Abstract
Identification of B-cell epitopes in target antigens is a critical step in epitope-driven vaccine design, immunodiagnostic tests, and antibody production. B-cell epitopes could be linear, i.e., a contiguous amino acid sequence fragment of an antigen, or conformational, i.e., amino acids that are often not contiguous in the primary sequence but appear in close proximity within the folded 3D antigen structure. Numerous computational methods have been proposed for predicting both types of B-cell epitopes. However, the development of tools for reliably predicting B-cell epitopes remains a major challenge in immunoinformatics.
Classifier ensembles a promising approach for combining a set of classifiers such that the overall performance of the resulting ensemble is better than the predictive performance of the best individual classifier. In this chapter, we show how to build a classifier ensemble for improved prediction of linear B-cell epitopes. The method can be easily adapted to build classifier ensembles for predicting conformational epitopes.
Keywords: B-cell epitope prediction, Classifiers ensemble, Random forest, Epitope prediction toolkit
1 Introduction
Antigen-antibody interactions play a crucial role in the humoral immune response. Antibodies, a family of structurally related glycoproteins produced in membrane-bound or secreted form by B lymphocytes, serve as mediators of specific humoral immunity by engaging various effector mechanisms that serve to eliminate the bound antigens [1]. The part of the antigen recognized by antibodies is called B-cell epitope. B-cell epitopes often classified into two categories: (1) linear (continuous) B-cell epitopes consist of amino acid residues that are sequential in the primary structure of the protein and (2) conformational (discontinuous) B-cell epitopes consist of residues that are not sequential in the protein primary structure but come together in the protein 3D structure. Conformational B-cell epitopes form the majority of B-cell epitopes. Several experimental procedures for mapping both types of B-cell epitopes have been presented [2]. However, in silico methods for identifying B-cell epitopes have the potential to dramatically decrease the cost and the time associated with the experimental mapping of B-cell epitopes [3].
Several computational methods have been proposed for predicting either linear or conformational B-cell epitopes [3–5]. Methods for predicting linear B-cell epitopes range from simple propensity scale profiling methods [6–9] to methods based on state-of-the-art machine learning predictors (e.g., [10–14]). Methods for predicting conformational B-cell epitopes (e.g., [15–19]) utilize some structure and physicochemical features derived from antigen-antibody complexes that could be correlated with antigenicity [3]. Despite the large number of B-cell epitope prediction methods proposed in literature, the performance of existing methods leaves significant room for improvement [4].
One of the promising approaches for improving the predictive performance of computational B-cell epitope prediction tools is to combine multiple classifiers. This approach is motivated by the observation that no single predictor outperforms all other predictors and that predictors often complement each other [20].
Against this background, we present a framework for developing classifier ensembles [21] and explain the procedure for building several variants of classifier ensembles based on the framework. Specifically, we describe a procedure for building classifier ensembles for predicting linear B-cell epitopes using Epitopes Toolkit (EpiT) [22]. We also show how to adapt the procedure for building classifier ensembles for predicting conformational B-cell epitopes (see Note 1). The procedures described in this chapter can be adapted for any other machine learning benchmark.
2 Materials
2.1 Data Set
We used the FBCPRED data set [11], a homology-reduced data set of variable-length linear B-cell epitopes extracted from Bcipep database [23]. The data set has 934 epitopes and non-epitopes (respectively) such that the length distribution of epitopes and non-epitopes is preserved.
2.2 Epitopes Toolkit (EpiT)
WEKA [24] is a machine learning workbench that is widely used by bioinformatics developers for developing prediction tools. Unfortunately, the vast majority of WEKA-implemented algorithms do not accept amino acid sequences as input. Hence, developers have to preprocess their sequence data for extracting useful features before using WEKA classification algorithms. Alternatively, developers of epitope prediction tools can use the Epitopes Toolkit (EpiT) [22] which is built on top of WEKA and provides a specialized set of useful data preprocessors (e.g., filters) and classification algorithms for developing B-cell epitope prediction tools. A java implementation of EpiT is freely available at the project website, http://ailab.ist.psu.edu/epit. More information about how to install and use EpiT is provided in the project documentation.
3 Methods
In this section, we show how to use EpiT to build individual and classifier ensembles for predicting linear B-cell epitopes. The procedure can be easily adapted for any other machine learning workbench (e.g., RapidMiner [25] and KNIME [26]).
3.1 Building a Single Classifier with EpiT
Here, we show how to build a single predictor using FlexLenBCPred. nr80.arff, FBCPRED data in WEKA format available at http://ailab.ist.pdu.edu/red/bcell/FBCPred.zip, and a Random Forest classifier [27] with 50 trees (RF50).
Run EpiT.
Go to Application menu and select model builder application.
In the model builder window (WEKA explorer augmented with EpiT filters and prediction methods) click open and select the file fbcprednr80.arff.
Click classify tab.
In the classifier panel, click choose and browse for weka.meta.FilteredClassifier. The FilteredClassifier is a WEKA class for running an arbitrary classifier on data that has been passed through arbitrary filter.
Click on the FilteredClassifier in the classifier panel and specify the following classifier and filter. For the classifier, choose weka.classifiers.trees.RandomForest and set numTrees to 50. For the filter, choose epit.filters.unsupervised.attribute.AAP. The AAP filter implements the amino acid propensity scale features proposed in [28].
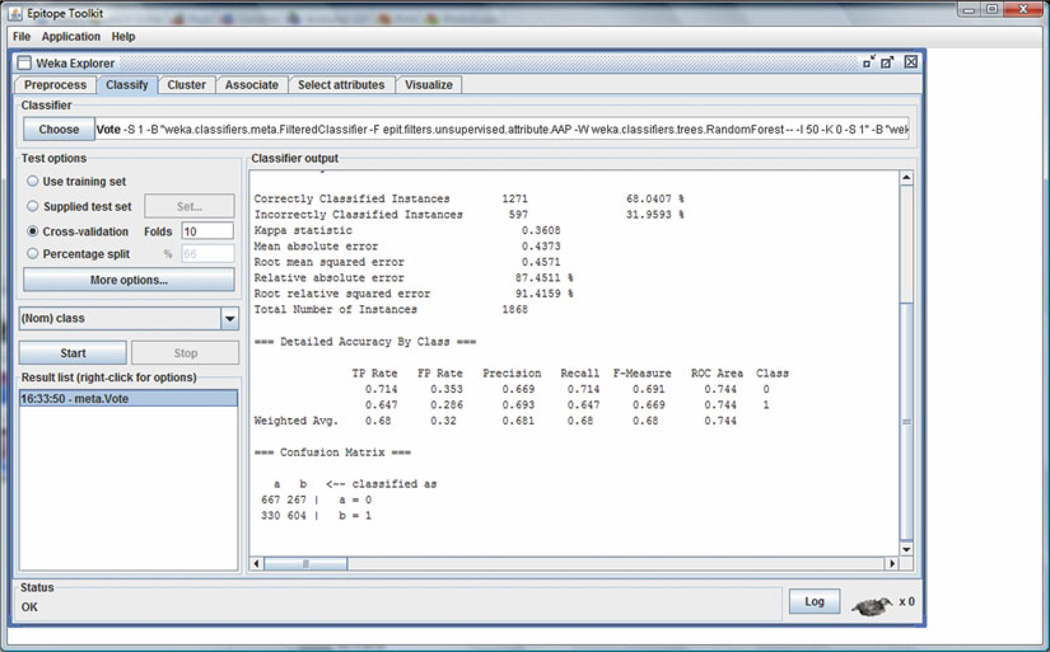
Having both the data set and the classification algorithm specified, we are ready to build the model and evaluate it using ten-fold cross-validation (see Note 2). Just click start button and wait for the ten-fold cross-validation procedure to finish. The classifier output panel shows several statistical estimates of the classifier using ten-fold cross-validation (see Fig. 1).
Fig. 1.
Output statistics for ten-fold cross-validation experiment evaluating Vote classifier with average (AVG) of probabilities combination rule
3.2 Building a Classifier Ensemble with EpiT
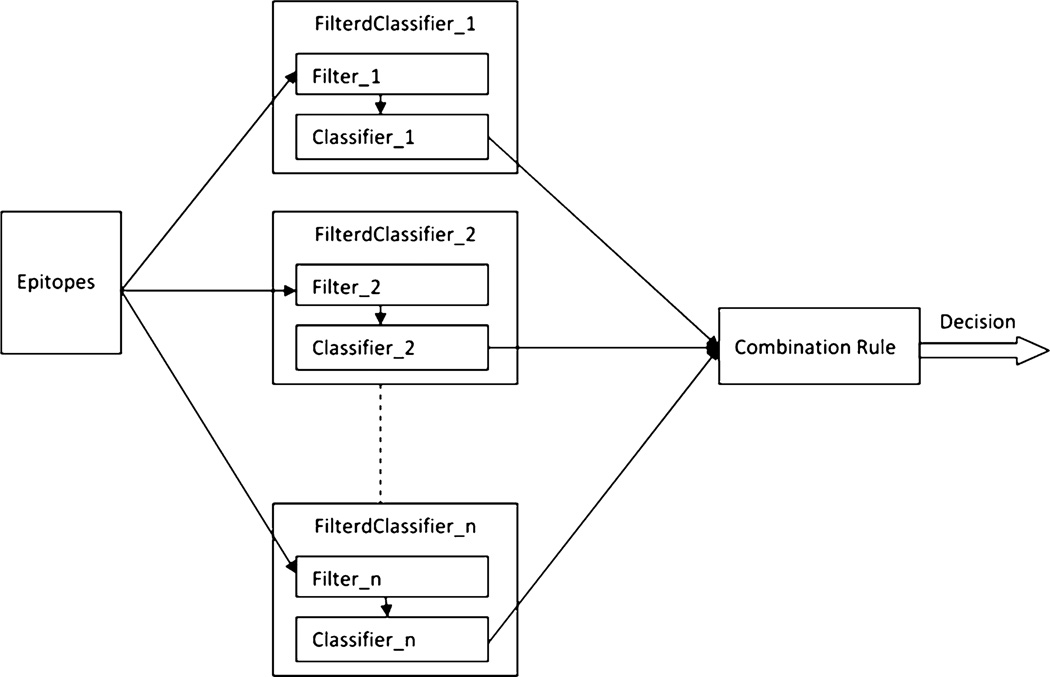
A classifier ensemble consists of a collection of individual (or base) classifiers that work together using a suitably designed fusion method (e.g., combination rule or second-level classifier) for optimally combining the outputs of the individual classifiers. This design process involves two basic steps: (1) design a set of complementary or diverse base classifiers: diversity of classifiers could be ensured by manipulating the classifiers’ inputs, outputs, or the training algorithms [21] (see Notes 3 and 4); (2) design a combination rule that exploits the behaviors of the individual classifiers to optimally combine them. Figure 2 shows a framework for constructing classifier ensembles using EpiT. In this framework, different classifier ensembles can be developed by using different combinations of choices of filters, base classifiers, and combination rules. In this example, we fix the base classifier to RF50 and use different filters for each individual classifier. We also experiment with different combination rules. To build a classifier ensemble for predicting flexible-length linear B-cell epitopes using EpiT, follow the following procedure:
Run EpiT.
Go to Application menu and select the model builder application.
In the model builder window (WEKA explorer augmented with EpiT filters and prediction methods) click open and select the file fbcprednr80.arff.
Click classify tab.
In the classifier panel, click choose and browse for weka.meta.Vote. The Vote classifier is a WEKA class for combining classifiers. Different combinations of probability estimates for classification are available.
Click on classifiers and enter four FilteredClassifiers. Set the classifier parameter for each FilteredClassifier to RF50 and set the filter parameter to AAP, CTD, SequenceComposition, and SequenceDiCompositions, respectively.
Select one of the available combination rule options. In our experiment we used the WEKA default setting for this parameter, average of probabilities.
Click start button to start a ten-fold cross-validation experiment and wait for the output results (see Fig. 1).
Fig. 2.
Framework for building classifier ensembles using EpiT tool
A more sophisticated way for combining multiple classifiers according to the framework in Fig. 2 is to replace the simple combination rule used with Vote classifier with a meta-predictor, a second-stage classifier. The procedure for building such a classifier ensemble is as follows:
Run EpiT.
Go to Application menu and select the model builder application.
In the model builder window (WEKA explorer augmented with EpiT filters and prediction methods) click open and select the file fbcprednr80.arff.
Click classify tab.
In the classifier panel, click choose and browse for weka.meta.Stacking. The Stacking classifier is a WEKA class for combining several classifiers using the stacking method [29].
Click on classifiers and enter four FilteredClassifiers. Set the classifier parameter for each FilteredClassifier to RF50 and set the filter parameter to AAP, CTD, SequenceComposition, and SequenceDiCompositions, respectively.
Click on metaclassifier and choose the naïve Bayes (NB) classifier, weka.classifiers.bayes.NaiveBayes.
Set numFolds to 3. This parameter sets the number of folds used for cross-validation experiment performed for training the meta-classifier. Click OK.
Click start button to start a ten-fold cross-validation experiment.
Table 1 compares the performance (in terms of AUC scores (see Note 5)) of two classifiers, NB and RF50, using four sets of input features: (1) amino acid pair (AA) propensities [28]; (2) composition-transition-distribution (CTD) [30]; (3) amino acid composition (AAC); and (4) dipeptide composition (DC). Table 2 compares the performance of a classifier ensemble that combines four NB classifiers trained using the four sets of input features (AAP, CTD, AAC, DC) and a classifier ensemble that combines four RF50 constructed using the four sets of input features. Four simple combination rules have been evaluated: AVG, PROD, MIN, and MAX which represent average, product, minimum, and maximum estimated probabilities from the four base classifiers for each input instance. Table 3 compares the performance of the NB- and RF50-based classifier ensembles (reported in Table 2) when the simple combination rule is replaced with a meta-classifier (second-stage classifier).
Table 1.
AUC values for naïve Bayes (NB) and Random Forest (RF50) classifiers using four different sets of input features
| Features | NB | RF50 |
|---|---|---|
| AAP | 0.67 | 0.72 |
| CTD | 0.65 | 0.65 |
| AAC | 0.66 | 0.71 |
| DC | 0.63 | 0.72 |
Table 2.
AUC values for a classifier ensemble that combines four NB classifiers trained using the four sets of input features (AAP, CTD, AAC, DC) and a classifier ensemble that combines four RF50 constructed using the four sets of input features
| Combination rule | NB | RF50 |
|---|---|---|
| AVG | 0.69 | 0.74 |
| PROD | 0.65 | 0.75 |
| MIN | 0.64 | 0.75 |
| MAX | 0.68 | 0.74 |
The classifier ensembles are obtained using the same base classifiers but different combination rules
Table 3.
AUC values for a classifier ensemble that combines four NB classifiers trained using the four sets of input features (AAP, CTD, AAC, DC) and a classifier ensemble that combines four RF50 constructed using the four sets of input features
| Meta-predictor | NB | RF50 |
|---|---|---|
| NB | 0.69 | 0.75 |
| Logistic | 0.69 | 0.75 |
The classifier ensembles are obtained using the same base classifiers but different meta-predictors
Table 1 shows that the predictive performance of each classifier seems to be highly dependent on the input features. For example, AUC scores of RF50 range from 0.65 to 0.72 for different choices of input features. Tables 2 and 3 show that combining individual classifiers constructed with different input features and using the same classification algorithm (e.g., NB and RF50) not only eliminate the dependency on the input features but also yields a classifier ensemble with performance higher than the best individual classifier performance obtained in Table 1.
It should be noted that the RF50 classifier, treated in our experiments as an individual classifier, is itself an ensemble of 50 different decision tree classifiers. The performance of RF50 might be improved using several approaches including (1) increasing the number of trees, (2) selecting a subset of the 50 trees using some criteria for eliminating redundant and poor tree predictors [31], and (3) building a multiple classifier system in which RF50 is treated as a base classifier.
Acknowledgments
This work was supported in part by a grant from the National Institutes of Health (NIH GM066387) and by Edward Frymoyer Chair of Information Sciences and Technology at Pennsylvania State University.
Footnotes
The current implementation of EpiT does not support the extraction of evolutionary or structure-based features since most of these features require running third-party programs (e.g., BLAST [32]). Building classifier ensemble that uses such features requires preprocessing the training data such that each epitope in the original data is represented with a combined set of extracted features (each set of features might be extracted using one or more third-party program (s)). The resulting combined set of features are used as inputs and the filter for each FilteredClassifier will select a range of attribute indices (corresponding to a set of features) to pass to the base classifier.
In ten-fold cross-validation experiments, the data set is randomly partitioned into ten equal subsets such that the relative proportion of epitopes to non-epitopes in each subset is preserved. Nine of the subsets are used for training the classifier and the remaining subset is used for testing the classifier. This procedure is repeated ten times, each time setting aside a different subset of the data for testing. The estimated performance of the classifier corresponds to an average of the results from the ten cross-validation runs.
Classifier ensembles can be developed using a single set of features and a single classification algorithm by training each base classifier with different training data (i.e., sampled instances or sampled subspace of the original training data). WEKA provides built-in classification algorithms for building such ensemble of classifiers (e.g., Bagging [33] and AdaBoost [34]).
For unbalanced data, an ensemble of classifiers system can be created by training each single classifier using all training instances from the minority class and an equal number of training instances (selected at random) from the majority class [21]. Such base classifiers can be created using EpiT Balanced Classifier (for more details please refer to EpiT documentation). The classifiers can be combined using a combination rule via Vote class or using a meta-classifier via Stacking class.
The receiver operating characteristic (ROC) curve is obtained by plotting the true positive rate as a function of the false-positive rate as the discrimination threshold of the binary classifier is varied. A widely used measure of classifier performance is the area under ROC curve (AUC). A perfect classifier will have an AUC = 1, while a random guessing classifier will have an AUC = 0.5, and any classifier performing better than random will have an AUC value that lies between these two values.
References
- 1.Abbas AK, Lichtman AH, Pillai S. Cellular and molecular immunology. 6th edn. Philadelphia: Saunders Elsevier; 2007. [Google Scholar]
- 2.Reineke U, Schutkowski M. Methods in molecular biology. 2nd edn. Vol. 524. New York: Humana Press; 2009. Epitope mapping protocols. [PubMed] [Google Scholar]
- 3.Ansari HR, Raghava GP. In silico models for B-cell epitope recognition and signaling. Methods Mol Biol. 2013;993:129–138. doi: 10.1007/978-1-62703-342-8_9. [DOI] [PubMed] [Google Scholar]
- 4.El-Manzalawy Y, Honavar V. Recent advances in B-cell epitope prediction methods. Immunome Res. 2010;6(Suppl 2):S2. doi: 10.1186/1745-7580-6-S2-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yao B, Zheng D, Liang S, et al. Conformational B-cell epitope prediction on antigen protein structures: a review of current algorithms and comparison with common binding site prediction methods. PLoS One. 2013;8(4):e62249. doi: 10.1371/journal.pone.0062249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Emini EA, Hughes JV, Perlow D, et al. Induction of hepatitis A virus-neutralizing antibody by a virus-specific synthetic peptide. J Virol. 1985;55(3):836–839. doi: 10.1128/jvi.55.3.836-839.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karplus P, Schulz G. Prediction of chain flexibility in proteins. Naturwissenschaften. 1985;72(4):212–213. [Google Scholar]
- 8.Parker JM, Guo D, Hodges RS. New hydrophilicity scale derived from high-performance liquid chromatography peptide retention data: correlation of predicted surface residues with antigenicity and X-ray-derived accessible sites. Biochemistry. 1986;25(19):5425–5432. doi: 10.1021/bi00367a013. [DOI] [PubMed] [Google Scholar]
- 9.Pellequer J-L, Westhof E, Van Regenmortel MH. Correlation between the location of antigenic sites and the prediction of turns in proteins. Immunol Lett. 1993;36(1):83–99. doi: 10.1016/0165-2478(93)90072-a. [DOI] [PubMed] [Google Scholar]
- 10.El-Manzalawy Y, Dobbs D, Honavar V. Predicting linear B-cell epitopes using string kernels. J Mol Recognit. 2008;21(4):243–255. doi: 10.1002/jmr.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.El-Manzalawy Y, Dobbs D. Honavar V (3400678) Predicting flexible length linear B-cell epitopes. Comput Syst Bioinformatics. 2008:121–132. [PMC free article] [PubMed] [Google Scholar]
- 12.Larsen JE, Lund O, Nielsen M. Improved method for predicting linear B-cell epitopes. Immunome Res. 2006;2:2. doi: 10.1186/1745-7580-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saha S, Raghava GP. Prediction of continuous B-cell epitopes in an antigen using recurrent neural network. Proteins. 2006;65(1):40–48. doi: 10.1002/prot.21078. [DOI] [PubMed] [Google Scholar]
- 14.Sweredoski MJ, Baldi P. COBEpro: a novel system for predicting continuous B-cell epitopes. Protein Eng Des Sel. 2009;22(3):113–120. doi: 10.1093/protein/gzn075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haste Andersen P, Nielsen M, Lund O. Prediction of residues in discontinuous B-cell epitopes using protein 3D structures. Protein Sci. 2006;15(11):2558–2567. doi: 10.1110/ps.062405906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kringelum JV, Lundegaard C, Lund O, et al. Reliable B cell epitope predictions: impacts of method development and improved benchmarking. PLoS Comput Biol. 2012;8(12):e1002829. doi: 10.1371/journal.pcbi.1002829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ponomarenko J, Bui H-H, Li W, et al. ElliPro: a new structure-based tool for the prediction of antibody epitopes. BMC Bioinformatics. 2008;9(1):514. doi: 10.1186/1471-2105-9-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun J, Wu D, Xu T, et al. SEPPA: a computational server for spatial epitope prediction of protein antigens. Nucleic Acids Res. 2009;37(suppl 2):W612–W616. doi: 10.1093/nar/gkp417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sweredoski MJ, Baldi P. PEPITO: improved discontinuous B-cell epitope prediction using multiple distance thresholds and half sphere exposure. Bioinformatics. 2008;24(12):1459–1460. doi: 10.1093/bioinformatics/btn199. [DOI] [PubMed] [Google Scholar]
- 20.Resende DM, Rezende AM, Oliveira NJ, et al. An assessment on epitope prediction methods for protozoa genomes. BMC Bioinformatics. 2012;13:309. doi: 10.1186/1471-2105-13-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wozniak M. Hybrid Classifiers: Methods of Data, Knowledge, and Classifier Combination. Vol. 519. Springer Heidelberg London: Studies in Computational Intelligence; 2013. [Google Scholar]
- 22.El-Manzalawy Y. Honavar V A framework for developing epitope prediction tools. Proceedings of the First ACM International conference on bioinformatics and computational biology; ACM; 2010. pp. 660–662. [Google Scholar]
- 23.Saha S, Bhasin M, Raghava GP. Bcipep: a database of B-cell epitopes. BMC Genomics. 2005;6:79. doi: 10.1186/1471-2164-6-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frank E, Hall M, Holmes G, Kirkby R, Pfahringer B, Witten IH, Trigg L. Data Mining and Knowledge Discovery Handbook. US: Springer; 2005. Weka: A machine learning workbench for data mining; pp. 1305–1314. [Google Scholar]
- 25.Jungermann F. Information extraction with rapidminer; Proceedings of the GSCL Symposium’ Sprachtechnologie und eHumanities; 2009. pp. 50–61. [Google Scholar]
- 26.Berthold MR, Cebron N, Dill F, et al. Data Analysis. Springer Berlin Heidelberg: Machine Learning and Applications; 2008. KNIME: The Konstanz information miner; pp. 319–326. [Google Scholar]
- 27.Breiman L. Random forests. Mach Learn. 2001;45(1):5–32. [Google Scholar]
- 28.Chen J, Liu H, Yang J, et al. Prediction of linear B-cell epitopes using amino acid pair antigenicity scale. Amino Acids. 2007;33(3):423–428. doi: 10.1007/s00726-006-0485-9. [DOI] [PubMed] [Google Scholar]
- 29.Wolpert DH. Stacked generalization. Neural Netw. 1992;5(2):241–259. [Google Scholar]
- 30.Cai C, Han L, Ji ZL, et al. SVM-Prot: web-based support vector machine software for functional classification of a protein from its primary sequence. Nucleic Acids Res. 2003;31(13):3692–3697. doi: 10.1093/nar/gkg600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bernard S, Heutte L, Adam S. Emerging Intelligent Computing Technology and Applications. Springer: With Aspects of Artificial Intelligence; 2009. Towards a better understanding of random forests through the study of strength and correlation; pp. 536–545. [Google Scholar]
- 32.Altschul SF, Madden TL, Schäffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Breiman L. Bagging predictors. Mach Learn. 1996;24(2):123–140. [Google Scholar]
- 34.Freund Y. Schapire RE Experiments with a new boosting algorithm. ICML. 1996:148–156. [Google Scholar]