Abstract
Endospores of Bacillus subtilis are encased in a thick, proteinaceous shell known as the coat, which is composed of a large number of different proteins. Here we report the identification of three previously uncharacterized coat-associated proteins, YabP, YheD, and YutH, and their patterns of subcellular localization during the process of sporulation, obtained by using fusions of the proteins to the green fluorescent protein (GFP). YabP-GFP was found to form both a shell and a ring around the center of the forespore across the short axis of the sporangium. YheD-GFP, in contrast, formed two rings around the forespore that were offset from its midpoint, before it eventually redistributed to form a shell around the developing spore. Finally, YutH-GFP initially localized to a focus at one end of the forespore, which then underwent transformation into a ring that was located adjacent to the forespore. Next, the ring became a cap at the mother cell pole of the forespore that eventually spread around the entire developing spore. Thus, each protein exhibited its own distinct pattern of subcellular localization during the course of coat morphogenesis. We concluded that spore coat assembly is a dynamic process involving diverse patterns of protein assembly and localization.
Endospores of the bacterium Bacillus subtilis are encased in a thick, proteinaceous shell known as the coat that helps to protect the dormant cell from hazardous environmental agents. The coat is composed of as many as 60 different proteins (11), but the detailed architecture of the coat and the mechanism of its assembly are poorly understood (3). Two proteins that are known to play critical morphogenetic roles in coat formation are SpoIVA, which is needed for directing assembly of the coat to its proper location around the outer surface of the developing spore (18), and CotE, which is responsible for the assembly of the outer layer of the coat (24).
Sporulation takes place in a two-chamber sporangium consisting of a smaller cell called the forespore, which ultimately becomes the spore, and a larger cell called the mother cell, which nurtures the developing spore. Coat proteins are produced in the mother cell and are then deposited around the outer surface of the forespore, which at the time of coat assembly is entirely contained within the mother cell (as a cell within a cell) (14). Coat proteins are produced under the control of the mother-cell-specific RNA polymerase sigma factors σE and σK (14) and the DNA-binding proteins SpoIIID (10) and GerE (13, 23). Recently, additional genes in the mother cell line of gene expression have been identified by gene microarray analysis (5). In ongoing work, we have created fusions of the coding sequence for the green fluorescent protein (GFP) to large numbers of genes identified in this manner. Here we report on the subcellular localization of three previously uncharacterized proteins produced under the control of σE. These proteins are YabP, which is encoded within an operon that also encodes the previously studied YabQ protein (2), YheD, and YutH. We show here that all three of these proteins localize to the assembling coat, but the detailed pattern of subcellular localization is different for each protein and differs from that previously described for other coat proteins. We concluded that protein localization during the process of spore coat assembly is more intricate than previously appreciated.
MATERIALS AND METHODS
General methods.
All cloning steps were performed by utilizing Escherichia coli strain DH5α. Plasmids used for single-recombinant integration were isolated from E. coli strain TG1, which allows for isolation of concatenated plasmids, which provide a higher transformation frequency for single-recombination integration. The parent strain for all Bacillus subtilis strains was PY79 (22).
Plasmid construction.
pCVO119 was synthesized by placing the multiple cloning site of pBluescript into pKL147 (12), which was done as follows. pBluescript was digested with SacI and then treated with T4 DNA polymerase as described previously (19) to produce blunt ends. The vector was then digested with XhoI, and the released 85-bp fragment was purified. pKL147 was digested with EcoRI and treated with T4 DNA polymerase to produce filled-in, blunt ends. The vector was then digested with XhoI and gel purified. The 85-bp fragment was then ligated to the pKL147 backbone to produce pCVO119.
pCVO122 was constructed by amplifying yabP and 200 bp of its promoter sequence by PCR from PY79 genomic DNA with the oligonucleotides YabP5′-3 (GGACGGATCCCGGCCAAAAGCTTGTAACGG) and YabP1-3′-3 (GGACCTCGAGTTTAAACAACTTGCTAAAAAACCC). The resulting DNA fragment was digested with BamHI and XhoI (restriction sites introduced into oligonucleotides are underlined in the sequences) and cloned in frame upstream of the gfp gene in the similarly digested vector pCVO119, forming a yabP-gfp fusion with a five-codon linker.
pCVO124 was constructed by amplifying yabQ by PCR from PY79 genomic DNA with primers YabQ5′-1 (GGACTCTAGAATGACGCTGACGACACAATTC) and YabQ3′-1 (GGACCTCGAGTCTCTTCAAAAAACGTG) by using PY79 genomic DNA as the template. The resulting fragment was digested with XbaI and XhoI and cloned in frame upstream of the gfp gene in pCVO119 digested with SpeI and XhoI to produce a yabQ-gfp fusion with a five-codon linker.
pCVO151 was constructed by amplifying the yabPQ-gfp operon, including 200 bp of upstream, promoter-containing DNA, by PCR from genomic DNA of strain CVO1026 by using primers yabP5′-3 and jd833 (GGCACGGGATCCTTATTTGTATAGTTCATCCATGC). The resulting fragment was digested with BamHI and HindIII and cloned into similarly digested amyE integration vector pDG364 (8).
pCVO152 was constructed by amplifying the yabP-gfp fusion by PCR by using oligonucleotides yabP5′-3 and jd833 from genomic DNA of strain CVO1024. The resulting fragment was digested with BamHI and HindIII and cloned into similarly digested amyE integration vector pDG364, resulting in pCVO152.
pCVO139 was constructed by cloning a BamHI-HindIII fragment from pCVO122 containing the yabP-gfp fusion into similarly digested pDG364.
pCVO197 was constructed by amplifying the coding region of yabP by PCR with oligonucleotide spoVM-RBS (GGACGCATGCGGAGGGGACAAAAATGAATTCATATTATGATCAAAAAGGTTC), which contains the sequence of the ribosome-binding site of the spoVM gene (boldface type) upstream of the yabP start codon, and oligonucleotide GFP3′-3 (GGACAAGCTTTTATTTGTATAGTTCATCCATGC). The resulting fragment was digested with HindIII and SphI and cloned into similarly digested pDR111, an amyE integration vector that contains a copy of the isopropyl-β-d-1-thiogalactopyranoside (IPTG)-inducible Phyperspank promoter (a gift from D. Rudner), to create a Phyperspank-yabP-gfp transcriptional fusion.
pCVO293 was constructed by amplifying yheD by PCR from PY79 genomic DNA by using primers yheD5′-1 (GACGGATCCCGCTGAAGCAAGACGGACTTTTG) and yheD3′-1 (GACCTCGAGCGAAGGCCACAATGCTTCCG). The resulting DNA fragment was digested with BamHI and XhoI and cloned in frame upstream of the gfp gene into similarly digested pCVO119.
pPE61 was constructed by amplifying 569 bp of the 3′ end of yutH by PCR from PY79 genomic DNA by using primers PE691 (CGTATCCCGGGATCCCCTCCGCATGAGCCATTCGATAAA) and PE692 (CGTCTAGCCCTCGAGCCTTGAGCTGCCTTTGCCGAGCCA). The resulting DNA fragment was digested with BamHI and XhoI and cloned in frame upstream into similarly digested pCVO119.
Strain construction.
CVO1024 (yabP-gfp) was produced by transforming PY79 with pCVO122. Transformants were selected for resistance to spectinomycin (Spcr). CVO1026 (yabQ-gfp) was produced by transforming PY79 with pCVO124. Transformants were selected for Spcr. CVO1084 (amyE::yabP-gfp) was produced by transforming PY79 with pCVO152 that had been linearized with KpnI. Transformants were selected for resistance to chloramphenicol (Cmr) and tested for loss of amylase activity. CVO1088 (amyE::yabPQ-gfp) was produced by transforming PY79 with pCVO151 that had been linearized with KpnI. Transformants were selected for Cmr and tested for loss of amylase activity. CVO1111 (ΔyabP amyE::yabPQ-gfp) was constructed by transforming strain RL2243 (yabP::spc) (6) with genomic DNA of strain CVO1088. Transformants were selected for Cmr and tested for loss of amylase activity. CVO1183 (amyE::yabP-gfp) was constructed by transforming PY79 with linearized pCVO139 and selecting transformants for Cmr. CVO1202 was constructed by transforming RL2244 (yabQ::tet) (6) with genomic DNA of strain CVO1183. Transformants were selected for Cmr and tested for resistance to tetracycline (Tcr) and loss of amylase activity. CVO1219 (amyE::PhyperspankyabP-gfp) was constructed by transforming PY79 with pCVO197 that had been linearized with BglII. Transformants were selected for Spcr and tested for loss of amylase activity. Strain CVO1233 (spoIIGB::erm amyE::PhyperspankyabP-gfp) was produced by transforming RL1061 (spoIIGB::erm) (9) with genomic DNA of CVO1219. Transformants were selected for Spcr and tested for resistance to erythromycin (Emr) and loss of amylase activity. Strain CVO1728 (yheD-gfp) was constructed by transforming PY79 with linearized pCVO293. Transformants were selected for Spcr. Chromosomal DNA from CVO1728 was used to transform RL1397 (spoIVA::neo) and RL322 (cotE::cat) (4) to Spcr to create PE556 and PE557, respectively.
PE479 was obtained by transformation of PY79 with pPE61, followed by selection for Spcr. Chromosomal DNA was prepared from strain PE479 and used to transform RL1397 (spoIVA::neo) and RL322 (cotE::cat) (4) to Spcr to create PE547 and PE500, respectively.
Measuring sporulation efficiency.
Sporulation efficiency was determined with a heat resistance assay, as described previously (7). Briefly, cells were grown at 37°C in Difco sporulation medium for 24 to 30 h. The culture was serially diluted 10-fold in T base supplemented with 1 mM MgSO4 six times. Aliquots (100 μl) of the dilutions were plated on Difco sporulation medium agar plates. The dilutions were then heated at 80°C for 20 min, and 100-μl aliquots of the heat-treated dilutions were plated on agar plates containing Difco sporulation medium. The numbers of CFU were determined after overnight incubation at 37°C.
Microscopy.
To prepare a culture for microscopy, a strain was grown overnight at 25°C in growth medium to an optical density at 600 nm of 0.5 to 0.7. Sporulation was induced by resuspension of the cells in Sterlini-Mandelstam medium (20) unless indicated otherwise; this time point represented h 0 of sporulation. For strains that carried a gene under the control of an inducible promoter, IPTG (Sigma, St. Louis, Mo.) was added to a concentration of 1 mM to induce gene expression at this time. Alternatively, where indicated below, 1 ml of Difco sporulation medium was inoculated with a single colony of the strain, which was grown for 16 to 18 h at 25 or 37°C (7). Cells were prepared for microscopy by centrifuging a 200-μl aliquot for about 2 min in a microcentrifuge at the times indicated below. The cells were resuspended in 10 μl of phosphate-buffered saline containing 1.5 μg of FM4-64 per ml. Three microliters was placed on a microscope slide and covered with a coverslip that had been treated for approximately 30 s with poly-l-lysine (Sigma). Cells were observed with an Olympus BX60 fluorescence microscope. Typical acquisition times ranged from 400 to 1,000 ms for GFP and were 1,000 ms for FM4-64. Images were captured and cropped by using METAMORPH software. Some additional adjustments were made with Adobe Photoshop.
Deconvolution microscopy was performed as follows. Cells of strain CVO1024 were induced to sporulate and prepared for microscopy as described above. The microscopy was carried out with an inverted DeltaVision microscope. Twenty-five images of optical sections showing fluorescence from GFP were collected with a spacing of 0.1 μm. The images were deconvolved through 15 iterations by using the DeltaVision deconvolution software.
RESULTS AND DISCUSSION
Fusions of GFP to YabP and YabQ.
To study the subcellular localization of YabP and YabQ, we fused the coding sequence for GFP in frame to the 3′ terminus of the gene for each protein. The resulting fusions, yabP-gfp and yabQ-gfp, were introduced into the chromosome of B. subtilis strain PY79 by double (marker replacement) recombination at the amyE locus, producing strains CVO1084 and CVO1088, respectively. Both strains were merodiploid, containing one copy of the wild-type gene at its original locus and one copy of the fusion at the amyE locus. When introduced into a strain with a deletion of the yabQ gene (creating strain CVO1111), the yabQ-gfp fusion was able to rescue the block in sporulation caused by the null mutation; cells with a deletion of the yabQ gene sporulated at a 104-fold-lower frequency (as judged by the production of heat-resistant spores) than the wild type sporulated, whereas CVO1111 produced spores at a frequency similar to that of the wild type. We concluded that the YabQ-GFP fusion protein was largely if not fully functional. Nonpolar deletion of yabP did not cause a detectable phenotype (2), and so it was not possible to assess whether the corresponding fusion protein was functional.
YabQ-GFP forms a shell around the forespore.
Strains CVO1084 and CVO1088 were treated with the vital membrane stain FM4-64 and examined by fluorescence microscopy. Figure 1B shows the results for strain CVO1088. The YabQ-GFP fusion protein was localized to the septum in sporangia that had undergone asymmetric division (data not shown), to the membrane migrating around the forespore in sporangia that were undergoing engulfment (Fig. 1B, cell on the right indicated by an arrow), and to the outer membrane surrounding the forespore in sporangia in which the forespore was completely pinched off as a free protoplast within the mother cell (Fig. 1B, cell on the left indicated by an arrow). The latter could be recognized by the absence of staining by FM4-64, which is membrane impermeable and hence unable to gain access to the outer forespore membrane when it is topologically isolated from the cytoplasmic membrane. Our results also show that YabQ-GFP is maintained on the spore coat during maturation and release of the mature spore (Fig. 1B, bottom panels). The localization of YabQ was investigated previously by Asai et al. (2), and our findings are in agreement with their findings.
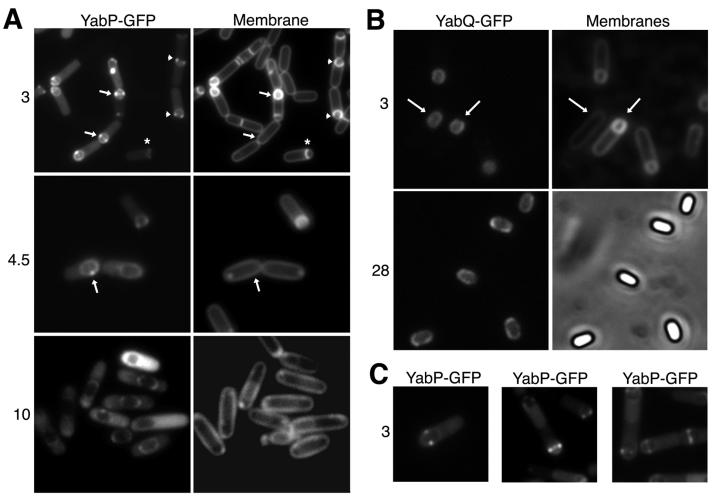
FIG. 1.
Subcellular localization of YabP-GFP and YabQ-GFP. (A) Localization of YabP-GFP. Cells of strain CVO1084 (amyE::yabP-gfp) were induced to sporulate, stained with the membrane dye FM4-64, and prepared for microscopy at the times (in hours) indicated on the left as described in Materials and Methods. YabP-GFP fluorescence is shown in the left panels, and the membrane staining is shown in the right panels. The arrows and arrowheads indicate corresponding cells in adjacent panels that have almost completed engulfment and are in the process of engulfing the forespore, respectively. The asterisks indicate a cell in which engulfment of the forespore is in a very early stage but in which YabP-GFP is nonetheless already concentrated on the sporulation septum. (B) Localization of YabQ-GFP. Cells of strain CVO1088 (amyE::yabPQ-gfp) were induced to sporulate and observed at the times (in hours) indicated on the left. The cells viewed after 3 h of sporulation were additionally stained with the membrane dye FM4-64. The panels on the left show the YabQ-GFP fluorescence. The panels on the right show the membrane dye fluorescence (top) or phase microscopy (bottom) of corresponding cells. The arrows indicate corresponding cells in the left and right panels. YabQ-GFP was concentrated on the outer forespore membrane in all cells examined. (C) Localization of YabP-GFP in the absence of YabQ. Cells of strain CVO1202 (yabQ::tet amyE::yabP-gfp) were induced to sporulate and observed after 3 h. Note the bright staining of the dots and the greatly decreased staining of the outer forespore membrane compared to the staining of YabP-GFP in a wild-type background (as shown in panel A).
YabP-GFP forms a ring around the forespore.
The localization pattern of YabP-GFP was more complex and dynamic than the pattern observed for YabQ-GFP. Like YabQ-GFP, YabP-GFP was initially concentrated at the sporulation septum (for example, see the cell labeled with an asterisk in the h 3 panels of Fig. 1A; since at this stage of sporulation the yabP-gfp gene had been transcribed for only a short time, YabP-GFP staining in this cell was weak). Then as the mother cell started to engulf the forespore, YabP-GFP became enriched at the leading edge of the engulfment membrane (the region where the septal membrane touches the cytoplasmic membrane; arrowheads in the h 3 panels of Fig. 1A). After the forespore was completely engulfed by the mother cell, YabP-GFP became concentrated in two bright dots on opposite sides of the forespore along the short axis of the sporangium (arrows in h 3 panels of Fig. 1A). In addition, we also detected a low level of YabP-GFP staining around the entire forespore membrane; YabP-GFP formed a shell around the forespore, similar to that observed for YabQ-GFP. Thus, there were two patterns of localization of YabP-GFP, one in which YabP-GFP decorated the entire outer forespore membrane and another in which YabP-GFP was concentrated in two dots on the sides of the forespore.
Deconvolution microscopy of sporulating cells of strain CVO1084, which allowed us to view thin (∼0.1-μm) focal planes without the interfering unfocused light from other focal planes, revealed that the dots on the side of the forespore represented a ring that encircled the entire forespore (Fig. 2). YabP-GFP was detected as a single dot at focal planes that represented the region where the cell was in contact with the glass slide (Fig. 2, panel 1). At the focal planes corresponding to the middle portion of the cell (Fig. 2, panels 2 to 5), YabP-GFP staining separated into two dots that gradually spread further apart as the focal plane was moved up through the cell. At the top of the cell YabP-GFP was once again detected as a single dot (Fig. 2, panels 6 to 8).
FIG. 2.
Deconvolution microscopy of sporulating cells producing YabP-GFP. Cells of strain CVO1084 (amyE::yabP-gfp) were prepared for deconvolution microscopy as described in Materials and Methods. Panel 1 shows the focal plane closest to the slide, and each consecutive image is ∼0.1 μm farther from the slide. Note the increase in the distance between the dots of YabP-GFP fluorescence as the focal plane moved up through the first five images. The last three images show that the distance between the dots decreased until they combined to form a single dot in panel 8, which shows the region of the cell farthest from the slide.
The ring of YabP-GFP staining disappeared from the outer forespore membrane as sporulation progressed. In many cells the YabP-GFP staining initially became concentrated in a single bright dot on one side of the engulfed forespore (arrow in h 4.5 panels of Fig. 1A), which we assumed represented an intermediate in the breakdown of the ring structure. During late stages of sporulation we did not detect any specific YabP-GFP staining at the outer forespore membrane, and most of the YabP-GFP was detected in the cytoplasm (Fig. 1A, h 10 panels). No YabP-GFP was detected in the released spore (data not shown). Interestingly, in other work we have found that this dispersal of YabP-GFP from the outer forespore membrane later in sporulation is blocked in the absence of SpoIVA (data not shown). SpoIVA is known to play a critical role in coat formation, forming a shell around the outer forespore membrane, which is required for proper localization of the assembling coat around the outer surface of the developing spore (18). Conceivably, SpoIVA, or a coat-associated protein(s) recruited by SpoIVA, displaces YabP from the outer forespore membrane, causing it to be released into the cytoplasm.
Ring formation by YabP does not depend on YabQ.
Because yabP and yabQ are in the same operon, it seemed possible that the localization of YabP might depend in whole or in part on YabQ. To investigate this, we introduced the yabP-gfp fusion into a strain with a deletion of the yabQ gene. The localization of YabP-GFP in the resulting strain (CVO1202) was subtly altered. Ring formation by YabP-GFP was unimpaired, but the staining around the entire outer forespore membrane was greatly diminished (Fig. 1C). We concluded that YabP-GFP is recruited to the outer forespore membrane by YabQ but that the assembly of the protein into a ring occurs in a YabQ-independent manner.
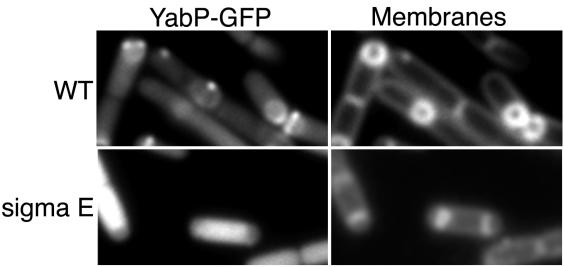
The localization pattern of several previously investigated proteins (SpoVM and the SpoIVFA-SpoIVFB-BofA complex) whose production in the mother cell is under the control of σE is known to depend on an as-yet-unidentified σE-controlled gene product(s) (15, 21). To test if the localization of YabP-GFP was similarly dependent on σE, we placed yabP-gfp under the control of the IPTG-inducible promoter Phyperspank (16) so that the protein could be synthesized in a σE-independent manner. In a strain harboring the Phyperspank-yabP-gfp construct (CVO1219), the fusion protein exhibited a normal pattern of localization (Fig. 3, top panels). However, when synthesis of YabP-GFP was induced in a strain lacking σE (CVO1233), YabP-GFP failed to localize in a specific manner (Fig. 3, bottom panels). Instead, the fusion protein exhibited a uniform pattern of fluorescence throughout the mother cell cytoplasm. Thus, at least two gene products influence the distribution of YabP-GFP: YabQ, which is required for the adherence of the fusion protein around the entire outer forespore membrane, and another, unidentified, gene product produced under the control of σE.
FIG. 3.
Subcellular localization of YabP-GFP requires σE-directed gene expression. Transcription of yabP-gfp was induced by addition of IPTG in strains carrying the Phyperspank-yabP-gfp translational fusion in backgrounds with and without σE activity (strains CVO1219 and CVO1233, respectively). Cells were prepared for microscopy 3 h after the induction of sporulation, as described in the legend to Fig. 1. The YabP-GFP fusion was recruited to the outer forespore membrane normally when it was expressed in the wild-type (WT) background from the inducible promoter (top panels). In the absence of σE, YabP was not recruited to the sporulation septum, and the protein was detected throughout the cytoplasm (bottom panels).
YheD-GFP forms two rings around the forespore.
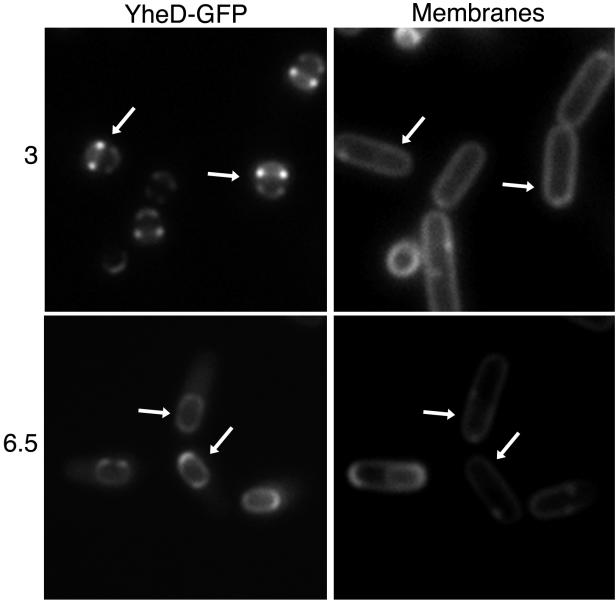
Next, we investigated the subcellular localization of the product of the σE-controlled gene yheD. The gene for GFP was joined to the 3′ end of the 429-codon yheD open reading frame. (Because a yheD mutation did not exhibit a conspicuous phenotype, we were unable to assess the functionality of the fusion protein.) Plasmid DNA containing the yheD-gfp fusion was integrated into the chromosome by single-reciprocal recombination at yheD, yielding strain CVO1728 in which gfp was fused to a full-length copy of yheD. YheD-GFP displayed a striking localization pattern; the fusion protein was concentrated in four dots that were juxtaposed on opposite sides of the engulfed forespore across the short axis of the sporangium (Fig. 4, top panels). We interpreted these images to indicate that YheD-GFP forms two rings that encircle the forespore. (The two rings were in every case examined in a plane perpendicular to the cytoplasmic membrane, which appeared to rule out the possibility that the two rings were part of a spiral.) In almost all sporangia examined the ring on the mother cell proximal side of the forespore was significantly brighter than the ring nearest the mother cell distal pole. Interestingly, the location of the YheD-GFP rings did not overlap the location of the YabP-GFP ring. Whereas YabP-GFP rings were located at the middle of the forespore, at its widest point, the YheD-GFP rings were offset from the middle. At later stages of sporulation, the YheD-GFP rings disappeared and YheD-GFP was redistributed to form a complete shell around the forespore (Fig. 4, bottom panels).
FIG. 4.
Subcellular localization of YheD-GFP. Cells of strain CVO1728 (yheD-gfp) were induced to sporulate, stained with the membrane dye FM4-64, and prepared for microscopy as described in Materials and Methods. GFP fluorescence is shown in the left panels, and membrane staining is shown in the right panels. Note the distinct subcellular localization pattern of YheD-GFP around the outer forespore membrane at 3 h after induction of sporulation (top panels). Engulfment had been completed in these cells, which greatly decreased the staining of the forespore. At 6.5 h after the induction of sporulation (bottom panels), YheD-GFP was redistributed to form a complete shell around the outer forespore membrane. The arrows indicate corresponding cells in the left and right panels.
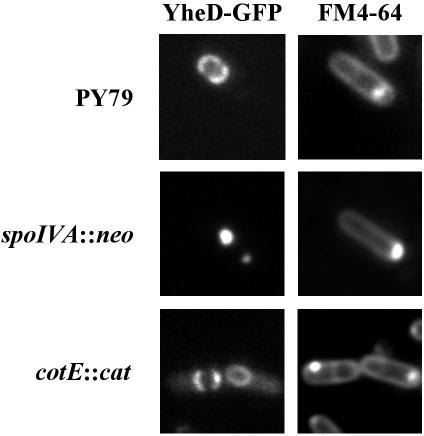
As a further indication that YheD is a coat protein, localization around the forespore was found to be dependent on SpoIVA, which, as indicated above, is known to anchor the coat to the outer surface of the engulfed forespore (18) (Fig. 5). Localization of YheD was not, however, affected by the absence of CotE, which is known to be needed for the assembly of the outermost layer of the spore coat (24) (Fig. 5). This result indicates that YheD-GFP was associated with the inner layer of the spore coat. This is in contrast to the localization of YabP-GFP, which is recruited to the outer forespore membrane in the absence of SpoIVA and thus also in the absence of CotE.
FIG. 5.
Subcellular localization of YheD-GFP depends on SpoIVA. Cells harboring a yheD-gfp fusion were grown for 18 h in Difco sporulation medium at 37°C, stained with the membrane dye FM4-64, and prepared for microscopy as described in Materials and Methods. YheD-GFP was visualized in a otherwise wild-type derivative of strain PY79 (strain CVO1728), in a spoIVA mutant (PE556), and in a cotE mutant (PE557), as indicated on the left. Representative sporangia are shown. Staining with the membrane dye FM4-64 is shown in the right panels, and GFP fluorescence is shown in the left panels.
Dynamic localization pattern of the coat protein YutH-GFP.
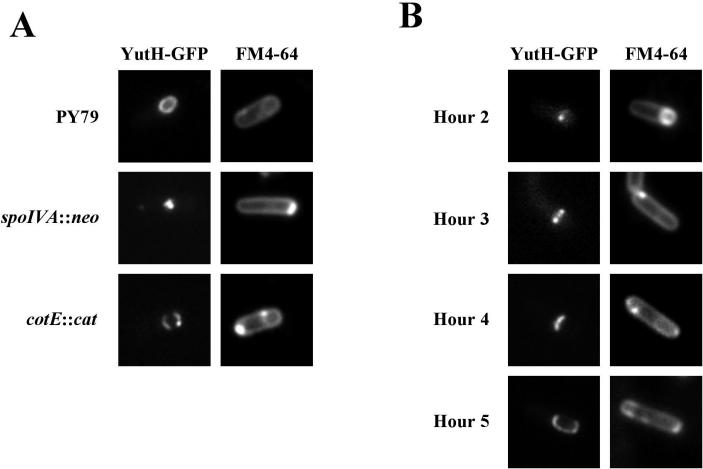
Finally, we investigated the localization of YutH, which is paralogous to the previously identified coat proteins CotI (11) and CotS (1). A plasmid harboring a yutH-gfp fusion was integrated into the chromosome at yutH by single-recombination integration. Sporulating cells harboring the fusion displayed a ring of fluorescence around the engulfed forespore, a pattern consistent with the pattern expected for a coat protein (Fig. 6A). As a further indication that YutH is a coat protein, localization of YutH-GFP around the forespore was dependent on SpoIVA but not on CotE (Fig. 6A). Thus, as in the case of YheD-GFP (see above), we inferred that YutH-GFP was associated with the inner layer of the spore coat. (As in the cases described above, a mutation in yutH did not result in a conspicuous phenotype, and hence the functionality of the YutH-GFP fusion could not be determined.)
FIG. 6.
Subcellular localization of YutH-GFP. Cells harboring a yutHΩyutH-gfp fusion were induced to sporulate, stained with the membrane dye FM4-64, and prepared for microscopy as described in Materials and Methods. (A) Fluorescence micrographs of representative cells producing YutH-GFP after growth for 18 h in Difco sporulation medium at 37°C. YutH-GFP was visualized in an otherwise wild-type derivative of strain PY79 (strain PE479), in a spoIVA mutant (PE547), and in a cotE mutant (PE500), as indicated on the left. Staining with the membrane dye FM4-64 is shown in the right panels, and GFP fluorescence is shown in the left panels. (B) Time course of YutH-GFP localization during sporulation. Cells of strain PE479 were sporulated by suspension in Sterlini-Mandelstam medium at 37°C, and samples were collected at the times indicated and analyzed by fluorescence microscopy. Only representative cells are shown. Staining with the membrane dye FM4-64 is shown in the right panels, and GFP fluorescence is shown in the left panels.
Time course experiments revealed a novel series of intermediates in the incorporation of YutH-GFP into the coat (Fig. 6B). Shortly after activation of σE, 2 h after suspension in sporulation medium, a single dot of YutH-GFP fluorescence was observed close to the engulfing forespore. Once engulfment was completed at h 3, two dots were observed along the short axis of the sporangium, suggesting that YutH-GFP had formed a ring. However, in contrast to the YabP-GFP and YheD-GFP rings, which were located at or near the center of the forespore, the YutH ring was at the extreme mother-cell-proximal end of the forespore and did not completely encircle it. By h 4, the ring had become a polar cap on the mother-cell-proximal side of the forespore. By h 5, the cap had spread to encircle most of the forespore.
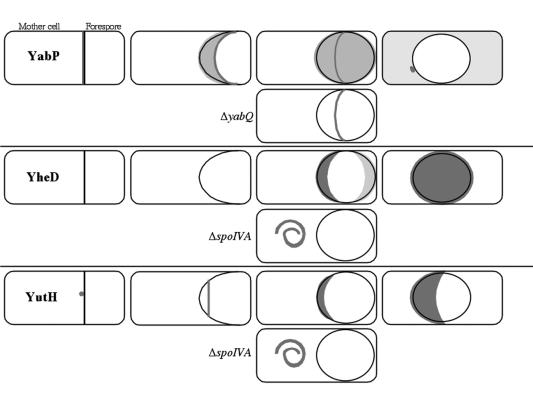
A model summarizing the results of the subcellular localization studies in this investigation is presented in Fig. 7. We concluded that protein localization during sporulation is richer and more intricate than previously recognized. Not only do proteins coalesce into shell-like structures that encase the developing spore, but, as we now see, some proteins (i.e., YabP, YheD and YutH) (Fig. 7) assemble into distinct rings that encircle the forespore during the process of coat assembly. Presumably, these rings contribute to the assembly of the spore coat, but how they do so is not yet apparent. A similar dynamic pattern of localization has been detected for the germination protein GerQ, a spore coat protein that is also dependent on SpoIVA for proper localization. In initial stages of coat formation GerQ is detected as a single focus on the mother cell side of the forespore, and at later stages it is distributed in a homogeneous shell around the forespore (17). A principal challenge for the future is to elucidate the nature of the spatial cues that dictate the characteristic positioning of YabQ, YabP, YheD, YutH, GerQ, and other development-specific proteins during the course of spore coat morphogenesis.
FIG. 7.
Model of localization of YabP, YheD, and YutH during sporulation. The cartoons show the following progressive stages of sporulation (from left to right): septation, engulfment in progress, engulfment completed, and maturing forespore. YabP initially associates with the sporulation septum and then migrates with the mother cell membrane that is engulfing the forespore, where it is concentrated at its leading edge. After engulfment is complete, YabP is concentrated in a ring that surrounds the forespore at its widest point but is also present as a shell around the outer forespore membrane. The latter pattern of localization is dependent on YabQ, as shown in the cartoon depicting the YabP localization in a yabQ mutant cell. At later stages of sporulation YabP is found primarily in the cytosol of the mother cell and occasionally as a small dot on the mother cell side of the forespore. YheD is detected initially in two caps at the poles of the forespore. Ultimately, it forms a complete shell around the forespore. Recruitment of YheD to the outer forespore depends on SpoIVA. YutH is initially detected as a thin cap at the mother cell side of the forespore, which gradually covers the outer forespore membrane as sporulation progresses. Localization of YutH on the outer forespore membrane is dependent on SpoIVA. Proteins are indicated by grey shading, and membranes are indicated by black lines; the intensity of shading indicates the relative amount of protein as detected by fluorescence microscopy.
Acknowledgments
We thank J. Kemp for assistance with the deconvolution microscopy, D. Rudner for the gift of pDR111, and A. Driks for helpful advice on the manuscript.
This work was supported by National Institutes of Health National Research Service Award GM20165 to C.V.O., a Swiss National Science Foundation postdoctoral fellowship and a Merck Core Educational Support Program to P.E., and National Institutes of Health grant GM18568 to R.L.
REFERENCES
- 1.Abe, A., H. Koide, T. Kohne, and K. Watabe. 1995. A Bacillus subtilis spore coat polypeptide gene, cotS. Microbiology 141:1433-1454. [DOI] [PubMed] [Google Scholar]
- 2.Asai, K., H. Takamatsu, M. Iwano, T. Kodama, K. Watabe, and N. Ogasawara. 2001. The Bacillus subtilis yabQ gene is essential for formation of the spore cortex. Microbiology 147:919-927. [DOI] [PubMed] [Google Scholar]
- 3.Driks, A. 2002. Proteins of the spore core and coat, p. 527-536. In A. Sonenshein, J. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives, ASM Press, Washington, D.C.
- 4.Driks, A., S. Roels, B. Beall, C. P. Moran, and R. Losick. 1994. Subcellular localization of proteins involved in the assembly of the spore coat of Bacillus subtilis. Genes Dev. 8:234-244. [DOI] [PubMed] [Google Scholar]
- 5.Eichenberger, P., S. Jensen, E. Conlon, C. van Ooij, J. Silvaggi, J. Gonzalez-Pastor, M. Fujita, S. Ben-Yehuda, P. Stragier, J. Liu, and R. Losick. 2003. The sigma E regulon and the identification of additional sporulation genes in Bacillus subtilis. J. Mol. Biol. 327:945-972. [DOI] [PubMed] [Google Scholar]
- 6.Fawcett, P., P. Eichenberger, R. Losick, and P. Youngman. 2000. The transcriptional profile of early to middle sporulation. Proc. Natl. Acad. Sci. USA 97:8063-8068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harwood, C. R., and S. M. Cutting. 1990. Molecular biological methods for Bacillus. John Wiley and Sons, New York, N.Y.
- 8.Karmazyn-Campelli, C., L. Fluss, T. Leighton, and P. Stragier. 1992. The spoIIN279(ts) mutation affects the FtsA protein of Bacillus subtilis. Biochimie 74:689-694. [DOI] [PubMed] [Google Scholar]
- 9.Kenney, T. J., and C. P. Moran. 1987. Organization and regulation of an operon that encodes a sporulation-essential sigma factor in Bacillus subtilis. J. Bacteriol. 169:33329-33339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kunkel, B., L. Kroos, H. Poth, P. Youngman, and R. Losick. 1989. Temporal and spatial control of the mother-cell regulatory gene spoIIID of Bacillus subtilis. Genes Dev. 3:1735-1744. [DOI] [PubMed] [Google Scholar]
- 11.Lai, E. M., N. Phadke, M. Kachman, R. Giorno, S. Vazquez, J. A. Vazquez, J. R. Maddock, and A. Driks. 2003. Proteomic analysis of the spore coats of Bacillus subtilis and Bacillus anthracis. J. Bacteriol. 185:1443-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lemon, K. P., and A. D. Grossman. 1998. Localization of bacterial DNA polymerase: evidence for a factory model of replication. Science 282:1516-1519. [DOI] [PubMed] [Google Scholar]
- 13.Moir, A., E. Lafferty, and D. A. Smith. 1979. Genetic analysis of spore germination mutants of Bacillus subtilis 168: the correlation of phenotype with map location. J. Gen. Microbiol. 111:165-180. [DOI] [PubMed] [Google Scholar]
- 14.Piggot, P. J., and R. Losick. 2002. Sporulation genes and intercompartmental regulation, p. 483-518. In A. Sonenshein, J. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives. ASM Press, Washington, D.C.
- 15.Price, K. D., and R. Losick. 1999. A four-dimensional view of assembly of a morphogenetic protein during sporulation in Bacillus subtilis. J. Bacteriol. 181:781-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quisel, J. D., W. F. Burkholder, and A. D. Grossman. 2001. In vivo effects of sporulation kinases on mutant Spo0A proteins in Bacillus subtilis. J. Bacteriol. 183:6573-6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ragkousi, K., P. Eichenberger, C. van Ooij, and P. Setlow. 2003. Identification of a new gene essential for germination of Bacillus subtilis spores with Ca2+-dipicolinate. J. Bacteriol. 185:2315-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roels, S., A. Driks, and R. Losick. 1992. Characterization of spoIVA, a sporulation gene involved in coat morphogenesis. J. Bacteriol. 174:575-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1990. Molecular cloning: a laboratory manual, vol. 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 20.Sterlini, J. M., and J. Mandelstam. 1969. Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem. J. 113:29-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Ooij, C., and R. Losick. 2003. Subcellular localization of a small sporulation protein in Bacillus subtilis. J. Bacteriol. 185:1391-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Youngman, P., J. B. Perkins, and R. Losick. 1984. Construction of a cloning site near one end of Tn917 into which foreign DNA may be inserted without affecting transposition in Bacillus subtilis or expression of the transposon-borne erm gene. Plasmid 12:1-9. [DOI] [PubMed] [Google Scholar]
- 23.Zheng, L., R. Halberg, S. Roels, H. Ichikawa, L. Kroos, and R. Losick. 1992. Sporulation regulatory protein GerE from Bacillus subtilis binds to and can activate or repress transcription from promoters for mother-cell-specific genes. J. Mol. Biol. 226:1037-1050. [DOI] [PubMed] [Google Scholar]
- 24.Zheng, L. B., W. P. Donovan, P. C. Fitz-James, and R. Losick. 1988. Gene encoding a morphogenic protein required in the assembly of the outer coat of Bacillus subtilis endospore. Genes Dev. 2:1047-1054. [DOI] [PubMed] [Google Scholar]