Abstract
The bacteriophage Mu genome contains a centrally located strong gyrase site (SGS) that is required for efficient prophage replication. To aid in studying the unusual properties of the SGS, we sought other gyrase sites that might be able to substitute for the SGS in Mu replication. Five candidate sites were obtained by PCR from Mu-like prophage sequences present in Escherichia coli O157:H7 Sakai, Haemophilus influenzae Rd, Salmonella enterica serovar Typhi CT18, and two strains of Neisseria meningitidis. Each of the sites was used to replace the natural Mu SGS to form recombinant prophages, and the effects on Mu replication and host lysis were determined. The site from the E. coli prophage supported markedly enhanced replication and host lysis over that observed with a Mu derivative lacking the SGS, those from the N. meningitidis prophages allowed a small enhancement, and the sites from the Haemophilus and Salmonella prophages gave none. Each of the candidate sites was cleaved specifically by E. coli DNA gyrase both in vitro and in vivo. Supercoiling assays performed in vitro, with the five sites or the Mu SGS individually cloned into a pUC19 reporter plasmid, showed that the Mu SGS and the E. coli or N. meningitidis sequences allowed an enhancement of processive, gyrase-dependent supercoiling, whereas the H. influenzae or Salmonella serovar Typhi sequences did not. While consistent with a requirement for enhanced processivity of supercoiling for a site to function in Mu replication, these data suggest that other factors are also important. The relevance of these observations to an understanding of the function of the SGS is discussed.
Bacteriophage Mu has proved to be an invaluable tool for dissecting the mechanism of replicative transposition. By virtue of the Mu life cycle, whereby the phage integrates into the host chromosome and replicates via multiple transposition events during the lytic cycle, this process can be studied en masse in a bacterial population (25). Upon infection of E. coli, the 37-kb linear Mu genome—albeit with its ends bound together by the coinjected Mu N protein—enters the cytoplasm and integrates at an essentially random location within the host chromosome, or nucleoid. During the lytic cycle, 100 or so copies of Mu are generated by replicative transposition, leading to the accumulation of genome units spread throughout the chromosome. These are then excised and packaged prior to host lysis.
Details of the transposition reaction at the molecular level have been supplied by extensive in vitro studies (3). The obligatory first steps in Mu replication are the formation of a series of synaptic complexes comprising the left and right ends of the integrated genome bound to copies of pA, the phage-encoded Mu A transposase. Transposase monomers bind and synapse the Mu ends and an internal enhancer located near the left end, initially forming the unstable LER (left end-enhancer-right end) complex. Rearrangement of transposase monomers into a stable tetramer and the transposase-bound ends into a plectonemically interwound supercoiled form results in formation of the type 0 complex and the trapping of five negative supercoils (24). Single-strand nicks or cleavages are introduced at the Mu ends forming a type 1 complex, with the nicks introduced by transposase in trans, i.e., pA bound at the Mu left end cleaves at the right end and vice versa. A concerted attack of the exposed 3′ hydroxyls at the Mu ends on a target site bound by the Mu B protein leads to strand transfer and formation of the type 2 complex. To complete a round of replicative transposition following the strand transfer reaction, the firmly bound pA tetramer is remodeled by the action of ClpX protease and other factors in preparation for binding of the host replication machinery.
Given the large size (37 kb) of a Mu prophage and the topological complexity involved in properly aligning the Mu ends within the confines of the bacterial nucleoid, we proposed that Mu has evolved a novel and active mechanism to overcome these barriers to synapsis, one that exploits the supercoiling activity of the host enzyme DNA gyrase (29). The model proposes that gyrase binds at the center of the Mu genome and by efficient processive supercoiling leads to the extrusion of a novel Mu-containing domain in the nucleoid. A centrally located gyrase, acting in this fashion, would direct a natural alignment of the Mu ends, thus facilitating the rapid formation of the LER complex, consequent DNA replication, and subsequent host lysis. In support of this model, a site near the center of the genome, within the 0.2-kb intergenic region between the Mu G and I genes, was identified as a strong gyrase site (SGS) (29). Deletion of the SGS leads to a very significant delay in Mu replication. Complementary studies have revealed a similar delay to replication of wild-type Mu when the host gyrase activity is disabled (33). More recently, we have demonstrated that the SGS sequence alone (when present in a reporter pUC19 plasmid) is able to modulate the activity of DNA gyrase in vitro, where it imparts a raised efficiency and processivity to the supercoiling reaction (20).
Several observations suggest that the function of the SGS is unusual or unique and that it represents a novel and subversive mechanism whereby gyrase activity is exploited by Mu. No other gyrase site that we have studied—including the strong gyrase sites from pBR322 or pSC101 (5, 12, 19, 36) or BIME elements (4)—can fully supply the function(s) of the SGS in vivo (28). Lysogens carrying a chimeric prophage, with the SGS replaced by other gyrase sites, exhibit large delays to host lysis, equivalent in most cases to that observed with deletion of the SGS. To understand the features of the SGS that are responsible for its unusual properties, it would be extremely useful to have other examples of gyrase sites that share these features. The availability of numerous bacterial genome sequences has revealed Mu-like prophage sequences present in various bacterial species, which could be potential sources of Mu SGS homologues (6, 8, 11, 14, 16, 22, 23, 35). We have obtained such candidate sequences from five Mu-like prophages by PCR and characterized the extent to which these sequences are able to supply the function of the Mu SGS in vivo and the effects of each of these sequences on DNA gyrase activity.
MATERIALS AND METHODS
Proteins, enzymes, and drugs.
DNA gyrase was kindly supplied by A. J. Howells of the John Innes Centre (Norwich, United Kingdom) and purified as described previously (15). Throughout this report, DNA gyrase concentrations are given in terms of the A2B2 tetramer. Enoxacin (a gift from N. P. Higgins) was dissolved in equimolar NaOH and stored at −20°C. Chloroquine phosphate was freshly dissolved in gel running buffer and used immediately.
Construction of DNA substrates.
Genomic DNA samples were generously supplied by the following sources: Haemophilus influenzae Rd KW20 from R. Redfield (University of British Columbia, Vancouver, Canada), Neisseria meningitidis serogroup A strain Z2491 from M. Achtman (Max Planck Institute, Berlin, Germany), Escherichia coli O157:K7 Sakai from K. Makino (Osaka University, Osaka, Japan), and Salmonella enterica serovar Typhi CT18 from D. Packard (Imperial College, London, United Kingdom). Oligonucleotide primers for PCR all contained an 8-bp leader sequence carrying a BamHI site for cloning purposes. Primers HI+ (5′ CGG GAT CCG GCG TCA GCG AAA TTA AAG C) and HI− (5′ CGG GAT CCC GGT GAG TAC CGC TAA AGA GG) amplified nucleotides 1,577,480 to 1,577,806 of the H. influenzae Rd KW20 sequence (6). Similarly, Nm3+ (5′ CGG GAT CCG ACA AAC AGG CTT TGA TGG ACG) and Nm3− (5′ CGG GAT CCG GCT GCA CCT CGA AAC TGC) gave DNA from nucleotides 1,226,322 to 1,226,662 of the N. meningitidis A Z2491 sequence (22), while Nm8+ (5′ CGG GAT CCG GCG GAA TAT CTG GCA CAG G) and Nm8− (5′ CGG GAT CCT CCA TAG CCC GCA T) gave the region from 1,787,998 to 1,788,246 of the same genome. ECs+ (5′ CGG GAT CCG GCA GAG CAG GAG ATC ATG G) and ECs− (5′ CGG GAT CCG GCA TTC AGA ATG GCA TAA GC) yielded the region 5,059,042 to 5,059,329 of E. coli O157:K7 Sakai (8), and finally Sty+ (5′ CGG GAT CCG TCC GCC ATC TTG AAT CAG C) and Sty− (5′ CGG GAT CCG CCG TGC ATA TCG GTA TGA G) gave nucleotides 1,554,709 to 1,555,006 of the Salmonella serovar Typhi CT18 chromosome (23). PCRs were performed by standard methods using Taq polymerase and approximately 1 ng of purified genomic DNA as a template. PCR products were purified from agarose gels and were sequenced to verify that no mutations had been introduced. Singly radiolabeled molecules were obtained by including one 5′ end-labeled primer (generated from pretreatment with polynucleotide kinase and [γ-32P]ATP) in the PCR. Three-kilobase substrates were obtained by ligating each PCR product into HincII-cut pUC19. Supercoiled DNA forms were purified from cesium chloride-ethidium bromide gradients; relaxed forms were obtained by subsequent treatment with calf thymus topoisomerase I and linear forms by cleavage with AhdI.
Construction of chimeric prophages.
The parent strain for all lysogens was E. coli AB1157 recB recC sbcB malF::Mu cts62 (29). The PCR fragments described above were cleaved with BamHI and cloned into the BglII site of plasmid pMP1856 (28): this carries a large central fragment of the Mu genome (BamHI-EcoRI; 17.2 to 23.7 kb), a deletion of the SGS, and an oligonucleotide inserted at the site of the deletion that contains a BglII site. Plasmid DNA was linearized and electroporated into strain MP1928 (28), containing a Mu prophage (in the malF locus) with the SGS replaced with a 3.5-kb fragment bearing the sacBR genes and a kanamycin resistance marker. A fraction of the pMP1856 constructs recombined with the MP1928 Mu prophage and double recombinants were identified by selection for sucrose resistance and screening for kanamycin sensitivity.
Growth, lysis, and replication assays.
Cultures of lysogens were grown in L broth at 30°C to a density of about 1 × 108 cells/ml, diluted twofold with L broth, and induced by transferring the culture to 42°C. Culture density was monitored with Klett readings to monitor growth and lysis. Replication was monitored using a semiquantitative PCR procedure (28). Pairs of PCR primers were used to simultaneously amplify a fragment of Mu DNA and a fragment of chromosomal DNA (from the malK gene adjacent to the prophage at malF) in PCRs designed to yield results in a linear range. The amounts of the two PCR fragments were determined after separation on agarose gels, and the data were expressed as the ratio of Mu to malK DNA. The ratio is reproducibly close to 1.0 in uninduced cells and increases after the onset of Mu DNA replication.
Gyrase cleavage in vivo.
Gyrase cleavage assays in vivo were performed as described previously (28), following a modified version of an established protocol (32). Cultures of lysogens were incubated with 200 μg of enoxacin/ml for 5 min and rapidly lysed at 80°C in 0.25% sodium dodecyl sulfate (SDS). After treatment with proteinase K, DNA was purified and digested with an appropriate restriction enzyme. A 32P 5′ end-labeled oligonucleotide primer, complementary to a chromosomal sequence such that the gyrase site falls between the primer site and the restriction site, was used for 30 cycles of iterative primer extension. The extension products thus terminated either at the cleaved gyrase site (for templates cleaved by gyrase in vivo) or at the cleaved restriction site (for templates not cleaved by gyrase). The resulting labeled fragments were separated on an 8% polyacrylamide sequencing gel, and the percentage of cleaved molecules in the population was quantified with a phosphorimager.
Gyrase assays in vitro.
Cleavage reactions (final volume, 10 μl) of 0.3-kb radiolabeled DNA molecules contained 35 mM Tris-HCl (pH 7.5), 20 mM KCl, 5 mM MgCl2, 1.8 mM spermidine, 0.1 mg of bovine serum albumin/ml, 5 mM dithiothreitol, 2.5% (vol/vol) glycerol, 150 μM enoxacin, 1 to 2 nM each PCR fragment, and increasing concentrations of DNA gyrase (typically up to a 20-fold molar excess); reactions were incubated at 37°C for 20 min. SDS to 0.1% and proteinase K to 0.1 mg/ml were added, followed by a further 15-min incubation at 37°C. Samples were resolved on 8% polyacrylamide gels. Gyrase cleavage sites were mapped at the nucleotide level by resolving the cleavage reaction products on denaturing 6% polyacrylamide gels, alongside appropriate di-deoxy sequencing ladders obtained using the Promega fmol DNA cycle sequencing kit. Cleavage reactions with 10 nM linear 3-kb plasmid DNA and up to 200 nM DNA gyrase were performed similarly: these reactions were prepared either as above or with 5 mM CaCl2 and 1.5 mM ATP replacing 5 mM MgCl2 and 150 μM enoxacin, respectively. Supercoiling reactions comprised 35 mM Tris-HCl (pH 7.5), 24 mM KCl, 6 mM MgCl2, 1.8 mM spermidine, 5 mM dithiothreitol, 6% (vol/vol) glycerol, 0.5 mg of bovine serum albumin/ml, 10 μg of tRNA/ml, 1.5 mM ATP, 10 nM relaxed plasmid DNA, and various concentrations of DNA gyrase (final volume, 20 μl). Reactions were incubated at 37°C for 30 min, followed by addition of SDS to 0.1%, EDTA to 20 mM, and proteinase K to 0.1 mg/ml. Incubation was continued an additional 30 min at 37°C. DNA products were resolved on 1.1% agarose-chloroquine gels (30). The running buffer comprised 80 mM Tris-HCl (pH 7.8), 25 mM phosphoric acid, 8 mM EDTA, with up to 45 μg of chloroquine/ml (freshly dissolved in the running buffer). Gels were developed at 4°C at 3 V/cm for 16 h, with constant buffer recircularization (∼1 tank volume/h). Gels were washed extensively with several changes of water (2 h) and 2 mM MgSO4 (30 min) and stained in 0.5-μg/ml ethidium bromide and 2 mM EDTA solutions (3 h). The final destaining wash comprised 2 mM MgSO4 (30 min). DNA bands were visualized by transillumination.
RESULTS
Sources of candidate SGS homologues.
The Mu SGS is the only gyrase site we have thus far identified that has the properties required for promoting efficient replication of a Mu prophage. To determine whether the SGS is unique or whether other sites exist that share this unusual property, and to unravel the molecular mechanism responsible for this activity, it would be extremely useful to isolate additional SGS-like sites. Therefore, as an approach to search for sequences that might substitute for the Mu SGS, we used DNA and protein BLAST searches to scan the sequence databases for homologues of the Mu G and I genes, those that flank the SGS in the Mu genome. This approach identified several bacterial species that contain homologues of the G and I proteins, with the corresponding intergenic regions being around 0.2 kb, as summarized in Table 1. One candidate SGS-like site was present in H. influenzae Rd (6) (this 0.2-kb sequence herein abbreviated as HI), one in E. coli O157:H7 Sakai (8) (abbreviated ECs), two in N. meningitidis serogroup A strain Z2491 (22) (abbreviated NMA1 and NMA2, the former identical to a prophage sequence from serogroup B strain MC58) (35), and the fifth in Salmonella serovar Typhi CT18 (23) (abbreviated Sty).
TABLE 1.
Homologues of Mu pG and pI and the G-I intergenic regiona
| Organism | pG (aa) | G-I (bp) | pI (aa) |
|---|---|---|---|
| E. coli K12 (Mu G-I) | 156 | 196 | 361 |
| E. coli O157:H7 (ECs 4972-4973) | 157 (3e-29) | 176 | 374 (1e-75) |
| H. influenzae (HI 1503-1504) | 146 (2e-12) | 236 | 355 (1e-75) |
| Salmonella serovar Typhi (Sty 1618-1619) | 152 (2e-04) | 214 | 371b |
| N. meningitidis (NMA 1316-1317) | 138 (0.014) | 206 | Truncatedc |
| N. meningitidis (NMA 1849-1850) | 165 (0.017) | 208 | 354 (5e-26) |
| N. meningitidis (NMB 1097-1098) | 138 (0.014) | 206 | 353 (1e-15) |
Numbers in brackets following each organism name are the gene names or numbers from the relevant annotated genomes. Numbers in the second and fourth columns give the sizes of the G and I protein homologues in amino acid (aa) residues. The figures in parentheses are the BLAST e-values for comparison with the corresponding Mu proteins. The sizes of the intergenic regions are given in the third column.
No e-value was obtained for pI in Salmonella serovar Typhi, which may not be homologue of Mu pI.
The NMA1317 pI homologue is truncated by a frameshift mutation, although the sequence for the G homologue, the 206-bp intergenic region, and the extant portion of the truncated pI are identical to the corresponding regions of NMB1097-1098.
In four out of five cases (the exception being the Sty prophage) the corresponding open reading frames for each G and I homologue resided in large (approximately 35- to 40-kb) chromosomal regions which have been assigned to the Mu-like family of prophage sequences, based on gene order alignments (2). The four G-I intergenic regions are located near the center of the encompassing prophage sequences, despite the existence of numerous insertions and deletions relative to the Mu sequence. In addition, each prophage carries an open reading frame for a protein with similarity to Mu A transposase. These four sequences were thus obvious candidates to test as sites that might have an effect on Mu replication. The case for the Sty sequence is less clear, as the downstream gene may not be an I homologue (Table 1) and there is much less overall resemblance to the Mu prophage. Nevertheless, we chose to include it in our studies. Primers were designed to amplify from the genomic DNA in all five cases a region from within the 3′ end of the G homologue, the intergenic region in its entirety, followed by a 5′ fragment of the I homologue. This yielded five DNA fragments around 0.3 kb in length (Table 2) which were used in the following experiments.
TABLE 2.
G-I intergenic regions
| Region | Sequencea |
|---|---|
| Mu G-I | aaacgcgtcagcgccgctctgaggcaataaACAGAATCAGGCATAAAATCAGCCGCACAGATTTTTTAAAACGCGCCACGGGATTTTTAAACCGGTATTTAACGGTGTATGAATCCCGTTTTATCTTCCTTTCACTTTCTTTCTCCAGTACTCAAATAGCATAACCCCAGATTTTCCCGCACCTCCCGCAAACTGACTGCTCACAAACCATGATGAGCAGCAGGACatgaaaaaac |
| E. coli O157 Sakai (ECs 4972-4973) | cgctttaaaaaggccacagaaacgccttaaTTGTTTAAGGTATGCAAATGCATTACCTTACCCCCTTCAGGCGCGTGTCGTGATTTTCTAACCTGTATTTAACGGGCTTTAAAATCTGCGTGAAGCGCCTTTGTATTTTTCCCCTGGAAGCCACTTCCTGAAATCCCGCAAAACCCGCCCTGAAAATTCCCGGCCTATGCTGCCGGAatgaagacg |
| H. influenzae (HI 1503-1504) | aaccgagagttgtcggaattaactcaagaaTAGGCTAAAATCGCAAATATCGCCACAGAATCACGCTAAGGGCGTTTGAAAATCAATTTATATCATTTTACGTCTCAAAAAATTTAAACGTGCTTAAACGCATTTAAACGGCATTTAAACGCTATTCTAATTTAAACCTTTCTCTAATTTTCCACCCCTGACTTTCAGCGTTTTATTTTAGGCTGATATATCACAAATCCGCCCTTCCCTCCCTTTACCGCAAAATAGCGGTTATGatgaaagcag |
| Salmonella serovar Typhi (Sty 1618-1619) | cgccatcttgaatcagcggcccgtcgctgaGTTTTCTCTCTTCAGGTGGGTGATTTATCATTGCCAGCGAATGAGGGGCTGTATTACCTTTATAAAGGCTTTACAGCCTCTGTTTTATAACCGCCTCCGGTTCACCGCATTGCTTTCCCTGTCCTTCTCCCCTGATGTTTTCTAAAGCAGATTAAAATCGCCGGGCCTGCATTTCTCACAAACTGTCTCCGACAACATAACGCGGGACAGCAAAatgtcagcca |
| N. meningitidis (NMA 1316-1317) | taggattatttttcgggtctgataccgtgaATTTATAAAACCCTCAAAACGCGCTTTTTAGCGCGTTTTTTTATGCGGGTAATACAAACCCCTGCCCAAGATATAAAAATCAATCCTAGACGCTTCTAAAAAGCCCCTGAAAAGATTAATGTGTATCGCGCGGACAGGTTTTAAAAAAATGGCGGGAGGGTTTGAAGCACGCCTACTCTTTGTTGTTTTTTCAAATAGGCAAAatgtccaaaa |
| N. meningitidis (NMA 1849-1850) | tatctggcacaggccattcaggggcggtaaTCATGCCTGCCTGAAAAATGCCGAAAAACAGCCCGAAAACGCCCTAACCCATACTTACCCCTACCCATCGCCTGAAAATCAATCCTGCGCGCGTTTGAACACCTTTTGAACACTATCCCACGCGTATCTCTGCAAGTACATTTCCCTTCCCTAGAAATTCATCTGTCTGACATAGGCCAAGCTTTGACGAGGCGCGGCTGCCGCACAatgcgggcta |
Each sequence is shown 5′ to 3′. The end of the coding sequences for the G gene homologues and the start of the coding sequences for the I gene homologues are given in lowercase letters. The intergenic regions are given in uppercase letters. In these regions, underlined letters indicate the locations of the DNA gyrase cleavage sites. The E. coli O157 site shown comprises two overlapping gyrase sites, the first (site A) spanning the sequence GCAA and the second (site B) spanning AAAT. The Salmonella serovar Typhi sequence contains two separated sites, with site A lying closer to the G gene.
Effects of candidate SGS homologues on Mu replication and host lysis.
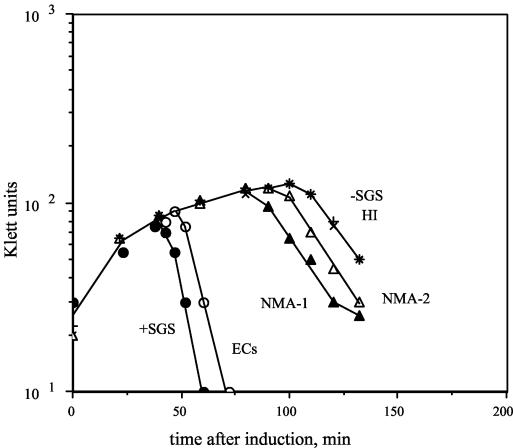
We wished to determine whether the candidate sequences are capable of supplying the functions required for promoting efficient prophage replication and host lysis. Therefore, to determine the biological effect(s) of the five DNA sequences, each fragment was used to replace the natural SGS in a Mu cts62 prophage, yielding five novel chimeric prophages. Cultures of lysogens carrying a Mu prophage, a mutant lacking the SGS, or chimeric prophages with the SGS replaced with each of the individual candidate sites were thermoinduced, and lysis profiles of the cultures and the kinetics of prophage DNA replication were determined. Representative lysis curves are shown in Fig. 1, and the corresponding lysis times are listed in Table 3. Mu-dependent lysis of the host was rapid with an intact SGS; however, in the absence of the SGS, lysis was delayed by over an hour. Replacement of the SGS with three of the candidate gyrase sites allowed more rapid lysis than that observed in the absence of the SGS. The ECs site allowed substantial improvement, the NMA1 site allowed intermediate improvement, and the NMA2 sites allowed a minimal improvement in lysis (Table 3). Prophages with the HI and Sty sites yielded lysis curves that were indistinguishable from that of the prophage lacking the SGS.
FIG. 1.
Host lysis induced by Mu and chimeric Mu prophages. Cultures of lysogens carrying either Mu cts62 or one of six recombinant prophages were grown in L broth at 30°C to a density of about 1 × 108 cells ml−1, diluted twofold in L broth, and then induced by a temperature shift to 42°C. The recombinant prophages either lacked the SGS altogether (−SGS) or carried one of five SGS homologue sequences in place of the natural SGS. Growth and lysis were monitored with Klett readings.
TABLE 3.
Estimated lysis times of lysogens bearing chimeric prophages
| SGS homologue | Time (min) |
|---|---|
| Mu + SGS | 45 |
| ECs | 55 |
| NMA1 | 80 |
| NMA2 | 95 |
| HI | 110 |
| Sty | 110 |
| Mu − SGS | 110 |
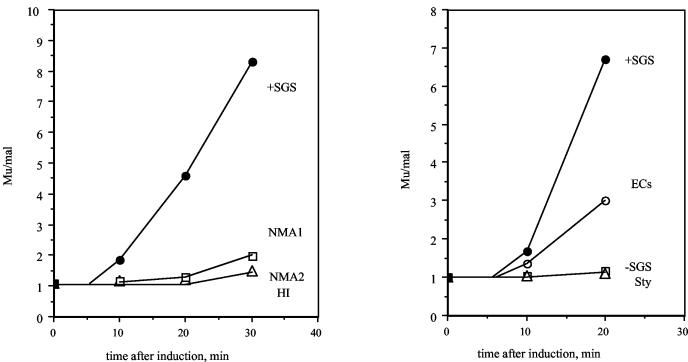
Replication of prophage DNA was examined with a semiquantitative PCR assay (28) that assesses the relative amounts of a sequence in Mu and a sequence in malK, adjacent to the prophage insertion site. Figure 2 shows that the kinetics of early replication are consistent with the lysis data (Fig. 1). During the initial 20 to 30 min after induction, the prophage with the ECs site showed extensive replication, although less than that with the wild-type SGS. The prophage with the NMA1 site showed a small increase in early replication compared to the prophage lacking the SGS, while, at these early time points, none of the other prophages showed increases in DNA replication.
FIG. 2.
Replication of Mu and chimeric Mu prophages. Cultures of lysogens were grown and induced as described for Fig. 1. At indicated times after induction, the cultures were assayed for the relative amounts of prophage DNA normalized to a control mal chromosomal locus by a semiquantitative PCR approach, as described in Materials and Methods. Two representative experiments are shown, encompassing data from the positive- and negative-control prophage sequences as well as the five recombinants.
Candidate SGS homologues are cleaved specifically by DNA gyrase in vitro.
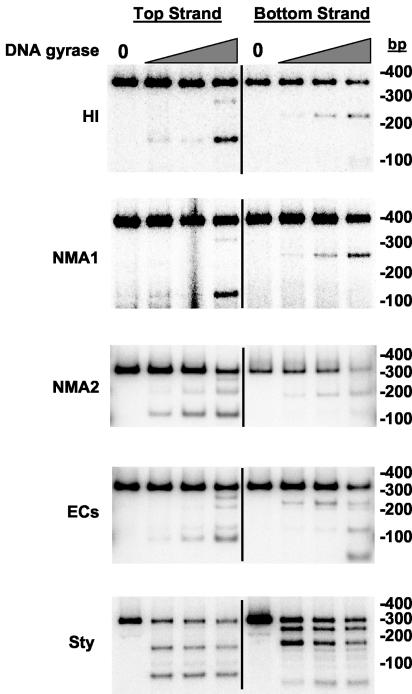
We wished to determine whether the five sequences indeed contained specific gyrase sites, particularly as three of the five showed some ability to supply the Mu SGS function in vivo. Therefore, we characterized the interaction of all five sequences with E. coli DNA gyrase by the following in vitro assays. During the supercoiling reaction gyrase binds a DNA segment, catalyzes the transient formation of a double-strand break in the bound DNA, or G segment, and passes a separate portion of DNA—the T segment—through the gap before religation of the break (10). The cleaved DNA intermediate can be trapped by quinolone drugs such as enoxacin (7, 34), and gyrase-dependent DNA cutting induced by these agents reveals the preferred sites of enzyme action on the DNA substrate. To determine the propensity of the sites to interact with DNA gyrase in vitro, each 0.3-kb PCR fragment was radiolabeled separately at the 5′ end of either the top or bottom strands and incubated with enoxacin and increasing concentrations of gyrase, to induce DNA cleavage. Figure 3 presents typical results. Both the HI and NMA1 fragments gave a single pair of bands at low gyrase levels, indicating a single preferred point of cleavage. At higher enzyme levels, a weaker secondary site was apparent. The NMA2 and ECs fragments gave a slightly more complex pattern, again with each possessing a primary cut site, but in addition each showed a greater tendency to be cleaved at other positions. The primary cut sites in these four sequences lie much closer to the 3′ end of the (putative) G gene than the start of the I gene (Fig. 3), a feature common to the location of the SGS primary cut site (29). Finally, the Sty DNA showed evidence of two similarly preferred points of cleavage. The gyrase cleavage sites in each fragment were subsequently mapped at the nucleotide level (Table 2). These in vitro studies provided confirmation that all five sites exhibited the 4-bp stagger characteristic of the gyrase cleavage reaction (17). A slightly more complex pattern was observed with the ECs fragment at the primary site of cleavage; however, here two closely overlapping cut sites (sites A and B) were apparent, separated by 2 bp (Table 2).
FIG. 3.
Enoxacin-induced cleavage of 0.3-kb PCR fragments by DNA gyrase. PCR fragments (ca. 2 nM) radiolabeled on either the top or bottom strands (indicated above each set of reactions in the topmost gel) were cleaved by DNA gyrase in the presence of enoxacin as described in Materials and Methods. DNA products were resolved on 8% polyacrylamide gels, and dried gels were exposed to phosphorimager screens. Control reactions lacking DNA gyrase are at the left of each set of reactions and indicated by a “0” above the relevant lanes. Increasing concentrations of DNA gyrase are represented by the grey triangles; each substrate is shown on the left of the gels and sizes of marker bands (in basepairs) on the right of the gels.
To determine whether gyrase would still cut at the five sites specifically even if presented with much longer flanking sequences, the 0.3-kb PCR fragments were cloned into HincII-cut pUC19, and 3-kb recombinant plasmids were obtained. Linear forms of these DNA molecules were similarly treated with gyrase in the presence of either enoxacin or calcium and ATP, the latter being conditions also able to promote gyrase-dependent DNA cutting (31). As in the case of equivalent experiments (20), discrete fragments were observed in all cases, once the reaction products were resolved on agarose gels (data not shown). Each plasmid gave rise to two pairs of bands, the exact intensities of which varied somewhat between each substrate or between each set of cleavage conditions. The first pair of subfragments arose from gyrase cleavage at the natural pUC19 site (20); the remaining bands were produced by specific cutting at the cloned sites. No other bands were produced. Thus, gyrase preferentially cut the DNA only at these locations, despite the availability of around 3 kb of plasmid DNA as a potential substrate. The results of this section, taken together, establish that each one of the sites forms a specific and efficient site for gyrase interaction.
In vivo gyrase cleavage of candidate SGS homologues.
While it is important to delineate gyrase sites with in vitro experiments as above, the biological properties of a site are determined by the in vivo interaction with gyrase. Therefore, we determined the location and extent of cleavage at the relevant sites in vivo after lysogens carrying the chimeric prophages were exposed to enoxacin for 5 min (29). Each site was cleaved at one or a few discrete locations in vivo, and these locations exactly matched the positions of gyrase cleavage in vitro for each of the five sequences. The calculated efficiency of cleavage at each of the sites is shown in Table 4. As observed previously, the wild-type Mu SGS was cleaved at a level of around 40% (28). By contrast, each of the candidate sites was cleaved less efficiently than the Mu SGS.
TABLE 4.
Efficiency of enoxacin-induced in vivo cleavage of SGS homologues
| SGS homologue | Molecules cleaved (% of total in culture)
|
|
|---|---|---|
| Expt 1 | Expt 2 | |
| Mu SGS | 42.5 | 43.0 |
| E. coli O157 (site A + B)a | 3.7a | 6.6a |
| H. influenzae | 3.3 | 2.6 |
| Salmonella serovar Typhi (site A) | 1.3 | 0.8 |
| Salmonella serovar Typhi (site B) | 4.3 | 4.5 |
| N. meningitidis NMA1 | 7.7 | 12.9 |
| N. meningitidis NMA2 | 0.4 | 2.0 |
Approximately equal amounts of cleavage at the ECs sites A and B were observed.
Effects of candidate SGS homologues on gyrase supercoiling.
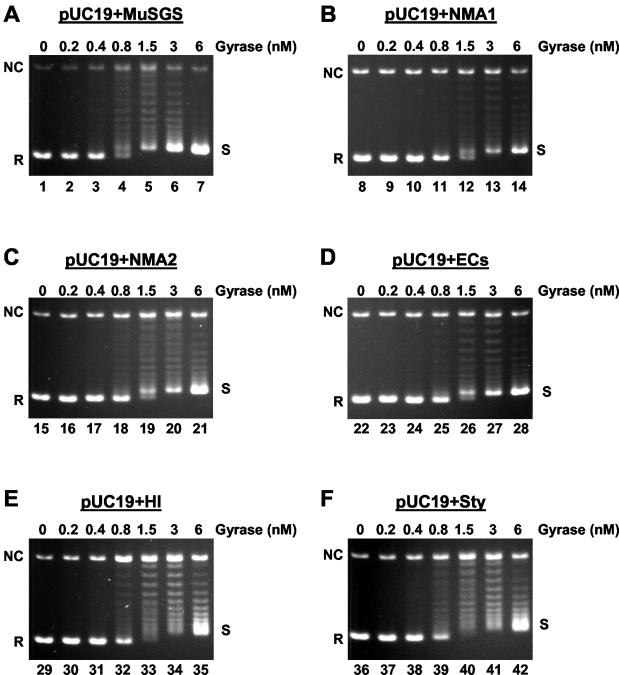
In previous work, we compared the properties of the Mu SGS and two well-studied gyrase sites from the plasmids pBR322 and pSC101 and observed that only the Mu SGS allowed highly efficient, processive supercoiling (20). We proposed that such an activity would be characteristic of any site that can promote efficient Mu replication. Therefore, we tested this prediction on the five candidate sites. To determine the effects of the candidate SGS homologues on gyrase supercoiling, recombinant pUC19 plasmids that contained the SGS homologues (along with a pUC19 plus Mu SGS construct and native pUC19 as controls) were obtained in relaxed form and used as substrates in gyrase supercoiling assays. Chloroquine-containing agarose gels were used to analyze the reaction products in all cases. On such gels, the relaxed substrate molecules run with high mobility due to intercalation of chloroquine. As the gyrase supercoiling reaction proceeds, the DNA molecules become more negatively supercoiled: this is seen as an initial decrease in the mobility of individual topoisomers under the gel conditions employed here. Once past a certain superhelical density, the plasmids run once more with increasing mobility, until at very high levels of supercoiling the topoisomers run as an unresolved band, almost but not quite with the mobility of the relaxed substrate.
Figure 4 shows results when equal amounts of each plasmid were incubated with increasing concentrations of DNA gyrase. The most efficient substrate for the enzyme was the SGS construct (Fig. 4A), in that less enzyme was needed to achieve comparable levels of supercoiling than was needed with the other substrates (Fig. 4, compare lane 4 with lanes 12, 19, and 26, for example). In addition, the majority of the pUC19 plus SGS DNA molecules were in one of two forms in this reaction even at low gyrase levels, either fully relaxed or fully supercoiled, and these two forms gave rise to a clear doublet of bands on the gels (e.g., Fig. 4A lane 4). This is a clear indication of processive supercoiling by the enzyme, where once bound, many supercoils are introduced into a substrate per binding event (18). A similar pattern was apparent with the three plasmids carrying the ECs, NMA1, or NMA2 sequences; once again, the greater part of the DNA was either relaxed or fully supercoiled (Fig. 4B to D). These are the three sites that could partially restore the biological function of the SGS.
FIG. 4.
DNA gyrase-catalyzed supercoiling of 3-kb plasmids. Each 0.3-kb SGS homologue, along with the Mu SGS as a control, was cloned into pUC19 to generate six 3-kb substrates as indicated above each panel. Relaxed forms of these molecules (ca. 10 nM) were incubated as described in Materials and Methods, with increasing concentrations of DNA gyrase. The products were resolved on 1.1% agarose gels containing 45 μg of chloroquine/ml. On the gels presented, relaxed forms of the substrates run with the greatest mobility, indicated by R on the left of each panel. NC shows the position of nicked circular molecules, and the fully supercoiled topoisomers, shown as S on the right of each panel, run as an unresolved band closely behind the relaxed substrate.
The plasmids with the HI and Sty sites, by contrast, gave a different pattern of topoisomers (Fig. 4E and F). In these cases most of the DNA was present in an intermediately supercoiled state at intermediate gyrase levels (Fig. 4E, lanes 33 and 34, and F, lanes 40 and 41). Such a pattern is indicative of a more distributive mode of supercoiling by gyrase. A similar pattern of distributive supercoiling was seen in control reactions with pUC19 alone (data not shown) or with plasmids carrying the pBR322 or pSC101 gyrase sites (20).
DISCUSSION
Gyrase sites from Mu-like prophages.
The presence of Mu-like prophage sequences in various bacterial strains and the availability of the corresponding genome sequences have enabled us to identify candidate homologues of the Mu SGS. These have been obtained by PCR, and a characterization of their biological effects in a Mu prophage and their interactions with DNA gyrase has been undertaken.
Two important points must be made initially about the candidate SGS homologues. First, the sites are present in more-or-less intact prophages, yet none has to date been shown capable of forming a viable phage. Therefore, the SGS-like sites may have degenerated and might not be fully functional. Second, our analyses of the sites from different bacterial strains were done with E. coli DNA gyrase. While gyrase function is highly conserved, e.g., mycobacterial gyrase cleaves the major pBR322 site at the same nucleotides as does E. coli gyrase (13), important differences may nevertheless exist. We cannot at present rule out the possibility that the experiments presented here have overlooked some subtlety of the interaction that each prophage sequence has with its cognate (i.e., host) gyrase. However, this issue is further complicated by the possibility that the prophages may be recent acquisitions by the various host strains. For example, the HI prophage has a GC content of about 50%, whereas the Haemophilus genomic DNA is about 38% GC (6, 16).
Given the above caveats, five candidate SGS-like sites have been identified and studied. All five sequences functioned as specific sites for DNA gyrase both in vitro and in vivo, although the extent of in vivo quinolone-induced cleavage at all five sites was significantly less than that at the authentic Mu SGS. The E. coli O157 Sakai sequence, when substituted for the SGS in a Mu prophage, allowed nearly normal levels of Mu replication and lysis of the induced lysogen, and the two sites from N. meningitidis species gave partial restoration of biological function. Mu prophages carrying the sites from H. influenzae and Salmonella serovar Typhi were indistinguishable from a prophage lacking the central SGS. In parallel supercoiling assays, only the three sites capable of (partial) substitution of SGS function gave an enhancement of processive supercoiling by gyrase. These results have allowed us to address questions relating to the function and singular nature of the Mu SGS and how it interacts with DNA gyrase to support efficient Mu replication.
Biological implications.
Recent studies have provided evidence that all double-stranded DNA bacteriophages share a common but very ancient origin (2). Extensive shuffling of genes appears to have occurred since, and now a mosaic pattern of genes is apparent between families of transposing and nontransposing phages, and within these families themselves (1, 2, 9). Four of the five sequences examined (the exception being the Sty sequence) are present on readily identifiable Mu-like prophages, although these differ, in part, by the presence of various deletions and insertions (2). The Sty sequence may be present on a prophage, but one with much less resemblance to Mu. Rather, it appears to possess Mu-like gene modules within an extensive mosaic pattern.
While we have no evidence that any of the other prophages examined here currently are capable of active transposition, they all probably confront, or once confronted, the problems faced by Mu during replicative transposition, i.e., possession of a large genome whose prophage ends must be efficiently synapsed with a defined topological orientation and within the confines of the bacterial nucleoid. It is then pertinent to ask if these share a similar gyrase-dependent mechanism to facilitate prophage DNA replication. No other transposing phage has been shown to exploit gyrase in the way that Mu does—except for D108, a phage so closely related to Mu itself that the example is trivial (28). However, we have now identified three novel prophage sites, the ECs, NMA1, and NMA2 sequences, which provide some ability to substitute for the Mu SGS. These sites may thus indicate that the Mu SGS is not unique and that other transposing phages utilize (or once utilized) a host topoisomerase to facilitate prophage replication.
Functional implications.
The complete mechanism by which Mu prophage termini are rapidly synapsed in the lytic cycle is still not fully known. We have proposed that highly processive supercoiling by gyrase bound at the SGS in the center of a Mu prophage promotes the formation of a plectonemically interwound, supercoiled loop, with the SGS at the apex and the prophage ends to be synapsed at the base (20, 27). One obvious point to make is that in addition to the action of gyrase at the SGS there could be additional host factors involved. While we cannot formally rule out such a possibility, the results of two lines of research suggest to us that the function of gyrase is of primary importance. Mutations in gyrase or the use of gyrase inhibitors are sufficient to eliminate efficient Mu replication (33), and the sequence of a given gyrase site can significantly affect the extent and the processivity of supercoiling in in vitro reactions with purified gyrase alone (20). In what follows, therefore, we focus on the role of the SGS in terms of its interaction with gyrase.
We recently proposed that any SGS-like site that can promote efficient Mu replication would impart enhanced processivity to the supercoiling activity of gyrase (20). The data presented here are consistent with this proposal, as only the three sites that gave this effect (ECs, NMA1, and NMA2) allowed observable decreases in the lysis times of the corresponding lysogens. However, while the extent of the enhancement to in vitro supercoiling by the ECs and the NMA sites was similar (Fig. 4), the sites had very different effects on lysis times (Table 3). Therefore, other aspects of the interaction of gyrase with the SGS-like sites are likely also of importance.
A second factor in determining the effectiveness of a site in supporting Mu replication and cell lysis is the efficiency of binding of gyrase to the different sites. This is particularly important as the affinity of the enzyme for supercoiled DNA is lower than for relaxed DNA and is anticipated to decrease as the superhelical density increases (17). Markedly different efficiencies of in vivo cleavage were observed with the different sites (Table 4), which might indicate differences in the efficiency of binding of gyrase. While the Mu SGS showed 40% cleavage under the conditions used, all the other sites showed significantly lower levels. These differences could help explain why the Mu SGS is the most efficient site in the biological assays. Similarly, the weak cleavage observed at the NMA2 site could help explain why this site, substituted for the SGS in a prophage, allowed only a minimal decrease of lysis time, even though it did allow increased processivity in the supercoiling reaction. However, while it is true to state that a high level of cleavage is indicative of efficient binding, it does not follow that a weak cleavage necessarily implies a weak binding. Cleavage involves an incompletely understood interaction between gyrase, DNA, and the quinolone (or other agent) such that the overall level of cleavage will be determined by the extent of binding, the particular cleavage agent, and the precise sequence of the site in question. For example, on a linear plasmid DNA the ECs site is cleaved less efficiently in vitro by gyrase than the other four candidate sites in the presence of enoxacin but with about equal efficiency with the use of Ca2+ (our unpublished data). Therefore, cleavage efficiency may not necessarily be an accurate indicator of binding efficiency. A determination of the exact occupancy by the enzyme at each site in vivo by means other than enoxacin-induced cleavage would be needed before we could conclude unequivocally that the five sites examined here are bound more weakly than the SGS in vivo by gyrase.
It is unlikely that there is a single biochemical property that can wholly account for the function of the SGS. We currently envision a scenario in which the overall biological activity of a given SGS-like sequence in Mu is the net result of several contributing biochemical effects. The efficiency of binding of gyrase and the extent of processivity of supercoiling imparted by a given site will likely be critical factors. Work in progress on quantifying the processivity effect and studies on binding of gyrase to supercoiled substrates of different superhelical densities may allow further dissection of the differences between the various sites and the relative contribution that each parameter makes to the SGS function.
Sequence analysis.
One motivation for seeking other SGS-like sites was to be able to compare the nucleotide sequences between sites sharing similar biological function, as a way to learn more about possible gyrase-DNA interactions important for a particular effect. Irrespective of the true biological function of each DNA sequence in its natural bacterial host, we have shown that each one forms a bona fide site for E. coli gyrase both in vitro and in vivo. It is meaningful, therefore, to compare the sites with the SGS at the sequence level to try and shed fresh light on the DNA-protein interactions with gyrase that might be responsible for imparting the particular function of each sequence.
Studies from several laboratories, primarily from footprinting analyses, have shown that gyrase sites are composed of a core of approximately 20 bp that bind gyrase most strongly, centered on the sites of cleavage by the enzyme, flanked by “arms” of about 50 to 60 bp that wrap around the enzyme (5, 17, 21). Our previous attempts to determine the features of the SGS that distinguish it from other gyrase sites have focused on a critical role of what we call the right arm of the gyrase site (26). Deletions into the right arm, or into the core of the SGS, inhibited replication of prophages with the deleted sites, while deletions into the left arm were tolerated. Additionally, replacing either arm of the pSC101 gyrase site (unable in itself to fully substitute SGS function) with just the SGS right arm yielded a hybrid site that is fully proficient in Mu replication.
We can now complement these functional dissections of the Mu SGS with sequence analyses. Listed below (5′ to 3′) are the central 20-bp regions, arbitrarily defined as cores (with the 4-bp gyrase cut site in bold): Mu, AGGCATAAAATCAGCCGCAC; ECs (site A), AGGTATGCAAATGCATTACC; HI, ATTTATATCATTTTACGTCT; Sty (site A), CAGGTGGGTGATTTATCATT; NMA1, GGTAATACAAACCCCTGCCC;NMA2, ACCCATACTTACCCCTACCC. The following are the adjacent 60-bp regions, towards the I gene in all cases, defined as right arms: Mu, AGATTTTTTAAAACGCGCCACGGGATTTTtAAACCGGTATTTaACGGTGTATGAATCCCG; ECs A, TTACCCCCTTCAGGCGCGTGTCGTGATTTTCTAACCTGTATTTAACGGGCTTTAAAATCT; HI, CAAAAAATTTAAACGTGCTTAAACGCATTTAAACGGCATTTAAACGCTATTCTAATTTAA; Sty A, CCAGCGAATGAGGGGCTGTATTACCTTTATAAAGGCTTTACAGCCTCTGTTTTATAACCG; NMA1, AAGATATAAAAATCAATCCTAGACGCTTCTAAAAAGCCCCTGAAAACGATTAATTGTGTA; NMA2, ATCGCCTGAAAATCAATCCTGCGCGCGTTTGAACACCTTTTGAACACTATCCCACGCGTA.
Firstly, the core sequences are highly diverse at the sequence level, although they show at least moderate agreement with the degenerate consensus sequence for gyrase cleavage sites (12) (i.e., RNNNRNRKGRYCKYNYNNNY, where R is A or G, K is G or T, Y is C or T, N is any nucleotide, and the 4-bp cleavage site is given in bold). Of greater interest is a comparison of the SGS and the ECs site, as the latter most closely reproduces the properties of the SGS. When the ECs right arm is shifted by one base relative to that of the Mu site, extensive sequence identity (15/16 bp) is observed in a region of the right arm (underlined) that is distal to the core (bp 32 to 47 from the 5′ end of the right arm). Evidence from earlier deletion studies also implicated this region as important for SGS function (26). A deletion of the SGS from the 3′ end through to bp 43 (lowercase “a”) partially disrupted Mu replication, while a deletion through to bp 30 (lower case “t”) completely inhibited replication. Structural studies on the interaction of these DNA molecules with gyrase and construction of new hybrid sites, e.g., the pSC101 gyrase site with a substitution of only the 16-bp distal portion of the SGS right arm highlighted by the above sequence analysis, should allow us to define more accurately the key sequence elements required for normal SGS function.
Acknowledgments
We thank R. Redfield, M. Achtman, K. Makino, and D. Packard for generous gifts of bacterial genomic DNA samples, A. J. Howells for DNA gyrase and topoisomerase I enzymes, and A. Maxwell and D. Marra-Oram for critical reading and comments on the manuscript.
This work was supported by the National Science Foundation (grant MCB-0090898).
REFERENCES
- 1.Brussow, H., and R. W. Hendrix. 2002. Phage genomics: small is beautiful. Cell 108:13-16. [DOI] [PubMed] [Google Scholar]
- 2.Casjens, S. 2003. Prophages and bacterial genomics: what have we learned so far? Mol. Microbiol. 49:277-300. [DOI] [PubMed] [Google Scholar]
- 3.Chaconas, G., and R. M. Harshey. 2002. Transcription of phage Mu DNA, p. 384-402. In R. Craigie (ed.), Mobile DNA II. ASM Press, Washington, D.C.
- 4.Espeli, O., and F. Boccard. 1997. In vivo cleavage of Escherichia coli BIME-2 repeats by DNA gyrase: genetic characterization of the target and identification of the cut site. Mol. Microbiol. 26:767-777. [DOI] [PubMed] [Google Scholar]
- 5.Fisher, L. M., K. Mizuuchi, M. H. O'Dea, H. Ohmori, and M. Gellert. 1981. Site-specific interaction of DNA gyrase with DNA. Proc. Natl. Acad. Sci. USA 78:4165-4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J. F. Tomb, B. A. Dougherty, J. M. Merrick, et al. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496-512. [DOI] [PubMed] [Google Scholar]
- 7.Gellert, M., K. Mizuuchi, M. H. O'Dea, T. Itoh, and J. I. Tomizawa. 1977. Nalidixic acid resistance: a second genetic character involved in DNA gyrase activity. Proc. Natl. Acad. Sci. USA 74:4772-4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11-22. [DOI] [PubMed] [Google Scholar]
- 9.Hendrix, R. W., M. C. Smith, R. N. Burns, M. E. Ford, and G. F. Hatfull. 1999. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc. Natl. Acad. Sci. USA 96:2192-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kampranis, S. C., A. D. Bates, and A. Maxwell. 1999. A model for the mechanism of strand passage by DNA gyrase. Proc. Natl. Acad. Sci. USA 96:8414-8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klee, S. R., X. Nassif, B. Kusecek, P. Merker, J. L. Beretti, M. Achtman, and C. R. Tinsley. 2000. Molecular and biological analysis of eight genetic islands that distinguish Neisseria meningitidis from the closely related pathogen Neisseria gonorrhoeae. Infect. Immun. 68:2082-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lockshon, D., and D. R. Morris. 1985. Sites of reaction of Escherichia coli DNA gyrase on pBR322 in vivo as revealed by oxolinic acid-induced plasmid linearization. J. Mol. Biol. 181:63-74. [DOI] [PubMed] [Google Scholar]
- 13.Manjunatha, U. H., M. Dalal, M. Chatterji, D. R. Radha, S. S. Visweswariah, and V. Nagaraja. 2002. Functional characterisation of mycobacterial DNA gyrase: an efficient decatenase. Nucleic Acids Res. 30:2144-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Masignani, V., M. M. Giuliani, H. Tettelin, M. Comanducci, R. Rappuoli, and V. Scarlato. 2001. Mu-like prophage in serogroup B Neisseria meningitidis coding for surface-exposed antigens. Infect. Immun. 69:2580-2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maxwell, A., and A. J. Howells. 1999. Overexpression and purification of bacterial DNA gyrase, p. 135-144. In M. A. Bjornsti and N. Osheroff (ed.), DNA topoisomerase protocols, vol. I. DNA topology and enzymes, vol. 94. Humana Press, Totowa, N.J. [DOI] [PubMed]
- 16.Morgan, G. J., G. F. Hatfull, S. Casjens, and R. W. Hendrix. 2002. Bacteriophage Mu genome sequence: analysis and comparison with Mu-like prophages in Haemophilus, Neisseria and Deinococcus. J. Mol. Biol. 317:337-359. [DOI] [PubMed] [Google Scholar]
- 17.Morrison, A., and N. R. Cozzarelli. 1979. Site-specific cleavage of DNA by E. coli DNA gyrase. Cell 17:175-184. [DOI] [PubMed] [Google Scholar]
- 18.Morrison, A., N. P. Higgins, and N. R. Cozzarelli. 1980. Interaction between DNA gyrase and its cleavage site on DNA. J. Biol. Chem. 255:2211-2219. [PubMed] [Google Scholar]
- 19.O'Connor, M. B., and M. H. Malamy. 1985. Mapping of DNA gyrase cleavage sites in vivo: oxolinic acid induced cleavages in plasmid pBR322. J. Mol. Biol. 181:545-550. [DOI] [PubMed] [Google Scholar]
- 20.Oram, M., A. J. Howells, A. Maxwell, and M. L. Pato. 2003. A biochemical analysis of the interaction of DNA gyrase with the bacteriophage Mu, pSC101 and pBR322 strong gyrase sites: the role of DNA sequence in modulating gyrase supercoiling and biological activity. Mol. Microbiol. 50:333-347. [DOI] [PubMed] [Google Scholar]
- 21.Orphanides, G., and A. Maxwell. 1994. Evidence for a conformational change in the DNA gyrase-DNA complex from hydroxyl radical footprinting. Nucleic Acids Res. 22:1567-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parkhill, J., M. Achtman, K. D. James, S. D. Bentley, C. Churcher, S. R. Klee, G. Morelli, D. Basham, D. Brown, T. Chillingworth, R. M. Davies, P. Davis, K. Devlin, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, S. Leather, S. Moule, K. Mungall, M. A. Quail, M. A. Rajandream, K. M. Rutherford, M. Simmonds, J. Skelton, S. Whitehead, B. G. Spratt, and B. G. Barrell. 2000. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature 404:502-506. [DOI] [PubMed] [Google Scholar]
- 23.Parkhill, J., G. Dougan, K. D. James, N. R. Thomson, D. Pickard, J. Wain, C. Churcher, K. L. Mungall, S. D. Bentley, M. T. Holden, M. Sebaihia, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, A. Krogh, T. S. Larsen, S. Leather, S. Moule, P. O'Gaora, C. Parry, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848-852. [DOI] [PubMed] [Google Scholar]
- 24.Pathania, S., M. Jayaram, and R. M. Harshey. 2002. Path of DNA within the Mu transpososome: transposase interactions bridging two Mu ends and the enhancer trap five DNA supercoils. Cell 109:425-436. [DOI] [PubMed] [Google Scholar]
- 25.Pato, M. L. 1989. Bacteriophage Mu, p. 23-52. In D. Berg and M. M. Howe (ed.), Mobile DNA. American Society for Microbiology, Washington, D.C.
- 26.Pato, M. L., and M. Banerjee. 2000. Genetic analysis of the strong gyrase site (SGS) of bacteriophage Mu: localization of determinants required for promoting Mu replication. Mol. Microbiol. 37:800-810. [DOI] [PubMed] [Google Scholar]
- 27.Pato, M. L., and M. Banerjee. 1996. The Mu strong gyrase-binding site promotes efficient synapsis of the prophage termini. Mol. Microbiol. 22:283-292. [DOI] [PubMed] [Google Scholar]
- 28.Pato, M. L., and M. Banerjee. 1999. Replacement of the bacteriophage Mu strong gyrase site and effect on Mu DNA replication. J. Bacteriol. 181:5783-5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pato, M. L., M. M. Howe, and N. P. Higgins. 1990. A DNA gyrase-binding site at the center of the bacteriophage Mu genome is required for efficient replicative transposition. Proc. Natl. Acad. Sci. USA 87:8716-8720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pruss, G. J. 1985. DNA topoisomerase I mutants. Increased heterogeneity in linking number and other replicon-dependent changes in DNA supercoiling. J. Mol. Biol. 185:51-63. [DOI] [PubMed] [Google Scholar]
- 31.Reece, R. J., and A. Maxwell. 1989. Tryptic fragments of the Escherichia coli DNA gyrase A protein. J. Biol. Chem. 264:19648-19653. [PubMed] [Google Scholar]
- 32.Scheirer, K. E., and N. P. Higgins. 1997. The DNA cleavage reaction of DNA gyrase. Comparison of stable ternary complexes formed with enoxacin and CcdB protein. J. Biol. Chem. 272:27202-27209. [DOI] [PubMed] [Google Scholar]
- 33.Sokolsky, T. D., and T. A. Baker. 2003. DNA gyrase requirements distinguish the alternate pathways of Mu transposition. Mol. Microbiol. 47:397-409. [DOI] [PubMed] [Google Scholar]
- 34.Sugino, A., C. L. Peebles, K. N. Kreuzer, and N. R. Cozzarelli. 1977. Mechanism of action of nalidixic acid: purification of Escherichia coli nalA gene product and its relationship to DNA gyrase and a novel nicking-closing enzyme. Proc. Natl. Acad. Sci. USA 74:4767-4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tettelin, H., N. J. Saunders, J. Heidelberg, A. C. Jeffries, K. E. Nelson, J. A. Eisen, K. A. Ketchum, D. W. Hood, J. F. Peden, R. J. Dodson, W. C. Nelson, M. L. Gwinn, R. DeBoy, J. D. Peterson, E. K. Hickey, D. H. Haft, S. L. Salzberg, O. White, R. D. Fleischmann, B. A. Dougherty, T. Mason, A. Ciecko, D. S. Parksey, E. Blair, H. Cittone, E. B. Clark, M. D. Cotton, T. R. Utterback, H. Khouri, H. Qin, J. Vamathevan, J. Gill, V. Scarlato, V. Masignani, M. Pizza, G. Grandi, L. Sun, H. O. Smith, C. M. Fraser, E. R. Moxon, R. Rappuoli, and J. C. Venter. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287:1809-1815. [DOI] [PubMed] [Google Scholar]
- 36.Wahle, E., and A. Kornberg. 1988. The partition locus of plasmid pSC101 is a specific binding site for DNA gyrase. EMBO J. 7:1889-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]