Abstract
Background and Objectives
Methicillin-resistant Staphylococcus aureus (MRSA) is a well-known pathogen with a worldwide distribution. Given the increasing rate of MRSA infections, implementing of reliable, accurate and rapid testing for diagnosis of MRSA is necessary. The aim of this study was to compare four diagnostic methods for detection of MRSA isolates.
Materials and Methods
From December 2012 to April 2014, 120 S. aureus isolates were collected from three hospitals affiliated with Tehran University of Medical Sciences. MRSA isolates were detected by four different methods including cefoxitin disc diffusion test, oxacillin disc diffusion test, minimum inhibitory concentration (MIC) of oxacillin as determined by MIC test strip, and mecA detection by PCR.
Results
Out of 120 S. aureus isolates, cefoxitin disc diffusion test, oxacillin disc diffusion test and MIC test strip identified 60 (50%), 48 (40%), 55 (45.83%) isolates as MRSA, respectively. The sensitivity and specificity for oxacillin disc diffusion, cefoxitin disc diffusion and MIC of oxacillin were 80% and 100%, 100% and 100%, and 91.6% and 100%, respectively.
Conclusion
Cefoxitin disc diffusion test is reliable substitute for detection of MRSA in clinical laboratory where MIC detection and molecular methods are not accessible.
Keywords: Staphylococcus aureus, Methicillin resistance, Microbial sensitivity tests
INTRODUCTION
Methicillin-resistant Staphylococcus aureus (MRSA) is considered as a major pathogen both in hospita land community settings (1, 2). MRSA is endemic in many hospitals causing excess mortality and economic burden compared to methicillin-susceptible isolates (3). MRSA strains are resistant to nearly all of the beta-lactam antibiotics by producing an alternative penicillin-binding protein known as PBP2a. This protein is encoded by the mecA gene and has a low affinity to many beta-lactam antibiotics (4). MRSA strains are not only resistant to beta-lactams and cephalosporins, but also often show resistance to a wide range of antibiotics (5).
Due to high prevalence of MRSA infections among hospitalized patients, rapid and accurate identification of MRSA is needed to initiate appropriate antimicrobial therapy and prevent the spread of MRSA infections. Usually, molecular methods such as detection of the mecA gene are preferred for this task because of high sensitivity and specificity. The results of molecular methods are also usually available faster than that of phenotypic methods (7).
Different phenotypic methods are available in clinical laboratories such as oxacillin and cefoxitin disc diffusion test, oxacillin agar screening test, and determination of minimum inhibitory concentration (MIC) for oxacillin and cefoxitin. However, the expression of resistance is affected in variant conditions such as difference in temperature, medium, inoculum size and NaCl concentration in the medium (8). In this study, we aimed to compare PCR of the mecA gene with three phenotypic methods including cefoxitin disc diffusion test, oxacillin disc diffusion test and MIC of oxacillin for detection of MRSA.
MATERIALS AND METHODS
Isolation and identification of S. aureus
From December 2012 to April 2014, we collected 120 S. aureus isolates from three hospitals affiliated with Tehran University of Medical Sciences. Samples were identified and confirmed by conventional biochemical tests (9). Control strains for methicillin-resistant and –susceptible S. aureus were COL and ATCC 8325-4, respectively.
Phenotype identification of MRSA
We performed phenotypic methods for detection of MRSA strains according to clinical and laboratory standards Institute (CLSI) guideline as follows:
Cefoxitin and oxacillindisc diffusion tests were done using cefoxitin (30μg) and oxacillin (1μg) discs purchased from MAST Company (UK). Müeller-Hinton agar (MHA) plates containing 2% NaCl were inoculated with broth suspension equivalent to 0.5McFarland. Discs were applied on to the plates and incubated at 35°C for 24h. The zone inhibitions were measured and interpreted according to CLSI guideline (10).
For oxacillin MIC strip test, isolates were cultured on MHA containing 2% NaCl and oxacillin strip was placed on the medium and incubated for 24h at 35°C. After incubation, inhibitory concentration studied and interpreted according to the CLSI criteria (10).
Genotype identification of MRSA by PCR
We extracted genomic DNA using Viogene kit (UK) based on manufacturer’s instructions. We used the extracted genomic DNA as template for PCR of the mecA gene. Forward (5’-TCC AGA TTA CAA CTT CAC CAG G-3’) and reverse (5’-CCA CTT CAT ATC TTG TAA CG-3’) primers were used to amplify the 162 bp mecA gene of MRSA as described previously (Fig. 1) (11). Each PCR mixture was composed of 2 μl DNA template, 0.5 μl of each primer (10 μM), 12.5 μl master mix (SinaClon, Iran), and 9.5 μl sterile distilled water. PCR program began with an initial denaturation step at 97°C for 6 min followed by 30 cycles of 92°C for 30 seconds, 55°C for 30 seconds, and 72°C for 45 second, and ended with a final extension step at 72°C for 10 min. The mecA-positive strain COL and the mecA-negative ATCC8325-4 were included as positive and negative controls, respectively. The amplified PCR products were electrophoresed in 1% agarose gel at 120 V for 1h, stained with KBC (0.5 μg/ml) (Kawsar,Iran), and photographed under UV light.
Fig. 1.
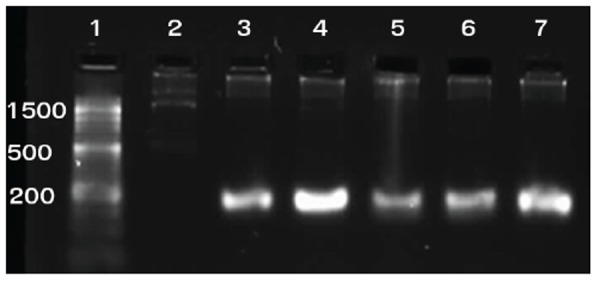
1%Agarose gel electrophoresis of the PCR-amplified mecA methicillin resistance gene. Lanes: 1: 50-bp ladder; 2: Negative control (S. aureus ATCC 8325-4); 3: Positive control (S. aureus strain COL); 4-7: S. aureus isolates showing 162 bpmecAamplicon
RESULTS
All S. aureus isolates were subjected to MRSA detection by four phenotypic methods. The PCR assay targeting mecA gene and cefoxitin disc diffusion identified 60 (50%) isolates as MRSA. Forty-eight MRSA (40%) were identified by oxacillin disc diffusion method, including the intermediate zones (Table 1). MIC test strip found 55 (45.83%) MRSA phenotype with MIC between 8-16 μg/ml. The remaining five MRSA strains were only identified by cefoxitin disc diffusion and had an MIC of oxacillin between 0.125-0.5 μg/ml (Table 2).
Table1.
Comparison of various laboratory methods for detection of methicillin-resistant Staphylococcus aureus isolates in this study
| Method | Specificity | Sensitivity | PPV | NPV |
|---|---|---|---|---|
| mecA gene | 100% | 100% | 100% | 100% |
| Cefoxitin disc | 100% | 100% | 100% | 100% |
| Oxacillin disc | 100% | 80% | 100% | 83% |
| Oxacillin strip | 100% | 91.6% | 100% | 92% |
PPV= Positive Predictive Value; NPV= Negative Predictive Value
Table 2.
Discordant results of minimum inhibitory concentration of oxacillin and PCR of mecA
| Isolate number | MIC (μg/dL) of Oxacillin | mecA |
|---|---|---|
| TTH-3-25 | 0.5 | + |
| TTH-1-17 | 0.25 | + |
| TTH-3-11 | 0.25 | + |
| TTH-3-8 | 0.25 | + |
| TTH-3-3 | 0.125 | + |
TTH: Tehran Teaching Hospital; MIC: Minimum Inhibitory Concentration
DISCUSSION
In recent years, detection of mecA by PCR is considered as the gold standard for identification of MRSA. In this study, we evaluated other methods as alternatives to PCR (12). Cefoxitin disc diffusion test was perceived to be the most sensitive method for detection of mecA-mediated resistance. CLSI has also recently substituted the oxacillin disc with cefoxitin disc for detection of MRSA (13). Numerous studies including the current one have informed that the results of the cefoxitin disc diffusion test correlates better with the presence of mecA compared with those of the oxacillin disc diffusion test (14-16).
Our results about cefoxitin disc diffusion method are consistent with previous report (15). However, Broekeme et al., reported the sensitivity and specificity of this method 97.3% and 100%, respectively among 1,611 S. aureus isolates (16).
In current study, MIC strip test showed the sensitivity and specificity about 91.6% and 100%, respectively. In the study of Rahbaret al., sensitivity and specificity were both 100% (17). Five isolates in our study showed discordant results for MIC of oxacillin and PCR. This can be probably explained by the fact that not all S. aureus isolates express their mecA gene (18). In our study, the sensitivity and specificity of oxacillin disc diffusion test were 80% and 100%, respectively. In the study of Farahani et al., the sensitivity and specificity of the oxacillin disc diffusion method was 100 and 73.6%, respectively (19). In previous study that performed by Pillaiet al., the sensitivity and specificity were reported 93.5% and 83.5%, respectively (20).
In conclusion, the present study showed that cefoxitin disc diffusion has both high sensitivity and specificity as compared with mecA PCR. Therefore, itcan be a good alternative to molecular methods due to its low cost for clinical laboratories.
Acknowledgments
The authors wish to express their gratitude to research council of Tehran University of Medical Sciences, Iran, for financial support (Grant number 26590).
References
- 1.Mediavilla JR, Chen L, Mathema B, Kreiswirth BN. Global epidemiology of community-associated methicillin resistant Staphylococcus aureus (CA-MRSA) Curr Oppin Microbial. 2012;15:588–595. doi: 10.1016/j.mib.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 2.Stafani S, Chung DR, Lindsay JA, Friedrich AW, Kearns AW, Westh H, et al. Meticillin-Resistant Staphylococcus aureus (MRSA): Global epidemiology and harmonisation of typing methods. Int J Antimicrobial Agent. 2012;39:273–283. doi: 10.1016/j.ijantimicag.2011.09.030. [DOI] [PubMed] [Google Scholar]
- 3.Cosqrove SE, Qi Y, Kays KS, Herbarth S, Karchmar AW, Carmeli Y. The impact of methicillin resistance in Staphylococcus aureus bacteremia on patient outcomes: mortality, length of stay, and hospital charges. Infect ontrol Hosp Epidemiol. 2005;56:166–174. doi: 10.1086/502522. [DOI] [PubMed] [Google Scholar]
- 4.Deurenberg R, Vink C, Kalanic S, Fariedrich AW, Bruggeman CA, Stobberingh EE. The molecular evolution of methicillin-resistant Staphylococcus aureus. Clin Microbiol Infect. 2007;13:222–235. doi: 10.1111/j.1469-0691.2006.01573.x. [DOI] [PubMed] [Google Scholar]
- 5.Grundmann H, Aires-de-Sousa M, Boyce J, Tiemersma E. Emergence and resurgence of meticillin-resistant Staphylococcus aureusas a public-health threat. Lancet. 2006;368:874–85. doi: 10.1016/S0140-6736(06)68853-3. [DOI] [PubMed] [Google Scholar]
- 6.Kaier K, Hagist C, Frank U, Conrad A, Meyer E. Two time-series analyses of the impact of antibiotic consumption and alcohol-based hand disinfection on the incidences of nosocomial methicillin-resistant Staphylococcus aureus infection and Clostridium difficile infection. Infect Control Hosp Epidemiol. 2009;30:346–53. doi: 10.1086/596605. [DOI] [PubMed] [Google Scholar]
- 7.Barski P, Piechowicz L, Galinski J, Kur J. Rapid assay for detection of methicillin-resistant Staphylococcus aureus using multiplex PCR. Mol Cell Probes. 1996;10:471–475. doi: 10.1006/mcpr.1996.0066. [DOI] [PubMed] [Google Scholar]
- 8.Afrough P, Pourmand MR, Zeinalinia N, Yousefi M, Abdossamadi Z, Yazdchi S. Molecular typing of clinical and nasal carriage isolates of staphylococcus aureus by spa gene patterns. J Mazand Univ Med. 2012;22:28–34. [Google Scholar]
- 9.Clinical and Laboratory Standards Institute. 23rd informational supplement M100-S23. CLSI; Wayne, PA: 2013. Performance standards for antimicrobial susceptibility testing. [Google Scholar]
- 10.Pourmand MR, Abdossamadi Z, Salari MH, Hosseini M. Slime layer formation and the prevalence of mecA and aap genes in Staphylococcus epidermidis isolates. J Infect Dev Ctries. 2011;5:34–40. doi: 10.3855/jidc.984. [DOI] [PubMed] [Google Scholar]
- 11.Vogelaers D. MRSA: total war or tolerance? Nephrol Dial Transplant. 2006;21:837–8. doi: 10.1093/ndt/gfl058. [DOI] [PubMed] [Google Scholar]
- 12.Bhutia KO, Singh TS, Biswas S, Adhikari L. Evaluation of phenotypic with genotypic methods for species identification and detection of methicillin resistant in Staphylococcus aureus. Int J Appl Basic Med Res. 2012;2:84–91. doi: 10.4103/2229-516X.106348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adaleti R, Nakipoglu Y, Karahan ZC, Tasdemir C, Kaya F. Comparison of polymerase chain reaction and conventional methods in detecting methicillin-resistant Staphylococcus aureus. J Infect Dev Ctries. 2008;2:46–50. doi: 10.3855/jidc.321. [DOI] [PubMed] [Google Scholar]
- 14.Cauwelier B, Gordts B, Descheemaecker P, Landuyt H. Evaluation of a disk diffusion method with cefoxitin (30 μg) for detection of methicillin-resistant Staphylococcus aureus. Eur J Clin Microbiol Infect Dis. 2004;23:389–92. doi: 10.1007/s10096-004-1130-8. [DOI] [PubMed] [Google Scholar]
- 15.Ananl K, Agrawal P, Kumar S, Kapila K. Comparison of cefoxitin disc diffusion test, oxacillin screen agar, and PCR for mecA gene for detection of MRSA. Indian J Med Microbiol. 2009;27:27–29. [PubMed] [Google Scholar]
- 16.Broekema NM, Van TT, Monson TA, Marshal SA, Warshauer DM. Comparison of cefoxitin and oxacillin disk diffusion methods for detection of mecA-mediated resistance in Staphylococcus aureus in a large-scale study. J Clin Microbiol. 2009;47:217–219. doi: 10.1128/JCM.01506-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rahbar M, Yaghoobi M, Fattahi A. Comparison of different laboratory methods for detection of methicillin resistant Staphylococcus aureus. Pak J Med Sci. 2006;22:442–445. [Google Scholar]
- 18.Ryffel C, Kayser FH, Berger Bachi B. Correlation between regulation of mecA transcription and expression of methicillin resistance in staphylococci. Antimicrobial Agent Chemother. 1992;36:25–31. doi: 10.1128/aac.36.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farahani A, Mohajeri P, Gholamine B, Rezaei M, Abbasi H. Comparison of different phenotypic and genotypic methods for the detection of methicillin-resistant Staphylococcus aureus. N Am J Med Sci. 2013;5:637–640. doi: 10.4103/1947-2714.122305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pillai MM, Latha R, Sarkar G. Detection of methicillin resistance in Staphylococcus aureus by polymerase chain reaction and conventional methods: A comparative study. J Lab Physicians. 2012;4:83–88. doi: 10.4103/0974-2727.105587. [DOI] [PMC free article] [PubMed] [Google Scholar]


