Abstract
This study describes a novel approach to identify unique genomic DNA sequences from the unsequenced strain C. jejuni ATCC 43431 by comparison with the sequenced strain C. jejuni NCTC 11168. A shotgun DNA microarray was constructed by arraying 9,600 individual DNA fragments from a C. jejuni ATCC 43431 genomic library onto a glass slide. DNA fragments unique to C. jejuni ATCC 43431 were identified by competitive hybridization to the array with genomic DNA of C. jejuni NCTC 11168. The plasmids containing unique DNA fragments were sequenced, allowing the identification of up to 130 complete and incomplete genes. Potential biological roles were assigned to 66% of the unique open reading frames. The mean G+C content of these unique genes (26%) differs significantly from the G+C content of the entire C. jejuni genome (30.6%). This suggests that they may have been acquired through horizontal gene transfer from an organism with a G+C content lower than that of C. jejuni. Because the two C. jejuni strains differ by Penner serotype, a large proportion of the unique ATCC 43431 genes encode proteins involved in lipooligosaccharide and capsular biosynthesis, as expected. Several unique open reading frames encode enzymes which may contribute to genetic variability, i.e., restriction-modification systems and integrases. Interestingly, many of the unique C. jejuni ATCC 43431 genes show identity with a possible pathogenicity island from Helicobacter hepaticus and components of a potential type IV secretion system. In conclusion, this study provides a valuable resource to further investigate Campylobacter diversity and pathogenesis.
Since the publication of the complete genomic sequence of C. jejuni NCTC 11168 in February 2000, evidence that Campylobacter jejuni strains exhibit high genomic sequence diversity has been accumulating (34). This genetic variability was demonstrated with a battery of different technologies, including microarray-based genotyping (9, 27, 35), multiple locus sequence typing (38), restriction fragment length polymorphism (44), amplified fragment length polymorphism (40), pulsed-field gel electrophoresis (36, 52), and subtractive hybridization (1). C. jejuni genotypic diversity was localized within distinct areas of the genome, particularly regions harboring genes involved mainly in the biosynthesis and modification of surface structures such as lipooligosaccharide (LOS) and capsule biosynthesis and flagellum posttranslational modification, iron acquisition, and DNA restriction and modification (9, 27, 35). This genetic diversity might, in part, explain the large spectrum of C. jejuni disease outcomes, ranging from asymptomatic colonization to acute inflammatory diarrhea and to induction of the Guillain-Barré syndrome (43).
While the genome sequence of C. jejuni NCTC 11168 provided many insights into Campylobacter physiology, the relevance of this particular genome sequence to more virulent isolates has been questioned (1, 3). Epithelial cell invasion is thought to be an essential step in Campylobacter infection (16, 25). This is supported by both in vitro tissue culture models and in vivo animal and human clinical pathologies (26, 37, 50). Yet C. jejuni NCTC 11168 has been shown to be poorly invasive in human embryonic intestinal INT407 cells (3). C. jejuni's ability to bind to, invade, and replicate in intestinal epithelial cells has been studied extensively in in vitro tissue culture models (26). Invasion studies with intestinal epithelial cells as well as other cell lines revealed that the relative ability of different C. jejuni isolates to invade cultured cells is strain dependent (10, 24, 31). Furthermore, a statistically significant correlation was observed between C. jejuni's ability to invade cultured cells and the clinical symptoms of infection (10). Noninvasive strains were isolated from patients with noninflammatory disease, while invasive strains were isolated from patients with inflammatory diarrhea (10). This observation suggests that different Campylobacter isolates vary in their virulence properties and that these virulence properties are correlated, at least in part, with the ability to invade intestinal epithelial cells. Consequently, while C. jejuni NCTC 11168 might be valuable to study the development of noninflammatory watery diarrhea, the use of a more invasive isolate is required to investigate inflammatory bloody diarrhea.
Recently, the invasion ability of C. jejuni 81-176 was shown to be associated with the presence of a plasmid named pVir (3). Mutations introduced into several genes from this plasmid affected the invasion abilities of isogenic mutant strains, which resulted in reduced virulence in the ferret diarrheal disease model for some of the strains (4). Moreover, the presence of this plasmid was detected in a subset of fresh clinical isolates (3). Taken together, these observations suggest a role for the genes encoded by this plasmid in Campylobacter virulence. The sequence of the complete plasmid has been reported and revealed the presence of genes encoding proteins involved in the formation of type IV secretion systems, which are absent in the C. jejuni NCTC 11168 genome (4). Intriguingly, many clinical isolates, including C. jejuni ATCC 43431 (TGH 9011), are known to be highly invasive and yet do not harbor any plasmids (17, 18). Therefore, the proteins involved in their mechanism of invasion are likely to be chromosomally encoded. Additionally, because C. jejuni strains differ significantly in their invasion abilities, this might reflect the use of different mechanisms for cell invasion. Consequently, the identification of new genomic sequences from invasive clinical isolates of C. jejuni is required to fully investigate the molecular mechanism(s) of Campylobacter invasion.
Recently, comparative genomic techniques such as microarray-based genotyping and subtractive hybridization have been used to investigate differences in the whole genome content of several C. jejuni isolates (1, 9, 27, 35). While comparative hybridization to C. jejuni NCTC 11168 microarrays allows the survey of the entire genetic content of any C. jejuni strain at single-gene resolution, it cannot identify new genomic sequences. On the other hand, although subtractive hybridization can identify new genomic information, this technology suffers from insensitivity resulting from failure to detect a proportion of unique genes as well as high background due to false positives. In addition, the screening procedures are labor intensive and time-consuming.
Here we present a method utilizing shotgun DNA microarray analysis to identify new genomic DNA sequences from a nonsequenced microbial strain by comparison with a related strain of known sequence. This approach could also be very useful for the identification of unique genes in important pathogens whose genome sequences are unavailable. In this study, we chose to identify unique genes from C. jejuni ATCC 43431, an invasive isolate, by comparison with the less invasive sequenced strain, C. jejuni NCTC 11168. This allowed us to identify strain-specific diversity which might be involved in virulence potential. These two C. jejuni strains differ by several phenotypic and genotypic features: (i) their Penner serotype: ATCC 43431 (TGH 9011) is Penner serotype O:3, while NCTC 11168 is Penner serotype O:2 (49); (ii) their invasion ability: NCTC 11168 is poorly invasive (3), while ATCC 43431 appears to be highly invasive (17, 18); (iii) their genome size: the genome of ATCC 43431 appears to be larger than that of NCTC 11168 (21, 34); and (iv) their gene order: gene positions seem to differ considerably between the two strains (21, 22, 34). Since invasion ability has been shown to correlate with epidemiological outcomes (10), the genes unique to ATCC 43431 will provide a valuable resource to dissect the C. jejuni features that affect pathogenic potential.
MATERIALS AND METHODS
Bacterial strain, growth conditions, and cell culture.
C. jejuni ATCC 43431 (TGH 9011) and NCTC 11168 were acquired from the American Type Culture Collection and the National Collection of Type Cultures. Bacterial strains were routinely grown at 37°C on Mueller-Hinton (MH) agar plates under microaerophilic conditions (83% N2, 4% H2, 8% O2 and 5% CO2) with the MACS-VA500 variable-atmosphere workstation (Don Whitley, West Yorkshire, England). Human INT407 embryonic intestinal cells were obtained from the American Type Culture Collection and routinely maintained in minimal essential medium alpha (MEM-α, Gibco) supplemented with 10% fetal bovine serum (Gibco). Cells were grown in an incubator at 37°C and 10% CO2.
Bacterial binding and invasion assays.
The invasion assay was performed by coincubating mid-log-phase C. jejuni (grown in biphasic MH medium) with semiconfluent INT407 cells (2 × 105 cells per well) at an infection ratio (cell-bacteria) of about 1:30. After 3 h of incubation at 37°C in the presence of 10% CO2, the MEMα was removed, and the monolayers were washed twice with fresh medium. For the invasion assay, the infected monolayers were incubated for an additional 2 h in fresh medium containing 100 μg of gentamicin per ml. Then the monolayers were washed three times with Hanks' balanced salt solution (Sigma) and lysed with 0.01% Triton X-100 for 30 min at room temperature. Finally, the released bacteria were enumerated by serial dilution and plate counting on MH agar plates. Invasion efficiency was expressed as the percentage of the bacterial inoculum surviving gentamicin treatment. For the binding assay, infected INT407 cells were lysed with 0.01% Triton X-100 for 30 min at room temperature, the suspensions were serially diluted, and the number of bacteria was determined by counting on MH agar plates. Each binding and invasion assay was repeated 10 times, and the results shown are the mean ± standard error of the replicates.
Construction of C. jejuni NCTC 11168 microarray.
The construction of a C. jejuni NCTC 11168 microarray has been previously described (46, 47). This array harbors PCR amplified fragments from approximately 98% of the open-reading-frames from the complete genome of C. jejuni NCTC 11168.
Construction of C. jejuni ATCC 43431 shotgun microarray.
A genomic library of C. jejuni ATCC 43431 was constructed with the TOPO shotgun subcloning kit from Invitrogen, following the manufacturer's protocol. Briefly, C. jejuni chromosomal DNA was purified by the method of Sambrook et al. (39), resuspended in nebulization buffer (0.5 M Tris-HCl [pH 8], 0.15 M MgCl2, 50% glycerol), and placed at the bottom of the nebulizer; 50 μg of chromosomal DNA in 2 ml of nebulization buffer was mechanically sheared by nebulization (nitrogen pressure of 10 lb/in2 for 5 min) in order to obtain DNA fragments of 500 to 1,100 bp in size (average size, 800 bp). After nebulization, the DNA was checked on a 1% agarose gel to ensure the desired size. Next, the DNA fragments were precipitated by adding 80 μl of 3 M sodium acetate (pH 5.2), 4 μl of glycogen (20 mg/ml), and 700 μl of isopropanol, and centrifuging at 12,000 × g for 15 min. The DNAs were further purified with the Qiagen PCR kit (Qiagen, Valencia, Calif.) by the manufacturer's recommendations, and resuspended in 10 μl of water. The fragments were blunt ended with a mixture of T4 DNA polymerase and Klenow fragment and then dephosphorylated with calf intestinal phosphatase. The resulting fragments were cloned into the pCR4Blunt-TOPO vector. Finally, the cloning mix was further purified with the PCR purification kit and electroporated into electrocompetent Escherichia coli strain TOP10.
The transformation reaction was plated on LB agar plates containing ampicillin (100 μg/ml) and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal, 64 μg/ml). Individual clones from this unamplified library were immediately replicated into 96-well plates containing LB broth and ampicillin (100 μg/ml). Plasmids from 9,600 clones were purified with the 96-well filter plates Millipore MultiScreen (Millipore MAFB NOB 50) and Costar polyvinylidene difluoride (0.2-μm, Costar catalog number 3504) according to the manufacturer's instructions and as previously described. The quality and concentration of the purified plasmids were assessed on 1% agarose gels. Purified plasmids were diluted in 3× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) buffer at a concentration ranging from 50 to 150 ng/μl and rearrayed into a 384-well format. The plasmid samples were subsequently printed on aminosilane-coated glass microscopic slides (CMT GAPS-II; Corning Inc., Corning, N.Y.) with an arrayer robot (Molecular Dynamics) in repeating blocks of a 62 by 13 spot pattern. Within each 62 by 13 block, spots are approximately 250 μm in diameter and are spaced 300 μm apart (center to center). Finally, the DNA fragments were immobilized onto the slides by baking at 80°C for 4 h. The quality of the microarray was assessed by direct labeling of the spotted DNA with a fluorescent nucleic acid stain (POPO-3 iodide; Molecular Probes), following the manufacturer's protocol.
DNA labeling and microarray hybridization.
For each microarray hybridization, 2 μg of genomic DNA from the sequenced strain C. jejuni NCTC 11168 and the test strain C. jejuni ATCC 43431 was fluorescently labeled with indodicarbocyanine (Cy5) and indocarbocyanine (Cy3) fluors, respectively, as follows. First, genomic DNAs were sheared by nebulization in order to obtain DNA fragments of 700 to 1,300 bp in size as described above. Second, sheared genomic DNAs were labeled by random priming with the BioPrime DNA labeling system from Invitrogen. Briefly, 2 μg of sheared genomic DNA in 21 μl of water was mixed with 20 μl of the 2.5× random primer solution. After denaturation at 99°C for 5 min, the reaction mixture was brought up to 47 μl by adding 5 μl of a deoxynucleoside triphosphate-dUTP mix (1.2 mM each dGTP, dATP, and dCTP; 0.6 mM aminoallyl-dUTP; and 0.6 mM dTTP) and 1 μl of highly concentrated Klenow fragment. This suspension was incubated at 37°C for 2 h. Then, the aminoallyl-labeled genomic DNAs were purified with Microcon YM-30 filters (Millipore) and labeled with monoreactive FluoroLink Cy3 and Cy5 dyes (Amersham) as previously described (46, 47).
Fluorescently labeled reference and test DNAs were combined in a 36-μl hybridization solution containing 25% formamide, 5× SSC buffer, 0.1% sodium dodecyl sulfate (SDS) and 25 μg of salmon sperm DNA, denatured for 2 min at 99°C, and hybridized to one microarray slide that had been prehybridized in a 1% bovine serum albumin solution (25% formamide, 5× SSC buffer, 0.1% SDS, and 1% bovine serum albumin) under a coverslip (Grace Bio-labs) and placed in a humidified chamber (ArrayIt). Hybridizations were carried out for 16 h at 42°C. Slides were then washed consecutively in 2× SSC-0.1% SDS for 5 min at 42°C, then in 0.1× SSC-1% SDS for 10 min at room temperature, and four times in 0.1× SSC for 1 min at room temperature. The slides were finally washed with distilled water and dried by centrifugation.
Microarray data analysis.
Microarray slides were scanned at 532 nm (Cy3) and 635 nm (Cy5) with a laser-activated confocal scanner (ScanArray 3000) at 10-μm resolution and analyzed with the GenePix Pro 4 software (Axon Instruments, Foster City, Calif.). Spots with abnormalities were removed either manually or automatically by the GenePix software before data processing. To filter out unreliable data, spots with a signal intensity below 2 times the standard deviation of the background in the control channel, which reveals the genomic DNA printed on the slide, were discarded.
For the hybridization experiments with the C. jejuni NCTC 11168 microarray, the data were processed following the method described by Kim et al. (20) with the algorithm implemented into the GACK software (available at http://falkow.stanford.edu/). Briefly, this algorithm uses the shape of the signal distribution to estimate the probability that any given gene is conserved or divergent (present or absent) independently of any normalization and thus provides a level of confidence for the gene category assignment. This method has been shown to be more reliable than methods with empirical determination of cutoff value to predict the presence or absence of genes. Three independent microarray hybridizations were performed and averaged across the GACK datasets.
For the hybridization experiments with the shotgun C. jejuni ATCC 43431 microarray, the fluorescent intensity in each wavelength was normalized with a linear regression analysis. This methodology is based on a three-step iterative identification of genes present in both C. jejuni stains in order to use them for normalization. In the first step, a linear regression was applied to the scatter plot of the mean Cy3 versus the mean Cy5 intensities and used to transform the raw data. For each spot, the ratio of the transformed Cy3 and Cy5 intensities was calculated as m = Cy3/Cy5. In the second step, a rank selection method was used to remove the first 20 genes with the lowest m value. In the third step, the raw data for the remaining spots were extracted and retransformed by applying a second linear regression (similar to the first step). These three steps were repeated in an iterative process until the linear correlation coefficient r converged to 1. Finally, the last linear regression was used to normalize the complete raw microarray data. DNA sequencing of 48 plasmids with an m value equal to 1 confirmed that their inserts were present in both C. jejuni strains, validating our normalization approach. All mathematical transformations were performed with OriginPro 7 software (OriginLab Corporation) and in-house written scripts. Two criteria, m < 0.5 and 0.5 < m < 0.6, were arbitrarily used to select spots carrying a DNA fragment unique to the C. jejuni ATCC 43431 strain. Finally, the plasmids from these two groups of spots were used as a template for DNA sequencing reactions.
DNA sequencing and annotation.
DNA sequencing reactions were carried out directly from the purified plasmid in both forward and reverse directions with the M13 universal primers by the DNA Sequencing Facility of the Laboratory for Genomics and Bioinformatics at the Oklahoma University Health Science Center with an ABI Prism 3700 capillary sequencer. The vector clipping and the sequence assembly and editing were performed with the SeqManII module from Lasergene software (DNAstar, Inc.). The sequences obtained were compared to those in the C. jejuni NCTC 11168 genome database from the Sanger Center with the BLASTN program (http://www.sanger.ac.uk/cgi-bin/BLAST/submitblast/c_jejuni). Sequences with 100% identity over their entire length to C. jejuni NCTC 11168 were removed from further analysis. Open reading frames (ORFs) of at least 100 bp in length were identified with the Artemis software (http://www.sanger.ac.uk/Software/Artemis/). Further analysis of the ORFs was performed with the BLAST program against the NCBI nonredundant database. Potential biological roles were assigned to the unique ORFs with the classification schemes from the Gene Ontology consortium.
DNA preparation for PFGE analysis.
C. jejuni cultures were grown overnight at 42°C in 25 ml of Mueller-Hinton broth under microaerophilic conditions as described above. Cultures were pelleted by centrifugation (1,000 × g, 15 min), washed once in 1× phosphate-buffered saline (0.15 M NaCl, 2.71 mM Na2HPO4 · 7H2O, 1.54 mM KH2PO4 [pH 7.4]), and resuspended in PBS for optical density (OD) measurements. The OD readings at 600 nm were in the range from 0.58 to 0.7, as recommended (36). Adjusted cell suspensions were processed as described before (36) with slight modifications. Briefly, 400 μl of each suspension was transferred separately to 1.5-ml microcentrifuge tubes containing 50 μl of proteinase K (10-mg/ml stock; Boehringer Mannheim) and mixed gently. An equal volume of molten 1.0% low EEO agarose (Fisher Biotech) in TE (10 mM Tris, 1 mM EDTA, pH 8.0) was added to the cell suspensions, one sample at a time, and mixed gently by pipetting. The agarose-cell suspension mixtures were loaded immediately into the wells of reusable plug molds (Bio-Rad Laboratories, Hercules, Calif.). The agarose plugs were placed at 4°C to harden. Then, the plugs were transferred to 15-ml conical tubes (polypropylene; Sarstedt) containing 5 ml of cell lysis buffer (50 mM Tris, 50 mM EDTA [pH 8.0], 1% Sarcosyl, 0.1 mg of proteinase K/ml). Lysis was allowed to proceed for 30 min at 54°C in a shaking water bath. After lysis, the plugs were washed four times as described by Ribot et al. (36). Finally, washed plugs were stored at 4°C until use.
Restriction digestion for PFGE analysis.
Slices from each plug 1 to 2 mm wide were cut with a sterile spatula edge and transferred to a 1.5-ml microcentrifuge tube containing 750 μl 1× buffer 4 (New England Biolabs) for SmaI and 750 μl of 1× SalI buffer (New England Biolabs) plus 1× bovine serum albumin for SalI for 5 min at room temperature for SmaI and 37°C for SalI. This prerestriction buffer was then removed and replaced with 200 μl of fresh buffer plus 40 U of the appropriate restriction enzyme. SmaI digests were incubated at room temperature for 2 h, while SalI digests were incubated at 37°C for 2 h. Prior to loading plugs into the gel for electrophoresis, the restriction buffer was first diluted with 0.5× Tris-borate-EDTA (TBE), allowed to stand for 5 min, and then removed. Fresh 0.5× TBE was then added for an additional 5 min.
PFGE and analysis.
Restricted plug slices were loaded into the appropriate wells of a 1% chromosomal grade agarose gel (Bio-Rad) in 0.5× TBE. Also included were molecular weight plugs corresponding to the lambda 50-kb ladder (Bio-Rad). The electrophoresis conditions used for the SmaI digest were initial switch time of 6.8 s and final switch time of 38.4 s (gradient of 6 V/cm and an included angle of 120°) for 18 h with the CHEF-DR III system with circulating and cooled (14°C) 0.5× TBE (36). The voltage gradient, included angle, gel percentage, temperature of running, and TBE concentration were the same for all digestions analyzed, but the other running conditions had to be modified to allow adequate band separation. For SalI, the conditions suggested for the separation of the 50-kb lambda ladder were used (Bio-Rad CHEF-DR III manual), since each strain had a very large fragment which could not be resolved with the SmaI running conditions. Conditions for SalI were initial switch time of 50 s and final switch time of 90 c for 22 h. After the electrophoresis, gels were stained with 200 ml of a 50-μg/ml ethidium bromide solution and the band patterns were documented with the UV Doc It system (UVP). The data from the two restriction digestions were analyzed with the LabWorks image acquisition and analysis software and by comparing the migration of the lambda marker bands with the C. jejuni genomic bands. An average molecular weight was determined for each strain based on the results from the two digestions.
Nucleotide sequence accession numbers.
The DNA sequences described in this work have been deposited in the NCBI database under consecutive accession numbers AY501906 to AY501998.
RESULTS AND DISCUSSION
Invasion abilities of C. jejuni NCTC 11168 and ATCC 43431.
In order to confirm the previously reported invasion properties of the two C. jejuni strains (3, 18), they were subjected to an invasion assay with human INT407 embryonic intestinal cells. As expected, this assay revealed considerable differences between the two strains in their ability to invade INT407 host cells. While C. jejuni ATCC 43431(TGH 9011) was confirmed to be a proficiently invasive isolate, C. jejuni NCTC 11168 was found to be poorly invasive (0.36% ± 0.257% and 0.00054% ± 0.00027% invasion, respectively). The invasion ability of C. jejuni NCTC 11168 was not restored following passage in vivo in the rabbit ligated ileal loop model (data not shown and unpublished data) (7). This inability to restore its invasion potential suggests that this phenotype has a genetic basis.
Recently, the original clonal clinical isolate of C. jejuni NCTC 11168, named C. jejuni strain 11168-O, has been shown to be substantially more invasive in epithelial cells than the sequenced strain (12). Interestingly, this phenotypic difference was not attributed to a large genetic variability (e.g., gene loss or gain) between the two strains but rather to small genetic changes (e.g., single-nucleotide polymorphism). The invasion efficiency of the 11168-O isolate was found to be approximately 0.0035% as calculated by Gaynor et al. (12) and to exhibit a threefold difference in invasion with the sequenced NCTC 11168 strain after 2 h of invasion. In contrast, ATCC 43431 invades epithelial cells with an efficiency of 0.36% and exhibits a 667-fold difference in invasion with the NCTC 11168 strain (after 3 h of invasion). Given that the invasion assay used by Gaynor et al. differs slightly from the one used in this study (e.g., different incubation time and multiplicity of infection), a direct comparison would be necessary to determine the exact difference in invasion efficiencies between the ATCC 43431 and the 11168-O isolates. Nevertheless, C. jejuni ATCC 43431 appears to be significantly more invasive than 11168-O. Consequently, large genetic differences, in addition to subtle genetic changes, may contribute to and enhance the invasion potential of certain Campylobacter isolates. As mentioned previously, the invasion efficiency of C. jejuni 81-176 is related to the presence of a plasmid carrying genes absent from the C. jejuni NCTC 11168 genome (3, 4).
In this study, the genes which are different and common between the ATCC 43431 and NCTC 11168 strains were identified by whole genome microbial comparison with both a shotgun DNA microarray of C. jejuni ATCC 43431 and the previously constructed C. jejuni NCTC 11168 microarray. Because invasion ability has been shown to be tightly associated with Campylobacter virulence, investigation of the genotypic differences and similarities between these two strains could reveal novel targets for the study of Campylobacter invasion mechanisms and pathogenesis as well as provide new insights about the evolution of Campylobacter species.
Conserved genes between C. jejuni NCTC 11168 and ATCC 43431.
The genes conserved between C. jejuni NCTC 11168 and ATCC 43431 were identified with a C. jejuni NCTC 11168 microarray. This array carries PCR-amplified fragments of each annotated open reading frame from C. jejuni NCTC 11168, as previously described (46, 47). Each strain was hybridized to the microarray in triplicate. Genes were classified as conserved or divergent, with a cutoff for the intensity ratio dynamically generated by a microarray genomic analysis program called GACK (20). The complete data set on the presence and absence of each gene is represented in Fig. 1 (first row). The genes are organized with respect to their chromosomal location on the NCTC 11168 genome.
FIG. 1.
Conserved and divergent C. jejuni NCTC 111168 ORFs in C. jejuni ATCC 43431 identified by microarray analysis (row 1), DNA sequencing (row 2), and PCR analysis (row 3). ORFs are listed with respect to their location on the NCTC 11168 genome horizontally from left to right in 21 sections of 80 ORFs. It should be noticed that the gene order might be different in the genome of C. jejuni ATCC 43431. For the microarray data (row 1), the color scale is located at the bottom of the figure, with the brightest blue representing ORFs found to be present with high certainty, the brightest yellow indicating ORFs found to be absent with high certainty, and black corresponding to ORFs found to be uncertain or slightly divergent. For the sequencing data (row 2), sequenced ORFs are indicated in blue, while green indicates missing data. For the PCR analysis (row 3), ORFs successfully amplified are represented in blue, ORFs with failed PCR amplification are indicated in yellow, and untested ORFs are indicated in green.
Overall, 88 ORFs were found to be absent with high certainty in C. jejuni ATCC 43431 but present in the reference strain NCTC 11168, which equals approximately 5.3% of all ORFs. This result contrasts with the previous study from Dorrell et al. (9), who identified 117 genes absent in C. jejuni ATCC 43431 with a “low-cost” microarray technology. This microarray was constructed by arraying PCR amplicons from a pUC18 genomic library, and thus each element on the array could contain more than one gene (9). Consequently, the data generated by Dorrell et al. (9) are difficult to interpret and likely result in a higher rate of false-positives and false-negatives.
In order to validate our data, we assessed the absence of the 88 ORFs (identified by our microarray analysis) by PCR with the same primers as used for the construction of the C. jejuni NCTC 11168 microarray (third row, Fig. 1). In addition, we similarly assessed the presence of 328 individual ORFs (randomly chosen) by PCR. Moreover, the presence of 146 genes in the C. jejuni ATCC 43431 genome (with >80% identity to the counterpart C. jejuni NCTC 111168 genes) was corroborated by sequencing the DNA inserts from the C. jejuni ATCC 43431 genomic library (second row, Fig. 1). These 146 genes include the clones identified as carrying DNA inserts common to both strains and the false-positives obtained from the shotgun microarray analysis (see below).
Among the 88 ORFs found to be absent by the microarray analysis, 85 were confirmed absent by PCR analysis, whereas three were found to be present, indicating that at least 3.5% of the genes were falsely called absent. It should be noted that the absence of PCR amplification does not prove the absence of a gene. Indeed, a single base pair mismatch at the primer annealing site could prevent a successful PCR amplification. Similarly, this possibility may explain the failure of the PCR to amplify 25 genes found to be present with high certainty by the microarray analysis among the 328 genes tested (Fig. 1, row 3). While several of these genes were found to be present by DNA sequencing (Fig. 1, row 2), we cannot exclude that some of these 25 genes are false-positives. While apparently contradictory, four genes found to be absent by the microarray and PCR analysis were identified as present upon sequencing of several clones from the C. jejuni ATCC 43431 genomic library (Cj0629, Cj1678, Cj0809c, and Cj1422c). However, further analysis of the DNA sequences revealed that they correspond to a different portion of the gene than the one printed on the microarray slide (data not shown), suggesting the presence of orthologues of these genes in the genome of C. jejuni ATCC 43431.
Many of the dispensable genes in ATCC 43431 were found to colocalize in five major chromosomal regions, referred to as plasticity zones PZ1 to PZ5. These zones were previously characterized by Dorrell et al. and others as hypervariable plasticity regions among C. jejuni strains of diverse origins (9, 27, 35). PZ1 contains eight genes: Cj0258 (a possible regulator), pyrC (dihydroorotase), Cj0262c (putative methyl-accepting chemotaxis signal transduction protein), Cj0264c (molybdopterin-containing oxidoreductase), Cj0265c (putative cytochrome c-type heme-binding periplasmic protein), and three genes encoding proteins of unknown function (Cj0260c, Cj0261c, and Cj0263). While the absence of PyrC might suggest pyrimidine auxotrophy, it should be noticed that C. jejuni possesses a second dihydroorotase gene (pyrC2) which was found to be present in ATCC 43431 (Fig. 1).
Cj0624c is a molybdenum-containing enzyme that has been shown to catalyze the reduction of trimethylamine-N-oxide and dimethyl sulfoxide (42). Together with Cj0265c, Cj0264c allows Campylobacter to respire under limited oxygen conditions with N- or S-oxide as an alternative electron acceptor. The absence of these genes in C. jejuni ATCC43431 suggests its inability to utilize these electron acceptors and is in agreement with the absence of the molybdenum transporter genes modABC, which are part of plasticity zone 2 (PZ2). In addition to these transporter genes, PZ2 contains genes encoding a putative acyltransferase (Cj0295), a MoeB/ThiF family protein (Cj0294), a protein of unknown function (Cj0302c), a putative periplasmic beta-lactamase (Cj0299), and two proteins involved in biotin biosynthesis (BioC and BioF).
The three other zones of plasticity have been previously described as being variable in other strains of C. jejuni (9, 27, 35). These zones include genes encoding proteins involved in sugar modification and transport (PZ3), genes encoding proteins involved in lipooligosaccharide (LOS) biosynthesis (PZ4), and genes from the capsular polysaccharide locus (PZ5). The genes encoding the capsule export system (kps) and the inner core genes of the LOS (waa) are conserved between the two C. jejuni strains, as has been suggested in other strain comparisons (14, 19). Given that the Penner serotype recognizes the capsular and LOS structures, the divergence of the capsular and LOS biosynthesis genes is in agreement with the different serotypes of the C. jejuni NCTC 11168 and ATCC 43431 strains (49).
Genome size comparison between C. jejuni NCTC 11168 and ATCC 43431 by PFGE.
The size of the C. jejuni ATCC 43431 genome has been previously evaluated by PFGE analysis and reported to be 1,812.5 ± 43.9 kb (21). The comparison of this result with the known size of the C. jejuni NCTC 11168 genome (1,641 kb) obtained by whole-genome sequencing suggests that these two genomes might differ by up to 171 kb. However, this size difference is likely inaccurate and overestimated because the genome sizes were determined by two different technologies. Therefore, in order to directly compare these two genomes, their sizes were reevaluated side by side with the same pulsed-field gel.
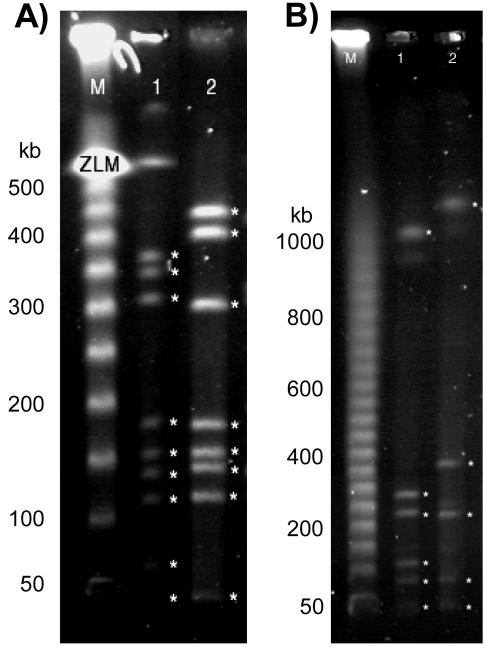
Given the low G+C content of C. jejuni and based on the previous study from Kim et al. (21), C. jejuni chromosomal DNA was digested with two different restriction endonucleases that recognize G- and C-rich cleavage sites. The restriction enzymes SmaI and SalI were used individually to digest the genome of both C. jejuni strains, and the restricted fragments were separated by PFGE (Fig. 2). The digestion of C. jejuni NCTC 11168 and ATCC 43431 chromosomal DNA with SmaI and SalI produced between 5 and 10 fragments which could be easily differentiated and sized. The sizes of C. jejuni NCTC 11168 and ATCC 43431 genomes were calculated to be 1,744 ± 29 kb and 1,811 ± 22 kb, respectively. While this result reflects the overestimation of the real genome size by PFGE analysis, it allows the direct comparison of the two genomes. Based on these data, these two genomes differ by only 66 ± 7 kb, with C. jejuni ATCC 43431 having the larger of the two genomes, as expected. Therefore, with the size of the sequenced C. jejuni NCTC 11168 genome as a reference, the size of C. jejuni ATCC 43431 can be estimated to be approximately 1,707 kb.
FIG. 2.
PFGE of SmaI-digested (A) and SalI-digested (B) C. jejuni chromosomal DNA. Lanes: M, lambda 50-kb markers; 1, restriction fragments obtained from digestion of C. jejuni NCTC 11168 DNA; 2, restriction fragments obtained from digestion of C. jejuni ATCC 43431 DNA. The ladder sizes are indicated on the left of each panel. For SalI, the gel running conditions allowed the entire 50-kb ladder (from 50 kb to 1 Mb) to be separated, whereas the SmaI conditions were optimized for separation up to 550 kb.
Construction of a shotgun DNA microarray of C. jejuni ATCC 43431.
The use of C. jejuni NCTC 11168 as a reference for the interspecies microarray hybridization obviously excluded genes missing in the genome of C. jejuni ATCC 43431. In order to identify genes unique to C. jejuni ATCC 43431, a shotgun microarray of this organism was constructed. As a prerequisite to the shotgun microarray construction, a library of the C. jejuni ATCC 43431 genome was produced with the pCR4Blunt-TOPO vector (Invitrogen) in E. coli strain TOP10 as described in the Materials and Methods section. Nine thousand six hundred individual clones from the unamplified library were immediately replicated into a 96-well plate format, and their plasmids were purified and analyzed by agarose gel electrophoresis. Given a genome size of approximately 1,707 kb, a mean DNA insert size of about 750 bp (determined upon sequencing of 624 DNA inserts), and 9,600 clones in the library, the probability of having each DNA sequence in our library is 0.98 (P = 1 − e(9,600 × ln[1 − 0.7/1,707])).
Initially, 96 clones were randomly chosen for microarray protocol development. Several printing conditions onto glass slides were evaluated: PCR-amplified inserts; untreated plasmids, plasmids cut with a restriction enzyme (EcoRI); plasmids treated with Plasmid-Safe ATP-dependent DNase (Epicentre); and denatured plasmids. The comparison of the hybridization efficiency and specificity of these different probes revealed that purified untreated plasmids produced microarrays of a quality similar to that of PCR products and suitable for microarray hybridization with genomic DNA (data not shown). Consequently, we constructed a shotgun DNA microarray by arraying the 9,600 purified whole plasmids from the C. jejuni ATCC 43431 genomic library onto a glass slide.
Identification of unique C. jejuni ATCC 43431 DNA fragments by competitive hybridization with the C. jejuni NCTC 11168 genome.
Genes unique to C. jejuni ATCC 43431 were identified by competitive hybridization on its array with genomic DNA from C. jejuni NCTC 11168. The fluorescent mean intensities in both channels 1 (Cy5) and 2 (Cy3) were normalized with an iterative linear regression analysis (as described in Materials and Methods). This normalization technique was validated by DNA sequencing of 48 clones identified as carrying DNA fragments common to both C. jejuni strains. The microarray hybridization was repeated three times by labeling the C. jejuni NCTC 11168 and ATCC 43431 genomes with the Cy5 and Cy3 fluors, respectively.
The geometric mean of the normalized [(Cy5 fluorescence intensity)/(Cy3 fluorescence intensity)] ratio was computed, and an initial cutoff below 0.5 for the identification of DNA fragments unique to C. jejuni ATCC 43431 was arbitrarily chosen. With this ratio threshold, 576 clones were selected. The insert from each individual clone was sequenced in one direction. The sequences obtained were compared to those in the C. jejuni NCTC 11168 genome database with the BLASTN program at the Sanger Center. Of the 576 sequences, 208 (36% of the total sequences) were false-positive, sharing 100% identity with genomic fragments from C. jejuni NCTC 11168. While the false-positives were not studied further, clones carrying DNA fragments unique to C. jejuni ATCC 43431 were sequenced in the opposite direction.
In order to ensure that every clone from the library containing a unique C. jejuni ATCC 43431 DNA fragment was selected with a ratio threshold of 0.5, 96 plasmids with a fluorescent intensity ratio between 0.5 and 0.6 were randomly picked and sequenced. The DNA sequencing revealed that all 96 vectors contained DNA inserts that had 100% identity with a chromosomal region of C. jejuni NCTC 11168 over their entire length. Therefore, we assume that all the clones carrying DNA fragments unique to C. jejuni ATCC 43431 and present in the genomic library have been sequenced. In total, 84 kb of new DNA was sequenced. Next, they were assembled into 93 contigs with the module SeqManII from Lasergene software (DNAstar). Each DNA fragment was sequenced from at least three different clones from the library, suggesting at least a threefold coverage of the genome. Among the 93 contigs, 9 contained a 150- to 700-bp DNA sequence at one of their extremities that exhibited 100% identity with regions from the C. jejuni NCTC 11168 genome, suggesting their possible location on the genome (data not shown).
General features of the unique C. jejuni ATCC 43431 DNA sequences.
The sum of the fragments unique to C. jejuni ATCC 43431 yields 83,909 bp of new DNA sequence. A total of 130 ORFs were identified by the presence of an uninterrupted coding region encoding a minimum of 40 amino acids with or without an initiation and/or stop codon. The G+C content of the ORFs ranged from 17.4 to 36.1%, with a mean of 26.2%. In contrast, the core set of genes (on the basis of the C. jejuni NCTC 11168 genome) has an average G+C content of 30.6%, suggesting that many of the C. jejuni ATCC 43431-specific genes have been acquired through horizontal gene transfer from an organism with a G+C content lower than that of C. jejuni.
BLAST searches allowed the assignment of potential biological roles to 66% of the unique ORFs (corresponding to a total of 86 ORFs) (Table 1). These unique ORFs of C. jejuni ATCC 43431 can be classified in eight functional categories: cell envelope and surface structures, restriction-modification, recombination, and repair (DNA modification), chemotaxis, other (bacteriophage sequence), transport, small-molecule metabolism, hypothetical proteins, and unknown. Genes encoding proteins involved in cell envelope and surface structure and genes encoding restriction modification systems and DNA recombination and repair constitute the two major functional groups of unique C. jejuni ATCC 43431 genes of known function.
TABLE 1.
C. jejuni ATCC 43431 ORFs absent from the genome of NCTC 11168a
| Category and ORF | Lengthb (bp) | %G+C content | Closest relationshipc | Accession no. | Identityd (%) | Notes | |
|---|---|---|---|---|---|---|---|
| Cell envelope and surface structures | |||||||
| Tgh001 | 774 | 23.5 | a) Unknown (C. jejuni NCTC 11828) | AAK12951 | 230/257 (89) | ||
| b) Lipooligosaccharide biosynthesis protein (Brucella melitensis) | NP_539335 | 54/207 (26) | |||||
| Tgh002 | 957 | 20.3 | a) Unknown (C. jejuni NCTC 11828) | AAK12950 | 306/318 (96) | ||
| b) Putative capsular polysaccharide synthesis protein (Bacteroides thetaiotaomicron) | NP_811784 | 88/257 (34) | |||||
| Tgh003 | 639 | 23.0 | a) Unknown (C. jejuni NCTC 11828) | AAK12949 | 196/211 (92) | ||
| b) HtrL (E. coli K-12) | NP_418075 | 60/189 (31) | |||||
| Tgh004 | 918 | 24.3 | a) Similar to C. jejuni unknown (C. jejuni NCTC 11828) | AAK12952 | 241/267 (90) | On the same contig (LOS biosynthesis locus)e | |
| b) β-1,4-N-Acetylgalactosaminyl transferase (C. jejuni O:4) | AAG43977 | 169/296 (57) | |||||
| Tgh011 | 700 | 22.4 | a) Unknown (C. jejuni NCTC 11828) | AAK12956 | 221/224 (98) | ||
| b) Glycosyltransferase (Nostoc sp. strain PCC 7120) | NP_486876 | 36/165 (21) | |||||
| Tgh020 | 357 | 23.2 | Unknown (C. jejuni NCTC 11828) | AAK12960 | 104/111 (94) | ||
| Tgh021 | 405 | 25.7 | Unknown (C. jejuni NCTC 11828) | AAK12959 | 122/134 (91) | ||
| Tgh022 | 301 | 26.2 | Putative acetyltransferase (C. jejuni NCTC 11828) | AAK12958 | 100/100 (100) | ||
| Tgh042 | 548 | 26.2 | Putative aminotransferase (C. jejuni NCTC 11828) | AAK12954 | 177/181 (97) | ||
| Tgh043 | 479 | 20.0 | a) Unknown (C. jejuni NCTC 11828) | AAK12953 | 155/159 (97) | ||
| b) Probable sugar transferase Cj1422c (C. jejuni NCTC 11168) | NP_282563 | 51/155 (32) | |||||
| Tgh101 | 462 | 26.2 | WaaV (C. jejuni NCTC 11828) | AAK12948 | 44/153 (94) | ||
| Tgh114 | 810 | 26.4 | rmlB (C. jejuni ATCC43431) | AAL06019 | 181/181 (100) | ||
| Tgh160 | 533 | 23.59 | Hypothetical protein HH0094 (Helicobacter hepaticus ATCC 51449) | AAP76691.1 | 83/162 (51) | ||
| Tgh006 | 1,7443′ | 24.6 | a) Hypothetical protein Cj1431c (C. jejuni NCTC 11168) | NP_282572 | 56/593 (92) | ||
| b) Probable sugar transferase Cj1432c (C. jejuni NCTC 11168) | NP_282573 | 36/108 (33) | On the same contig | ||||
| Tgh007 | 1305′ | 24.4 | α-2,3-Sialyltransferase (C. jejuni OH4384) | AAF13495 | 79/120 (65) | ||
| Tgh009 | 1,2443′,5′ | 24.7 | a) Unknown (Actinobacillus suis) | AAO65492 | 65/413 (39) | ||
| b) Glycosyltransferase (Nitrosomonas europaea ATCC 19718) | NP_841416 | 146/423 (34) | |||||
| Tgh010 | 6283′,5′ | 26.4 | Hypothetical membrane protein HH0255 (H. hepaticus ATCC 51449) | NP_859786 | 76/232 (32) | ||
| Tgh012 | 1,3775′ | 22.1 | a) Hypothetical protein Cj1431c (C. jejuni NCTC 11168) | NP_282572 | 112/436 (25) | ||
| b) Probable sugar transferase Cj1422c (C. jejuni NCTC 11168) | NP_282563 | 42/111 (37) | |||||
| Tgh036 | 8713′ | 20.0 | Glycosyl transferase (Nitrosomonas europaea ATCC 19718) | NP_841416 | 115/262 (43) | Linked to Tgh037 | |
| Tgh046 | 6963′ | 23.4 | Hypothetical membrane protein HH0255 (H. hepaticus ATCC 51449) | NP_859786 | 58/232 (25) | ||
| Tgh048 | 9445′ | 22.2 | Probable sugar transferase Cj1422c (C. jejuni NCTC 11168) | NP_282563 | 58/151 (38) | ||
| Tgh056 | 1,0083′,5′ | 25.8 | Hypothetical membrane protein HH0255 (H. hepaticus ATCC 51449) | NP_859786 | 90/247 (36) | ||
| Tgh059 | 1,0253′,5′ | 24.68 | Hypothetical membrane protein HH0255 (H. hepaticus ATCC 51449) | NP_859786 | 83/328 (25) | ||
| Tgh077 | 6045′ | 32.3 | a) Hypothetical protein HH0051 (H. hepaticus ATCC 51449) | NP_859582 | 79/192 (41) | ||
| b) Putative ankyrin 3-like protein (H. hepaticus) | AAL16685 | 39/82 (47) | |||||
| Tgh086 | 227 | 25.0 | Predicted periplasmic or secreted lipoprotein (Nostoc punctiforme) | ZP_00108381 | 24/78 (30) | Linked to Tgh084-88 | |
| Tgh120 | 8855′ | 25.4 | Probable sugar transferase Cj1422c (C. jejuni NCTC 11168) | NP_282563 | 183/292 (62) | ||
| Tgh126 | 2913′ | 29.2 | Probable fucose synthetase Cj1428c (C. jejuni NCTC 11168) | NP_28256 | 55/98 (56) | ||
| Tgh137 | 3043′,5′ | 28.5 | rmlA (C. jejuni ATCC43431) | AAL06018 | 100/100 (100) | ||
| Restriction-modification, recombination and repair (DNA modification) | |||||||
| Tgh014 | 4655′ | 30.3 | Type I restriction-modification system methyltransferase subunit (Trichodesmium erythraeum IMS101) | ZP_00071587 | 34/96 (35) | ||
| Tgh049 | 2,0435′ | 26.6 | a) Hypothetical protein jhp1271 (H. pylori strain J99) | NP_223990 | 312/712 (43) | ||
| b) Site-specific DNA-methyltransferase (H. pylori strain 26695) | NP_208146 | 219/481 (45) | Linked to Tgh050 | ||||
| Tgh076 | 7303′,5′ | 24.8 | a) Hypothetical protein jhp1271 (H. pylori strain J99) | G71827 | 97/197 (49) | ||
| b) Putative adenine-specific DNA methyltransferase (H. pylori 26695) | NP_208146 | 97/200 (48) | |||||
| Tgh084 | 1,3205′ | 26.9 | Putative integrase (Wolinella WS2030) | NP_908131 | 40/124 (32) | Linked to Tgh085-087 | |
| Tgh098 | 3943′,5′ | 27.9 | Putative type I specificity subunit HsdS (C. jejuni RM3200) | AAN33144 | 51/103 (49) | ||
| Tgh108 | 3245′ | 28.4 | Integrase (N. punctiforme) | ZP_00109546 | 35/99 (35) | Linked to Tgh109 | |
| Tgh117 | 5783′ | 21.9 | Integrase-recombinase protein XerCD family WS1221 (W. succinogenes DSMZ 1740) | NP_907402 | 44/119 (36) | ||
| Tgh124 | 3395′ | 25.1 | Probable type IIS restriction-modification enzyme, C-terminal half Cj0032 (C. jejuni NCTC11168) | NP_281254 | 65/101 (64) | Linked to Tgh123 | |
| Tgh128 | 3993′,5′ | 28.9 | Hypothetical protein, putative integrase WS2030 (W. succinogenes DSMZ 1740) | NP_908131 | 42/130 (32) | ||
| Tgh132 | 2833′,5′ | 28.1 | a) Conserved hypothetical protein RB1621 (Pirellula sp. strain 1) | NP_864512 | 20/65 (30) | ||
| b) ATPase involved in DNA repair (P. fluorescens PfO-1) | ZP_00085233 | ||||||
| Tgh133 | 15185′ | 22.3 | a) Conserved hypothetical protein (Rhizobium solanacearum) | NP_520740 | 87/372 (23) | ||
| b) ATPase involved in DNA repair (P. fluorescens PfO-1) | ZP_00085233 | 37/134 (27) | |||||
| Transport | |||||||
| Tgh005 | 9563′,5′ | 22.5 | Conserved hypothetical protein HH0252 (H. hepaticus ATCC 51449) | NP_859783 | 117/318 (36) | ||
| Tgh016 | 7685′ | 23.8 | a) Conserved hypothetical protein HH0252 (H. hepaticus ATCC 51449) | NP_859783 | 45/254 (57) | ||
| b) IcmF (Agrobacterium tumefaciens C58) | AH3088 | 58/206 (28) | |||||
| Tgh038 | 7383′,5′ | 24.0 | a) Conserved hypothetical protein HH0252 (H. hepaticus ATCC 51449) | NP_859783 | 101/241 (41) | ||
| b) IcmF-related protein (V. cholerae O1 biovar E Tor strain NI6961 | NP_232521 | 51/208 (24) | |||||
| Tgh051 | 6795′ | 27.8 | a) Conserved hypothetical protein HH0245 (H. hepaticus ATCC 51449) | NP_859776 | 97/227 (42) | ||
| b) ImpG involved in temperature-dependent protein secretion (R. leguminosarum bv. trifolii) | AAL17805 | 55/235 (23) | |||||
| Tgh073 | 6323′,5′ | 36.1 | TraG (N. gonorrhoeae) | AAK77139 | 46/170 (27) | ||
| Tgh103 | 9143′ | 24.2 | a) Conserved hypothetical protein HH0245 (H. hepaticus ATCC 51449) | NP_859776 | 137/301 (45) | ||
| b) ImpG (R. leguminosarum bv. trifolii) | AAL17805 | 76/303 (25) | |||||
| Tgh105 | 13065′ | 33.1 | a) Conserved hypothetical protein HH0247 (H. hepaticus ATCC 51449) | NP_859778 | 242/294 (82) | ||
| b) ImpC (R. leguminosarum bv. trifolii) | AAL17801 | 115/297 (38) | |||||
| Tgh119 | 3685′ | 27.1 | a) Conserved hypothetical protein HH0250 (H. hepaticus ATCC 51449) | NP_859781 | 62/119 (52) | ||
| b) ImpJ (R. leguminosarum bv. trifolii) | AF361470_2 | 28/108 (25) | |||||
| Chemotaxis | |||||||
| Tgh024 | 13293′ | 28.9 | Probable methyl-accepting chemotaxis signal transduction protein Cj1564 (C. jejuni NCTC 11168) | NP_282692 | 299/436 (68) | ||
| Other (bacteriophage sequence) | |||||||
| Tgh039 | 7545′ | 27.8 | Hypothetical protein y3088, possible phage protein (Yersinia pestis KIM) | NP_670387 | 36/117 (30) | ||
| Tgh088 | 401 | 23.4 | Similar to Bacillus subtilis YwlA (bacteriophage SPBc2) | NP_046626 | 19/78 (24) | Linked to Tgh089 | |
| Small-molecule metabolism | |||||||
| Tgh013 | 2823′,5′ | 17.4 | Tetrapyrrole methylases (Bacillus anthracis A2012) | NP_656000 | 16/41 (39) | ||
| Tgh017 | 8075′ | 27.5 | Hypothetical protein rodA_1 (H. pylori strain J99) | NP_223399 | 71/257 (27) | ||
| Tgh058 | 5235′ | 23.7 | a) Hypothetical protein HH0094 (H. hepaticus ATCC 51449) | AAP76691 | 84/167 (50) | ||
| b) Putative butyryltransferase (E. coli ECA95) | AAK60452 | 73/159 (45) | |||||
| Tgh106 | 9993′ | 25.2 | a) Conserved hypothetical protein HH0090 (H. hepaticus ATCC 51449) | NP_859621 | 171/327 (52) | Linked to Tgh107 | |
| b) Adenylating enzyme CmlK (Streptomyces venezuelae) | AAM01214 | 58/144 (40) | |||||
| Tgh107 | 3445′ | 31.7 | a) Conserved hypothetical protein HH0092 (H. hepaticus ATCC 51449) | NP_859623 | 102/123 (82) | Linked to Tgh106 | |
| b) Dehydrogenases (Rhodobacter sphaeroides) | ZP_00007120 | 73/109 (66) | |||||
| Hypothetical and unknown proteins | |||||||
| Tgh015 | 2513′ | 25.21 | Hypothetical protein HH0213 (H. hepaticus ATCC 51449) | NP_859744 | 31/76 (40) | ||
| Tgh018 | 18813′ | 25.7 | Hypothetical protein HH0242 (H. hepaticus ATCC 51449) | NP_859773 | 265/728 (36) | ||
| Tgh025 | 7835′ | 27.3 | Hypothetical protein HH0235 (H. hepaticus ATCC 51449) | NP_859766 | 50/155 (32) | Linked to Tgh026-028 | |
| Tgh029 | 5675′ | 22.4 | Hypothetical protein HH0253 (H. hepaticus ATCC 51449) | NP_859784 | 28/80 (35) | On the same contig | |
| Tgh030 | 993 | 31.3 | Hypothetical protein HH0254 (H. hepaticus ATCC 51449) | NP_859785 | 185/340 (54) | ||
| Tgh031 | 6773′,5′ | 28.5 | Hypothetical protein HH0383 (H. hepaticus ATCC 51449) | NP_859914 | 137/226 (60) | ||
| Tgh034 | 1965′ | 33.8 | Conserved hypothetical protein HH0243 (H. hepaticus ATCC 51449) | NP_859774 | 45/60 (75) | Linked to Tgh035 | |
| Tgh035 | 347 | 23.0 | Conserved hypothetical protein HH0251 (H. hepaticus ATCC 51449) | NP_859782 | 32/74 (43) | ||
| Tgh045 | 2273′ | 26.7 | Hypothetical protein HH1587 (H. hepaticus ATCC 51449) | NP_861118 | 43/67 (64) | Linked to Tgh044 | |
| Tgh047 | 4223′,5′ | 29.8 | Hypothetical protein HH0242 (H. hepaticus ATCC 51449) | NP_859773 | 37/88 (42) | ||
| Tgh050 | 3895′ | 28.7 | Hypothetical protein Cj0261c (C. jejuni NCTC 11168) | NP_281455 | 72/113 (63) | Linked to Tgh049 | |
| Tgh053 | 8833′,5′ | 23.1 | Hypothetical protein Cj1431c (C. jejuni NCTC 11168) | NP_282572 | 69/280 (24) | ||
| Tgh055 | 2515′ | 23.8 | Hypothetical protein (H. somnus 2336) | ZP_00132709 | 22/64 (34) | Linked to Tgh054 | |
| Tgh061 | 12743′ | 26.1 | Hypothetical protein HH0256 (H. hepaticus ATCC 51449) | NP_859787 | 149/439 (33) | ||
| Tgh071 | 464 | 24.7 | Hypothetical protein HH0212 (H. hepaticus ATCC 51449) | NP_859743 | 78/159 (49) | On the same contig and linked to Tgh070 | |
| Tgh072 | 3163′ | 27.3 | Hypothetical protein HH1374 (H. hepaticus ATCC 51449) | NP_860905 | 43/67 (64) | ||
| Tgh075 | 4913′ | 32.1 | Hypothetical protein HH0390 (H. hepaticus ATCC 51449) | NP_859921 | 57/155 (36) | ||
| Tgh079 | 11363′ | 23.9 | Hypothetical protein VCA0119 (V. cholerae O1 biovar El TOR strain N16961) | NP_232520 | 71/338 (21) | ||
| Tgh080 | 6023′ | 27.0 | Conserved hypothetical protein HH0250 (H. hepaticus ATCC 51449) | NP_859781 | 102/200 (51) | On the same contig | |
| Tgh081 | 335 | 31.2 | Conserved hypothetical protein HH0249 (H. hepaticus ATCC 51449) | NP_859780 | 46/104 (44) | ||
| Tgh082 | 4423′,5′ | 24.4 | Hypothetical protein Cj1431c (C. jejuni NCTC 11168) | NP_282572 | 37/106 (34) | ||
| Tgh083 | 5405′ | 31.1 | Hypothetical protein Chte2282 (C. thermocellum ATCC 27405) | ZP_00061855 | 25/94 (26) | ||
| Tgh085 | 449 | 25.6 | Conserved hypothetical protein HH0278 (H. hepaticus ATCC 51449) | NP_859809 | 50/108 (46) | On the same contig and linked to Tgh084 | |
| Tgh087 | 369 | 28.5 | Uncharacterized conserved protein Npun2806 (N. punctiforme) | ZP_00108380 | 33/111 (29) | ||
| Tgh089 | 305 | 30.1 | Hypothetical protein HH0264 (H. hepaticus ATCC 51449) | NP_859787 | 20/29 (68) | Linked to Tgh088 | |
| Tgh092 | 3033′ | 29.9 | Hypothetical protein HH0386 (H. hepaticus ATCC 51449) | NP_859917 | 50/102 (49) | Linked to Tgh091 | |
| Tgh093 | 287 | 27.8 | Hypothetical protein WS1132 (W. succinogenes DSMZ 1740) | NP_907321 | 47/84 (55) | Linked to Tgh095 | |
| Tgh104 | 392 | 25.7 | Conserved hypothetical protein HH0246 (H. hepaticus ATCC 51449) | NP_859777 | 43/130 (33) | ||
| Tgh115 | 196 | 30.4 | Hypothetical protein WS1734 (W. succinogenes DSMZ 1740) | NP_907859 | 25/66 (37) | Linked to Tgh116 | |
| Tgh121 | 557 | 30.1 | Hypothetical protein HH0550 (H. hepaticus ATCC 51449) | NP_860081 | 135/183 (73) | ||
| Tgh123 | 2283′ | 18.4 | Hypothetical protein ORF No. CR006 (Staphylococcus aureus MR108) | BAC67554 | 34/77 (44) | Linked to Tgh124 |
Only ORFs with significant protein matches in the NCBI database are reported.
The superscript number indicates a partial ORF with the stop codon (3′) and/or the start codon (5′) missing.
Only the first BLAST hit is reported if not otherwise noted as followed: a), first hit; b), second hit.
Identity refers to the protein identity in the region of homology identified by BLAST.
The complete LOS locus of ATCC 43431 was obtained by PCR and DNA sequencing of the junctions between six contigs identified by this study (Fig. 3).
Genes encoding proteins involved in cell envelope and surface structures.
Up to 20 ORFs show significant similarity to ORFs encoding proteins involved in the capsular polysaccharide and LOS biosynthesis pathways. The capsular polysaccharides and LOS have been shown to be the major determinants in the Penner and Hennessy serotyping system (19). Additionally, genes encoding proteins involved in their biosynthesis were shown to be highly divergent between strains of different Penner serotypes (9).
In particular, for the LOS gene cluster, only a few anchor loci such as the waaC gene (encoding heptosyltransferase I) and htrB (encoding lipid A biosynthesis acyltransferase) at one end of the cluster and the waaF gene (encoding heptosyltranserase II) at the other end of the cluster were determined to be conserved, and the genes between these loci were absent or highly divergent in non-serotype O2 strains (9). Subsequent studies by Gilbert et al. (14) determined that at least three conserved classes of LOS genes are present in C. jejuni strains. The three classes, described in detail as A, B, and C, contained 12 ORFs with high sequence similarity, with class C also containing three additional ORFs not present in the A and B classes and the A and B classes sharing an additional ORF absent from class C. Of special importance the three previously described LOS cluster classes, all contained genes required for the production of ganglioside mimics (14).
By microarray and genomic sequencing analysis, the capsule locus (kps) was anchored by genes homologous to E. coli group II capsule genes (51) designated kpsMTEDF at one end of the cluster and kpsCS at the other end, with intervening genes not detected in non-serotype O2 strains (9, 19). With the exception of conserved kps anchor loci (19), no additional class structure(s) has been described for C. jejuni kps clusters.
As described above, the LOS and capsule cluster genes were found to vary between C. jejuni ATCC 43431 and NCTC 11168, which belong to Penner serotypes O:3 and O:2, respectively (Fig. 1, regions PZ5 for the capsular polysaccharide locus and PZ4 for the LOS biosynthesis locus). Consequently, this identification of a new set of genes encoding proteins involved in capsule and LOS biosynthesis in C. jejuni ATCC 43431 is in agreement with its different Penner serotype. In addition, the observed gene variability in the LOS biosynthesis locus is supported by the difference in LOS structure between the two C. jejuni strains (49). In particular, ATCC 43431 lacks sialic acid in its LOS (2), while NCTC 11168 has sialylated LOS (30).
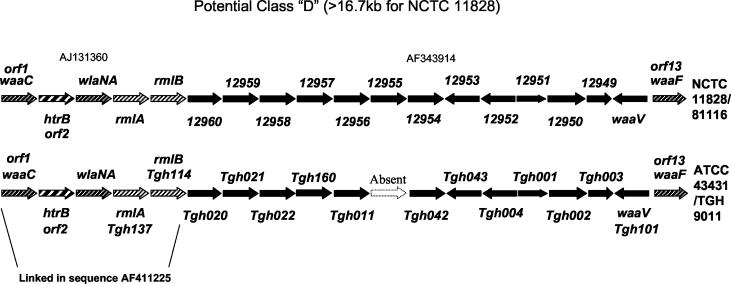
Interestingly, the LOS biosynthesis genes from ATCC 43431 appear highly similar to those of NCTC 11828 (Table 1), which suggests an additional unreported class (class D) of conserved LOS clusters is found in C. jejuni strains (Fig. 3). Of the 12 conserved ORFs described previously (35), the D class contains only the four anchor ORFs 1 and 2 (waaC and htrB) and 12 and 13 (waaV and waaF). Instead, this class shares genes similar to those required for dTDP-rhamnose biosynthesis in several bacteria (28, 29, 45), i.e., rmlA and rmlB homologues, and lacks the neuABC genes (required for sialic acid biosynthesis) (14, 15, 30) and the cst-encoded sialyltransferases involved with LOS sialylation (14). The lack of neu and cst genes thus correlates with the lack of sialic acid in the LOS of ATCC 43431, as has been reported previously (2). The absence of LOS sialylation machinery in class D clusters may have important implications for the pathogenesis of LOS class D strains. In particular, since LOS sialylation is considered an important prerequisite for Guillain-Barré induction, LOS class D strains may be unable to induce this condition.
FIG. 3.
Comparison of the LOS cluster from NCTC 11828/81116 and ATCC 43431 suggests the existence of a fourth class of LOS gene clusters, designated class D. The 11828 sequences from 12960 to waaF were from AF343914 (33). The 11828/81116 sequences from waaC to rmlB were from AJ131360 (11). The sequence data for ATCC 43431 from waaC to rmlB are from AF411225 (13). The gene order and direction in the LOS locus of ATCC 43431 were confirmed by PCR and DNA sequencing of the junctions between each contig identified by this study. The entire DNA sequence from the LOS cluster of this strain has been deposited in the NCBI database under accession number AY501976. This cluster of genes is anchored by waaC and waaF, similarly to the class A, B and C clusters (14), but differs substantially from these by lacking the neu genes and the cst-encoded sialyltransferase. Where relevant, both the suggested gene name and the ORF designations as described previously (14) are shown. The striking finding of numerous conserved genes in ATCC 43431 linked as in 81116 suggests that these two strains represent the same LOS cluster class. One of the 81116 genes was not found (absent) in the sequencing of unique ATCC 43431 ORFs. The glycosyltransferase designated wlaNA was also not identified in this study but was previously reported by Gilbert et al. (13) to be contiguous with waaC, htrB, and rmlAB.
Four ORFs (Tgh010, Tgh046, Tgh056, and Tgh059) show similarity to four different parts of a 3,897-bp hypothetical gene from Helicobacter hepaticus (HH0255), which encodes a predicted membrane-associated protein of unknown function. Interestingly, HH0255 is part of a possible H. hepaticus pathogenicity island named HHGI1 (48). While the relevance of this gene to pathogenesis remains to be demonstrated, HHGI1-negative H. hepaticus strains are affected in their virulence potential compared to HHGI1-carrying strains (48).
Genes encoding restriction modification systems and DNA recombination and repair.
A potentially very important group of unique ATCC 43431 genes contain five ORFs predicted to encode proteins involved in DNA restriction and modification as well as six ORFs likely to encode proteins involved in DNA repair, integration, and recombination. Since all of the DNA restriction and modification systems annotated in NCTC 11168 appear to be present in ATCC 43431 (Fig. 1), ATCC 43431 contains a larger complement of restriction-modification systems than NCTC 11168. While the biological significance of these additional restriction-modification systems remains to be elucidated, their lower G-C content (27%) compared to the remainder of the genome (30.6%) indicates that they may have been acquired through horizontal gene transfer. In support of this hypothesis, restriction-modification systems in other organisms have been proposed to have been transferred between genomes based on their different G-C content, their codon usage bias, their strain specificity, and their common association with mobile genetic elements (23). Despite the fact that a physical link to mobile genetic elements cannot be deduced from our data, the presence in ATCC 43431 of numerous possible integrase-recombinases (Table 1) and a TraG homologue (a protein required for DNA transfer in bacterial conjugation; Tgh073) is indicative of a conjugative lifestyle, which may have contributed to the acquisition of the unique set of genes.
Four ORFs (Tgh084, Tgh108, Tgh117, and Tgh128) exhibit significant homology to integrases that belong to the larger family of tyrosine recombinases. These enzymes are commonly found on transposons and catalyze the process of DNA recombination, promoting the integration of transposons into the host genome. The finding of sequences related to mobile elements such as bacteriophages, transposons, and inserted sequences in ATCC 43431 contrasts sharply with the lack of such sequences in the NCTC 11168 genome (34). Like the presence or absence of mobile elements, the restriction-modification systems in C. jejuni have been shown to be strain specific (9). Of note, the chick colonization strain C. jejuni NCTC 11828/81116 possesses a unique repertoire of restriction-modification systems compared to C. jejuni NCTC 11168, suggesting a potential role for restriction-modification proteins in host colonization (1). In Helicobacter pylori, the presence of certain restriction-modification systems has been shown to be associated with the extent of host response to the bacterial infection (5). While it is tempting to propose a similar function for the ATCC 43431 restriction-modification systems, their role in Campylobacter host colonization and/or virulence needs to be investigated further.
Genes encoding orthologues of Helicobacter hepaticus proteins.
Notably, 36 unique ATCC 43431 DNA fragments out of 130 (≈28% of the new sequences identified) showed high similarities with 28 proteins from H. hepaticus ATCC 51449, in particular with several proteins belonging to a possible pathogenicity island (HH0233 to HH0302). This H. hepaticus island contains 70 ORFs (48), 16 of which have orthologues in C. jejuni ATCC 43431 (Table 1). The G+C content of these genes is substantially different between the genomes, suggesting that both strains may have acquired these genes horizontally from a third common source. To support this hypothesis, both strains are naturally competent to take up DNA, and both strains inhabit the same intestinal niche. Alternatively, these genes may have been exchanged between these two bacterial species.
Interestingly, several C. jejuni orthologues of H. hepaticus genes encode proteins that also have homology to members of the Rhizobium leguminosarum Imp operon (ImpG, ImpC, and ImpJ; Table 1) (6). Highly similar Imp loci have been identified in various human and animal pathogens (such as Pseudomonas aeruginosa and Vibrio cholerae) (6). This locus has been proposed to constitute a new family of operons involved in modification of host responses (6). In R. leguminosarum, the Imp locus is involved in the temperature-dependent secretion of proteins and influences the level of infection in plants (6). The Imp proteins appear to govern host specificity. The interaction of the Imp proteins with a plant-specific complementary resistance gene leads to a hypersensitive response, while the same Imp proteins can act as virulence factors on susceptible host plants, resulting in infection (6). The Imp locus contains a homolog of the IcmF protein that is a component of a type IV secretion system. Interestingly, we have identified an orthologue of H. hepaticus HH0252 that also has similarity with IcmF of Vibrio cholerae, Agrobacterium tumefaciens, and Legionella pneumophila. In L. pneumophila, IcmF is required for human macrophage killing (41).
In summary, our data indicate that C. jejuni ATCC 43431 harbors several H. hepaticus orthologues that are also homologues to genes from the Imp locus. In H. hepaticus, these genes are part of a pathogenicity island that has been shown to be required for the induction of liver disease (48). Based on these observations, one could propose a similar role for these genes in the modulation of Campylobacter virulence. The function of these genes in C. jejuni ATCC 43431 is currently under investigation. The other C. jejuni orthologues of H. hepaticus genes encode proteins involved in small-molecule metabolism or hypothetical proteins of unknown function (Table 1).
Genes encoding other proteins of known and unknown function.
A noteworthy unique ORF of C. jejuni ATCC 43431 encodes a homologue of TraG from Neisseria gonorrhoeae. In other organisms, the TraG protein is an essential component of the type IV secretion apparatus and is required for DNA transfer in bacterial conjugation (8). Its potential nucleoside triphosphatase activity has been proposed to provide the energy required for substrate secretion. Type IV secretion systems have been identified in many pathogens and have been shown to play an important role in bacterial pathogenesis (8). In Helicobacter pylori, the type IV system is required for excretion of the cytotoxin CagA protein into gastric cells (32). Recently, sequencing of the pVir plasmid of C. jejuni 81-176 revealed the presence of genes encoding orthologues of type IV secretion proteins found in H. pylori (4). Mutations in these type IV secretion genes affected the ability to invade human epithelial cells in vitro and decreased virulence in the ferret diarrhea model (4). In summary, the identification of a TraG-like protein in C. jejuni ATCC 43431, together with the identification of IcmF and Imp-like proteins, suggests that this bacterium might be able to produce a type IV secretion system.
In addition to 33 DNA fragments encoding proteins with homology to hypothetical proteins from other organisms, an additional 44 ORFs were predicted (not shown in Table 1). These ORFs do not have significant homology to any known or hypothetical proteins currently in the databases.
Concluding remarks.
To our knowledge, this is the first report describing the successful construction and use of a whole-plasmid shotgun microarray to investigate bacterial diversity. This approach is particularly powerful to identify new genomic DNA sequences from a nonsequenced strain by comparison with a reference sequenced genome. Although the emphasis of this publication has been on the identification of genes unique to C. jejuni ATCC 43431, a similar approach could be used with any other organism. Undoubtedly, the combination of C. jejuni NCTC 11168 and ATCC 43431 microarrays provides valuable information on Campylobacter genetic diversity by identifying both common and unique sets of genes between these two strains. However, this method cannot detect point mutations, chromosomal rearrangements, differences in the number of multicopy genes, or physical location on the chromosome. Nevertheless, this identification of genes unique to C. jejuni ATCC 43431 should enable the construction of a more comprehensive Campylobacter microarray and thus further the investigation of genetic diversity. Of special interest is the identification of a probable fourth class of LOS cluster genes, since the variability of LOS structure is likely to feature importantly in the pathogenic process. Additionally, genes potentially involved with type IV secretion and a possible pathogenicity island are other novel findings.
One of our goals for this study was to identify potential genes encoding proteins that participate in the mechanism of epithelial cell invasion of C. jejuni ATCC 43431. From our data, we failed to identify ORFs with similarity to genes from the pVir plasmid of C. jejuni 81-176, which encodes proteins involved in the mechanism of human epithelial cell invasion. However, we did identify several ORFs encoding proteins with homology to structural components of a type IV secretion apparatus, which may play an important role in human epithelial cell invasion. In addition, we identified a large number of ORFs with homology to genes from a pathogenicity island in H. hepaticus. Future experiments will be necessary to determine the role of these unique genes in the mechanism of human epithelial cell invasion and C. jejuni pathogenesis.
Acknowledgments
We thank I. Turcot and J. Andrus for contributions to this manuscript. We are grateful to all the staff from the OSU and OU microarray core facilities and OU Health Science Center Laboratory for Genomics and Bioinformatics.
This work was supported by National Institutes of Health grants RO1-AI055612 and RR15564.
REFERENCES
- 1.Ahmed, I. H., G. Manning, T. M. Wassenaar, S. Cawthraw, and D. G. Newell. 2002. Identification of genetic differences between two Campylobacter jejuni strains with different colonization potentials. Microbiology 148:1203-1212. [DOI] [PubMed] [Google Scholar]
- 2.Aspinall, G. O., C. M. Lynch, H. Pang, R. T. Shaver, and A. P. Moran. 1995. Chemical structures of the core region of Campylobacter jejuni O:3 lipopolysaccharide and an associated polysaccharide. Eur. J. Biochem. 231:570-578. [PubMed] [Google Scholar]
- 3.Bacon, D. J., R. A. Alm, D. H. Burr, L. Hu, D. J. Kopecko, C. P. Ewing, T. J. Trust, and P. Guerry. 2000. Involvement of a plasmid in virulence of Campylobacter jejuni 81-176. Infect. Immun. 68:4384-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bacon, D. J., R. A. Alm, L. Hu, T. E. Hickey, C. P. Ewing, R. A. Batchelor, T. J. Trust, and P. Guerry. 2002. DNA sequence and mutational analyses of the pVir plasmid of Campylobacter jejuni 81-176. Infect. Immun. 70:6242-6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bjorkholm, B. M., J. L. Guruge, J. D. Oh, A. J. Syder, N. Salama, K. Guillemin, S. Falkow, C. Nilsson, P. G. Falk, L. Engstrand, and J. I. Gordon. 2002. Colonization of germ-free transgenic mice with genotyped Helicobacter pylori strains from a case-control study of gastric cancer reveals a correlation between host responses and HsdS components of type I restriction-modification systems. J. Biol. Chem. 277:34191-34197. [DOI] [PubMed] [Google Scholar]
- 6.Bladergroen, M. R., K. Badelt, and H. P. Spaink. 2003. Infection-blocking genes of a symbiotic Rhizobium leguminosarum strain that are involved in temperature-dependent protein secretion. Mol. Plant-Microbe Interact. 16:53-64. [DOI] [PubMed] [Google Scholar]
- 7.Caldwell, M. B., R. I. Walker, S. D. Stewart, and J. E. Rogers. 1983. Simple adult rabbit model for Campylobacter jejuni enteritis. Infect. Immun. 42:1176-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christie, P. J. 2001. Type IV secretion: intercellular transfer of macromolecules by systems ancestrally related to conjugation machines. Mol. Microbiol. 40:294-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorrell, N., J. A. Mangan, K. G. Laing, J. Hinds, D. Linton, H. Al-Ghusein, B. G. Barrell, J. Parkhill, N. G. Stoker, A. V. Karlyshev, P. D. Butcher, and B. W. Wren. 2001. Whole genome comparison of Campylobacter jejuni human isolates using a low-cost microarray reveals extensive genetic diversity. Genome Res. 11:1706-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Everest, P. H., H. Goossens, J. P. Butzler, D. Lloyd, S. Knutton, J. M. Ketley, and P. H. Williams. 1992. Differentiated Caco-2 cells as a model for enteric invasion by Campylobacter jejuni and C. coli. J. Med. Microbiol. 37:319-325. [DOI] [PubMed] [Google Scholar]
- 11.Fry, B. N., V. Kolorik, B. A. van der Zeijst, and P. J. Coloe. 1998. A gene cluster from Campylobacter jejuni involved in inner core and lipid A synthesis. NCBI accession number AJ131360. Applied Biology and Biotechnology, Royal Melbourne Institute of Technology University, Melbourne, Australia.
- 12.Gaynor, E. C., S. Cawthraw, G. Manning, J. K. MacKichan, S. Falkow, and D. G. Newell. 2004. The genome-sequenced variant of Campylobacter jejuni NCTC 11168 and the original clonal clinical isolate differ markedly in colonization, gene expression, and virulence-associated phenotypes. J. Bacteriol. 186:503-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilbert, M., J. Bellefeuille, M. F. Karwaski, A. M. Cunningham, and W. W. Wakarchuk. 2001. Partial sequence of the lipooligosaccharide biosynthesis locus of Campylobacter jejuni O:3. NCBI accession number AF411225. Institute for Biological Sciences, National Research Council of Canada, Montreal, Canada.
- 14.Gilbert, M., M. F. Karwaski, S. Bernatchez, N. M. Young, E. Taboada, J. Michniewicz, A. M. Cunningham, and W. W. Wakarchuk. 2002. The genetic bases for the variation in the lipo-oligosaccharide of the mucosal pathogen, Campylobacter jejuni. Biosynthesis of sialylated ganglioside mimics in the core oligosaccharide. J. Biol. Chem. 277:327-337. [DOI] [PubMed] [Google Scholar]
- 15.Guerry, P., C. P. Ewing, T. E. Hickey, M. M. Prendergast, and A. P. Moran. 2000. Sialylation of lipooligosaccharide cores affects immunogenicity and serum resistance of Campylobacter jejuni. Infect. Immun. 68:6656-6662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu, L., and D. Kopecko. 2000. Interactions of Campylobacter with eukaryotic cells: gut luminal colonization and mucosal invasion mechanism, p. 191-215. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. ASM Press, Washington, D.C.
- 17.Jin, S., Y. C. Song, A. Emili, P. M. Sherman, and V. L. Chan. 2003. JlpA of Campylobacter jejuni interacts with surface-exposed heat shock protein 90alpha and triggers signalling pathways leading to the activation of NF-kappaB and p38 MAP kinase in epithelial cells. Cell. Microbiol. 5:165-174. [DOI] [PubMed] [Google Scholar]
- 18.Jin, S., A. Joe, J. Lynett, E. K. Hani, P. Sherman, and V. L. Chan. 2001. JlpA, a novel surface-exposed lipoprotein specific to Campylobacter jejuni, mediates adherence to host epithelial cells. Mol. Microbiol. 39:1225-1236. [DOI] [PubMed] [Google Scholar]
- 19.Karlyshev, A. V., D. Linton, N. A. Gregson, A. J. Lastovica, and B. W. Wren. 2000. Genetic and biochemical evidence of a Campylobacter jejuni capsular polysaccharide that accounts for Penner serotype specificity. Mol. Microbiol. 35:529-541. [DOI] [PubMed] [Google Scholar]
- 20.Kim, C. C., E. A. Joyce, K. Chan, and S. Falkow. 2002. Improved analytical methods for microarray-based genome-composition analysis. Genome Biol. 3(11):1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim, N. W., H. Bingham, R. Khawaja, H. Louie, E. Hani, K. Neote, and V. L. Chan. 1992. Physical map of Campylobacter jejuni TGH9011 and localization of 10 genetic markers by use of pulsed-field gel electrophoresis. J. Bacteriol. 174:3494-3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim, N. W., R. Lombardi, H. Bingham, E. Hani, H. Louie, D. Ng, and V. L. Chan. 1993. Fine mapping of the three rRNA operons on the updated genomic map of Campylobacter jejuni TGH9011 (ATCC 43431). J. Bacteriol. 175:7468-7470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kobayashi, I. 2001. Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res. 29:3742-3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Konkel, M. E., and L. A. Joens. 1989. Adhesion to and invasion of HEp-2 cells by Campylobacter spp. Infect. Immun. 57:2984-2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Konkel, M. E., L. A. Joens, and P. F. Mixter. 2000. Molecular characterization of Campylobacter jejuni virulence determinants., p. 217-240. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. ASM Press, Washington, D.C.
- 26.Kopecko, D. J., L. Hu, and K. J. Zaal. 2001. Campylobacter jejuni microtubule-dependent invasion. Trends Microbiol. 9:389-396. [DOI] [PubMed] [Google Scholar]
- 27.Leonard, E. E., 2nd, T. Takata, M. J. Blaser, S. Falkow, L. S. Tompkins, and E. C. Gaynor. 2003. Use of an open-reading frame-specific Campylobacter jejuni DNA microarray as a new genotyping tool for studying epidemiologically related isolates. J. Infect. Dis. 187:691-694. [DOI] [PubMed] [Google Scholar]
- 28.Li, Q., and P. R. Reeves. 2000. Genetic variation of dTDP-L-rhamnose pathway genes in Salmonella enterica. Microbiology 146:2291-2307. [DOI] [PubMed] [Google Scholar]
- 29.Li, Q., M. Hobbs, and P. R. Reeves. 2003. The variation of dTDP-L-rhamnose pathway genes in Vibrio cholerae. Microbiology 149:2463-2474. [DOI] [PubMed] [Google Scholar]
- 30.Linton, D., M. Gilbert, P. G. Hitchen, A. Dell, H. R. Morris, W. W. Wakarchuk, N. A. Gregson, and B. W. Wren. 2000. Phase variation of a beta-1,3 galactosyltransferase involved in generation of the ganglioside GM1-like lipo-oligosaccharide of Campylobacter jejuni. Mol. Microbiol. 37:501-514. [DOI] [PubMed] [Google Scholar]
- 31.Newell, D. G., H. McBride, F. Saunders, Y. Dehele, and A. D. Pearson. 1985. The virulence of clinical and environmental isolates of Campylobacter jejuni. J. Hyg. (London) 94:45-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Odenbreit, S., J. Puls, B. Sedlmaier, E. Gerland, W. Fischer, and R. Haas. 2000. Translocation of Helicobacter pylori CagA into gastric epithelial cells by type IV secretion. Science 287:1497-1500. [DOI] [PubMed] [Google Scholar]
- 33.Oldfield, N. J., A. P. Moran, L. A. Millar, M. M. Prendergast, and J. M. Ketley. 2002. Characterization of the Campylobacter jejuni heptosyltransferase II gene, waaF, provides genetic evidence that extracellular polysaccharide is lipid A core independent. J. Bacteriol. 184:2100-2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M. A. Rajandream, K. M. Rutherford, A. H. van Vliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 35.Pearson, B. M., C. Pin, J. Wright, K. I'Anson, T. Humphrey, and J. M. Wells. 2003. Comparative genome analysis of Campylobacter jejuni using whole genome DNA microarrays. FEBS Lett. 554:224-230. [DOI] [PubMed] [Google Scholar]
- 36.Ribot, E. M., C. Fitzgerald, K. Kubota, B. Swaminathan, and T. J. Barrett. 2001. Rapid pulsed-field gel electrophoresis protocol for subtyping of Campylobacter jejuni. J. Clin. Microbiol. 39:1889-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Russell, R. G., M. O'Donnoghue, D. C. Blake, Jr., J. Zulty, and L. J. DeTolla. 1993. Early colonic damage and invasion of Campylobacter jejuni in experimentally challenged infant Macaca mulatta. J. Infect. Dis. 168:210-215. [DOI] [PubMed] [Google Scholar]
- 38.Sails, A. D., B. Swaminathan, and P. I. Fields. 2003. Clonal complexes of Campylobacter jejuni identified by multilocus sequence typing correlate with strain associations identified by multilocus enzyme electrophoresis. J. Clin. Microbiol. 41:4058-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 40.Schouls, L. M., S. Reulen, B. Duim, J. A. Wagenaar, R. J. Willems, K. E. Dingle, F. M. Colles, and J. D. Van Embden. 2003. Comparative genotyping of Campylobacter jejuni by amplified fragment length polymorphism, multilocus sequence typing, and short repeat sequencing: strain diversity, host range, and recombination. J. Clin. Microbiol. 41:15-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Segal, G., M. Purcell, and H. A. Shuman. 1998. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc. Natl. Acad. Sci. USA. 95:1669-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sellars, M. J., S. J. Hall, and D. J. Kelly. 2002. Growth of Campylobacter jejuni supported by respiration of fumarate, nitrate, nitrite, trimethylamine-N-oxide, or dimethyl sulfoxide requires oxygen. J. Bacteriol. 184:4187-4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Skirrow, M. B., and M. J. Blaser. 2000. Clinical aspects of Campylobacter infection, p. 69-88. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. American Society for Microbiology, Washington, D.C.
- 44.Steinhauserova, I., J. Ceskova, and M. Nebola. 2002. PCR/restriction fragment length polymorphism (RFLP) typing of human and poultry Campylobacter jejuni strains. Lett. Appl. Microbiol. 34:354-358. [DOI] [PubMed] [Google Scholar]
- 45.Stevenson, G., B. Neal, D. Liu, M. Hobbs, N. H. Packer, M. Batley, J. W. Redmond, L. Lindquist, and P. Reeves. 1994. Structure of the O antigen of Escherichia coli K-12 and the sequence of its rfb gene cluster. J. Bacteriol. 176:4144-4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stintzi, A. 2003. Gene expression profile of Campylobacter jejuni in response to growth temperature variation. J. Bacteriol. 185:2009-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stintzi, A., and L. Whitworth. 2003. Investigation of the Campylobacter jejuni cold-shock response by global transcript profiling. Genome Lett. 2:18-27. [Google Scholar]
- 48.Suerbaum, S., C. Josenhans, T. Sterzenbach, B. Drescher, P. Brandt, M. Bell, M. Droge, B. Fartmann, H. P. Fischer, Z. Ge, A. Horster, R. Holland, K. Klein, J. Konig, L. Macko, G. L. Mendz, G. Nyakatura, D. B. Schauer, Z. Shen, J. Weber, M. Frosch, and J. G. Fox. 2003. The complete genome sequence of the carcinogenic bacterium Helicobacter hepaticus. Proc. Natl. Acad. Sci. USA 100:7901-7906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Szymanski, C. M., F. S. Michael, H. C. Jarrell, J. Li, M. Gilbert, S. Larocque, E. Vinogradov, and J. R. Brisson. 2003. Detection of conserved N-linked glycans and phase-variable lipooligosaccharides and capsules from Campylobacter cells by mass spectrometry and high resolution magic angle spinning NMR spectroscopy. J. Biol. Chem. 278:24509-24520. [DOI] [PubMed] [Google Scholar]
- 50.van Spreeuwel, J. P., G. C. Duursma, C. J. Meijer, R. Bax, P. C. Rosekrans, and J. Lindeman. 1985. Campylobacter colitis: histological immunohisto-chemical and ultrastructural findings. Gut 26:945-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Whitfield, C., and I. S. Roberts. 1999. Structure, assembly and regulation of expression of capsules in Escherichia coli. Mol. Microbiol. 31:1307-1319. [DOI] [PubMed] [Google Scholar]
- 52.Yan, W., N. Chang, and D. E. Taylor. 1991. Pulsed-field gel electrophoresis of Campylobacter jejuni and Campylobacter coli genomic DNA and its epidemiologic application. J. Infect. Dis. 163:1068-1072. [DOI] [PubMed] [Google Scholar]