Abstract
Helicobacter pylori FlgR activates transcription with σ54-RNA polymerase holoenzyme (σ54-holoenzyme) from at least five flagellar operons. Activators of σ54-holoenzyme generally bind enhancer sequences located >70 bp upstream of the promoter and contact σ54-holoenzyme bound at the promoter through DNA looping to activate transcription. H. pylori FlgR lacks the carboxy-terminal DNA-binding domain present in most σ54-dependent activators. As little as 42 bp of DNA upstream of the flaB promoter and 26 bp of DNA sequence downstream of the transcriptional start site were sufficient for efficient FlgR-mediated expression from a flaB′-′xylE reporter gene in H. pylori, indicating that FlgR does not use an enhancer to activate transcription. Other examples of σ54-dependent activators that lack a DNA-binding domain include Chlamydia trachomatis CtcC and activators from the other Chlamydia spp. whose genomes have been sequenced. FlgR from Helicobacter hepaticus and Campylobacter jejuni, which are closely related to H. pylori, appear to have carboxy-terminal DNA-binding domains, suggesting that the loss of the DNA-binding domain from H. pylori FlgR occurred after the divergence of these bacterial species. Removal of the amino-terminal regulatory domain of FlgR resulted in a constitutively active form of the protein that activated transcription from σ54-dependent genes in Escherichia coli. The truncated FlgR protein also activated transcription with E. coli σ54-holoenzyme in an in vitro transcription assay.
Helicobacter pylori is a microaerophilic, motile bacterium that is the etiological agent of chronic gastritis in humans (10, 11). Colonization of the gastric mucosa by H. pylori is associated with development of peptic ulcers, gastric non-Hodgkin's lymphomas, and gastric mucosa-associated lymphoid tissue lymphoma (5). Motility in H. pylori is achieved through two to six polar flagella and is essential for colonization in gnotobiotic piglets (12, 13).
The H. pylori genome contains about 40 known flagellar genes scattered throughout the genome, which are organized into 25 or more transcriptional units (1, 58). Where the regulation of flagellar biogenesis has been studied in other bacteria, flagellar gene expression is under the control of a regulatory hierarchy in which genes encoding the basal body and protein export apparatus are expressed first, followed by the genes encoding components of the hook, and then finally the genes encoding filament proteins (31, 62).
Flagellar gene regulation in H. pylori is complex, involving all three sigma factors found in the bacterium. Genes encoding flagellar components required early in flagellar biogenesis are transcribed by σ80-RNA polymerase holoenzyme, the primary form of RNA polymerase holoenzyme in H. pylori, and are equivalent to the class II flagellar genes in Escherichia coli and Salmonella enterica serovar Typhimurium (31). Class III flagellar genes in H. pylori are transcribed by σ54-RNA polymerase holoenzyme (σ54-holoenzyme) and encode basal body rod proteins (flgBC), the hook protein (flgE), and a minor flagellin (flaB) (54, 56). Expression of the σ54-dependent flagellar genes requires a two-component system consisting of the sensor kinase HP0244 (3), which we refer to as FlgS, and the response regulator FlgR (54). The gene encoding FlgS appears to be within a class II operon (1, 58), which may provide a mechanism for controlling the hierarchical expression of the class III operons. The class IV flagellar genes in H. pylori, which include the major flagellin gene flaA, are transcribed by σ28-RNA polymerase holoenzyme (22, 29). Temporal control of class IV genes appears to be coordinated through the regulation of both the expression and activity of σ28. The gene encoding σ28, fliA, is part of a class II operon (1, 58), and the activity of σ28 is negatively regulated through interactions with the anti-σ28 factor FlgM (9, 19).
FlgR activates transcription with σ54-holoenzyme and belongs to a large family of activators that are widespread in bacteria and that are involved in regulation of diverse functions including nitrogen fixation, C4-dicarboxylic acid transport, degradation of aromatic compounds, hydrogen metabolism, flagellar biogenesis, and pilin formation (24, 64). To activate transcription, activators of σ54-holoenzyme typically bind to enhancer-like sequences located upstream of the promoter and contact σ54-holoenzyme bound at the promoter in a closed complex through DNA looping (7, 50, 51, 55). Productive interactions between the activators and σ54-holoenzyme lead to conversion of the closed promoter complex to an open complex in a reaction that is coupled to ATP hydrolysis by the activator (38, 47, 52, 61).
Activators of σ54-holoenzyme are modular in structure, generally consisting of an amino-terminal regulatory domain, a central domain responsible for transcriptional activation and ATP hydrolysis, and a carboxy-terminal DNA-binding domain (41, 64). The central activation domain belongs to the AAA+ superfamily of ATPases (ATPases associated with various cellular activities), the members of which are involved in diverse functions including transcription, DNA replication, protein folding and unfolding, proteolysis, and membrane fusion (39, 43, 65).
FlgR is unusual in that it lacks the DNA-binding domain found in other activators (Fig. 1); it consists of only an amino-terminal response regulator domain and an AAA+ domain. Chlamydia trachomatis CtcC was reported recently to lack the carboxy-terminal DNA-binding domain (23), indicating that FlgR is not unique in its unusual structural arrangement. Previous study on CtcC, however, did not address the issue of whether an enhancer is required for efficient CtcC-mediated transcriptional activation in C. trachomatis.
FIG. 1.
Comparison of a partial sequence of H. pylori FlgR with those of other σ54-dependent activators. The conserved Walker A, Walker B, and sensor II motifs within the AAA+ domains of selected σ54-dependent activators are indicated. X, any amino acid residue, with the number that follows indicating the spacing between the motifs. The sequences from σ54-dependent activators that are shown are those of S. meliloti DctD (Sm DctD); H. pylori FlgR (Hp FlgR); H. hepaticus FlgR (Hh FlgR); Campylobacter jejuni FlgR (Cj FlgR); C. trachomatis CtcC (Ct CtcC); and activators from C. muridarum (Cm act), C. pneumoniae (Cp act), and Chlamydophila caviae (Cc act). Helix 4 indicates the last helix of the α-helical subdomain of the DctD AAA+ domain, which was predicted by threading the DctD sequence onto the “A. aeolicus” NtrC1 AAA+ domain structure (27). The serine residue underlined in helix 4 of the DctD sequence is Ser-390, which is the carboxy terminus of the DctD AAA+ domain used in the studies described here. The predicted helix-turn-helix motifs of DctD, H. hepaticus FlgR, and C. jejuni FlgR are indicated. The four amino acid residues of the carboxy termini of the FlgR proteins are shown for comparison.
To address the possibility that an enhancer-binding activity needed for FlgR function resides on a separate polypeptide, flaB′-′xylE reporter genes that carried various amounts of DNA sequence upstream of the promoter were constructed and FlgR-mediated transcriptional activation from these reporters in H. pylori was monitored. As little as 42 bp of sequence upstream of the flaB promoter and 26 bp of sequence downstream of the transcriptional start site were needed for efficient expression in H. pylori, indicating that FlgR does not use an enhancer to activate transcription. The levels of FlgR in H. pylori were estimated by Western blotting and found to be somewhat higher than those reported for other σ54-dependent activators that have a carboxy-terminal DNA-binding domain. The AAA+ domain of FlgR was expressed in E. coli and purified. This truncated FlgR protein was constitutively active and was able to function with E. coli σ54-holoenzyme both in vivo and in vitro.
MATERIALS AND METHODS
Bacterial strains and media.
E. coli strain DH5α [φ80d lacZ ΔM15 recA1 gyrA96 thi-1 hsdR17(rK− mK+) supE44 relA1 deoR Δ(lacZYA-argF)U169] was used for cloning and was cultured in Luria-Bertani (LB) medium at 37°C. H. pylori strains ATCC 43504 and 26695 were grown on tryptic soy agar (TSA) supplemented with 5% horse serum at 37°C under microaerobic conditions, which consisted of an atmosphere of 4% O2, 5% CO2, and 91% N2. When included in the medium, antibiotics were used at the following concentrations: ampicillin, 100 μg/ml; chloramphenicol, 30 μg/ml; kanamycin, 30 μg/ml; bacitracin, 200 μg/ml; colistin, 15 μg/ml.
PCRs.
Genomic DNA used for PCR was isolated from bacterial strains with the Wizard genomic DNA purification kit (Promega). PCR primers were from Integrated DNA Technologies. PCR amplifications were done with either Taq (Promega) or Pfu (Stratagene) DNA polymerase. DNA was amplified in 30 cycles with the following temperature regimen: 94°C for 2 min, 49°C for 1.5 min, and 72°C for 3 min. PCR products were cloned into the cloning vector pGEM-T (Promega), and sequencing of the cloned PCR products was performed at the Molecular Genetics Instrumentation Facility at the University of Georgia.
Transformation of H. pylori.
Electrocompetent H. pylori cells were prepared and transformed as described previously (53). When transforming cells with derivatives of the shuttle vector pHel3, plasmid DNA was methylated with S-adenosylmethionine and H. pylori cell extract essentially as described previously to improve the transformation efficiency (44). Briefly, 25 μg of plasmid DNA was treated with H. pylori cell extract (3 mg/ml) in a 200-μl reaction mixture containing 20 mM Tris-HCl, 50 mM KCl (pH 7.9), 5 mM EDTA, and 2 μM S-adenosylmethionine (Sigma). Samples were incubated at 37°C for 30 min, after which time the DNA was purified with QIAGEN gel extraction columns. Treated plasmids were introduced into H. pylori by electroporation, and transformants were selected on TSA containing kanamycin.
Construction of H. pylori mutant strains with insertions in flgR or flgS.
Plasmid pGHPAY91 carries the flgR gene from H. pylori strain 26695 and was obtained from the American Type Culture Collection. A 1.4-kb EcoRI fragment from plasmid pHP1 (34) that carries the Campylobacter coli aphA3 cassette was cloned into a unique StuI site within flgR in plasmid pGHPAY91 to create a suicide vector that was introduced into H. pylori strain ATCC 43504 by electroporation. Transformants were selected on TSA containing kanamycin, and genomic DNA from some of these colonies was analyzed by PCR to confirm that the chromosomal copy of flgR had been inactivated with the aphA3 cassette. One of these strains was saved and named MGD1. An H. pylori flgR insertion mutant was similarly generated with a cassette bearing a chloramphenicol transacetylase (cat) gene from C. coli. The entire flgR from H. pylori strain 26695 was amplified by PCR and cloned in pGEM-T. A 1.3-kb EcoRI fragment from plasmid pSKAT4 (59) that carried the C. coli cat cassette was introduced into a unique Eco47III site within flgR in this plasmid, which then was transformed into H. pylori strain ATCC 43504 by electroporation. Transformants were selected on TSA containing chloramphenicol, and inactivation of the chromosomal copy of flgR by the cat cassette in these strains was confirmed by PCR. One flgR:cat mutant strain was saved and designated HP31.
The entire flgS was amplified from H. pylori strain ATCC 43504 by PCR and cloned into pGEM-T. The 1.4-kb EcoRI fragment carrying the C. coli aphA3 cassette was inserted into a unique HindIII site with flgS in this plasmid to create plasmid pMD5. Plasmid pMD5 was introduced into H. pylori strain ATCC 43504 by electroporation, and transformants were selected on TSA containing kanamycin. Genomic DNA from kanamycin-resistant colonies was analyzed by PCR to confirm that the chromosomal copy of flgS had been inactivated with the aphA3 cassette, and one flgS:aphA3 mutant strain was saved and designated MGD2. The 1.3-kb EcoRI fragment bearing the C. coli cat cassette was also inserted into the HindIII site within flgS in pMD5. This plasmid was introduced into H. pylori strain ATCC 43504 by electroporation, and transformants were selected on TSA containing chloramphenicol. Genomic DNA from chloramphenicol-resistant colonies was analyzed by PCR to confirm that the chromosomal copy of flgS had been inactivated with the cat cassette, and one of the flgS:cat strains was saved and named strain HP22.
Construction of xylE reporter genes.
For construction of all of the reporter genes described below, genomic DNA from H. pylori strain 26695 was used as a template. Cloned PCR products were sequenced to verify that no mutations had been introduced during DNA amplification. Reporter genes were constructed with a promoterless Pseudomonas putida catechol-2,3-dioxygenase (xylE) reporter (44). A region of DNA that corresponded to positions −67 to +26 relative to the transcriptional start site of H. pylori flaB, which was determined previously (54), was amplified by PCR and cloned upstream of the promoterless xylE reporter gene. The resulting flaB′-′xylE reporter gene (designated flaB1′-′xylE) was moved into the shuttle vector pHel3 (16) to create plasmid pPBHP21, which was introduced into H. pylori strains by electroporation. Similarly, a region of DNA corresponding to positions −393 to +26 relative to the transcriptional start site of flaB was amplified and cloned upstream of xylE to create the flaB2′-′xylE reporter gene, which was moved into the shuttle vector pHel3 to create plasmid pPBHP22.
For construction of the flaA′-′xylE reporter gene, DNA corresponding to positions −126 to +47 relative to the transcriptional start site of flaA (37) was amplified by PCR and then introduced upstream of xylE. The resulting flaA′-′xylE reporter gene was cloned into pHel3 to create plasmid pPBHP24. For construction of the flgI′-′xylE reporter gene, a DNA fragment corresponding to positions −225 to +24 relative to the translational start site of flgI was amplified and cloned upstream of xylE. The resulting flgI′-′xylE reporter gene was moved into the shuttle vector pHel3 to create plasmid pPBHP23.
Plasmid constructions for expression of FlgR proteins.
To produce a full-length histidine-tagged version of FlgR, flgR was amplified from pGHPAY91 and cloned into the expression vector pTrcHis-C (Invitrogen). The resulting plasmid, pMD20, introduced a sequence coding for a histidine tag to the 5′ end of flgR. To express a maltose-binding protein-FlgR fusion protein (MBP-FlgR), a DNA fragment bearing flgR was moved from pMD20 into a derivative of pMAL-c (New England Biolabs), resulting in plasmid pPBHP12. A plasmid that expressed the FlgR AAA+ domain (residues His-131 to Arg-381) with a histidine tag at the amino terminus was constructed by amplifying a 750-bp DNA fragment corresponding to this region of flgR with H. pylori 26695 genomic DNA as a template and cloning the PCR product into the expression vector pTrcHis-C to create plasmid pPBHP80. Plasmid pHX182, which expresses the Sinorhizobium meliloti DctD AAA+ domain (residues Leu-141 to Ser-390) linked to a histidine tag at the amino terminus, is a derivative of pTrcHis-C and was provided by Hao Xu (63).
Measurement of XylE activity.
XylE activities in whole cells were measured as described previously (44). H. pylori strains containing the xylE reporter plasmids were grown on TSA supplemented with kanamycin for 48 h and then resuspended in 50 mM phosphate buffer, pH 7.4, to a cell density of 1 optical density at 600 nm (OD600) unit, which corresponded to 109CFU/ml. Reactions were initiated by adding cells (50 to 100 μl) to reaction mixtures containing 10 mM catechol in 50 mM potassium phosphate, pH 7.4. Catechol oxidation to 2-hydroxymuconic semialdehyde was monitored continuously at 375 nm with a Beckman DU 640B recording spectrophotometer at room temperature. A unit of XylE activity corresponds to 1 μmol of catechol/min, and values were expressed as units per minute per 108 cells.
Purification of FlgR proteins.
Cultures of E. coli DH5α bearing plasmid pPBHP12 were grown in LB medium at 37°C to an OD650 of 0.5, at which point isopropyl-β-d-thiogalactopyranoside (IPTG) was added to the culture medium to a final concentration of 1 mM to induce the expression of MBP-FlgR. After an additional 3 to 4 h of incubation, cells were harvested by centrifugation and the resulting cell pellet was resuspended in a mixture containing 20 mM Tris-HCl (pH 7.4), 5% (vol/vol) glycerol, 1 mM dithiothreitol (DTT), 1 mM EDTA, 200 mM KCl, and 0.5 mM phenylmethylsulfonyl fluoride (buffer C). Cells were lysed in a French pressure cell at 9,000 lb/in2, and the resulting cell extract was clarified by centrifugation at 1,000 × g for 50 min. The supernatant was loaded onto an amylose-agarose (New England Biolabs) affinity column, and MBP-FlgR was eluted with buffer C plus 10 mM maltose. Protein fractions were pooled and dialyzed against 20 mM HEPES (pH 7.4)-5% (vol/vol) glycerol-1 mM DTT-100 mM potassium thiocyanate. Protein concentrations were determined by a bicinchoninic acid protein assay (Pierce) using bovine serum albumin as a standard.
The histidine-tagged FlgR AAA+ domain was expressed in E. coli DH5α from plasmid pPBHP80. Cells were grown at 37°C to an OD650 of 0.8, at which point IPTG was added to the medium to a final concentration of 1 mM. Cultures were incubated for an additional 3 to 4 h. Cells were harvested by centrifugation, resuspended in 50 mM Tris-acetate (pH 8.2)-200 mM KCl-1 mM EDTA-0.5 mM phenylmethylsulfonyl fluoride, and lysed in a French pressure cell at 9,000 lb/in2. The cell extract was clarified by centrifugation at 1,000 × g for 50 min, and the resulting supernatant was loaded onto a nickel-nitrilotriacetic acid resin column (QIAGEN). The histidine-tagged FlgR AAA+ domain was eluted in a buffer containing 300 mM NaCl, 50 mM sodium phosphate (pH 7.8), 250 mM imidazole, and 5% (vol/vol) glycerol. Fractions containing the FlgR AAA+ domain were pooled and dialyzed against 20 mM Tris-HCl (pH 8.8)-5% (vol/vol) glycerol-100 mM potassium thiocyanate-0.5 mM DTT (buffer A). The protein was then applied to a 5-ml HiTrapQ anion-exchange column (Pharmacia) and eluted in a linear gradient to 1 mM KCl in buffer A.
Assaying activity of FlgR proteins in E. coli.
Plasmids pMD20, pPBHP80, and pHX182 were introduced into an E. coli DH5α strain that contained plasmid pRKMAZ:+UAS, which bears an S. meliloti dctA′-′lacZ reporter gene (25). Cultures were grown in LB medium at 37°C to an OD650 of 0.8, at which time IPTG was added to a final concentration of 1 mM. Cultures were incubated an addition 7 h, at which point six independent sets of whole cell β-galactosidase assays were done in duplicate as described previously (2), with activities expressed as Miller units (36).
Western blot analysis.
H. pylori cells were lysed in sodium dodecyl sulfate loading buffer and then applied to a 10% polyacrylamide gel. Following electrophoresis, proteins were transferred to nitrocellulose membranes, which were probed with antiserum prepared in New Zealand White rabbits and directed against either full-length histidine-tagged FlgR (this study) or the H. pylori flagellum (kindly provided by Paul O'Toole, Massey University). A peroxidase-conjugated goat affinity-purified antibody against rabbit immunoglobulin G was used as the secondary antibody (ICN/Cappel). Cross-reactive protein bands were visualized by luminescence with an ECL kit (Amersham).
In vitro transcription assay.
Single-round transcription assays were performed as described previously (21). Plasmid pJES534 (48) yields an ∼155-nucleotide uracilless transcript from the S. enterica serovar Typhimurium glnA promoter and was used as a DNA template. Reaction mixtures contained 0.1 to 10 μM FlgR AAA+ domain protein, 1 U of E. coli RNA polymerase (Epicentre), 200 nM S. enterica serovar Typhimurium σ54, 10 nM plasmid DNA, 4 mM ATP, 400 μM GTP, 5 μM CTP, and 7.5 μCi of [α-32P]CTP (3,000 Ci/mmol; Amersham). Proteins were incubated with the DNA template at 37°C for 10 min, after which time ATP was added to stimulate open-complex formation. After 10 min, the remaining nucleotides were added to allow synthesis of the transcripts along with 0.1 mg of heparin/ml to prevent further open-complex formation. Reactions were stopped after 10 min, and transcripts were visualized on a denaturing polyacrylamide gel, followed by exposure to X-ray film.
Glutamine synthetase assays.
Glutamine synthetase activities were determined by the γ-glutamyltransferase assay as described previously (4). Cultures of DH5α that carried plasmids pPBHP80 or pHX182 were grown in LB medium to mid-log phase and harvested by centrifugation. Cells were permeabilized by including hexadecyltrimethylammonium bromide in the assay buffer as described previously (4). Glutamine synthetase units were expressed as nanomoles of γ-glutamyl hydroxymate produced per minute and normalized to a cell density of 1 OD650 unit. All assays were done at least six times.
RESULTS
FlgR and FlgS are required for motility and expression of σ54-dependent flagellar genes.
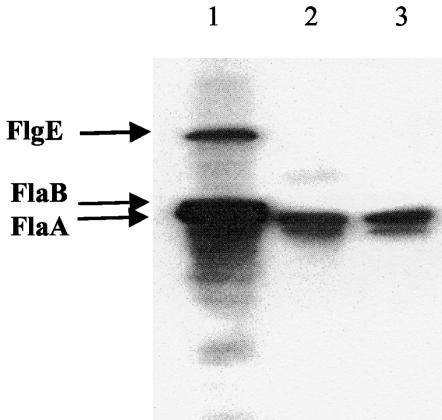
FlgR is an activator of σ54- holoenzyme that was shown previously to be required for motility and transcription of five σ54-dependent flagellar operons (flaB, flgE, orf0906, flgBC, and orf1120-flgK) in H. pylori strain G27 (54). The gene encoding FlgR is at the 3′ end of a putative operon that includes genes encoding diacylglycerol kinase (dgkA), the α subunit of DNA gyrase (gyrA), and three hypothetical proteins of unknown function (1, 58). Similar to the previous observations with H. pylori strain G27, inactivation of flgR in H. pylori strain ATCC 43504 with either an aphA3 or cat cassette resulted in loss of motility and expression of FlgE and FlaB (Fig. 2; data only shown for the flgR:aphA3 mutant strain).
FIG. 2.
Immunoblot of cell extracts of wild-type H. pylori and flgR and flgS mutant strains. Each lane contained cell extracts (50 μg of protein) from one of the H. pylori strains. The blot was probed with antiserum directed against the H. pylori flagellum. Lane 1, wild-type H. pylori ATCC 43504; lane 2, MGD1 (flgR:aphA3 mutant strain); lane 3, MGD2 (flgS:aphA3 mutant strain). FlgE, FlaB, and FlaA bands are indicated.
The flgR mutant strain expressed the major flagellin, FlaA, which is dependent on σ28 for its expression (29). This observation was consistent with the previous report for H. pylori strain G27 that flgR was not required for expression of flaA (54). This finding also illustrates a difference in the mechanisms for controlling the expression of σ28-dependent flagellar genes in S. enterica serovar Typhimurium and H. pylori. In S. enterica serovar Typhimurium, σ28 function is inhibited by the anti-σ28 factor FlgM until the hook-basal body complex is completed, at which point FlgM is translocated out of the cell by the flagellar protein export apparatus (31). Although H. pylori possesses a FlgM homolog that negatively regulates the function of σ28 (9, 19), alleviation of the inhibitory effect of FlgM on σ28 does not involve formation of the hook-basal body complex since H. pylori strains with mutations in the hook protein gene do not produce flagellar filaments yet they still accumulate FlaA (46).
The flgS gene (orf0244) encodes the cognate sensor kinase of FlgR and is at the 3′ end of a potential operon that includes flgI and orf0245, which encode the flagellar basal body P-ring protein and a hypothetical protein of unknown function, respectively (1, 58). Inactivation of flgS in H. pylori strain G27 resulted in loss of motility and expression of σ54-dependent flagellar genes, as assessed by two-dimensional gel electrophoresis analysis of cell extracts of the mutant strain (3). As in these earlier observations, it was found that inactivation of flgS in H. pylori strain ATCC 43504 resulted in loss of motility and expression of FlgE and FlaB (Fig. 2). In contrast to the previous report with H. pylori strain G27, however, inactivation of flgS in H. pylori strain ATCC 43504 did not eliminate expression of FlaA. This apparent discrepancy could be due to differences between the two H. pylori strains. Alternatively, since FlaA would not localize correctly in the absence of FlgE and FlgB, it may not accumulate to wild-type levels in the flgS mutant strain. Therefore, our ability to detect FlaA in the flgS mutant strain may simply reflect the fact that the sensitivity of the Western blotting procedure that we used was higher than that of the two-dimensional gel electrophoresis method used in the previous study for detecting expression of FlaA.
FlgR does not require upstream activation sequences for efficient transcriptional activation from the flaB promoter.
Activators of σ54-holoenzyme generally bind to DNA sequences located relatively far from the promoter and contact the closed promoter complex through DNA looping to activate transcription. These activator binding sites are typically located >100 bp upstream of the transcriptional start site since shorter distances hinder the ability of the activator to interact with σ54-holoenzyme through DNA looping due to constraints in DNA flexibility (64). As illustrated in Fig. 1, H. pylori FlgR lacks the carboxy-terminal DNA-binding domain found in most other σ54-dependent activators, suggesting that FlgR does not bind DNA to activate transcription. Consistent with the lack of a DNA-binding domain, purified full-length FlgR failed to bind a DNA fragment that bore the flaB promoter regulatory region in gel mobility shift assays (data not shown). We wanted to examine the possibility, however, that a DNA-binding activity required for FlgR function resides on a separate polypeptide.
To determine if upstream activation sequences are required for expression of σ54-dependent flagellar genes in H. pylori, we constructed flaB′-′xylE reporter genes that had either 393 or 67 bp of DNA sequence upstream of the transcriptional start site of flaB, which we designated flaB2′-′xylE and flaB1′-′xylE, respectively. Both reporter genes contained 26 bp of DNA sequence downstream of the transcriptional start site of flaB, which corresponded to the untranslated region of the flaB transcript. These reporter genes were placed on the shuttle vector pHel3 and introduced into H. pylori strain ATCC 43504. Levels of expression from the two flaB′-′xylE reporter genes were similar and dependent on both FlgR and FlgS (Table 1). Low-level expression from the flaB2′-′xylE reporter gene was observed in the flgR and flgS mutant strains, suggesting that σ80-holoenzyme or σ28-holoenzyme might initiate transcription weakly from a sequence located upstream of the flaB promoter between positions −67 and −393.
TABLE 1.
Expression of flaB′-′xylE, flaA′-′xylE, and flgI′-′xylE reporter genes in various H. pylori strains
| H. pylori strain | Relevant genotype | XylE activity (U/min/108 cells) for reporter genea:
|
|||
|---|---|---|---|---|---|
| flaB2′-′xylE | flaB1′-′xylE | flaA′-′xylE | flgI′-′xylE | ||
| ATCC 43504 | Wild type | 12.4 ± 4.9 | 9.0 ± 2.1 | 21.5 ± 10.9 | 5.2 ± 2.3 |
| HP31 | flgR:cat | 2.6 ± 1.9 | 0.3 ± 0.3 | 17.2 ± 6.3 | 2.2 ± 0.4 |
| HP22 | flgS:cat | 4.1 ± 1.9 | 0.1 ± 0.2 | 19.2 ± 7.9 | 5.6 ± 2.9 |
Values represent the averages of at least six replicates ± standard deviations.
Expression from a flaA′-′xylE reporter gene, which is dependent on σ28-holoenzyme, and a flgI′-′xylE reporter gene, which is dependent on σ80-holoenzyme, was compared with that from the flaB′-′xylE reporter genes (Table 1). Expression from the flaA′-′xylE reporter gene was independent of FlgR and FlgS, while expression from the flgI′-′xylE reporter gene was approximately twofold lower in the flgR mutant strain but not in the flgS mutant strain (Table 1). We do not understand why disruption of flgR resulted in lower expression from the flgI′-′xylE reporter gene, but it does not seem likely that it was due to the failure of the mutant strain to express the σ54-dependent flagellar genes, since this would also require FlgS. The XylE activities observed for the flagellar reporter genes were consistent with the expected relative amounts of the products of these genes associated with the H. pylori flagellum (i.e., FlaA > FlaB > FlgI). Taken together, these data indicate that FlgR does not use an upstream activation sequence or enhancer to elicit its function on the flaB promoter.
FlgR levels in H. pylori are slightly higher than levels of σ54-dependent activators in other bacteria.
Transcriptional activation in the absence of DNA binding has been reported previously for other σ54-dependent activators, including NtrC (42), DctD (18, 60), NifA (17), and PspF (20). In these previous studies, the DNA-binding motifs of the activators were either deleted or mutated to eliminate DNA-binding activity or the upstream activation sequence of the target gene was removed. Transcriptional activation under these conditions required that the activator be present at higher-than-normal levels. We wished to estimate the levels of FlgR in H. pylori to determine if it was expressed at levels that were higher than those for σ54-dependent activators that function by binding to an upstream activation sequence or enhancer.
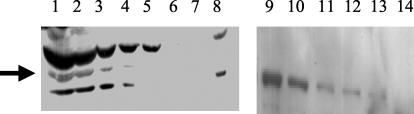
Levels of NtrC in E. coli range from about 10 molecules of monomeric protein per cell when cells are grown in a medium with excess nitrogen to approximately 140 molecules per cell following starvation of cells for nitrogen (32, 49). We estimated FlgR levels in H. pylori strain ATCC 43504 by Western blotting using antiserum directed against the full-length histidine-tagged FlgR. For the Western blot, purified MBP-FlgR was used as a standard to avoid underestimating the FlgR concentration due to antibodies that may have recognized the histidine tag. Various amounts of H. pylori cells were lysed and loaded directly onto the sodium dodecyl sulfate-polyacrylamide gel for the Western blot assay. Under the assay conditions the detection limit for MBP-FlgR was ∼12.5 ng, or approximately 0.14 pmol of MBP-FlgR monomer (Fig. 3). FlgR could be detected by the Western blot assay when as few as 108 cells were lysed and loaded on the gel, which corresponded to ∼800 FlgR monomers per cell. Thus, assuming that the intracellular volumes of H. pylori and E. coli are similar, the level of FlgR in H. pylori appears to be about sixfold higher than that of NtrC in E. coli under conditions where these proteins are activating transcription.
FIG. 3.
Immunoblot of FlgR in H. pylori cell extracts. The immunoblot was probed with an antibody directed against the histidine-tagged FlgR protein. Arrow, band corresponding to FlgR. Different amounts of cell extract of wild-type H. pylori were loaded onto lanes 1 to 7 (lane 1, 1 × 109 cells; lane 2, 5 × 108 cells; lane 3, 2 × 108 cells; lane 4, 1 × 108 cells; lane 5, 5 × 107 cells; lane 6, 2 × 107 cells; lane 7, 1 × 107 cells). In lane 8, 109 cells of the mutant H. pylori flgR:cat strain were loaded. Different amounts of purified MBP-FlgR were loaded onto lanes 9 to 14 (lane 9, 200 ng; lane 10, 100 ng; lane 11, 50 ng; lane 12, 25 ng; lane 13, 12.5 ng; lane 14, 6 ng).
FlgR functions with E. coli σ54-holoenzyme.
We wished to examine the function of FlgR in an in vitro transcription assay. H. pylori σ54-holoenzyme has not been purified in an active form, so we used E. coli σ54-holoenzyme for these experiments. Previous work in the laboratory showed that H. pylori rpoN, which encodes σ54, failed to complement an S. enterica serovar Typhimurium rpoN mutant strain (M. Dashti, unpublished data), so it was unclear if FlgR could function with E. coli σ54-holoenzyme.
To examine H. pylori FlgR function in E. coli, flgR was cloned into the expression vector pTrc-HisC, which introduced a sequence coding for a histidine tag at the 5′ end of the gene. Removal of the amino-terminal receiver domain of S. meliloti DctD had been shown previously to result in a constitutively active form of the protein (26), and we wished to determine if removal of the receiver domain of FlgR would similarly result in constitutive activity. Therefore, a truncated flgR allele that encoded residues His-131 through Arg-381, which is the carboxy-terminal amino acid residue of native FlgR, was cloned into pTrc-HisC.
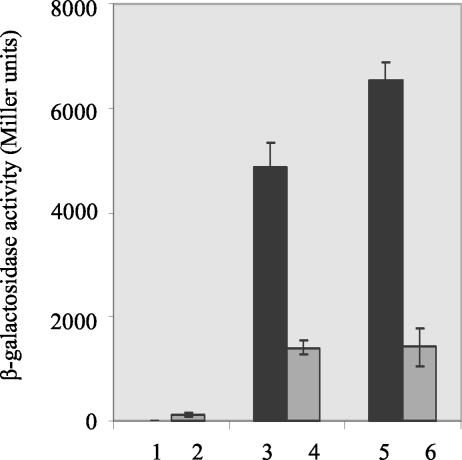
Expression of both the full-length and truncated FlgR proteins was inducible with IPTG to about the same level (data not shown). Activity of the FlgR proteins was monitored in E. coli with an S. meliloti dctA′-′lacZ reporter gene, the expression from which is dependent on σ54-holoenzyme. This reporter gene was used because S. enterica serovar Typhimurium σ54 (and also presumably E. coli σ54) has a low affinity for the H. pylori flaB promoter (L. Pereira and T. R. Hoover, unpublished data). Full-length FlgR activated transcription from the dctA′-′lacZ reporter gene very weakly (approximately threefold above background) when its expression was induced with IPTG but failed to activate transcription above background levels when its expression was not induced (Fig. 4). Since FlgR presumably needs to be phosphorylated to activate transcription, the low-level activity observed with the full-length FlgR in E. coli suggested that the protein could be phosphorylated by another sensor kinase or a small phosphor donor, as occurs with other response regulators (30, 40).
FIG. 4.
In vivo activity of AAA+ domains of FlgR and DctD. Activity of full-length FlgR, the FlgR AAA+ domain, and the DctD AAA+ domain on a dctA′-′lacZ reporter gene in E. coli. Gray bars, activity for culture in which the expression of the FlgR or DctD proteins was induced with IPTG; black bars, activity for cultures in which the expression of the proteins was not induced with IPTG. Lanes 1 and 2, activity for full-length FlgR; lanes 3 and 4, activity for the FlgR AAA+ domain; lanes 5 and 6, activity for the DctD AAA+ domain.
In contrast to the results with the full-length protein, the FlgR AAA+ domain activated transcription from the dctA′-′lacZ reporter gene >120-fold above background levels when its expression was not induced with IPTG. The activity of the FlgR AAA+ domain in E. coli compared favorably with that of the DctD AAA+ domain, indicating that the FlgR AAA+ domain was able to function effectively with E. coli σ54-holoenzyme. The activities of both the FlgR AAA+ domain and the DctD AAA+ domain decreased upon induction of these proteins with IPTG. This decrease in activity was not likely due to aggregation since these proteins were in the soluble fraction when we overexpressed them for purification. Since σ54-dependent activators can bind σ54 (8), the FlgR and DctD AAA+ domains may have sequestered σ54 and prevented it from binding core RNA polymerase to form the holoenzyme.
To determine if the FlgR and DctD AAA+ domains influenced expression of other σ54-dependent genes in E. coli, we examined the effect of these proteins on expression of glnA, which encodes glutamine synthetase. Expression of glnA is dependent on σ54 and NtrC, which binds to several sites located upstream of glnAp2, the σ54-dependent glnA promoter (50). The presence of the FlgR AAA+ domain or the DctD AAA+ domain resulted in an approximately threefold increase in glutamine synthetase activity (Table 2), suggesting that these activators stimulated transcription from glnAp2. Overexpression of these proteins, however, did not inhibit glnA expression, as observed with the dctA′-′lacZ reporter gene. We do not know the reason for this, but one possibility is that σ54-holoenzyme has a higher affinity for glnAp2 than it does for the dctA promoter and therefore might be less sensitive to decreased levels of free σ54 in the cell.
TABLE 2.
Glutamine synthetase activities in E. coli DH5α strains that express the FlgR AAA+ domain or the DctD AAA+ domain
| Plasmid | Protein expressed | Glutamine synthetase activity (nmol of γ-glutamyl hydroxymate/min/A650 unit)a
|
|
|---|---|---|---|
| −IPTG | +IPTG | ||
| None | None | 5.46 ± 1.22 | 6.38 ± 2.00 |
| pPBHP80 | FlgR AAA+ domain | 17.4 ± 5.42 | 15.4 ± 3.14 |
| pHX182 | DctD AAA+ domain | 16.5 ± 5.07 | 9.62 ± 3.40 |
Values represent the averages of at least six replicates ± standard deviations. −IPTG, without IPTG induction; +IPTG, with IPTG induction.
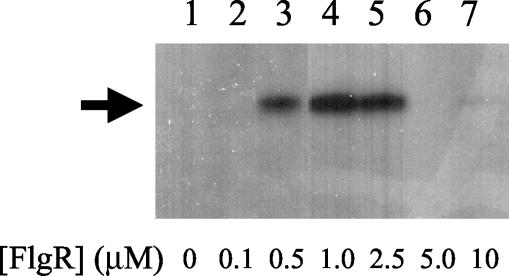
The FlgR AAA+ domain was purified, and its activity was examined in vitro. The purified FlgR AAA+ domain hydrolyzed ATP and activated transcription with E. coli RNA polymerase from a DNA template that carried S. enterica serovar Typhimurium glnAp2 in an in vitro transcription assay (Fig. 5). Transcripts were detected with as little as 0.5 μM FlgR AAA+ domain monomer. As observed with the dctA′-′lacZ reporter gene in vivo, but in contrast to the results for glnAp2, high concentrations of the FlgR AAA+ domain inhibited transcription initiation from glnAp2 in the in vitro transcription assay. The reason for the discrepancy in the in vivo and in vitro results with glnAp2 might be the smaller amounts of σ54-holoenzyme in the in vitro system. Alternatively, conditions in the in vitro system may not be optimized for efficient transcription, making the in vitro assay more sensitive to perturbations. Regardless of the reason, the results of the in vivo and in vitro transcription assays suggest further that FlgR does not bind DNA to activate transcription, nor does it require another DNA-binding protein to do so since it is unlikely that the promoter regulatory regions of dctA, glnA, and flaB have a common upstream activation sequence.
FIG. 5.
In vitro transcriptional activation with the FlgR AAA+ domain. Reaction mixtures contained FlgR AAA+ domain (monomer) at the indicated concentrations. Arrow, transcript of the expected size (∼155 nucleotides) from the glnA promoter.
DISCUSSION
Most bacterial transcriptional activators bind to specific sites within the promoter regulator regions of their target genes to recruit RNA polymerase to the promoter or to stimulate a step in transcription initiation that occurs after the initial binding of RNA polymerase to the promoter. One notable exception is the bacteriophage N4 single-stranded DNA binding protein (N4SSB), which activates transcription with E. coli σ70-holoenzyme at N4 late promoters without binding DNA (35). N4SSB interacts with the carboxy terminus of the β′ subunit of RNA polymerase and appears to stimulate a step that follows the initial binding of RNA polymerase to the promoter (35). E. coli MarA and the closely related SoxS protein are other examples of transcriptional activators that function somewhat differently from most bacterial activators. Although MarA and SoxS are DNA-binding proteins, they appear to bind DNA after interacting with RNA polymerase in solution (33). MarA and SoxS interact with the α subunit of RNA polymerase in the absence of DNA to form binary complexes that are thought to scan chromosomal DNA for target promoters.
Activators of σ54-holoenzyme generally bind to sites that are located relatively far from the promoter and contact the closed complex through DNA looping to activate transcription (64). H. pylori FlgR lacks the DNA-binding domain associated with most other σ54-dependent activators, and the results presented here demonstrate that FlgR does not require an enhancer or upstream activation sequence to activate transcription. We infer that FlgR binds σ54-holoenzyme directly, either before or after formation of the closed promoter complex, to activate transcription. While mutant forms of other σ54-dependent activators have been reported to activate transcription in the absence of DNA binding (17, 18, 20, 42), FlgR is unusual in that it represents a naturally occurring enhancer-independent activator of σ54-holoenzyme. FlgR appears to be present at concentrations in the cell that are higher than those of NtrC in E. coli. The glnA enhancer facilitates oligomerization of NtrC, which is required for transcriptional activation from glnAp2 (48). Thus, higher concentrations of FlgR may be needed to compensate for the absence of enhancer binding and to allow oligomerization of the protein.
C. trachomatis CtcC was described recently as an activator of σ54-holoenzyme that lacks a DNA-binding domain (23), and we infer that, like FlgR, CtcC does not require sequences upstream of the promoter to activate transcription. Search of the databases for additional σ54-dependent activators that lack the DNA-binding domain identified activators from Chlamydia pneumoniae, Chlamydia muridarum, and Chlamydophila caviae with this unusual structural property (Fig. 1). Two other potential σ54-dependent activators that appear to lack the carboxy-terminal DNA-binding domain were found in database searches, one in Xanthomonas campestris pv. campestris ATCC 33913 (gene designated pilR in the database) and another in P. putida KT2440 (designated PP5166 in the database). There is some doubt, however, as to whether the sequences of these activators are correct or if they encode σ54-dependent activators. The carboxy-terminal end of the deduced amino acid sequence of X. campestris PilR corresponds to a sequence located 28 amino acids before that of H. pylori FlgR, placing it within the conserved sensor II motif of the AAA+ domain. Since this motif is important for function of other σ54-dependent activators, it is unlikely that such a truncation would result in an active protein. The carboxy-terminal end of the deduced amino acid sequence of P. putida PP5166 extends 3 residues beyond that of H. pylori FlgR. P. putida PP5166, however, has a poor match for the highly conserved GAFTGA motif in the C3 region of the protein (deduced amino acid sequence of this motif in PP5166 is GSHGGT). The GAFTGA motif, which is diagnostic of σ54-dependent activators (45) functions in contacting σ54 and coupling ATP hydrolysis to open-complex formation, and substitutions within this motif often result in loss of activity (6, 8, 14, 28, 60, 61). Thus, PP5166 may not be an activator of σ54-holoenzyme but may function with another form of RNA polymerase holoenzyme.
A potential advantage to the cell for using an activator of σ54-holoenzyme that does not bind DNA is that extensive regulatory regions upstream of target promoters which harbor binding sites for activators and auxiliary proteins involved in transcriptional activation, such as the integration host factor, are dispensable. A major drawback, however, is that the cell may be limited to a single σ54-dependent activator dedicated for a particular cellular function since there is no obvious mechanism for preventing activation from all of the σ54-dependent genes within the genome. Indeed, FlgR, CtcC, and the other chlamydial activators are the sole σ54-dependent activators in their respective bacteria, and, at least for FlgR, the activator appears to be dedicated for a specific cellular activity. The metabolic savings gained by employing activators that function without binding DNA seems small given the sacrifice in regulatory potential that accompanies the use of such activators. Therefore, we expect activators of σ54-holoenzyme that do not bind DNA to be restricted to bacteria with relatively limited needs for regulatory potential. Consistent with this hypothesis, such σ54-dependent activators have been found only in pathogens that have limited biosynthetic capability.
All of the chlamydial genomes that have been sequenced to date include an open reading frame that encodes a potential σ54-dependent activator that lacks the DNA-binding domain. In contrast, Helicobacter hepaticus and Campylobacter jejuni, which are closely related to H. pylori, each have a single σ54-dependent activator but these activators each have a potential DNA-binding domain (Fig. 1). This is somewhat surprising given that these activators appear to have the same role in flagellar biogenesis as H. pylori FlgR (15, 54, 57). We infer from this observation that loss of the DNA-binding domain in the chlamydial σ54-dependent activators and its loss in H. pylori FlgR occurred independently of each other.
The crystal structure of the AAA+ domain of NtrC1, a σ54-dependent activator from “Aquifex aeolicus,” was recently reported (27). Alignment of the FlgR, CtcC, and chlamydial activator sequences with that of NtrC1 indicated that the carboxy termini of these proteins correspond to residues within the last helix of the α-helical subdomain of the NtrC1 AAA+ domain. Thus, these σ54-dependent activators appear to have lost the entire DNA-binding domain during the course of evolution. It is unclear if the loss of the entire DNA-binding domain reflects an economic benefit by removing as much of the protein as possible or if there is a structural or functional basis for the proteins to terminate at this point.
Acknowledgments
This work was funded by award MCB-9974558 to T.R.H. from the National Science Foundation.
We thank Paul O'Toole for providing antiserum directed against the H. pylori flagellum.
REFERENCES
- 1.Alm, R. A., L.-S. L. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smithe, B. Noonon, B. D. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, J. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. R. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 2.Ashraf, S. I., M. T. Kelly, Y.-K. Wang, and T. R. Hoover. 1997. Genetic analysis of the Rhizobium meliloti nifH promoter, using the P22 challenge phage system. J. Bacteriol. 179:2356-2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beier, D., and R. Frank. 2000. Molecular characterization of two-component systems of Helicobacter pylori. J. Bacteriol. 182:2068-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bender, R. A., A. D. Janssen, A. D. Resnick, M. Blumenberg, F. Foor, and B. Magasanik. 1977. Biochemical parameters of glutamine synthetase from Klebsiella aerogenes. J. Bacteriol. 129:1001-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blaser, M. J. 1998. Helicobacter pylori and gastric diseases. Biomed. J. 316:1507-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bordes, P., S. R. Wigneshweraraj, J. Schumacher, X. Zhang, M. Chaney, and M. Buck. 2003. The ATP hydrolyzing transcription activator phage shock protein F of Escherichia coli: identifying a surface that binds σ54. Proc. Natl. Acad. Sci. USA 100:2278-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buck, M., S. Miller, M. Drummond, and R. Dixon. 1986. Upstream activator sequences are present in the promoters of nitrogen fixation genes. Nature (London) 320:374-378. [Google Scholar]
- 8.Chaney, M., R. Grande, S. R. Wigneshweraraj, W. Cannon, P. Casaz, M.-T. Gallegos, J. Schumacher, S. Jones, S. Elderkin, A. E. Dago, E. Morett, and M. Buck. 2001. Binding of transcriptional activators to sigma 54 in the presence of the transition state analog ADP-aluminum fluoride: insights into activator mechanochemical action. Genes Dev. 15:2282-2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colland, F., J.-C. Rain, P. Gounon, A. Labigne, P. Legrain, and H. De Reuse. 2001. Identification of the Helicobacter pylori anti-σ28 factor. Mol. Microbiol. 41:477-487. [DOI] [PubMed] [Google Scholar]
- 10.Cover, T. L., and M. J. Blaser. 1992. Helicobacter pylori and gastroduodenal disease. Annu. Rev. Med. 43:135-145. [DOI] [PubMed] [Google Scholar]
- 11.Dick, J. D. 1990. Helicobacter (Campylobacter) pylori: a twist on an old disease. Annu. Rev. Microbiol. 108:70-90. [DOI] [PubMed] [Google Scholar]
- 12.Eaton, K. A., D. R. Morgan, and S. Krakowka. 1989. Campylobacter pylori virulence factors in gnotobiotic piglets. Infect. Immun. 57:1119-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eaton, K. A., D. R. Morgan, and S. Krakowka. 1992. Motility as a factor in the colonisation of gnotobiotic piglets by Helicobacter pylori. J. Med. Microbiol. 37:123-127. [DOI] [PubMed] [Google Scholar]
- 14.Gonzalez, V., L. Olvera, X. Soberon, and E. Morett. 1998. In vivo studies on the positive control function of NifA: a conserved hydrophobic amino acid patch at the central domain involved in transcriptional activation. Mol. Microbiol. 28:55-67. [DOI] [PubMed] [Google Scholar]
- 15.Hendrixson, D. R., B. J. Akerley, and V. J. DiRita. 2001. Transposon mutagenesis of Campylobacter jejuni identifies a bipartite energy taxis system required for motility. Mol. Microbiol. 40:214-224. [DOI] [PubMed] [Google Scholar]
- 16.Heuermann, D., and R. Haas. 1998. A stable shuttle vector system for efficient genetic complementation of Helicobacter pylori strains by transformation and conjugation. Mol. Gen. Genet. 257:519-528. [DOI] [PubMed] [Google Scholar]
- 17.Huala, E., and E. M. Ausubel. 1989. The central domain of Rhizobium meliloti NifA is sufficient to activate transcription from the R. meliloti nifH promoter. J. Bacteriol. 171:3354-3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huala, E., J. Stigter, and F. M. Ausubel. 1992. The central domain of Rhizobium leguminosarum DCTD functions independently to activate transcription. J. Bacteriol. 174:1428-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Josenhans, C., E. Niehus, S. Amersbach, A. Horster, C. Betz, B. Drescher, K. T. Hughes, and S. Suerbaum. 2002. Functional characterization of the antagonistic flagellar late regulators FliA and FlgM of Helicobacter pylori and their effects on the H. pylori transcriptome. Mol. Microbiol. 43:307-322. [DOI] [PubMed] [Google Scholar]
- 20.Jovanovic, G., J. Rakonjac, and P. Model. 1999. In vivo and in vitro activities of the Escherichia coli σ54 transcription activator, PspF, and its DNA-binding mutant, PspFΔHTH. J. Mol. Biol. 285:469-483. [DOI] [PubMed] [Google Scholar]
- 21.Kelly, M. T., J. A. Ferguson III, and T. R. Hoover. 2000. Transcription initiation-defective forms of σ54 that differ in ability to function with a heteroduplex DNA template. J. Bacteriol. 182:6503-6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim, J. S., J. H. Chang, S. I. Chung, and J. S. Yum. 1999. Molecular cloning and characterization of the Helicobacter pylori fliP gene, an essential factor in flagellar structure and motility. J. Bacteriol. 181:6969-6976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koo, I. S., and R. S. Stephens. 2003. A developmentally regulated two-component signal transduction system in Chlamydia. J. Biol. Chem. 278:17314-17319. [DOI] [PubMed] [Google Scholar]
- 24.Kustu, S., E. Santero, D. Popham, D. Weiss, and J. Keener. 1989. Expression of σ54 (ntrA)-dependent genes is probably united by a common mechanism. Microbiol. Rev. 53:367-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ledebur, H., and B. T. Nixon. 1992. Tandem DctD-binding sites of the Rhizobium meliloti dctA upstream activating sequence are essential for optimal function despite a 50- to 100-fold difference in affinity for DctD. Mol. Microbiol. 6:3479-3492. [DOI] [PubMed] [Google Scholar]
- 26.Lee, J. H., D. Scholl, B. T. Nixon, and T. R. Hoover. 1994. Constitutive ATP hydrolysis and transcriptional activation by a stable, truncated form of Rhizobium meliloti DCTD, a σ54-dependent transcriptional activator. J. Biol. Chem. 269:20401-20409. [PubMed] [Google Scholar]
- 27.Lee, S.-K., A. De La Torre, D. Yan, S. Kustu, T. Nixon, and D. E. Wemmer. 2003. Regulation of the transcriptional activator NtrC1: structural studies of the regulatory and AAA+ ATPase domains. Genes Dev. 17:2552-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lew, C. M., and J. D. Gralla. 2002. New roles for conserved regions within a σ54-dependent enhancer-binding protein. J. Biol. Chem. 277:41517-41524. [DOI] [PubMed] [Google Scholar]
- 29.Leying, H., S. Suerbaum, G. Geis, and R. Haas. 1992. Cloning and genetic characterization of a Helicobacter pylori flagellin gene. Mol. Microbiol. 6:2863-2874. [DOI] [PubMed] [Google Scholar]
- 30.Lukat, G. S., W. R. McCleary, A. M. Stock, and J. B. Stock. 1992. Phosphorylation of bacterial response regulator proteins by low molecular weight phospho-donors. Proc. Natl. Acad. Sci. USA 89:718-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Macnab, R. M. (ed.). 1996. Flagella and motility, 2nd ed. ASM Press, Washington, D.C.
- 32.Magasanik, B. 1996. Regulation of nitrogen utilization, p. 1344-1356. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 33.Martin, R. G., W. K. Gillette, N. I. Martin, and J. L. Rosner. 2002. Complex formation between activator and RNA polymerase as the basis for transcriptional activation by MarA and SoxS in Escherichia coli. Mol. Microbiol. 43:355-370. [DOI] [PubMed] [Google Scholar]
- 34.McGee, D. J., F. J. Radcliff, R. L. Mendz, R. L. Ferrero, and H. L. Mobley. 1999. Helicobacter pylori rocF is required for arginase activity and acid protection in vitro but is not essential for colonization of mice or for urease activity. J. Bacteriol. 181:7314-7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller, A., D. Wood, R. H. Ebright, and L. B. Rothman-Denes. 1997. RNA polymerase β′ subunit: a target of DNA binding-independent activation. Science 275:1655-1657. [DOI] [PubMed] [Google Scholar]
- 36.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Mobley, H. L. T., and S. L. Hazell. 2001. Regulation of virulence genes, p. 400-450. In H. L. T. Mobley (ed.), Helicobacter pylori: physiology and genetics. ASM Press, Washington, D.C. [PubMed]
- 38.Morett, E., and M. Buck. 1989. In vivo studies on the interaction of RNA polymerase-σ54 with the Klebsiella pneumoniae and Rhizobium meliloti nifH promoters: the role of NIFA in the formation of an open promoter complex. J. Mol. Biol. 210:65-77. [DOI] [PubMed] [Google Scholar]
- 39.Neuwald, A. F., L. Aravind, J. L. Spouge, and E. V. Koonin. 1999. AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 9:27-43. [PubMed] [Google Scholar]
- 40.Ninfa, A. J., E. G. Ninfa, A. N. Lupas, A. M. Stock, B. Magasanik, and J. Stock. 1988. Crosstalk between bacterial chemotaxis signal transduction proteins and regulators of transcription of the Ntr regulon: evidence that nitrogen assimilation and chemotaxis are controlled by a common phosphotransfer mechanism. Proc. Natl. Acad. Sci. USA 85:5492-5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.North, A. K., K. E. Klose, K. M. Stedman, and S. Kustu. 1993. Prokaryotic enhancer-binding proteins reflect eukaryotic-like modularity: the puzzle of nitrogen regulatory protein C. J. Bacteriol. 175:4267-4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.North, A. K., and S. Kustu. 1997. Mutant forms of the enhancer-binding protein NtrC can activate transcription from solution. J. Mol. Biol. 267:17-36. [DOI] [PubMed] [Google Scholar]
- 43.Ogura, T., and A. J. Wilkinson. 2001. AAA+ superfamily ATPases: common structure-diverse function. Genes Cells 6:575-597. [DOI] [PubMed] [Google Scholar]
- 44.Olczak, A. A., J. W. Olson, and R. J. Maier. 2002. Oxidative-stress resistance mutants of Helicobacter pylori. J. Bacteriol. 184:3186-3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Osuna, J., X. Soberon, and E. Morett. 1997. A proposed architecture for the central domain of the bacterial enhancer-binding proteins based on secondary structure prediction and fold recognition. Protein Sci. 6:543-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O'Toole, P. W., M. Kostrzynska, and T. J. Trust. 1994. Non-motile mutants of Helicobacter pylori and Helicobacter mustelae defective in flagellar hook production. Mol. Microbiol. 14:691-703. [DOI] [PubMed] [Google Scholar]
- 47.Popham, D., D. Szeto, J. Keener, and S. Kustu. 1989. Function of a bacterial activator protein that binds to transcriptional enhancers. Science 243:629-635. [DOI] [PubMed] [Google Scholar]
- 48.Porter, S. C., A. K. North, A. B. Wedel, and S. Kustu. 1993. Oligomerization of NTRC at the glnA enhancer is required for transcriptional activation. Genes Dev. 7:2258-2272. [DOI] [PubMed] [Google Scholar]
- 49.Reitzer, L. J., and B. Magasanik. 1983. Isolation of the nitrogen assimilation regulator NRI, the product of the glnG gene of Escherichia coli. Proc. Natl. Acad. Sci. USA 80:5554-5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reitzer, L. J., and B. Magasanik. 1986. Transcription at glnA of E. coli is stimulated by activator bound to sites far from the promoter. Cell 45:785-792. [DOI] [PubMed] [Google Scholar]
- 51.Rippe, K., M. Guthold, P. H. von Hippel, and C. Bustamante. 1997. Transcriptional activation via DNA-looping: visualization of intermediates in the activation pathway of E. coli RNA polymerase σ54 holoenzyme by scanning force microscopy. J. Mol. Biol. 270:125-138. [DOI] [PubMed] [Google Scholar]
- 52.Ssse-Dwight, S., and J. D. Gralla. 1988. Probing the Escherichia coli glnALG upstream activation mechanism in vivo. Proc. Natl. Acad. Sci. USA 85:8934-8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seyler, R. W., Jr., J. W. Olson, and R. J. Maier. 2001. Superoxide dismutase-deficient mutants of Helicobacter pylori are hypersensitive to oxidative stress and defective in host colonization. Infect. Immun. 69:4034-4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spohn, G., and V. Scarlato. 1999. Motility of Helicobacter pylori is coordinately regulated by the transcriptional activator FlgR, an NtrC homolog. J. Bacteriol. 181:593-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Su, W., S. Porter, S. Kustu, and H. Echols. 1990. DNA-looping and enhancer activity: association between DNA-bound NTRC activator and RNA polymerase at the bacterial glnA promoter. Proc. Natl. Acad. Sci. USA 87:5504-5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Suerbaum, S., C. Josenhans, and A. Labigne. 1993. Cloning and genetic characterization of the Helicobacter pylori and Helicobacter mustelae flaB flagellin genes and construction of H. pylori flaA- and flaB-negative mutants by electroporation-mediated allelic exchange. J. Bacteriol. 175:3278-3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suerbaum, S., C. Josenhans, T. Sterzenbach, B. Drescher, P. Brandt, M. Bell, M. Droge, B. Fartmann, H.-P. Fischer, Z. Ge, A. Horster, R. Holland, K. Klein, J. Konig, L. Macko, G. L. Mendez, G. Nyakatura, D. B. Schauer, Z. Shen, J. Weber, M. Frosch, and J. G. Fox. 2003. The complete genome sequence of the carcinogenic bacterium Helicobacter hepaticus. Proc. Natl. Acad. Sci. USA 100:7901-7906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tomb, J.-F., O. White, A. R. Keflavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, E. K. Hickey, D. E. Berg, J. D. Gocayne, T. R. Utterback, J. D. Peterson, J. M. Kelley, M. D. Cotton, J. M. Weidman, C. Fujii, C. Bowman, L. Watthey, E. Wallin, W. S. Hayes, M. Borodovsky, P. D. Karp, H. O. Smith, C. M. Fraser, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 59.Wang, Y., and D. E. Taylor. 1990. Chloramphenicol resistance in Campylobacter coli: nucleotide sequence, expression, and cloning vector. Gene 94:23-28. [DOI] [PubMed] [Google Scholar]
- 60.Wang, Y.-K., J. H. Lee, J. M. Brewer, and T. R. Hoover. 1997. A conserved region in the σ54-dependent activator DctD is involved in both binding to RNA polymerase and coupling ATP hydrolysis to activation. Mol. Microbiol. 26:373-386. [DOI] [PubMed] [Google Scholar]
- 61.Weiss, D. S., J. Batut, K. E. Klose, J. Keener, and S. Kustu. 1991. The phosphorylated form of the enhancer-binding protein NTRC has an ATPase activity that is essential for activation of transcription. Cell 67:155-167. [DOI] [PubMed] [Google Scholar]
- 62.Wu, J., and A. Newton. 1997. Regulation of the Caulobacter flagellar gene hierarchy; not just for motility. Mol. Microbiol. 24:233-239. [DOI] [PubMed] [Google Scholar]
- 63.Xu, H. 2003. Purification and characterization of the AAA+ domain of Sinorhizobium meliloti DctD, a sigma54-dependent activator. Ph.D. thesis. University of Georgia, Athens.
- 64.Xu, H., and T. R. Hoover. 2001. Transcriptional regulation at a distance in bacteria. Curr. Opin. Microbiol. 4:138-144. [DOI] [PubMed] [Google Scholar]
- 65.Zhang, X., M. Chaney, S. R. Wigneshweraraj, J. Schumacher, P. Bordes, W. Cannon, and M. Buck. 2002. Mechanochemical ATPases and transcriptional activation. Mol. Microbiol. 45:895-903. [DOI] [PubMed] [Google Scholar]