2 patients treated with 36 or 48-wk pegylated interferon-α plus ribavirin combination therapy
| SVR | non-SVR |
SVR vs non-SVR (1: non-SVR/2: SVR) |
||||
| Univaritate analysis |
Multivaritate analysis |
|||||
| P value | OR | 95%CI | P value | |||
| Baseline factors | ||||||
| Demographic data | ||||||
| Number of patients | 50 | 19 | ||||
| Gender (1:male/2:female) | 20/30 | 11/8 | 0.2873 | |||
| Age (yr) | 60.5 (18-80) | 53 (39-73) | 0.2341 | |||
| Body weight (kg) | 55.0 (37.0-101.0) | 64.0 (44.0-104.0) | 0.1563 | |||
| Body mass index (kg/m2) | 22.8 (15.2-32.8) | 22.8 (18.8-34.8) | 0.2426 | |||
| Histological fibrosis of liver (F0/1/2/3/4/ND)1 | 1/18/6/6/10/9 | 1/6/3/0/7/2 | ||||
| (F0-3/F4) | 31/10 | 10/7 | 0.2050 | |||
| Prior interferon and ribavirin treatment response | ||||||
| Naïve/Relapse/Non response | 46/3/1 | 18/0/1 | 0.8933 | |||
| Laboratory data | ||||||
| Genetic variation at rs8099917 (TG or GG/TT) | 6/44 | 8/11 | 0.0146 | 5.87 (1; TG or GG/ 2; TT) | 1.62-21.22 | 0.0059 |
| HCV-RNA (Log10IU/mL) | 6.3 (5.0-7.3) | 6.4 (5.0-7.2) | 0.6064 | |||
| HCV Genotype (2a/2b/2a + 2b/ND) | 21/24/2/3 | 8/10/0/1 | 0.8156 | |||
| White blood cells (/μL) | 4700 (2800-7800) | 5550 (2000-10200) | 0.3001 | |||
| Hemoglobin (g/dL) | 13.8 (11.0-16.9) | 13.75 (11.0-16.8) | 0.9279 | |||
| Platelets (× 104/μL) | 16.6 (6.0-34.1) | 16.55 (6.1-29.7) | 0.9834 | |||
| Aspartate aminotransferase (IU/L) | 54 (15-455) | 32 (13-264) | 0.0751 | |||
| Alanine aminotransferase (IU/L) | 59 (12-356) | 36.5 (9-231) | 0.1866 | |||
| Gamma-glutamyl-transpeptitase (IU/L) | 49 (9-292) | 47 (11-331) | 0.9237 | |||
| Albumin (g/dL) | 4.0 (2.9-4.8) | 3.9 (3.1-4.7) | 0.5738 | |||
| Fasting total cholesterol (mg/dL) | 172 (108-256) | 178 (106-270) | 0.6894 | |||
| Fasting low density lipoprotein-cholesterol (mg/dL) | 102 (54-177) | 110 (40-181) | 0.9647 | |||
| Fasting plasma glucose (mg/dL) | 98.5 (76-320) | 103.5 (93-158) | 0.1554 | |||
| Homeostasis model assessment-Insulin Resistance | 1.36 (0.78-6.78) | 1.44 (1.19-5.39) | 0.3151 | |||
| Alpha-fetoprotein (ng/mL) | 5.0 (1.0-96.0) | 5.0 (2.0-130) | 0.8593 | |||
| On-treatment factors | ||||||
| Treatment | ||||||
| Adherence of Peg-IFN (%)2 | 77.1 (62.5-100.0) | 91.4 (66.0-104.6) | 0.0936 | |||
| Adherence of RBV (%)2 | 75.0 (56.8-100.0) | 86.1 (86.1-94.4) | 0.0457 | |||
| Duration of treatment (wk) | 37 (36-48) | 48 (36-48) | 0.1449 | |||
Classified by METAVIR score;
Calculated on the basis of 48-wk treatment. Note: Data are expressed as number of patients or median (range); ND: Not done; SVR: Sustained virologic response; HCV: Hepatitis C virus; Peg-IFN: Pegylated interferon; RBV: Ribavirin.
Among the on-treatment factors, adherence to RBV was significantly higher in non-SVR patients than in SVR patients (P = 0.0457) and adherence to Peg-IFN was only marginally higher in non-SVR than in SVR patients (P = 0.0936), indicating that these adherence factors did not influence SVR. The duration of Peg-IFNα/RBV combination therapy (36 wk or 48 wk) did not affect the outcome of treatment (Table 2).
SVR rates according to RVR, G2 subtype, and IFNL3 SNPs
The SVR rates were not statistically different between patients with HCV G2a and G2b in either the IFNL3 genotype TT patient group (90.7% vs 88.6%) or the non-TT group (82.6% vs 61.5%). The remaining 21 patients (11.7%) were not found to have G2a or G2b. Twenty of 21 patients achieved SVR and the remaining patient (who did not achieve RVR) showed relapse. In the RVR patient group, the SVR rates in HCV G2a patients were comparable to those observed with HCV G2b patients, regardless of the IFNL3 genotype (Figure 3A).
Figure 3.
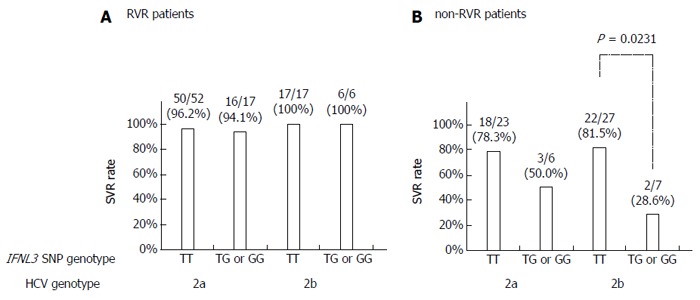
Sustained virologic response rates according to interferon-λ3 single nucleotide polymorphism genotype and hepatitis C virus sub-genotype. A: Rapid virologic response (RVR); and B: Non-RVR patient groups. HCV: Hepatitis C virus; RVR: Rapid virologic response; SNP: Single nucleotide polymorphism; IFNL3: Interferon-λ3.
Among non-RVR patients, the SVR rate in HCV G2b patients with the IFNL3 TT genotype was significantly higher than in those with the non-TT genotype (81.5% vs 28.6%, P = 0.0231; Figure 3B). The SVR rate in G2a patients with the TT genotype was higher than in those with the non-TT genotype (78.3% vs 50.0%), though the difference was not statistically significant.
DISCUSSION
IFNL3 SNPs, such as rs8099917 and rs12979860, have a strong impact on virologic responses in chronically HCV G1-infected patients to Peg-IFNα/RBV combination therapy[7-9]. However, more potent antiviral treatments, including direct-acting antiviral agents (DAAs), would attenuate the value of IFNL3 SNPs as a predictor of treatment outcome, because they could markedly improve the SVR rate. In countries/areas where DAAs are not available, Peg-IFNα/RBV combination therapy is still the standard of care for HCV G2 patients. Therefore, IFNL3 SNPs still have prognostic value in such settings[10-13].
In a previous study from Japan, IFNL3 SNPs were found to be an independent predictive factor for SVR (but not RVR) in patients infected with HCV subtype G2b, but not G2a[11]. However, the study analysis included both RVR patients and non-RVR patients in whom the treatment duration was limited to 24 wk and not extended to 36 or 48 wk. Another study conducted in the United States showed that the IFNL3 rs12979860 genotype was associated with SVR to 24-wk Peg-IFNα/RBV combination therapy in HCV-2/3 patients who did not achieve RVR[12]. Our findings were in partial concordance with these results; the IFNL3 SNP significantly influenced the achievement of SVR in patients who did not attain RVR, but did not affect SVR in RVR patients. Therefore, IFNL3 SNP genotyping is valuable for predicting SVR only in non-RVR patients, irrespective of G2 subtype, even if Peg-IFNα/RBV combination therapy is extended to 36 or 48 wk. Conversely, neither IFNL3 SNPs nor G2 subtypes are associated with SVR in RVR patients. However, the relatively small number of patients in our study may limit the conclusions that can be drawn, and these results should be verified in a larger study cohort.
The SVR rate for HCV G2 patients in our study was similar to those reported in previous studies[17,18]. As the SVR rate was very high (96.4%) in patients who achieved RVR and were treated with standard 24-wk Peg-IFNα/RBV combination therapy, the treatment period of 24 wk is sufficient and could be abbreviated without reducing the SVR rate. Conversely, the SVR rates following 24-wk Peg-IFNα/RBV combination therapy were reported to be fairly low in non-RVR patients[6,19], and response-guided extension to 36 or 48 wk has been used to improve treatment efficacy[5,6,20]. However, our previous study[6] and the present study reveal that there are no distinct differences in the SVR rates of non-RVR patients who receive either 36 or 48 wk of therapy, and that the SVR rate is significantly lower in non-RVR patients (treated for 36 wk or 48 wk) than in RVR patients (treated for 24 wk). These findings suggest that there are limitations to prolonged treatment duration in non-RVR patients. Specifically, this study highlights the low SVR rates in non-RVR patients with unfavorable IFNL3 genotypes.
In the near future, DAA-based combination therapy will be used worldwide as the first-line therapy for treating chronic HCV G2 infection because extremely high SVR rates can be attained with shorter treatment durations and without distinctive side effects[21-23]. In many countries/areas, however, Peg-IFNα/RBV combination therapy will still be the standard of care before DAAs are approved and available. Until then, response-guided therapy based on RVR to Peg-IFNα/RBV combination therapy is useful in yielding high SVR rates for RVR patients and reducing economic and physical burdens by prematurely discontinuing unnecessary treatment for non-RVR patients. Alternatively, to make a decision to continue treatment in non-RVR patients, IFNL3 genotyping may be valuable in predicting the probability of achieving SVR.
In conclusion, neither the IFNL3 SNP genotype nor the G2 subtype influenced the probability of achieving SVR in RVR patients treated with response-guided Peg-IFNα/RBV combination therapy. However, the SVR rate in non-RVR patients was higher in those with the IFNL3 TT genotype compared to those with the non-TT genotype, irrespective of G2 subtype, even if therapy was extended to 36 or 48 wk, indicating that the IFNL3 SNP has a significant impact only on the achievement of SVR in non-RVR patients.
ACKNOWLEDGMENTS
We thank the participating physicians and staff at the Jikei University Katsushika Medical Center and Kashiwa Hospital and the Shinmatsudo Central General Hospital for their assistance. We also thank Ms. Rie Agata and Ms. Yoko Yumoto (ICMR, Jikei University School of Medicine) for providing excellent technical support.
COMMENTS
Background
Genotype 2 (G2) hepatitis C virus (HCV) is the second-most frequent HCV genotype and accounts for approximately 30% of chronic HCV infections in Japan. Most HCV G2 patients who achieve rapid virologic response (RVR) in 24-wk response-guided pegylated interferon-α plus ribavirin (Peg-IFNα/RBV) combination therapy achieve sustained virologic response, whereas a fraction of patients who do not achieve RVR may remain uncured even when therapy is extended for 36 or 48 wk. The impact of interferon-λ3 (IFNL3) single nucleotide polymorphisms (SNPs) on Peg-IFNα/RBV combination therapy for HCV G1 has been firmly established. However, it remains controversial whether the IFNL3 genotype is useful in predicting virologic responses of HCV G2 patients to Peg-IFNα/RBV therapy.
Research frontiers
IFNL3 genotyping is advantageous in clinical practice for patients who do not achieve RVR. The results of this study provide a strong rationale for the use of IFNL3 SNPs testing to personalize antiviral therapy.
Innovations and breakthroughs
This work aims at emphasizing the role of IFNL3 SNPs in HCV G2 patients who received Peg-IFNα/RBV combination therapy. In non-RVR patients, the evaluation of the IFNL3 SNPs still holds significance to establish the therapeutic schedule.
Applications
In patients with IFNL3 non-TT genotypes and non-RVR, clinicians should not extend treatment with combination therapy. The relevance of this approach is cost-effective at the time of DAA therapy.
Terminology
IFNL3, located 8 kb upstream of the interleukin-28B gene, is a cytokine that plays a role in HCV clearance.
Peer-review
The authors describe associations of IFNL3 genotypes with Peg-IFNα/RBV treatment outcome in HCV G2 patients who do not achieve RVR. The data are interesting, in that a role for IFNL3 genotype in treatment outcome for HCV G2 patients is demonstrated only in patients not achieving RVR. These data contribute to the IFNL3 literature and thus merit reporting.
Footnotes
Ethics approval: The study was reviewed and approved by the Jikei University School of Medicine Institutional Review Board.
Informed consent: All study participants, or their legal guardian, provided informed written consent prior to study enrollment.
Conflict-of-interest: All authors declare no conflicts of interest.
Data sharing: No additional data are available.
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Peer-review started: September 19, 2014
First decision: November 5, 2014
Article in press: January 16, 2015
P- Reviewer: Meissner EG, Kitson MT S- Editor: Yu J L- Editor: AmEditor E- Editor: Liu XM
References
- 1.Tsubota A, Kumada H, Chayama K, Arase Y, Saitoh S, Koida I, Murashima N, Suzuki Y, Kobayashi M, Takagi K, et al. Relationship between pretreatment viremia level and response to interferon-alpha therapy in chronic hepatitis C differs in viral type 1 and 2 infections. Dig Dis Sci. 1996;41:1925–1932. doi: 10.1007/BF02093591. [DOI] [PubMed] [Google Scholar]
- 2.Tsubota A, Chayama K, Arase Y, Koida I, Saitoh S, Ikeda K, Iwasaki S, Matsumoto T, Kobayashi M, Kumada H. Factors useful in predicting the response to interferon therapy in chronic hepatitis C. J Gastroenterol Hepatol. 1993;8:535–539. doi: 10.1111/j.1440-1746.1993.tb01648.x. [DOI] [PubMed] [Google Scholar]
- 3.Manns MP, McHutchison JG, Gordon SC, Rustgi VK, Shiffman M, Reindollar R, Goodman ZD, Koury K, Ling M, Albrecht JK. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet. 2001;358:958–965. doi: 10.1016/s0140-6736(01)06102-5. [DOI] [PubMed] [Google Scholar]
- 4.Fried MW, Shiffman ML, Reddy KR, Smith C, Marinos G, Gonçales FL, Häussinger D, Diago M, Carosi G, Dhumeaux D, et al. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med. 2002;347:975–982. doi: 10.1056/NEJMoa020047. [DOI] [PubMed] [Google Scholar]
- 5.Sato K, Hashizume H, Yamazaki Y, Horiguchi N, Kakizaki S, Takagi H, Mori M. Response-guided peginterferon-alpha-2b plus ribavirin therapy for chronic hepatitis C patients with genotype 2 and high viral loads. Hepatol Res. 2012;42:854–863. doi: 10.1111/j.1872-034X.2012.00997.x. [DOI] [PubMed] [Google Scholar]
- 6.Abe H, Aida Y, Ishiguro H, Yoshizawa K, Seki N, Miyazaki T, Itagaki M, Sutoh S, Ika M, Kato K, et al. New proposal for response-guided peg-interferon-plus-ribavirin combination therapy for chronic hepatitis C virus genotype 2 infection. J Med Virol. 2013;85:1523–1533. doi: 10.1002/jmv.23626. [DOI] [PubMed] [Google Scholar]
- 7.Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K, Sakamoto N, Nakagawa M, Korenaga M, Hino K, Hige S, et al. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41:1105–1109. doi: 10.1038/ng.449. [DOI] [PubMed] [Google Scholar]
- 8.Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, Heinzen EL, Qiu P, Bertelsen AH, Muir AJ, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 9.Tsubota A, Shimada N, Yoshizawa K, Furihata T, Agata R, Yumoto Y, Abe H, Ika M, Namiki Y, Chiba K, et al. Contribution of ribavirin transporter gene polymorphism to treatment response in peginterferon plus ribavirin therapy for HCV genotype 1b patients. Liver Int. 2012;32:826–836. doi: 10.1111/j.1478-3231.2011.02727.x. [DOI] [PubMed] [Google Scholar]
- 10.Rauch A, Kutalik Z, Descombes P, Cai T, Di Iulio J, Mueller T, Bochud M, Battegay M, Bernasconi E, Borovicka J, et al. Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology. 2010;138:1338–1345, 1345.e1-e7. doi: 10.1053/j.gastro.2009.12.056. [DOI] [PubMed] [Google Scholar]
- 11.Kawaoka T, Hayes CN, Ohishi W, Ochi H, Maekawa T, Abe H, Tsuge M, Mitsui F, Hiraga N, Imamura M, et al. Predictive value of the IL28B polymorphism on the effect of interferon therapy in chronic hepatitis C patients with genotypes 2a and 2b. J Hepatol. 2011;54:408–414. doi: 10.1016/j.jhep.2010.07.032. [DOI] [PubMed] [Google Scholar]
- 12.Mangia A, Thompson AJ, Santoro R, Piazzolla V, Tillmann HL, Patel K, Shianna KV, Mottola L, Petruzzellis D, Bacca D, et al. An IL28B polymorphism determines treatment response of hepatitis C virus genotype 2 or 3 patients who do not achieve a rapid virologic response. Gastroenterology. 2010;139:821–827, 827.e1. doi: 10.1053/j.gastro.2010.05.079. [DOI] [PubMed] [Google Scholar]
- 13.Sakamoto N, Nakagawa M, Tanaka Y, Sekine-Osajima Y, Ueyama M, Kurosaki M, Nishida N, Tamori A, Yuki NS, Itsui Y, et al. Association of IL28B variants with response to pegylated-interferon alpha plus ribavirin combination therapy reveals intersubgenotypic differences between genotypes 2a and 2b. J Med Virol. 2011;83:871–878. doi: 10.1002/jmv.22038. [DOI] [PubMed] [Google Scholar]
- 14.Ohno O, Mizokami M, Wu RR, Saleh MG, Ohba K, Orito E, Mukaide M, Williams R, Lau JY. New hepatitis C virus (HCV) genotyping system that allows for identification of HCV genotypes 1a, 1b, 2a, 2b, 3a, 3b, 4, 5a, and 6a. J Clin Microbiol. 1997;35:201–207. doi: 10.1128/jcm.35.1.201-207.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanaka T, Tsukiyama-Kohara K, Yamaguchi K, Yagi S, Tanaka S, Hasegawa A, Ohta Y, Hattori N, Kohara M. Significance of specific antibody assay for genotyping of hepatitis C virus. Hepatology. 1994;19:1347–1353. [PubMed] [Google Scholar]
- 16.Bedossa P, Poynard T. An algorithm for the grading of activity in chronic hepatitis C. The METAVIR Cooperative Study Group. Hepatology. 1996;24:289–293. doi: 10.1002/hep.510240201. [DOI] [PubMed] [Google Scholar]
- 17.Toyoda H, Kumada T, Kiriyama S, Sone Y, Tanikawa M, Hisanaga Y, Kanamori A, Atsumi H, Nakano S, Arakawa T. Eight-week regimen of antiviral combination therapy with peginterferon and ribavirin for patients with chronic hepatitis C with hepatitis C virus genotype 2 and a rapid virological response. Liver Int. 2009;29:120–125. doi: 10.1111/j.1478-3231.2008.01736.x. [DOI] [PubMed] [Google Scholar]
- 18.Inoue Y, Hiramatsu N, Oze T, Yakushijin T, Mochizuki K, Hagiwara H, Oshita M, Mita E, Fukui H, Inada M, et al. Factors affecting efficacy in patients with genotype 2 chronic hepatitis C treated by pegylated interferon alpha-2b and ribavirin: reducing drug doses has no impact on rapid and sustained virological responses. J Viral Hepat. 2010;17:336–344. doi: 10.1111/j.1365-2893.2009.01182.x. [DOI] [PubMed] [Google Scholar]
- 19.Andriulli A, Mangia A, Iacobellis A, Ippolito A, Leandro G, Zeuzem S. Meta-analysis: the outcome of anti-viral therapy in HCV genotype 2 and genotype 3 infected patients with chronic hepatitis. Aliment Pharmacol Ther. 2008;28:397–404. doi: 10.1111/j.1365-2036.2008.03763.x. [DOI] [PubMed] [Google Scholar]
- 20.Yamaguchi Y, Tamori A, Tanaka Y, Iwai S, Kobayashi S, Fujii H, Morikawa H, Hagihara A, Enomoto M, Kawada N. Response-guided therapy for patients with chronic hepatitis who have high viral loads of hepatitis C virus genotype 2. Hepatol Res. 2012;42:549–557. doi: 10.1111/j.1872-034X.2011.00956.x. [DOI] [PubMed] [Google Scholar]
- 21.Lawitz E, Mangia A, Wyles D, Rodriguez-Torres M, Hassanein T, Gordon SC, Schultz M, Davis MN, Kayali Z, Reddy KR, et al. Sofosbuvir for previously untreated chronic hepatitis C infection. N Engl J Med. 2013;368:1878–1887. doi: 10.1056/NEJMoa1214853. [DOI] [PubMed] [Google Scholar]
- 22.Jacobson I, Yoshida EM, Sulkowski M, Nelson DR, Svarovskaia E, An D, McNally J, Brainard DM, Symonds WT, McHutchison JG, et al. Treatment with sofosbuvir ribavirin for 12 weeks achieves svr12 of 78% in GT2/3 interferon-ineligible, -intolerant, or -unwilling patients: results of the phase 3 POSITRON trial. J Hepatol. 2013;58:S28. [Google Scholar]
- 23.Dore GJ, Lawitz E, H’ezode C, Shafran S, Ramji A, Tatum H, Taliani G, Tran A, Brunetto M, Zaltron S, et al. Daclatasvir combined with peginterferon alfa-2a and ribavirin for 12 or 16 weeks in patients with HCV genotype 2 or 3 infection: COMMAND GT2/3 study. J Hepatol. 2013;58:S570–S571. [Google Scholar]


