Abstract
Despite the importance of the oxidative pentose phosphate (PP) pathway as a major source of reducing power and metabolic intermediates for biosynthetic processes, almost no direct genetic or biochemical evidence is available for Bacillus subtilis. Using a combination of knockout mutations in known and putative genes of the oxidative PP pathway and 13C-labeling experiments, we demonstrated that yqjI encodes the NADP+-dependent 6-P-gluconate dehydrogenase, as was hypothesized previously from sequence similarities. Moreover, YqjI was the predominant isoenzyme during glucose and gluconate catabolism, and its role in the oxidative PP pathway could not be played by either of two homologues, GntZ and YqeC. This conclusion is in contrast to the generally held view that GntZ is the relevant isoform; hence, we propose a new designation for yqjI, gndA, the monocistronic gene encoding the principal 6-P-gluconate dehydrogenase. Although we demonstrated the NAD+-dependent 6-P-gluconate dehydrogenase activity of GntZ, gntZ mutants exhibited no detectable phenotype on glucose, and GntZ did not contribute to PP pathway fluxes during growth on glucose. Since gntZ mutants grew normally on gluconate, the functional role of GntZ remains obscure, as does the role of the third homologue, YqeC. Knockout of the glucose-6-P dehydrogenase-encoding zwf gene was primarily compensated for by increased glycolytic fluxes, but about 5% of the catabolic flux was rerouted through the gluconate bypass with glucose dehydrogenase as the key enzyme.
The carbon-rearranging transaldolase and transketolase reactions in the nonoxidative branch of the pentose phosphate (PP) pathway constitute the exclusive route for catabolism of pentoses. During growth on hexoses, the PP pathway becomes a major source of pentose phosphates for nucleotide biosynthesis and of the anabolic redox cofactor NADPH, the reducing equivalent for biosynthesis reactions (21, 22). For this purpose, two consecutive NADP+-dependent dehydrogenase reactions convert glucose-6-P into ribulose-5-P in the oxidative branch of the PP pathway. Because of its important role in central metabolism, the PP pathway has been investigated in great biochemical and genetic detail (18, 19), and more recently it has also been investigated from a metabolic systems perspective (5, 26, 41) in the gram-negative model bacterium Escherichia coli.
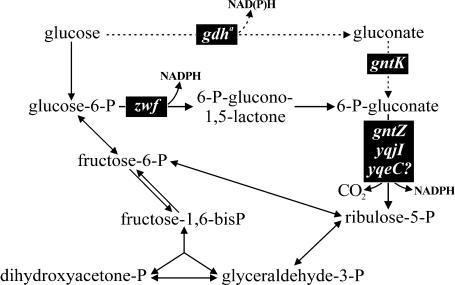
The PP pathway in the gram-positive model bacterium Bacillus subtilis, in contrast, has received very little attention, and most evidence has been indirectly inferred by comparison to E. coli (17). In particular, no biochemical data are available on the enzymes of the oxidative PP pathway, glucose-6-P dehydrogenase and 6-P-gluconate dehydrogenase, and there is no genetic evidence for the gene(s) encoding the 6-P-gluconate dehydrogenase. Based on sequence similarity, the distal gntZ gene of the gluconate operon was classified as a 6-P-gluconate dehydrogenase gene (38). Despite the presence of three homologues in the genome (35) and the homology-based suggestion that B. subtilis contains two 6-P-gluconate dehydrogenases with different cofactor specificities (47), gntZ has been considered the relevant gene (4, 35). Additionally, a P-gluconolactonase may be involved in the oxidative PP pathway of B. subtilis, but no gene is known to date and the reaction may also proceed by spontaneous hydrolysis (30) (Fig. 1).
FIG. 1.
Oxidative PP pathway and related reactions of B. subtilis. Relevant genes are indicated in black boxes. At least six more genes show significant similarity to gdh (35).
In recent years, there has been a resurgence of interest in the oxidative PP pathway that has been driven largely by the ability to estimate the metabolic flux through this pathway from novel 13C-labeling experiments (8, 40, 49, 50). For glucose-grown B. subtilis, such flux analyses have revealed highly variable fluxes through the oxidative PP pathway, ranging from an almost complete absence during slow growth under nitrogen limitation (12) or when glucose and intermediates of the tricarboxylic acid cycle are cometabolized (11) to PP pathway fluxes that may exceed the glycolytic flux under phosphate limitation (12) or in certain riboflavin-producing strains (42). Generally, the flux through the PP pathway does not appear to be regulated by the cellular demand for NADPH and/or pentoses in B. subtilis but rather is determined by the kinetic properties of the enzymes at the glucose-6-P branch point (11), as has been shown for Corynebacterium glutamicum (33, 34). During standard batch growth in minimal medium, around 30 to 40% of the consumed glucose is catabolized through the oxidative PP pathway flux in B. subtilis (52), which is more than the amount catabolized in E. coli (16) or other bacilli (9).
To identify the physiological function of enzymes in the oxidative PP pathway, we constructed B. subtilis knockout mutants with mutations in the zwf gene encoding the glucose-6-P dehydrogenase and in three homologues of the 6-P-gluconate dehydrogenase gene (gntZ, yqjI, and yqeC). In addition to physiological characterization and in vitro enzyme assays, metabolic flux ratio analysis by gas chromatography (GC)-mass spectrometry (MS) was used to determine the in vivo role of these enzymes by quantifying the ratio of glycolysis to oxidative PP pathway flux at the glucose-6-P branch point by performing [1-13C]glucose experiments (15).
MATERIALS AND METHODS
Bacterial strains, growth conditions, and media.
The B. subtilis wild-type strain used, a close relative of strain 168, and mutants derived from it are listed in Table 1. Chromosomal genes were inactivated by replacement with neomycin (27) and spectinomycin resistance cassettes (23) or by using the integrative vector pMUTIN that bears an erythromycin resistance marker (48). Both zwf and yqjI were removed from the start codon to the stop codon, and bp 45 to 1391 of the gntZ open reading frame (bp 1 to 1404) was removed. For all physiological and 13C-labeling experiments, frozen stocks were used to inoculate 5 ml of Luria-Bertani broth (24) that contained neomycin (final concentration, 5 mg liter−1), spectinomycin (final concentration, 100 mg liter−1), or erythromycin (final concentration, 0.5 mg liter−1). After 8 h of shaking at 37°C, 250-μl portions of precultures were used to inoculate 50-ml portions of M9 minimal medium (24) with 10 g of glucose liter−1 in 500-ml baffled shake flasks. After about 12 h, 250 μl was withdrawn from each flask and used to inoculate 50 ml of M9 medium with 5 g of glucose liter−1 for an actual growth experiment. All cultures were incubated at 37°C on a gyratory shaker at 250 rpm. For 13C-labeling experiments, glucose was added as the 1-13C-labeled isotope isomer (Euriso-Top, Gif-sur-Yvette, France). Leucine and methionine were each added to M9 medium cultures at a final concentration of 50 mg liter−1.
TABLE 1.
B. subtilis strains used in this study
| Strain | Relevant genotype | Reference or source |
|---|---|---|
| 1012 wild type | leuA8 metB5 | 39 |
| 1012 zwf | leuA8 metB5 zwf::spc | This study |
| 1012 gntZ | leuA8 metB5 gntZ::spc | This study |
| 1012 zwf gntZ | leuA8 metB5 zwf::neo gntZ::spc | This study |
| 1012 yqjI | leuA8 metB5 yqjI::neo | This study |
| 1012 yqjI* | leuA8 metB5 yqjI::neo; adapted to grow on glucose as the sole carbon source | This study |
| 1012 yqjI gntZ | leuA8 metB5 yqjI::neo gntZ::spc | This study |
| 1012 yqjI* gntZ | leuA8 metB5 yqjI::neo gntZ::spc; adapted to grow on glucose as the sole carbon source | This study |
| 168 yqeC | trpC2 yqeC::pMUTIN | S. Aymerich |
Analytical techniques.
Cell growth was monitored by measuring the optical density with a Klett-Summerson colorimeter (Bel-Art, Pequannock, N.J.) with a green filter (520 to 580 nm). The glucose and acetate concentrations in the culture supernatant were determined enzymatically with commercially available kits (Beckman, Palo Alto, Calif.), and the acetoin concentrations were determined by GC analysis by using a Carbowax MD-10 column (Macherey-Nagel). Specific consumption and production rates were calculated as described previously (44) by using an experimentally determined correlation curve for cellular dry weight and Klett units.
To prepare crude cell extracts for the 6-P-gluconate dehydrogenase activity assay, cultures were grown in 50 ml of either M9 or VY medium containing 25 g of veal infusion broth liter−1, 5 g of yeast extract liter−1, and 15 g of glucose liter−1. Cultures were centrifuged at 3,500 × g for 10 min, washed once with 0.9% (wt/vol) NaCl-10 mM MgSO4, and frozen for future use or resuspended in assay buffer containing 100 mM triethanolamine (pH 7.6), 4 mM MgCl2, 4 mM phenylmethylsulfonyl fluoride, and 1 mM dithiothreitol (3). Cells were disrupted by passage through a French press (SLM Aminco, SLM Instruments) and centrifuged at 14,000 × g and 4°C for 30 min. In vitro 6-P-gluconate dehydrogenase activity in the crude cell extracts was assayed within 1 h at a 1:10 dilution. Background activity was determined at 340 nm for a few minutes after addition of either NAD+ or NADP+ (ɛ = 6.2 mM−1 cm−1) to a final concentration of 1 mM. The reaction assay then was started with 6-P-gluconate at a final concentration of 1 mM and was monitored for at least 3 min. The protein concentrations in the samples were determined colorimetrically (Beckman).
Determination of metabolic flux ratios.
Culture aliquots (2 ml) were harvested at about 200 Klett units (equivalent to an optical density at 600 nm of 1.5 to 2.0) by centrifugation at 14,500 × g for 5 min, washed at least twice with double-distilled H2O, and hydrolyzed in 1.5 ml of 6 M HCl at 105°C for 24 h in sealed microtubes. The hydrolysate was dried at 60°C and derivatized for 1 h at 85°C in a solution containing 50 μl of dimethylformamide and 50 μl of N-(tert-butyldimethylsilyl)-N-methyl-trifluoroacetamide (15). Derivatized amino acids were measured with a series 8000 GC combined with an MD 800 mass spectrometer (Fisons Instruments, Beverly, Mass.) as described previously (15). The mass distributions in the amino acids were corrected for naturally occurring stable isotopes to obtain the mass distribution vectors (MDV), which, in turn, were used for metabolic flux ratio analysis to determine the split ratio between the fluxes through glycolysis and the PP pathway at the glucose-6-P branch point (15, 52). In particular, the following equation was used to determined the fraction (f) of serine derived through the PP pathway (15):
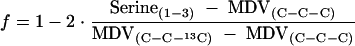 |
(1) |
where MDV(C-C-C) and MDV(C-C-13C) are the mass distributions of natural and 3-13C-labeled C3 fragments, respectively, and Serine(1-3) is the MDV of the C3 backbone of serine. Since MDV are vectors, f represents the least-squares solution. To correct for the nonproportional withdrawal of 13C label in dihydroxyacetone-P for the biosynthesis of phosphatidylglycerol, Serine(1-3) in equation 1 was replaced by the MDV of the triose-P pool [Triose-P(1-3)] estimated by using the following equation:
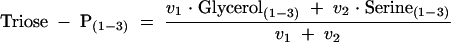 |
(2) |
where v1 is the flux from dihydroxyacetone-P to glycerol (in this case exclusively for phosphatidylglycerol biosynthesis) and v2 is the flux from glyceraldehyde-3-P to 1,3-di-P-glycerate. At a growth rate of 0.4 h−1 and a biomass yield of 0.3 g/g (as seen for the zwf gntZ mutant), v1 is only about 2% of v2 (10). This correction is more important for METAFoR analysis of organisms that secrete glycerol into the medium.
RESULTS
Knockout of glucose-6-P dehydrogenase.
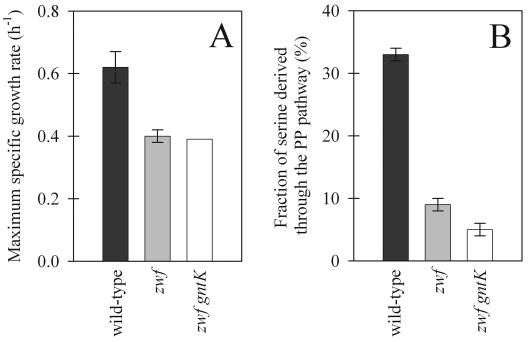
As the first enzyme of the oxidative PP pathway, the zwf-encoded 6-P-glucose dehydrogenase plays a pivotal role in the branching between glycolysis and the PP pathway during glucose catabolism. To quantify the intracellular carbon flux distribution in the absence of a functional oxidative PP pathway, we deleted zwf in B. subtilis wild-type 1012 by partial replacement with a spectinomycin resistance cassette; successful deletion was verified by the complete absence of in vitro 6-P-glucose dehydrogenase activity (data not shown). Since analysis of the surrounding chromosomal sequence indicated that zwf is monocistronic, polar effects of the deletion are unlikely. Similar to the results obtained for E. coli (41), the maximum specific growth rate of the zwf mutant was about 35% lower than that of the parent on glucose (Fig. 2A).
FIG. 2.
Maximum specific growth rates (A) and fractions of serine derived through the PP pathway (B) for wild-type B. subtilis and its isogenic zwf and zwf gntK mutants during growth on glucose. After correction for the labeling pattern in glycerol, the percentages of serine derived through the PP pathway were 32%, 8%, and 4% ± 1%, respectively. The growth rate error bars indicate the deviations based on duplicate experiments. The flux ratio error bars indicate the experimental measurement errors based on a single analysis, assuming that there was a standard error of 1% in the MS signals.
The metabolic impact of the mutation was then detected by metabolic flux ratio analysis by GC-MS (15) with batch cultures grown on 100% [1-13C]glucose. This isotopic tracer is optimally suited to determine the flux splitting between glycolysis and the oxidative PP pathway (7, 16); glycolytic breakdown of [1-13C]glucose yields 50% unlabeled triose-P and 50% [3-13C]triose-P, while catabolism via the oxidative PP pathway yields unlabeled trioses exclusively, because the label is lost as CO2 in the P-gluconate dehydrogenase reaction. Specifically, we determined the split ratio between glycolysis and the PP pathway from the fractional labeling of serine (15), whose C3 is derived from the C3 of the triose-P pool (32). A value of 0% serine derived through the PP pathway corresponded to 100% catabolic flux through glycolysis, as was expected for the zwf mutant. The relative oxidative PP pathway flux in the mutant, however, was 9% serine derived through the PP pathway, compared to 33% serine in the parent (Fig. 2B). Since no glucose-6-P dehydrogenase isoenzymes or zwf homologues are known and since no in vitro activity was detected in mutant extracts (data not shown), this residual flux could potentially be catalyzed by the direct oxidation of glucose to gluconate via the gdh-encoded glucose dehydrogenase or homologues of this enzyme (Fig. 1) (17, 29, 35). To test this possibility, we blocked the potential bypass by introducing a gluconate kinase (gntK) mutation. In the zwf gntK double mutant, the glycolysis-to-PP pathway split ratio was further reduced to 5% ± 1%, revealing the small but not negligible contribution of the gluconate bypass to glucose catabolism in the zwf mutant.
The calculated remaining fraction of about 5% serine derived through the PP pathway in the zwf gntK mutant was based on a higher fraction of unlabeled serine than was expected from exclusive breakdown of [1-13C]glucose via glycolysis (15). Unexpectedly, MS analysis of glycerol, another triose-3-P-derived compound in the total-cell hydrolysates, revealed a higher fraction of 13C label than the fraction in serine (Table 2). Since glycerol is synthesized from dihydroxyacetone-P and serine is synthesized from glyceraldehyde-3-P, the two triose-3-P pools were apparently not fully equilibrated via the rapid triose-3-P isomerase reaction. Because the exchange of trioses via isomerase is a rapid reaction, this is not overly surprising because the net glycolytic flux is from dihydroxyacetone-P to glycerol-3-P. As a consequence, the PP pathway contribution to serine synthesis was slightly overestimated because glycolysis produces [3-13C]dihydroxyacetone-P and unlabeled glyceraldehyde-3-P from [1-13C]glucose. To correct for the small withdrawal of [3-13C]dihydroxyacetone-P for glycerol biosynthesis that was not seen in serine, we considered the net carbon fluxes from dihydroxyacetone-P to glycerol-3-P and from glyceraldehyde-3-P into glycolysis. Since glycerol-3-P is required only for the biosynthesis of phosphatidylglycerol in B. subtilis (14, 43), this biosynthetic flux was very small (10), and the corrected values for serine derived through the PP pathway were only 1% lower than the values shown in Fig. 2B. The remaining small, but not negligible fraction of 4% serine derived through the PP pathway in the zwf gntK mutant may thus be explained by (i) minor secretion of glycerol into the medium and/or (ii) the gluconate bypass if another, GntK-independent kinase phosphorylates gluconate.
TABLE 2.
Mass distributions in serine and glycerol of whole-cell hydrolysates from wild-type B. subtilis and the zwf gntK mutant during growth on [1-13C]glucose
| Compound | Relative mass distributiona
|
|||
|---|---|---|---|---|
| m0 | m1 | m2 | m3 | |
| Wild-type strain | ||||
| Serine | 0.663 | 0.333 | 0.003 | 0.001 |
| Glycerol | 0.532 | 0.470 | 0.003 | 0.000 |
| zwf gntK mutant | ||||
| Serine | 0.518 | 0.471 | 0.012 | 0.000 |
| Glycerol | 0.368 | 0.625 | 0.007 | 0.000 |
The m0 isotope isomer consists exclusively of 12C, and the subscripts in the other isotope isomers indicate the numbers of 13C atoms.
Knockout of 6-P-gluconate dehydrogenase.
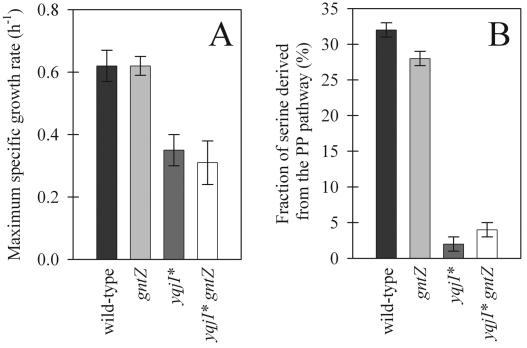
In contrast to the first reaction of the oxidative PP pathway, the second major reaction, catalyzed by 6-P-gluconate dehydrogenase, cannot be bypassed via the gluconate pathway. The major isoform of this enzyme is generally considered to be encoded by gntZ, the distal gene in the catabolic gluconate operon that is weakly induced in the presence of glucose (4, 35, 51). A second homologue is encoded by the monocistronic yqjI gene that is adjacent to zwf. Based on sequence homology, YqjI has been suggested to encode an NADP+-dependent 6-P-gluconate dehydrogenase (47), but this function is not generally recognized (4, 35, 45). To identify the catabolic roles of both homologues, we grew single gntZ and yqjI mutants in minimal medium with glucose as the sole carbon source. While the gntZ mutant had no detectable physiological phenotype (Fig. 3A), the maximum specific growth rate of the yqjI mutant was significantly lower than that of the parent (Fig. 3A), suggesting that YqjI is the major isoenzyme of the 6-P-gluconate dehydrogenase in B. subtilis. When it was first cultivated in minimal medium with glucose, the yqjI mutant exhibited an unusually long lag phase (at least 24 h), and this lag phase was seen again upon subcultivation in complex media. On glucose plates, few yqjI clones appeared after about 24 h, but we were unable to isolate stable suppressor mutants. Thus, it appears that an adaptation (or an unstable suppressor) is necessary to enable growth of the mutant on glucose as the sole carbon source, and such adapted yqjI cultures are indicated below with an asterisk.
FIG. 3.
Maximum specific growth rates (A) and (glycerol-corrected) fractions of serine derived through the PP pathway (B) for wild-type B. subtilis and 6-P-gluconate dehydrogenase mutants. The growth rate error bars indicate the deviations based on duplicate experiments. The flux ratio error bars indicate the experimental measurement error based on a single analysis, assuming that there was a standard error of 1% in the MS signals. Asterisks indicate yqjI mutants that were adapted for growth on glucose alone.
Consistent with the growth phenotype, disruption of gntZ had only a marginal effect on the flux partitioning at the glucose-6-P branch point, but yqjI* cultures did not use the PP pathway at all (Fig. 3B). Notably, both the maximum growth rate and the glycolysis-to-PP pathway split ratio of the yqjI* knockout were comparable to those of the zwf gntK complete PP pathway knockout mutant (compare Fig. 2B and 3B); hence, the undefined adaptation did not involve increased PP pathway fluxes. Combining both mutations in the yqjI* gntZ double-knockout mutant had no further detectable impact on either the phenotype or the PP pathway flux (Fig. 3), providing further evidence for the conclusion that yqjI encodes the major 6-P-gluconate dehydrogenase during growth on glucose. Since GntZ apparently has no function during growth on glucose, we investigated the phenotype on gluconate plates. Like the results for growth on glucose, the gntZ mutant and the glucose-adapted yqjI* mutant were indistinguishable from the wild type. The unadapted yqjI mutant, in contrast, formed only very small colonies within 24 h, and there were no rapidly growing clones such as those seen on glucose plates. This residual growth of a yqjI mutant was independent of GntZ because the yqjI gntZ double knockout was indistinguishable from the single yqjI deletion. Thus, YqjI is also the major 6-P-gluconate dehydrogenase isoenzyme during growth on gluconate. The slow growth of the yqjI gntZ mutant on gluconate may be explained either by catabolic flux to riboses catalyzed by YqeC or by catabolic flux to glucose via the reverse gluconate bypass (Fig. 1).
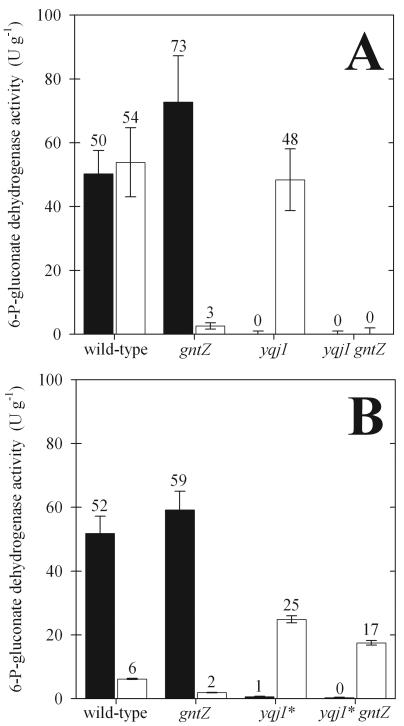
Next, we wondered whether both isoenzymes were differentially expressed and to what extent they contributed to NADPH generation, the major function of the oxidative PP pathway. Therefore, we assayed 6-P-gluconate dehydrogenase activity in crude cell extracts of B. subtilis mutants grown in complex and minimal media (Fig. 4). Unlike most bacteria, wild-type B. subtilis had a high activity with either NADP+ or NAD+ as the electrons acceptor in complex media (Fig. 4A). In minimal medium with glucose, however, the NAD+-dependent activity was low (Fig. 4B). Under both conditions, knockout of gntZ and knockout of yqjI eliminated the NAD+- and NADP+-dependent 6-P-gluconate dehydrogenase activities, respectively. Thus, YqjI appears to be the exclusive NADPH-producing isoform in B. subtilis, as was hypothesized previously based on sequence comparison (47). While GntZ appears to be the exclusive NADH-producing isoform in the wild type, the significant NAD+-dependent 6-P-gluconate dehydrogenase activity in the yqjI* gntZ double mutant in the presence of glucose was probably related to YqeC (Fig. 4B). Although NAD+-dependent activity was clearly present in the yqjI mutant, it could not compensate for the yqjI mutation during growth on glucose. The necessary adaptation of yqjI* cultures was not related to altered regulation or activity of GntZ or YqeC because the growth rates of yqjI* gntZ and yqjI* mutants on glucose were identical (Fig. 3A) and the oxidative PP pathway flux was virtually zero during growth on glucose (Fig. 3B).
FIG. 4.
NADP+-dependent (solid bars) and NAD+-dependent (open bars) 6-P-gluconate dehydrogenase activities in crude cell extracts of B. subtilis 1012 gntZ and yqjI mutants. The error bars indicate standard deviations based on triplicate experiments.
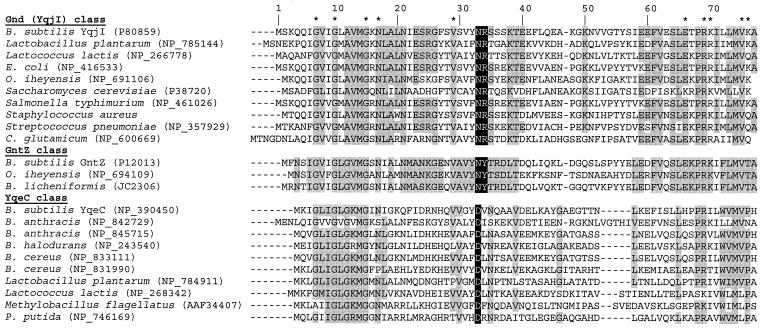
Multispecies alignment of 6-P-gluconate dehydrogenases.
The presence of three 6-P-gluconate dehydrogenases with different cofactor specificities and hence potentially different functions in the PP pathway of B. subtilis motivated us to investigate the distribution of orthologues in microbes. A conserved arginine residue (R-34 in YqjI) is generally necessary to bind and stabilize the 2′-phosphate of NADP+ in the NADP+-dependent 6-P-gluconate dehydrogenase homologues (1, 47). In NAD+-dependent GntZ-like isoforms, this arginine residue is replaced by a tyrosine (Fig. 5) that is specific for NAD+-dependent isoforms (25). Among the presently available prokaryotic and eukaryotic protein sequences in the GenBank database (2), only two other tyrosine-34 orthologues were identified; they were found in Bacillus licheniformis and Oceanobacillus iheyensis, and the latter also contains a YqjI homologue (Fig. 5). Unlike the findings for B. subtilis, we detected only the NADP+-dependent 6-P-gluconate dehydrogenase activity in crude cell extracts of O. iheyensis (data not shown).
FIG. 5.
Alignment of N-terminal dinucleotide binding domains of putative 6-P-gluconate dehydrogenases. The GenBank accession numbers are indicated in parentheses. Conserved residues at positions 33 and 34 are indicated by white type on a black background. Residues conserved in at least 70% of the sequences in a class are indicated by shading. Asterisks indicate residues conserved in all three classes.
B. subtilis YqeC is the prototype of a third class of less conserved homologues (Fig. 5). In contrast to the YqjI and GntZ classes, the conserved basic residue at position 33 is replaced by an acidic aspartate. The YqeC homologues are about one-third shorter than members of the other two classes, and they appear to lack key residues for binding to the phosphate group of 6-P-gluconate at the C terminus. Mainly for this reason, they were previously hypothesized to encode 3-hydroxyacid dehydrogenases (25, 47). More recently, however, it was demonstrated that the YqeC orthologue of Methylobacillus flagellatus is essential for the NAD+-dependent oxidation of 6-P-gluconate (6). Further evidence for the 6-P-gluconate dehydrogenase function of YqeC homologues comes from Pseudomonas species, in which a 6-P-gluconate dehydrogenase is active and neither YqjI nor GntZ is present but YqeC homologues are encoded in the genome (Table 3). B. subtilis YqeC also probably has a 6-P-gluconate dehydrogenase function because 6-P-gluconate dehydrogenase activity was detected in the yqjI* gntZ mutant during growth on glucose alone but not in complex media (Fig. 4). The absence of PP pathway flux in yqjI* cultures (Fig. 3B) demonstrates, however, that YqeC does not participate in the PP pathway of B. subtilis under the conditions tested here. This conclusion was confirmed by the lack of a detectable phenotype and an unaltered glycolysis-to-PP pathway split ratio in a B. subtilis yqeC mutant (data not shown).
TABLE 3.
Occurrence of B. subtilis Gnd, GntZ, and YqeC homologues with at least 40% identity in selected microorganismsa
| Organism | Gram stain | Genbank accession no. of homologues
|
||
|---|---|---|---|---|
| Gnd (YqjI) | GntZ | YqeC | ||
| Bacillus subtilis | + | P80859 | P12013 | NP_390450 |
| Bacillus halodurans | + | NP_243540 | ||
| Bacillus cereus | + | NP_831990, NP_833111 | ||
| Bacillus anthracis | + | NP_845715, NP_842729 | ||
| Bacillus licheniformisb | + | JC2306 | ||
| Oceanobacillus iheyensis | + | NP_691106 | NP_694109 | |
| Lactobacillus plantarum | + | NP_785144 | NP_784911 | |
| Lactococcus lactis | + | NP_266778 | NP_268342 | |
| Streptomyces coelicolor | + | NP_625271 | NP_628064, NP_630733 | |
| Staphylococcus aureus | + | NP_374625 | ||
| Escherichia coli | − | NP_416533 | ||
| Salmonella enterica serovar Typhimurium | − | NP_461026 | ||
| Corynebacterium glutamicum | − | NP_600669 | ||
| Corynebacterium diphtheriae | − | CAE49740 | ||
| Pseudomonas putida | − | NP_746169 | ||
The homologues were identified by Blast searches and were classified based on Clustal W alignments of the cofactor binding domains.
The complete genome sequence was not available, and thus there may be additional homologues.
DISCUSSION
The two major functions of the oxidative PP pathway are considered to be supply of biosynthetic precursors and supply of the anabolic redox cofactor NADPH (21). As in E. coli (41), neither function was essential for B. subtilis during growth on glucose because mutants with mutations in key enzymes without oxidative PP pathway fluxes grew rapidly, although the growth was about one-third slower than that of the parent. Using isotopic tracer experiments, we demonstrated that glycolysis was the primary pathway for glucose catabolism in glucose-6-P dehydrogenase mutants but that about 5% of the catabolic flux was catalyzed through the gluconate bypass that is typically found in pseudomonads (31). A similar in vivo flux observation was made with E. coli zwf mutants (15, 41), although in vitro gluconate bypass activities were described to be below the level of detection in a related E. coli strain (53). Knockout of the second major oxidative PP pathway enzyme, 6-P-gluconate dehydrogenase, was exclusively compensated for by flux rerouting through glycolysis, which is in contrast to what happens in E. coli, in which the rerouting occurs to a large extent via the Entner-Doudoroff pathway (28). In B. subtilis, we could exclude the possibility that there was a functional Entner-Doudoroff pathway under the conditions investigated because we never observed [1-13C]alanine in experiments with [1-13C]glucose (15), even in the yqjI mutant. Although B. subtilis contains a homologue of the second pathway enzyme, 2-keto-3-deoxygluconate-6-P aldolase (37), the absence of a 6-P-gluconate dehydratase homologue and our results suggest that the pathway does not exist in B. subtilis.
Based on 13C-labeling data from isogenic knockouts, we demonstrated that the major 6-P-gluconate dehydrogenase isoenzyme is encoded by the yqjI gene in B. subtilis. This conclusion contrasts with the generally held perception of the oxidative PP pathway in B. subtilis (4, 35, 45); hence, we propose a new designation for yqjI, gndA, the monocistronic gene encoding the principal 6-P-gluconate dehydrogenase. For growth on glucose, YqjI was important, but an unidentified adaptation permitted growth without an active oxidative PP pathway. The second isoform, GntZ, could not substitute for a knockout of yqjI, possibly because of different enzyme kinetics. Although in vitro enzyme data demonstrated the presence of GntZ on complex media, GntZ was seemingly not involved in the necessary adaptation of yqjI* cultures for growth on glucose because the oxidative PP pathway flux was at the level of detection. Moreover, a double yqjI* gntZ mutant had essentially the same phenotype as the yqjI* mutant. A role for the third, yqeC-encoded 6-P-gluconate dehydrogenase is unlikely because the PP pathway flux was at or below the detection limit in yqjI* cultures. Confirming previous observations (20, 45), GntZ was apparently not relevant even during gluconate catabolism because a gntZ mutant exhibited no phenotype and GntZ could not substitute for a knockout of yqjI that severely impaired growth on this substrate. Thus, the metabolic functions of GntZ and YqeC remain obscure.
By using in vitro enzyme assays with crude cell extracts of B. subtilis knockout mutants, the yqjI-encoded 6-P-gluconate dehydrogenase was shown to be NADP+ dependent, as was hypothesized previously from sequence comparisons (47), while the gntZ-encoded dehydrogenase was NAD+ dependent. Gram-negative genomes usually encode a single NADP+-dependent 6-P-gluconate dehydrogenase of the Gnd (or YqjI) class (Table 3). Pseudomonads and some bacilli (Fig. 5) appear to be an exception to this and rely exclusively on the truncated YqeC class. From the sequence, the preferred YqeC cofactor remains unclear because the acidic aspartate at position 33 favors binding of NAD+, while the basic arginine at position 34 stabilizes the phosphate group of NADP+, assuming that the same three-dimensional fold is adopted by the enzymes of the three classes and that the positions of residues 33 and 34 are the same in these proteins (Fig. 5). Indeed, the 6-P-gluconate dehydrogenase of Pseudomonas fluorescens was shown to be active with either NAD+ and NADP+ (46), and the described NAD+-dependent activity in Streptomyces and heterofermentative lactic acid bacteria (13, 36) probably originated from the YqeC homologue (Table 3). In contrast to pseudomonads, gram-positive bacteria exhibit a much wider spectrum of combinations. While many gram-positive bacteria contain a member of the NADP+-dependent Gnd (YqjI) class, most gram-positive organisms contain a member of the truncated YqeC class. At this time B. subtilis is unique because it contains a homologue of all three classes. Here, we demonstrated NAD+- and NADP+-dependent activities encoded by the gntZ and yqjI genes of B. subtilis, respectively. Furthermore, the first evidence that there is a yqeC-encoded NAD+-dependent activity in B. subtilis comes from the significant residual 6-P-gluconate dehydrogenase activity in the yqjI* gntZ mutant (Fig. 4B). In principle, the coexistence of NAD+- and NADP+-dependent isoenzymes would enable flexible adjustment of NADP+ or NAD+ reduction in the PP pathway to the overall metabolic requirement of the cell, a function that is performed in E. coli by transhydrogenases (41) that have not been identified in B. subtilis yet.
Acknowledgments
We thank Simon Tännler and Tobias Fuhrer for technical assistance.
REFERENCES
- 1.Adams, M. J., G. H. Ellis, S. Gover, C. E. Naylor, and C. Phillips. 1994. Crystallographic study of coenzyme, coenzyme analogue and substrate binding in 6-phosphogluconate dehydrogenase: implications for NADP specificity and the enzyme mechanism. Structure 2:651-668. [DOI] [PubMed] [Google Scholar]
- 2.Benson, D. A., I. Karsch-Mizrachi, D. J. Lipman, J. Ostell, and D. L. Wheeler. 2003. GenBank. Nucleic Acids Res. 31:23-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergmeyer, H. U. 1985. Methods of enzymatic analysis, vol. III. VCH Publishers, Deerfield Beach, Fla.
- 4.Blencke, H. M., G. Homuth, H. Ludwig, U. Mader, M. Hecker, and J. Stülke. 2003. Transcriptional profiling of gene expression in response to glucose in Bacillus subtilis: regulation of the central metabolic pathways. Metab. Eng. 5:133-149. [DOI] [PubMed] [Google Scholar]
- 5.Chassagnole, C., N. Noisommit-Rizzi, J. W. Schmid, K. Mauch, and M. Reuss. 2002. Dynamic modeling of the central carbon metabolism of Escherichia coli. Biotechnol. Bioeng. 79:53-73. [DOI] [PubMed] [Google Scholar]
- 6.Chistoserdova, L., L. Gomelsky, J. A. Vorholt, M. Gomelsky, Y. D. Tsygankov, and M. E. Lidstrom. 2000. Analysis of two formaldehyde oxidation pathways in Methylobacillus flagellatus KT, a ribulose monophosphate cycle methylotroph. Microbiology 146:233-238. [DOI] [PubMed] [Google Scholar]
- 7.Christensen, B., T. Christiansen, A. K. Gombert, and J. Nielsen. 2001. Simple and robust method for estimation of the split ratio between the oxidative pentose phosphate pathways and the Embden-Meyerhof-Parnas pathway in microorganisms. Biotechnol. Bioeng. 74:517-523. [DOI] [PubMed] [Google Scholar]
- 8.Christensen, B., and J. Nielsen. 1999. Metabolic network analysis. Adv. Biochem. Eng. Biotechnol. 66:209-231. [PubMed] [Google Scholar]
- 9.Christiansen, T., B. Christensen, and J. Nielsen. 2002. Metabolic network analysis of Bacillus clausii on minimal and semirich medium using 13C-labeled glucose. Metab. Eng. 4:159-169. [DOI] [PubMed] [Google Scholar]
- 10.Dauner, M., and U. Sauer. 2001. Stoichiometric growth model for riboflavin-producing Bacillus subtilis. Biotechnol. Bioeng. 76:132-143. [DOI] [PubMed] [Google Scholar]
- 11.Dauner, M., M. Sonderegger, M. Hochuli, T. Szyperski, K. Wüthrich, H.-P. Hohmann, U. Sauer, and J. E. Bailey. 2002. Intracellular carbon fluxes in riboflavin-producing Bacillus subtilis during growth on two-carbon substrate mixtures. Appl. Environ. Microbiol. 68:1760-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dauner, M., T. Storni, and U. Sauer. 2001. Bacillus subtilis metabolism and energetics in carbon-limited and carbon-excess chemostat cultures. J. Bacteriol. 183:7308-7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dekleva, M. L., and W. R. Strohl. 1988. Biosynthesis of ɛ-rhodomycinone from glucose by Streptomyces C5 and comparison with intermediary metabolism of other polyketide-producing streptomycetes. Can. J. Microbiol. 34:1235-1240. [DOI] [PubMed] [Google Scholar]
- 14.de Mendoza, D., R. Grau, and J. E. Cronan. 1993. Biosynthesis and function of membrane lipids, p. 411-424. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. American Society for Microbiology, Washington, D.C.
- 15.Fischer, E., and U. Sauer. 2003. Metabolic flux profiling of Escherichia coli mutants in central carbon metabolism using GC-MS. Eur. J. Biochem. 270:880-891. [DOI] [PubMed] [Google Scholar]
- 16.Fischer, E., N. Zamboni, and U. Sauer. 2004. High-throughput metabolic flux analysis based on gas chromatography-mass spectrometry derived 13C constraints. Anal. Biochem. 325:308-316. [DOI] [PubMed] [Google Scholar]
- 17.Fortnagel, P. 1993. Glycolysis, p. 171-180. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. American Society for Microbiology, Washington, D.C.
- 18.Fraenkel, D. G. 1996. Glycolysis, p. 189-198. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 19.Fraenkel, D. G. 1986. Mutants in glucose metabolism. Annu. Rev. Biochem. 55:317-337. [DOI] [PubMed] [Google Scholar]
- 20.Fujita, Y., and T. Fujita. 1989. Effect of mutations causing gluconate kinase or gluconate permease deficiency on expression of the Bacillus subtilis gnt operon. J. Bacteriol. 171:1751-1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garrett, R. H., and C. M. Grisham. 1999. Biochemistry, 2nd ed., p. 742-774. Saunders College Publishing, Fort Worth, Tex.
- 22.Gottschalk, G. 1986. Bacterial metabolism, 2nd ed. Springer-Verlag, New York, N.Y.
- 23.Guérout-Fleury, A. M., K. Shazand, N. Frandsen, and P. Stragier. 1995. Antibiotic-resistance cassettes for Bacillus subtilis. Gene 167:335-336. [DOI] [PubMed] [Google Scholar]
- 24.Harwood, C. R., and S. M. Cutting. 1990. Molecular biological methods for Bacillus. John Wiley & Sons Ltd., Chichester, England.
- 25.Hawes, J. W., E. T. Harper, D. W. Crabb, and R. A. Harris. 1996. Structural and mechanistic similarities of 6-phosphogluconate and 3-hydroxyisobutyrate dehydrogenases reveal a new enzyme family, the 3-hydroxyacid dehydrogenases. FEBS Lett. 389:263-267. [DOI] [PubMed] [Google Scholar]
- 26.Hua, Q., C. Yang, T. Baba, H. Mori, and K. Shimizu. 2003. Responses of the central carbon metabolism in Escherichia coli to phosphoglucose isomerase and glucose-6-phosphate dehydrogenase knockouts. J. Bacteriol. 185:7053-7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Itaya, M., K. Kondo, and T. Tanaka. 1989. A neomycin resistance gene cassette selectable in a single copy state in the Bacillus subtilis chromosome. Nucleic Acids Res. 17:4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiao, Z., T. Baba, H. Mori, and K. Shimizu. 2003. Analysis of metabolic and physiological responses to gnd knockout in Escherichia coli by using C-13 tracer experiment and enzyme activity measurements. FEMS Microbiol. Lett. 220:295-301. [DOI] [PubMed] [Google Scholar]
- 29.Kanehisa, M., S. Goto, S. Kawashima, and A. Nakaya. 2002. The KEGG databases at GenomeNet. Nucleic Acids Res. 30:42-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kupor, S. R., and D. G. Fraenkel. 1972. Glucose metabolism in 6-phosphogluconolactonase mutants of Escherichia coli. J. Biol. Chem. 247:1904-1910. [PubMed] [Google Scholar]
- 31.Lessie, T. G., and P. V. Phibbs, Jr. 1984. Alternative pathways of carbohydrate utilization in pseudomonads. Annu. Rev. Microbiol. 38:359-388. [DOI] [PubMed] [Google Scholar]
- 32.Michal, G. 1999. Biochemical pathways. Spektrum Akad. Verlag GmbH, Heidelberg, Germany.
- 33.Moritz, B., K. Striegel, A. A. de Graaf, and H. Sahm. 2002. Changes of pentose phosphate pathway flux in vivo in Corynebacterium glutamicum during leucine-limited batch cultivation as determined from intracellular metabolite concentration measurements. Metab. Eng. 4:295-305. [DOI] [PubMed] [Google Scholar]
- 34.Moritz, B., K. Striegel, A. A. de Graaf, and H. Sahm. 2000. Kinetic properties of the glucose-6-phosphate and 6-phosphogluconate dehydrogenases from Corynebacterium glutamicum and their application for predicting pentose phosphate pathways fluxes in vivo. Eur. J. Biochem. 267:3442-3452. [DOI] [PubMed] [Google Scholar]
- 35.Moszer, I., L. M. Jones, S. Moreira, C. Fabry, and A. Danchin. 2002. SubtiList: the reference database for the Bacillus subtilis genome. Nucleic Acids Res. 30:62-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ohara, H., K. Uchida, M. Yahata, and H. Kondo. 1996. NAD-specific 6-phosphogluconate dehydrogenase in lactic acid bacteria. Biosci. Biotechnol. Biochem. 60:692-693. [DOI] [PubMed] [Google Scholar]
- 37.Pujic, P., R. Dervyn, A. Sorokin, and S. D. Ehrlich. 1998. The kdgRKAT operon of Bacillus subtilis: detection of the transcript and regulation by the kdgR and ccpA genes. Microbiology 144:3111-3118. [DOI] [PubMed] [Google Scholar]
- 38.Reizer, A., J. Deutscher, M. H. Saier, Jr., and J. Reizer. 1991. Analysis of the gluconate (gnt) operon of Bacillus subtilis. Mol. Microbiol. 5:1081-1089. [DOI] [PubMed] [Google Scholar]
- 39.Saito, H., T. Shibata, and T. Ando. 1979. Mapping of genes determining nonpermissiveness and host-specific restriction to bacteriophages in Bacillus subtilis Marburg. Mol. Gen. Genet. 170:117-122. [DOI] [PubMed] [Google Scholar]
- 40.Sauer, U. 2004. High-throughput phenomics: experimental methods for mapping fluxomes. Curr. Opin. Biotechnol. 15:58-63. [DOI] [PubMed] [Google Scholar]
- 41.Sauer, U., F. Canonaco, S. Heri, A. Perrenoud, and E. Fischer. 2004. The soluble and membrane-bound transhydrogenases UdhA and PntAB have divergent functions in NADPH metabolism of Escherichia coli. J. Biol. Chem. 279:6613-6619. [DOI] [PubMed] [Google Scholar]
- 42.Sauer, U., V. Hatzimanikatis, J. E. Bailey, M. Hochuli, T. Szyperski, and K. Wüthrich. 1997. Metabolic fluxes in riboflavin-producing Bacillus subtilis. Nat. Biotechnol. 15:448-452. [DOI] [PubMed] [Google Scholar]
- 43.Sauer, U., V. Hatzimanikatis, H.-P. Hohmann, M. Manneberg, A. P. G. M. van Loon, and J. E. Bailey. 1996. Physiology and metabolic fluxes of wild-type and riboflavin-producing Bacillus subtilis. Appl. Environ. Microbiol. 62:3687-3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sauer, U., D. R. Lasko, J. Fiaux, M. Hochuli, R. Glaser, T. Szyperski, K. Wüthrich, and J. E. Bailey. 1999. Metabolic flux ratio analysis of genetic and environmental modulations of Escherichia coli central carbon metabolism. J. Bacteriol. 181:6679-6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Steinmetz, M. 1993. Carbohydrate catabolism: pathways, enzymes, genetic regulation, and evolution, p. 157-170. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. American Society for Microbiology, Washington, D.C.
- 46.Stournaras, C., P. Maurer, and G. Kurz. 1983. 6-Phospho-d-gluconate dehydrogenase from Pseudomonas fluorescens. Properties and subunit structure. Eur. J. Biochem. 130:391-396. [DOI] [PubMed] [Google Scholar]
- 47.Tetaud, E., S. Hanau, J. M. Wells, R. W. Le Page, M. J. Adams, S. Arkison, and M. P. Barrett. 1999. 6-Phosphogluconate dehydrogenase from Lactococcus lactis: a role for arginine residues in binding substrate and coenzyme. Biochem. J. 338:55-60. [PMC free article] [PubMed] [Google Scholar]
- 48.Vagner, V., E. Dervyn, and S. D. Ehrlich. 1998. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology 144:3097-3104. [DOI] [PubMed] [Google Scholar]
- 49.Wiechert, W. 2001. 13C metabolic flux analysis. Metab. Eng. 3:195-206. [DOI] [PubMed] [Google Scholar]
- 50.Wittmann, C. 2002. Metabolic flux analysis using mass spectroscopy. Adv. Biochem. Eng. Biotechnol. 74:39-64. [DOI] [PubMed] [Google Scholar]
- 51.Yoshida, K., I. Ishio, E. Nagakawa, Y. Yamamoto, M. Yamamoto, and Y. Fujita. 2000. Systematic study of gene expression and transcription organization in the gntZ-ywaA region of the Bacillus subtilis genome. Microbiology 146:573-579. [DOI] [PubMed] [Google Scholar]
- 52.Zamboni, N., and U. Sauer. 2003. Knockout of the high-coupling cytochrome aa3 oxidase reduces TCA cycle fluxes in Bacillus subtilis. FEMS Microbiol. Lett. 226:121-126. [DOI] [PubMed] [Google Scholar]
- 53.Zhao, J., T. Baba, H. Mori, and K. Shimizu. 2004. Global metabolic response of Escherichia coli to gnd or zwf gene-knockout, based on 13C-labeling experiments and the measurement of enzyme activities. Appl. Microbiol. Biotechnol. 64:91-98. [DOI] [PubMed] [Google Scholar]