Abstract
Background and Objective
Molecular epidemiological studies have shown that certain genotypes of Mycobacterium tuberculosis (MTB) are over-represented in limited geographical regions, suggesting of evolution of certain genotypes with increasing virulence and pathogenicity. Beijing strain of MTB was initially described by its potential to cause outbreaks worldwide and its association with drug resistance. Due to tuberculosis (TB)-related mortality which is associated with Beijing genotype, this study was designed with the aim to detect the MTB Beijing genotype in the region of study.
Materials and Methods
A total of 170 clinical isolates of MTB were collected from the TB reference laboratory of Khuzestan province, Iran, over one year period from February 2010 to February 2011. Phenotypic tests were used for preliminary detection of MTB. Culture positive MTB isolates were confirmed by multiplex PCR based on IS6110 gene with subsequent screening for resistance to isoniazid (INH), and rifampin (RIF) by PCR using relevant primers. Three set of primers were used to differentiate Beijing from non-Beijing strains by using Deletion- Targeted Multiplex (DTM) PCR.
Results
From 160 PCR-confirmed MTB isolates, 18 (11.25%) showed mutation in katG gene related to INH resistance and 20 (12.5%), associated with mutation in rpoB gene related to RIF resistance, and 8 (5%) were detected as Beijing strain using multiplex PCR. The majority of detected Beijing strains (6/8[75%]) comprised mutation in katG gene with the prevalent mutation specifically in codon 315. In 4 Beijing strains (2.5%), mutation in rpoB gene were also detected.
Conclusion
Using DTM- PCR, the rate of Beijing strains in the region of study was determined as 5%. Although for detection of MTB antimicrobial resistance, it is advised to use a combination of conventional antimicrobial susceptibility testing and molecular techniques, however for time saving, it seems that DTM-PCR, is a simple technique for use in areas of the world where Beijing strains are highly prevalent.
Keywords: M. tuberculosis, DTM PCR, Beijing strain, mutation
INTRODUCTION
Tuberculosis (TB) is a public health hazard in both developed and developing countries and the rapid emergence of drug resistance in the forms of multi-drug resistance (MDR) and extensively drug resistance (XDR), has become a major barrier in the management of TB all over the world. Molecular epidemiological studies have shown that certain genotypes of Mycobacterium tuberculosis (MTB), are over-represented in limited geographical regions, suggesting that certain genotypes have evolved unique properties increasing virulence and pathogenicity (1). Beijing strain of MTB is one such genotype of bacterium, which is pathogenic for human and it has been found to be the most frequently observed strain genotype of MTB to cause disease globally (2). It is most prevalent in Asia, but is also found in the former Soviet Union, Europe, Africa, and the United States. It was firstly identified in the Chinese capital by van Soolingen et al.(3), after analyzing MTB isolates to have shared multicopy IS6110 restriction fragment length polymorphism (RFLP) patterns that were collected between 1992-1994 from TB patients. Since the Beijing genotype strain’s origin in East Asia, it has spread to become endemic over much of Central, Northern and Western Asia and is known to have a global distribution now (3, 4).
Many studies have since reported that high emergence worldwide of the MTB Beijing strains is linked to its frequent association with MDR and TB-related mortality (2). This genotype is described by its potential to cause epidemics and its association with drug resistance (5, 6).
So far, most studies have focused on the Beijing genotype as it has been associated with numerous outbreaks worldwide (7). Rapid dissemination of Beijing strain may be originated from various characteristics of this genotype, including its interaction with the host immune system and ability to evasion and escape from BCG vaccine –induced protection, its high virulence and increase its ability to develop drug resistance (5, 8).
In the murine infection model, comparative survival studies have revealed that strains of the Beijing genotype had a higher level of virulence as compared to non-Beijing genotype strains (9, 10). In this investigation, we applied the Deletion- Targeted Multiplex (DTM) PCR technique for rapid detection of MTB Beijing strains in TB patients admitted to TB reference laboratory of Ahvaz, Iran.
MATERIALS AND METHODS
A total of 170 clinical specimens from suspected tuberculosis patients referred to TB reference laboratory of Khuzestan, Iran, over one year period from February 2010 to February 2011 were screened. These were included sputum, urine and bronchoalveolar Lavage (BAL) samples. For preliminary detection of MTB isolates, conventional acid fast staining, culture on Lowenstein-Jensen (LJ) and standard biochemical tests were performed (11). The MTB H37Rv strain was used as a reference strain for both traditional and molecular tests.
DNA extraction
For DNA extraction from MTB isolates, the simple boiling method was used as previously described (12). In brief, a few colonies harvested from surface of LJ medium, were dissolved in TE (Tris-EDTA) buffer and boiled at 100°C for 15 minutes with subsequent precipitation in a 1200 x g refrigerated centrifuge at 4°C for 3 min. The supernatant containing DNA was used for PCR amplification. Besides for extraction of DNA from culture negative samples, the extraction and purification kit (Roche, Germany) was used according to manufacturer’s instructions.
DTM- PCR assay
Multiplex PCR assay for confirmation of MTB strains was performed using the first set of primers of IS59 and IS60 amplified a 523-bp PCR product from IS6110 which is present in all MTB isolates, to serve as the internal PCR control (13). The genomic DNA which was also prepared from standard MTB H37Rv, used as a control positive in the amplification process. As previously determined, the region spanning genes Rv2816 to Rv2819 and part of Rv2820 is deleted in all Beijing strains of MTB (14).
So, the second set of primers of BjF and BjR producing a 129-bp PCR product containing the 3’ end of IS6110 and the 5’ End of the Rv2820 gene from the Beijing strains of MTB which reveals no deletion, were used for detection of Beijing strains in DTM- PCR (15). The third set of PCR primers, nBjF and nBjR producing a 95-bp PCR product from the Rv2819 gene were used for the detection of non-Beijing strains of MTB (16). The primers details are presented in Table 1.
Table 1.
Primers used for the amplification of IS59 and IS60 by multiplex PCR
| Genes | Primer sequence | Amplicon size (bp) |
|---|---|---|
| IS59 | 5’-GCGCCAGGCGCAGGTCGATGC-3’ | 523 |
| IS60 | 5’-GCGCCAGGCGCAGGTCGATGC-3’ | |
| BjF | 5’-CTCGGCAGCTTCCTCGAT-3’ | 129 |
| BjR | 5’-CGAACTCGAGGCTGCCTACTAC-3’ | |
| nBjF | 5’-AAGCATTCCCTT GACAGTCGAA-3’ | 95 |
| nBjR | 5’-GGCGCATGACTCGAAA GAAG-3’ |
The amplification reactions were performed in a final volume of 20 μl, containing 0.2 μg of genomic DNA, 20 pmol of each primer and 10 μl of Ampli Taq Gold master mix (ABI, USA). multiplex PCR was performed in an Eppendorf thermal cycler (Germany), using the following condition for amplification containing: Temperature cycling conditions included 96°C for 5 min, followed by 35 cycles of 96°C for 20 s, 55°C for 25 s, and 72°C for 30 s, and a final extension at 72°C for 7 min. The PCR products were analyzed by electrophoresis on a 2 % agarose gel. The molecular size of the PCR products were estimated by comparing the migration distances of DNA bands compared to a 100-bp DNA size marker.
PCR assay for detecting of antimicrobial resistance
All PCR confirmed strains were screened for resistance to isoniazid (INH) and rifampin (RIF) using the primers and PCR protocol which were described previously (17, 18).
Gene sequencing
PCR products of resistance strains were collected and sent for sequencing analysis at Bioneer Company, Korea.
Statistical analysis
This was done by SPSS package (SPSS Incorporation, Chicago, USA). Fisher exact test was used and P-values less than 0.05 were considered as statistically significant.
RESULTS
In this study a total of 170 samples from suspected TB patients referred to TB reference laboratory of Khuzestan, Iran, were examined. The samples included 126 sputum (74.1%), 34 urine (20%) and 10 BAL (5.9%). The samples were belonged to 72 (42.35%) male and 98 (57.64%) female patients with mean age of 47 ± 11.7. Using acid fast staining and direct microscopy 128 (75.3%) samples were positive for acid fast bacilli (AFB), while 156 (91.7%) samples were culture positive. By later application of PCR assay based on the IS6110 gene, a 523 bp fragment related to this gene were amplified in 160 (94.1%) of the isolates, which these isolates were confirmed as MTB. The overall results of traditional and molecular confirmatory tests are presented in Table 2.
Table 2.
Frequency of detected MTB among clinical samples by phenotypic and molecular methods
| Sample (No.) | acid-fast stain+ No. (%) | culture+ No. (%) | PCR+ No. (%) |
|---|---|---|---|
| Sputum (126) | 106(84.1) | 120(95.2) | 120(95.2) |
| Urine (34) | 16(47) | 28(82.3) | 30 (88.2) |
| BAL (10) | 6(60) | 8(80) | 10(100) |
| Total (170) | 128 (75.3) | 156 (91.7) | 160(94.1) |
The 160 PCR-confirmed MTB isolates, were screened for resistance to INH and RIF later. There were 18 tuberculosis patients (11.25%) whose isolate showed mutation in katG gene related to INH resistance, 20 (12.5%) associated with mutation in rpoB gene related to RIF resistance, and 8(5 %) were identified as MDR. The DNA extracts were then investigated for the presence of Beijing and non Beijing strains by DTM PCR (Fig. 1).
Fig. 1.
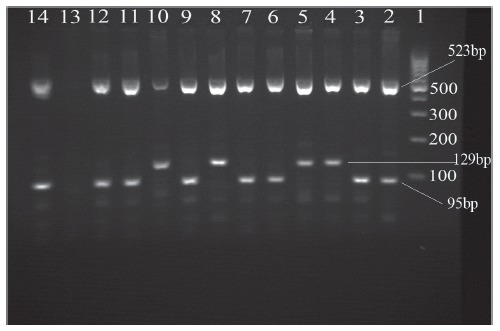
Multiplex polymerase chain reaction (PCR) products on a 2% agarose gel. Beijing strains generated 129-bp and 523-bp fragments. Non-Beijing strains generated 95-bp and 523-bp fragments.
Lanes 1: 100-bp DNA ladder;
2: Mycobacterium tuberculosis H37Rv;
3, 6, 7, 9, 11, 12 and 14: non-Beijing strain;
4, 5, 8 and10: Beijing strain; Lane 13: Distilled water blank control
The results revealed that 8 (5%) out of 160 MTB isolates belonged to Beijing strain. Sequencing data revealed that the majority of detected Beijing strains (6/8[75%]) comprised mutation in katG gene with the prevalent mutation specifically in codon 315. In 4 Beijing strains (50%) mutation in rpoB gene were also detected mainly in codons 527 and 531. Only 2 out of 8 detected MDR isolates in present study were confirmed as Beijing strains. Correlation between Beijing strains with age and sex of patients was not statistically significant, however the majority of Beijing strains (6/8) were detected from patients under retreatment for tuberculosis (n=46), which was statistically significant (P=0.069).
DISCUSSION
High emergence worldwide and frequent association with multi-drug resistance is the most important characteristic of the Beijing strains of MTB (5). In our study, we have used a simple and rapid multiplex PCR for analyzing the isolates for Beijing strain. This PCR-method requires only common PCR reagents and equipment, which is economical in both money and time and is suitable for high throughput analysis. Based on multiplex PCR assay from these samples, our study showed that in the city of Ahvaz, among 160 PCR-confirmed MTB isolates, 8 (5%) isolates were detected as Beijing genotype. These genotypes were mainly showed mutations in KatG (75%) with specifically codon 315 (Ser→Thr) and in rpoB gene (50%) [codon: 511, 527 and 531].
So, as shown, the frequency of Beijing genotype among kat G mutation (related to INH resistant strains) was significantly higher than rpoB mutation (related to RIF resistant strains). Besides, this study demonstrated that prevalence of this genotype among strains with mutation related with resistance was significantly higher than strains with no mutation in two genes (katG and rpoB) or susceptible strains. Moreover, katG mutation showed the most incorporation with Beijing genotype in TB patients in Iran according to study of Doustdar et al. (19). In their report, among 30 RIF resistant MTB isolates, by application of spoligotyping, IS6110-RFLP typing and sequencing methods, Beijing genotype were identified in 9 isolates that 6 isolates had mutation in rpoB codon 531(TCG →TGG (16.7%) and TCG →TTG (83.3%). While in our findings, among 20 mutations in rpoB gene related with RIF resistant strains, only 2 mutations were seen in codon S531L (TCG→TTG). In another study, the predominance of MDR, LAM and Beijing family strains were investigated in 87 MTB isolates, which 24 (77.4%) rpoB (S531L) mutation was reported among the 38 Beijing genotypes, higher than the rate we achieved (20). In our study, simultaneous mutation in rpoB and katG genes presenting the MDR strains were detected in 2 of 8 (25%) Beijing genotypes.
The correlation between the prevalence of Beijing strains and demographic information of the patients, revealed that distribution of these strains was identical in both genders, while the frequency of Beijing genotype was higher in patients with age ≤50 years compared to those with age ≥50 years. However, Fisher exact test showed that there was no significant correlation between age, sex and case definition of samples in TB patients with Beijing genotype. Previous studies from Iran, revealed the rate of 6.3% Beijing strain among Iranian and Afghans patients (21), and 7.1% Beijing strain in second study (22), which findings of both studies were close to our results. In a report from Pakistan (23), on characterization of mutations conferring extensive drug resistance to MTB isolates, 3 out of 4 Beijing strains, had mutation in rpoB codon S531L (TCG → TTG) and also, 3 isolates had mutation in katG codon S315T (AGC → ACC). In conclusion, using DTM- PCR, the rate of Beijing strains in the region of study was determined as 5%. Although for detection of MTB antimicrobial resistance, it is advised to use a combination of conventional antimicrobial susceptibility testing and molecular techniques. However, for time saving, it seems that DTM-PCR, is a simple technique for use in areas of the world where Beijing strains are highly prevalent.
Acknowledgments
This work is approved in Infectious and Tropical Diseases Research Center and was financially supported by Grant No. 88109, Research affairs, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran.
References
- 1.Hanekom M, van der Spuy GD, Streicher E, Ndabambi SL, McEvoy CR, Kidd M, et al. A recently evolved sublineage of the Mycobacterium tuberculosis Beijing strain family is associated with an increased ability to spread and cause disease. J Clin Microbiol. 2007;45:1483–1490. doi: 10.1128/JCM.02191-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parwati I, van Crevel R, van Soolingen D. Possible underlying mechanisms for successful emergence of the Mycobacterium tuberculosis Beijing genotype strains. Lancet Infect Dis. 2010;10:103–111. doi: 10.1016/S1473-3099(09)70330-5. [DOI] [PubMed] [Google Scholar]
- 3.Mokrousov I, Vyazovaya A, Zhuravlev V, Otten T, Millet J, Jiao WW, et al. Real-time PCR assay for rapid detection of epidemiologically and clinically significant Mycobacterium tuberculosis Beijing genotype. J Clin Microbiol. 2014;52:1691–1693. doi: 10.1128/JCM.03193-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bifani PJ, Mathema B, Kurepina NE, Kreiswirth BN. Global dissemination of the Mycobacterium tuberculosis W-Beijing family strains. Trends Microbiol. 2002;10:45–52. doi: 10.1016/s0966-842x(01)02277-6. [DOI] [PubMed] [Google Scholar]
- 5.Glynn JR. Beijing/W genotype Mycobacterium tuberculosis and drug resistance. Emerg Infect Dis. 2006;12:736–743. doi: 10.3201/eid1205.050400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanekom M, Gey van Pittius NC, McEvoy C, Victor TC, van Helden PD, Warren RM. Mycobacterium tuberculosis Beijing genotype: a template for success. Tuberculosis. 2011;91:510–523. doi: 10.1016/j.tube.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 7.Kremer K, Glynn JR, Lillebaek T, Niemann S, Kurepina NE, Kreiswirth BN, et al. Definition of the Beijing/W lineage of Mycobacterium tuberculosis on the basis of genetic markers. J Clin Microbiol. 2004;42:4040–4049. doi: 10.1128/JCM.42.9.4040-4049.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lasunskaia E, Ribeiro SC, Manicheva O, Gomes LL, Suffys PN, Mokrousov I, et al. Emerging multidrug resistant Mycobacterium tuberculosis strains of the Beijing genotype circulating in Russia express a pattern of biological properties associated with enhanced virulence. Microbes Infect. 2010;12:467–475. doi: 10.1016/j.micinf.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 9.Lopez B, Aguilar D, Orozco H, Burger M, Espitia C, Ritacco V, et al. A marked difference in pathogenesis and immuneresponse induced by different Mycobacterium tuberculosis genotypes. Clin Exp Immunol. 2003;133:30–37. doi: 10.1046/j.1365-2249.2003.02171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ordway D, Henao-Tamayo M, Harton M, Palanisamy G, Troudt J, Shanley C, et al. The hypervirulent Mycobacterium tuberculosis strain HN878 induces a potent TH1 response followed by rapid down-regulation. J Immunol. 2007;179:522–531. doi: 10.4049/jimmunol.179.1.522. [DOI] [PubMed] [Google Scholar]
- 11.Forbes BA, Sahm DF, Weissfeld AS. Bailey & Scott’s Diagnostic Microbiology. 12. St. Louis: Mosby; 2007. pp. 478–502. [Google Scholar]
- 12.Hosek J, Svastova P, Moravkova M, Pavlik I, Bartos M. Methods of mycobacterial DNA isolation from different biological material: A review. Vet Med. 2006;51:180–192. [Google Scholar]
- 13.Plikaytis BB, Marden JL, Crawford JT, Woodley CL, Butler WR, Shinnick TM. Multiplex PCR assay specific for the multidrug-resistant strain W of Mycobacterium tuberculosis. J Clin Microbiol. 2004;32:1542–1546. doi: 10.1128/jcm.32.6.1542-1546.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun JR, Lee SY, Dou HY, Lu JJ. Using a multiplex polymerase chain reaction for the identification of Beijing strains of Mycobacterium tuberculosis. Eur J Clin Microbiol Infect Dis. 2009;28:105–107. doi: 10.1007/s10096-008-0590-7. [DOI] [PubMed] [Google Scholar]
- 15.Warren RM, Victor TC, Streicher EM, Richardson M, Beyers N, Gey van Pittius NC, van Helden PD. Patients with active tuberculosis often have different strains in the same sputum specimen. Am J Respir Crit Care Med. 2004;169:610–614. doi: 10.1164/rccm.200305-714OC. [DOI] [PubMed] [Google Scholar]
- 16.Hillemann D, Warren R, Kubica T, Rüsch-Gerdes S, Niemann S. Rapid detection of Mycobacterium tuberculosis Beijing genotype strains by real-time PCR. J Clin Microbiol. 2006;44:302–306. doi: 10.1128/JCM.44.2.302-306.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hwang HY, Chang CH, Chang LL, Chang SF, Chang YH, Chen YJ. Characterization of Rifampin – resistant M. tuberculosis in Taiwan. J Med Microbiol. 2003;52:239–245. doi: 10.1099/jmm.0.05045-0. [DOI] [PubMed] [Google Scholar]
- 18.Baker LV, Brown TJ, Maxwell O, Gibson AL, Fang Z, Yates MD, Drobniewski FA. Molecular analysis of isoniasid – resistant M. tuberculosis isolate from England and Wales reveals the phylogenetic significance of the ahpc – 46A polymorphism. Antimicrob Agents Chemother. 2005;49:1445–1464. doi: 10.1128/AAC.49.4.1455-1464.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doustdar F, Khosravi AD, Farnia P, Bahrmand AR, Masjedi MR, Velayati AA. Mutations in rpoB Gene and genotypes of rifampin resistant Mycobacterium tuberculosis isolates in Iran. Tanaffos. 2008;7:11–17. [Google Scholar]
- 20.Ignatova A, Dubiley S, Stepanshina V, Shemyakin I. Predominance of multidrug resistant LAM and Beijing family strains among Mycobacterium tuberculosis isolates recovered from prison inmates in Tula Region, Russia. J Med Microbiol. 2006;55:1413–1418. doi: 10.1099/jmm.0.46575-0. [DOI] [PubMed] [Google Scholar]
- 21.Ramazanzadeh R, Farnia P, Amirmozafari N. characterization of Mycobacterium tuberculosis complex isolated from Iranian and Afghan patients by spoligotyping method. Braz J Microbiol. 2009;40:314–320. doi: 10.1590/S1517-838220090002000019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rohani M, Farnia P, Nasab MN, Moniri R, Torfeh M, Amiri MM. Beijing genotype and other predominant Mycobacterium tuberculosis spoligotypes observed in Mashhad city, Iran. Indian J Med Microbiol. 2009;27:306–310. doi: 10.4103/0255-0857.55441. [DOI] [PubMed] [Google Scholar]
- 23.Ali A, Hasan R, Jabeen K, Jabeen N, Qadeer E, Hasan Z. Characterization of mutations conferring extensive drug resistance to Mycobacterium tuberculosis isolates in Pakistan. Antimicrob Agents Chemother. 2011;55:5654–5659. doi: 10.1128/AAC.05101-11. [DOI] [PMC free article] [PubMed] [Google Scholar]


