Abstract
We have investigated whether DNA polymerase IV (Pol IV; the dinB gene product) contributes to the error rate of chromosomal DNA replication in Escherichia coli. We compared mutation frequencies in mismatch repair-defective strains that were either dinB positive or dinB deficient, using a series of mutational markers, including lac targets in both orientations on the chromosome. Virtually no contribution of Pol IV to the chromosomal mutation rate was observed. On the other hand, a significant effect of dinB was observed for reversion of a lac allele when the lac gene resided on an F′(pro-lac) episome.
Several mechanisms control the fidelity of the DNA replication process. These include correct base selection by the DNA polymerase, removal of base insertion errors by 3′-exonucleolytic proofreading, and correction by DNA mismatch repair (29). In Escherichia coli, base selection and proofreading are performed by the DNA polymerase III (Pol III) holoenzyme, the enzyme that replicates the bacterial chromosome. It is generally considered a highly accurate enzyme (29). Mismatch repair is performed by the mutHLS mismatch repair system (17). In combination, these three processes yield an error rate of 10−9 to 10−11 error per base pair replicated per cell division (6, 29).
In addition to Pol III, E. coli possesses four other DNA polymerases, Pol I, Pol II, Pol IV, and Pol V, whose precise functions are still being defined. Pol IV and Pol V belong to the recently described Y family of DNA polymerases (H. Ohmori, E. C. Friedberg, R. P. P. Fuchs, M. F. Goodman, F. Hanaoka, D. Hinkle, T. A. Kunkel, C. W. Lawrence, Z. Livneh, T. Nohmi, L. Prakash, S. Prakash, T. Todo, G. C. Walker, Z. Wang, and R. Woodgate, Letter, Mol. Cell 8:7-8, 2001). Pol IV and Pol V are generally considered low-fidelity DNA polymerases in view of their lack of proofreading activity and their high in vitro error rates (14, 37). Both enzymes are inducible as part of the SOS regulon, and under SOS-induced conditions they function in lesion bypass and mutagenesis (3, 14, 21, 22, 28, 33, 37, 39).
In contrast to Pol V, which is tightly controlled and essentially not present outside of SOS-induced conditions, Pol IV has a relatively high basal expression level (21), an estimated 250 molecules of Pol IV per cell (20) compared to about 30 molecules of Pol III and much less of the complete Pol III holoenzyme (25). It has been proposed (14) that in certain instances, such as at blocked DNA replication forks, Pol IV might gain access to the replication fork and perform remedial DNA synthesis. Thus, its abundance and potential access to the replication point raise the question of whether this low-fidelity enzyme may contribute to the replication error rate and spontaneous mutagenesis. Consistent with this potential role, overexpression of Pol IV was shown to result in a mutator phenotype (20, 40). Another instance where Pol IV has been implicated is mutagenesis in nondividing cells (adaptive or stationary-phase mutagenesis) (9). Adaptive mutations are routinely assessed through reversion of the lacI33ΩlacZ frameshift allele residing on an F′ episome (9). Loss of Pol IV significantly diminishes these mutations, implicating Pol IV in their production (9, 26, 38).
In this report we examine the possible role of Pol IV in contributing to the chromosomal mutation rate in dividing cells. We did this by carefully analyzing the level and specificity of spontaneous mutation in mismatch repair-defective mutL strains with or without Pol IV present. Mismatch repair-defective strains are most suitable for this purpose, as their mutation rates most directly reflect the error rate of ongoing DNA replication. We used a variety of mutational detection systems, including a system developed previously in our laboratory that permits measurement of reversion of a series of lacZ alleles residing in two orientations in the E. coli chromosome with respect to the direction of replication (7), causing lac sequences to be replicated either as a leading or a lagging strand during chromosomal replication. Thus, in addition to assessing the general role of Pol IV by comparing din+ to dinB strains, it may be possible to assess whether Pol IV might work differentially in one of the two strands, specifically. To inactivate Pol IV, we used the ΔdinB::kan allele, which also includes a partial deletion of the yafN gene (21).
Forward mutagenesis to rifampin and nalidixic acid resistance and reversion of the trpE9777 frameshift allele.
We first tested a series of chromosomal markers by using strain NR13145 [ara thi Δ(pro-lac) mutL::Tn10 trpE9777] and its ΔdinB::kan derivative, strain NR13146. The strains are derivatives of KA796 (32) into which the trpE9777 (30, 34), mutL::Tn10 (31), and ΔdinB::kan (21) alleles were introduced by P1 transduction. The strains allow measurement of trpE9777 frameshift reversion as well as forward mutagenesis to rifampin resistance (Rifr) and nalidixic acid resistance (Nalr). The trpE9777 allele contains an extra A · T base pair added to a run of five A · T base pairs (M. J. Bronson and C. Yanofsky, Letter, J. Mol. Biol. 88:913-915, 1974), and reversion occurs by loss of the extra pair (34). The data shown in Fig. 1 indicate that there were no significant differences in mutability between the dinB-positive and dinB-deficient strains for this marker. Likewise, no effects of dinB were observed for the Rifr or Nalr forward-mutagenesis systems, which allow scoring of base substitution mutagenesis at multiple sites in the rpoB or gyrA gene, respectively (11, 41). These experiments were performed multiple times, and no significant differences between dinB and dinB+ strains were observed in any of them (see Fig. 1).
FIG. 1.
Effect of dinB deletion on frequencies of mutants (per 108 cells) of mismatch repair-defective mutL strains. Light gray bars, dinB positive; dark gray bars, dinB deficient. The numbers above each bar indicate the average frequencies of mutants calculated for 30 independent cultures. This experiment was performed a total of four times with similar results. In no case was a significant difference for the dinB-to-dinB+ comparison found (P > 0.5). Error bars represent standard errors of the means.
Chromosomal lac system for assaying leading- versus lagging-strand mutagenesis.
Our laboratory has developed a system that allows comparison of the mutabilities of the lacZ gene when it is present in the two opposing orientations on the bacterial chromosome. Inversion of the target gene places lac sequences that were previously copied by leading-strand replication into the lagging strand and vice versa. Thus, a comparison of the mutabilities of the same target sequence in the two orientations can reveal differential mutabilities, if any, during leading- or lagging-strand replication. The two orientations of the lac operon with respect to the direction of replication through the target have been arbitrarily designated right (R) and left (L) (7). Using this system, it was previously deduced (7) that base substitutions likely occur more frequently during leading-strand replication. Likewise, the frequencies of frameshift mutations are not equal on the two DNA strands (12), although the complexity of frameshift mutagenesis (1) does not permit a ready strand assignment in this case (12). These sets of strains with lac alleles in the two orientations provide a potentially discriminating tool to assay the influence of Pol IV on chromosomal replication fidelity.
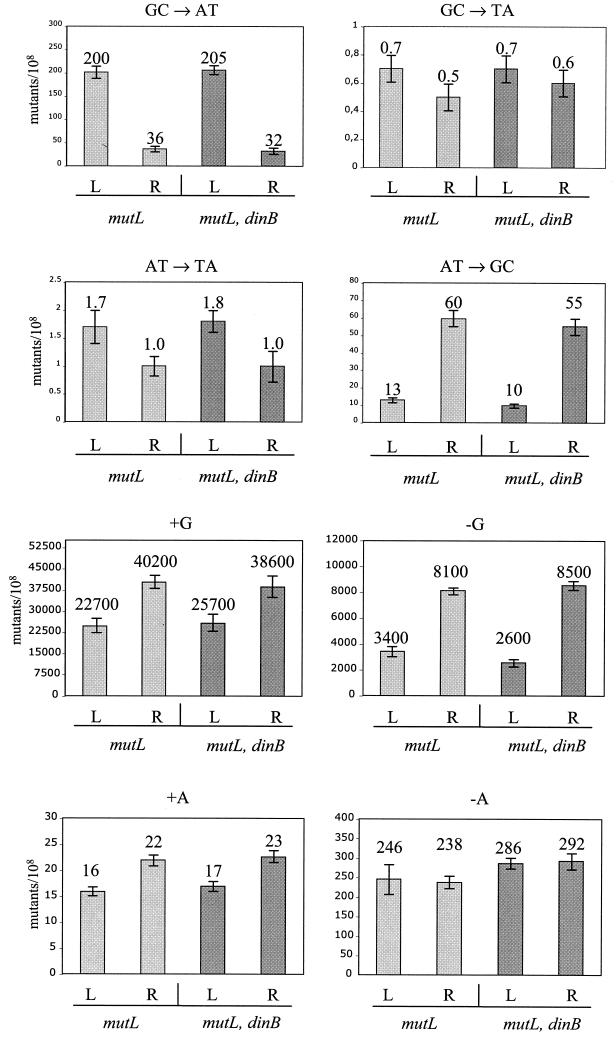
The strains used have been described previously (7, 12, 13). They are mutL::Tn5 derivatives of MC4100 (Δlac) carrying a series of defined lac alleles (4, 5) in the λ attachment site and having either a dinB+ or ΔdinB::kan genotype. For the lac alleles, we used four base substitution markers and four frameshift markers. The base substitution markers (5) were two transitions (G · C→A · T and A · T→G · C) and two transversions (G · C→T · A and A · T→T · A). The frameshift markers (4) were two (+1) frameshifts [addition of G · C to a (G · C)6 run or A · T to an (A · T)6 run] and two (−1) frameshifts [loss of G · C from (G · C)6 or A · T from (A · T)7].
The data in Fig. 2 show that, as before (7, 12, 13), significantly different frequencies of mutants were observed when the effect of orientation (R versus L) on all four base substitution alleles and three of the four frameshift alleles in the dinB+ control strains was tested. However, introduction of the dinB allele did not significantly affect the level of any of the observed mutations, regardless of the gene orientation (with one minor exception described below). The lack of differences is statistically significant (see legend to Fig. 2) and was observed in numerous repeated experiments (4 to 10 repeats for each lac allele). In the case of the −1G frameshift event only, a small but repeatable difference was observed between the dinB+ and dinB strains. In each repeated experiment with this allele, we observed a very moderate (∼20%) but consistent decrease in the mutability of the dinB derivative carrying the L-oriented lac operon. No difference was observed for the corresponding R-oriented strain. This suggests that for this lacZ allele, Pol IV may play a minor, strand-specific role.
FIG. 2.
Frequencies of base substitution or frameshift mutants (Lac+ revertants per 108 cells) of dinB+ and dinB pairs of strains containing the lac operon in two orientations. The strains were mismatch repair defective (mutL). The base substitution or frameshift measured in each case is indicated above the panel. Light gray bars indicate frequencies of mutants of the dinB+ strains; dark gray bars indicate frequencies of mutants of the dinB-deficient strains. L and R indicate the transcription orientation of the lac operon (7). The numbers above the bars are the average frequencies of mutants calculated for 30 independent cultures, and error bars represent standard errors of the means. For all of these experiments, the observed differences in lac reversion frequency between dinB+ and dinB pairs of strains are not statistically significant (P > 0.5), with the exception of a small (20%) effect for the −1G allele in the L orientation (P < 0.05).
Effects of dinB in F′(pro-lac) strains.
Lac reversion in nongrowing cells (stationary or adaptive mutagenesis) has been shown to be, to a large extent, dinB dependent (9, 26, 35, 38). These experiments have generally used a particular frameshift allele, lacI33ΩlacZ, residing on an F′(pro-lac) episome. Among other differences, such as those in growth and selection conditions, these experiments differ from ours in that they are generally conducted at higher basal Pol IV levels. The F′(pro-lac) episome used (F′128) also contains a copy of the dinB gene. Moreover, multiple copies of F′ are often present in the cell (10, 15). Thus, these experiments are generally conducted under conditions of enhanced Pol IV levels (21).
To complement our chromosomal experiments, we also conducted an experiment with a set of strains containing F′(pro-lac). We assayed for two chromosomal markers (Rifr and trpE9777 reversion) and one F′(pro-lac)-contained marker, the lacZ gene from strain CC104, which permits measurement of G · C→T · A transversions (5). The strains used were as described above for Fig. 1, except that the relevant F′ (either dinB positive or with the ΔdinB::kan deletion) was introduced by conjugation. The data in Table 1 show the results obtained for four different dinB configurations: fully positive (gene present on both chromosome and F′), semideficient (gene absent on either F′ or the chromosome), and fully deficient. The data show that the frequencies for the two chromosomal markers are unaffected or are little affected by the dinB copy number, whereas the episomal G · C→T · A marker is significantly affected. In four independent experiments, the average reductions due to the dinB defect were 1.9-fold (loss of the F′ copy), 1.3-fold (loss of the chromosomal copy), and 3.0-fold (loss of both copies). This result indicates that, in this system, Pol IV contributes significantly (∼70%) to the production of G · C→T · A transversions. The mutants on the Lac plates were counted after about 40 to 45 h of incubation, when Lac+ revertants first appeared. Thus, even though the lac G · C→T · A transversion allele has been shown to produce (modest amounts of) “adaptive” mutants upon prolonged incubation on the plate (16, 23), the observed mutants must be considered preexisting (growth dependent). Similar results were obtained in this experiment when the ΔdinB::kan allele was replaced by the nonpolar ΔdinB::zeo allele (2; data not shown).
TABLE 1.
Mutant frequencies in F′(pro-lac) strains with deletion of chromosomal and/or episomal dinB genes
| Straina |
dinB genotypeb
|
Mutation frequency (mean ± SEM)c
|
|||
|---|---|---|---|---|---|
| Chromosome | F′ | Rifr | Trp+ | Lac+ | |
| NR13153 | + | + | 440 ± 38 | 283 ± 18 | 7.3 ± 0.5 |
| NR13155 | + | − | 415 ± 48 | 253 ± 35 | 3.5 ± 0.5 |
| NR13157 | − | + | 485 ± 30 | 260 ± 20 | 5.3 ± 0.8 |
| NR13159 | − | − | 425 ± 40 | 243 ± 25 | 1.6 ± 0.3 |
The strains, which were created as described in the text, were mismatch repair defective (mutL::Tn10).
+, gene present; −, gene absent.
Presented are frequencies per 108 cells of Rifr, reversion of the trpE9777 frameshift, and reversion of the episomal lacZ G · C→T · A transversion. The numbers are the averages for 16 independent cultures.
Conclusions.
The present results indicate that a lack of functional Pol IV does not significantly lower mutation frequencies in growing mismatch repair-deficient E. coli cells. Therefore, we conclude that Pol IV does not contribute significantly to the normal chromosomal error rate. This does not mean that Pol IV does not participate in chromosomal activities; it means only that this involvement, if any, is generally error free compared to other error-producing mechanisms. Presumably, the access of Pol IV to the replication fork is carefully controlled, permitting only limited DNA synthesis by this error-prone enzyme.
Our experiments did show a small (∼20%) but consistent role for Pol IV in the reversion of the −1G frameshift lacZ allele, but only when the lacZ gene was in the L orientation (Fig. 2). This result suggests that Pol IV does contribute to errors in this configuration. Due to the complexity of frameshift mutagenesis (1), whether this role is an involvement in leading- or lagging-strand replication cannot be readily deduced. However, overexpression of Pol V preferentially increased base substitution mutagenesis in the lagging strand, presumably because of easier access (24). Preliminary data suggest that the same may be true for the mutator effect resulting from Pol IV overexpression (unpublished data). Overall, based on multiple repeats of the experiments shown in Fig. 2, we estimate that the 20% effect observed for the case of the −1G lacZ allele represents a general upper limit for the contribution of Pol IV in any of the other chromosomal tests reported here.
In contrast to the case with the chromosomal mutations, our data indicate that Pol IV can play a significant role in mutation production on the F′(pro-lac) episome, even in growing cells (Table 1). Presumably, one factor contributing to this mutation production is the increased dinB gene dosage in this experimental setting, as the F′ carries an additional copy of the gene and multiple F′ copies may be present. Overproduction of Pol IV has been demonstrated to lead to a mutator phenotype (20, 41). Interestingly, and consistent with the data of Kim et al. (21), who noted that the episomal dinB copy produced more Pol IV protein than the chromosomal copy, we noticed that the deletion of dinB from the episome decreased the frequency of mutants more than the deletion of dinB from the chromosome (1.3- versus 1.9-fold; average result of four experiments).
Interestingly, increased dinB copy number did not significantly increase the frequency of chromosomal Rifr mutations or trpE9777 frameshift reversions (Table 1). This finding suggests that location of the target gene on the F′ itself might also be necessary for enhanced mutagenesis. A rigorous demonstration of this will require comparison of the lacZ target genes on the chromosome and F′ under the various dinB gene dosage conditions. In the meantime, one might speculate that the same principles that underlie the production of growth-independent mutations (which are to a large extent dinB dependent) are also relevant to the growth-dependent mutations observed here. The duplications and amplifications of episomal lac sequences that have been shown to be prerequisites for the production of mutations under nongrowing conditions (19) are, in fact, produced during the preceding growth phase (19). These amplified sequences may include both lac and dinB (19, 35). Therefore, even in growing cells, conditions may be favorable for preferential mutagenesis of episomal lac sequences in a dinB-dependent manner. Although experiments reported here used the Δ(dinB-yafN) allele, for which polar effects on the yaf genes have been suggested to contribute to certain mutational end points (26, 27), we obtained identical results with the nonpolar ΔdinB::zeo allele (2; data not shown). Thus, we conclude that the mutator effect of Pol IV on F′(pro-lac) is due to Pol IV and not to other members of the yaf operon.
The question of the role of Pol IV in replication fidelity has been addressed before. Strauss et al. (36), using dnaE mutator strains containing an impaired Pol III, noted that about 75% of mutations in this background were dinB dependent, based on measurements of either lac reversion on F′ or chromosomal Rifr mutations. Interestingly, they also noted a corresponding 1.5- to 3-fold dinB-dependent reduction in the dnaE+ control strain, implying a role for Pol IV in creating these mutations. However, the effect of the ΔdinB::kan allele in the control strain was limited to the mismatch repair-proficient background. No effect was observed in the mismatch repair-deficient mutS background, in which replication errors are more directly viewed. Experiments in our laboratory with mut+ derivatives of the strains shown in Fig. 2 have not revealed any effect of dinB when chromosomal Rifr mutations or chromosomal G · C→T · A or −1G lac reversions are being scored (data not shown). This discrepancy remains to be explored.
McKenzie et al. (26) demonstrated the lack of an effect of the dinB10 allele on the mutability of a series of chromosomal markers. These experiments were performed in the mismatch repair-proficient background, and their results are therefore not directly comparable to those of our experiments, which focus on the production of replication errors. Nevertheless, the absence of an effect in both studies is consistent with the lack of a role for Pol IV in chromosomal replication fidelity. With regard to the effect of dinB on episomal genes, the experiments of McKenzie et al. indicated a lack of effect of dinB10 on the lacI33ΩlacZ allele (26), a result that appears to contradict our results with the episomal lac G · C→T · A transversion (Table 1). However, a reasonable explanation may be that DNA mismatch repair is efficient in removing most or all of the Pol IV-mediated frameshift revertants of lacI33ΩlacZ, masking the effect of Pol IV on replication errors. Efficient correction of the lacI33ΩlacZ revertants has been demonstrated (8, 9, 18). In a second study, McKenzie et al. (27) made a distinction between the nonpolar dinB alleles, such as dinB10, and the polar alleles, such as ΔdinB::kan, which exerts a polar effect on the distal members of the dinB-yafNOP operon. An effect (approximately twofold) was observed on lacI33ΩlacZ reversion with the polar ΔdinB::kan allele but not with the nonpolar alleles (27), leading the authors to conclude that the distal members of the operon might be involved. In our study, we observed an effect on the episomal lacZ G · C→T · A marker with both the polar ΔdinB::kan allele (Table 1) and the nonpolar ΔdinB::zeo allele, suggesting that only Pol IV is involved. These differences may reflect both the action of mismatch repair, which may dramatically influence the extent to which replication errors can be observed, and any mechanistic differences in the genesis of lacI33ΩlacZ frameshifts and lacZ G · C→T · A transversions. This issue requires further study.
In summary, no evidence has been found for a significant contribution of Pol IV to chromosomal mutagenesis in growing cells, although episomal mutagenesis on F′(pro-lac) may be enhanced by Pol IV action.
ADDENDUM IN PROOF
Recently, Wolff et al. (E. Wolff, M. Kim, K. Hu, H. Yang, and J. H. Miller, J. Bacteriol 186:2900-2905, 2004), based on an analysis of the spectral fingerprint of DNA polymerase IV-induced mutations, also concluded that Pol IV does not contribute significantly to mutations occurring during exponential growth.
Acknowledgments
We thank K. Bebenek and S. McCulloch of the National Institute of Environmental Health Sciences for careful review of this paper. We thank Takehiko Nohmi for providing ΔdinB::kan strain YG7207 and Roger Woodgate for providing the ΔdinB::zeo strain AR30.
This research was supported by grant 3PO4A04223 from the Polish Ministry of Scientific Research and Information Technology, State Committee for Scientific Research. D.G. was the recipient of a fellowship from the Kosciuszko Foundation.
REFERENCES
- 1.Bebenek, K., and T. A. Kunkel. 2000. Streisinger revisited: DNA synthesis errors mediated by substrate misalignments. Cold Spring Harbor Symp. Quant. Biol. 65:81-91. [DOI] [PubMed] [Google Scholar]
- 2.Borden, A., P. I. O'Grady, D. Vandewiele, A. R. Fernández de Henestrosa, C. W. Lawrence, and R. Woodgate. 2002. Escherichia coli DNA polymerase III can replicate efficiently past a T-T cis-syn cyclobutane dimer if DNA polymerase V and the 3′ to 5′ exonuclease proofreading function encoded by dnaQ are inactivated. J. Bacteriol. 184:2674-2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brotcorne-Lannoye, A., and G. Maenhaut-Michel. 1986. Role of RecA protein in untargeted UV mutagenesis of bacteriophage λ: evidence for the requirement for the dinB gene. Proc. Natl. Acad. Sci. USA 83:3904-3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cupples, C. G., M. Cabrera, C. Cruz, and J. H. Miller. 1990. A set of lacZ mutations in Escherichia coli that allow rapid detection of specific frameshift mutations. Genetics 125:275-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cupples, C. G., and J. H. Miller. 1989. A set of lacZ mutations in Escherichia coli that allow rapid detection of each of the six base substitutions. Proc. Natl. Acad. Sci. USA 86:5345-5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drake, J. W. 1991. A constant rate of spontaneous mutation in DNA-based microbes. Proc. Natl. Acad. Sci. USA 88:7160-7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fijalkowska, I. J., P. Jonczyk, M. Maliszewska-Tkaczyk, M. Bialoskorska, and R. M. Schaaper. 1998. Unequal fidelity of leading and lagging strand DNA replication on the Escherichia coli chromosome. Proc. Natl. Acad. Sci. USA 95:10020-10025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foster, P. L. 1999. Are adaptive mutations due to a decline in mismatch repair? The evidence is lacking. Mutat. Res. 436:179-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foster, P. L. 2000. Adaptive mutation in Escherichia coli. Cold Spring Harbor Symp. Quant. Biol. 65:21-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frame, R., and J. O. Bishop. 1971. The number of sex-factors per chromosome in Escherichia coli. Biochem. J. 121:93-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garibyan, L., T. Huang, M. Kim, E. Wolff, A. Nguyen, T. Nguyen, A. Diep, K. Hu, A. Iverson, H. Yang, and J. H. Miller. 2003. Use of the rpoB gene to determine the specificity of base substitution mutations on the Escherichia coli chromosome. DNA Repair 2:593-608. [DOI] [PubMed] [Google Scholar]
- 12.Gawel, D., M. Bialoskorska, P. Jonczyk, R. M. Schaaper, and I. J. Fijalkowska. 2002. Asymmetry of frameshift mutagenesis during leading and lagging strand replication in E. coli. Mutat. Res. 501:129-136. [DOI] [PubMed] [Google Scholar]
- 13.Gawel, D., M. Maliszewska-Tkaczyk, P. Jonczyk, R. M. Schaaper, and I. J. Fijalkowska. 2002. Lack of strand bias in UV-induced mutagenesis in Escherichia coli. J. Bacteriol. 184:4449-4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodman, M. F. 2002. Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu. Rev. Biochem. 71:17-50. [DOI] [PubMed] [Google Scholar]
- 15.Gordon, G. S., D. Sitnikov, C. D. Webb, A. Teleman, and A. Straight. 1997. Chromosome and low copy plasmid segregation in E. coli: visual evidence for distinct mechanisms. Cell 90:1113-1121. [DOI] [PubMed] [Google Scholar]
- 16.Hall, B. G. 1991. Spectrum of mutations that occur under selective and non-selective conditions in E. coli. Genetica 84:73-76. [DOI] [PubMed] [Google Scholar]
- 17.Harfe, B. D., and S. Jinks-Robertson. 2000. DNA mismatch repair and genetic instability. Annu. Rev. Genet. 34:359-399. [DOI] [PubMed] [Google Scholar]
- 18.Harris, R. S., H. J. Bull, and S. M. Rosenberg. 1997. A direct role for DNA polymerase III in adaptive reversion of a frameshift mutation in Escherichia coli. Mutat. Res. 375:19-24. [DOI] [PubMed] [Google Scholar]
- 19.Hendrickson, H., E. S. Slechta, U. Bergthorsson, D. I. Andersson, and J. R. Roth. 2002. Amplification-mutagenesis: evidence that “directed” adaptive mutation and general hypermutability result from growth with a selected gene amplification. Proc. Natl. Acad. Sci. USA 99:2164-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim, S.-R., G. Maenhaut-Michel, M. Yamada, Y. Yamamoto, K. Matsui, T. Sofuni, T. Nohmi, and H. Ohmori. 1997. Multiple pathways for SOS-induced mutagenesis in Escherichia coli: an SOS gene product (DinB/P) enhances frameshift mutations in the absence of any exogenous agents that damage DNA. Proc. Natl. Acad. Sci. USA 94:13792-13797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim, S.-R., K. Matsui, M. Yamada, P. Gruz, and T. Nohmi. 2001. Roles of chromosomal and episomal dinB genes encoding pol IV in targeted and untargeted mutagenesis in Escherichia coli. Mol. Genet. Genomics 266:207-215. [DOI] [PubMed] [Google Scholar]
- 22.Lenne-Samuel, N., R. Janel-Bintz, A. Kolbanovskiy, N. E. Geacintov, and R. P. P. Fuchs. 2000. The processing of a benzo(a)pyrene adduct into a frameshift or a base substitution mutation requires a different set of genes in Escherichia coli. Mol. Microbiol. 38:299-307. [DOI] [PubMed] [Google Scholar]
- 23.Mackay, W. J., S. Han, and L. D. Samson. 1994. DNA alkylation repair limits spontaneous base substitution mutations in Escherichia coli. J. Bacteriol. 176:3224-3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maliszewska-Tkaczyk, M., P. Jonczyk, M. Bialoskorska, R. M. Schaaper, and I. J. Fijalkowska. 2000. SOS mutator activity: unequal mutagenesis on leading and lagging strands. Proc. Natl. Acad. Sci. USA 97:12678-12683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McHenry, C., and A. Kornberg. 1977. DNA polymerase III holoenzyme of Escherichia coli. Purification and resolution into subunits. J. Biol. Chem. 252:6478-6484. [PubMed] [Google Scholar]
- 26.McKenzie, G. J., P. L. Lee, M.-J. Lombardo, P. J. Hastings, and S. M. Rosenberg. 2001. SOS mutator DNA polymerase IV functions in adaptive mutation and not adaptive amplification. Mol. Cell 7:571-579. (Erratum, 7:1119.) [DOI] [PubMed] [Google Scholar]
- 27.McKenzie, G. J., D. B. Magner, P. L. Lee, and S. M. Rosenberg. 2003. The dinB operon and spontaneous mutation in Escherichia coli. J. Bacteriol. 185:3972-3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Napolitano, R., R. Janel-Bintz, J. Wagner, and R. P. P. Fuchs. 2000. All three SOS-inducible DNA polymerases (Pol II, Pol IV, and Pol V) are involved in induced mutagenesis. EMBO J. 19:6259-6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schaaper, R. M. 1993. Base selection, proofreading, and mismatch repair during DNA replication in Escherichia coli. J. Biol. Chem. 268:23762-23765. [PubMed] [Google Scholar]
- 30.Schaaper, R. M. 1993. The mutational specificity of two Escherichia coli dnaE antimutator alleles as determined from lacI mutation spectra. Genetics 134:1031-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schaaper, R. M., B. I. Bond, and R. G. Fowler. 1989. A · T→C · G transversions and their prevention by the Escherichia coli mutT and mutHLS pathways. Mol. Gen. Genet. 219:256-262. [DOI] [PubMed] [Google Scholar]
- 32.Schaaper, R. M., B. N. Danforth, and B. W. Glickman. 1985. Rapid repeated cloning of mutant lac repressor genes. Gene 39:181-189. [DOI] [PubMed] [Google Scholar]
- 33.Shen, X., J. M. Sayer, H. Kroth, I. Ponten, M. O'Donnell, R. Woodgate, D. M. Jerina, and M. F. Goodman. 2002. Efficiency and accuracy of SOS-induced DNA polymerases replicating benzo[a]pyrene-7,8-diol 9,10-epoxide A and G adducts. J. Biol. Chem. 277:5265-5274. [DOI] [PubMed] [Google Scholar]
- 34.Siegel, E. C., and K. K. Vaccaro. 1978. The reversion of trp frameshift mutations in mut, polA, lig and dnaE mutant strains of Escherichia coli. Mutat. Res. 50:9-17. [Google Scholar]
- 35.Slechta, S. E., K. L. Bunny, E. Kugelberg, E. Kofoid, D. I. Andersson, and J. R. Roth. 2003. Adaptive mutation: general mutagenesis is not a programmed response to stress but results from rare coamplification of dinB with lac. Proc. Natl. Acad. Sci. USA 100:12847-12852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strauss, B. S., R. Roberts, L. Francis, and P. Pouryazdanparast. 2000. Role of the dinB gene product in spontaneous mutation in Escherichia coli with an impaired replicative polymerase. J. Bacteriol. 182:6742-6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang, M., P. Pham, X. Shen, J. S. Taylor, M. O'Donnell, R. Woodgate, and M. F. Goodman. 2000. Roles of E. coli DNA polymerases IV and V in lesion-targeted and untargeted SOS mutagenesis. Nature 404:1014-1018. [DOI] [PubMed] [Google Scholar]
- 38.Tompkins, J. D., J. L. Nelson, J. C. Hazel, S. L. Leugers, J. D. Stumpf, and P. L. Foster. 2003. Error-prone polymerase, DNA polymerase IV, is responsible for transient hypermutation during adaptive mutation in Escherichia coli. J. Bacteriol. 185:3469-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wagner, J., P. Grúz, S. R. Kim, M. Yamada, K. Matsui, R. P. P. Fuchs, and T. Nohmi. 1999. The dinB gene encodes a novel E. coli DNA polymerase, Pol IV, involved in mutagenesis. Mol. Cell 4:281-286. [DOI] [PubMed] [Google Scholar]
- 40.Wagner, J., and T. Nohmi. 2000. Escherichia coli DNA polymerase IV mutator activity: genetic requirements and mutational specificity. J. Bacteriol. 182:4587-4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoshida, H., M. Bogaki, M. Nakamura, and S. Nakamura. 1990. Quinolone resistance-determining region in the DNA gyrase gyrA gene of Escherichia coli. Antimicrob. Agents Chemother. 34:1271-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]