Abstract
Precise deletions of cell surface-exposed loops of FhuA resulted in mutants of Escherichia coli with distinct phenotypes. Deletion of loop 3 or 11 inactivated ferrichrome transport activity. Deletion of loop 8 inactivated receptor activity for colicin M and the phages T1, T5, and φ80. The loop 7 deletion mutant was colicin M resistant but fully phage sensitive. The loop 4 deletion mutant was resistant to the TonB-dependent phages T1 and φ80 but fully sensitive to the TonB-independent phage T5. The phenotypes of the deletion mutants revealed important sites for the multiple FhuA transport and receptor activities. The ligand binding sites are nonidentical and are distributed among the entire exposed surface. Presumably, FhuA evolved as a ferrichrome transporter and was subsequently used as a receptor by the phages and colicin M, which selected the same as well as distinct loops as receptor sites.
The crystal structures of the Escherichia coli TonB-dependent outer membrane transport proteins FhuA (15, 25), FepA (5), FecA (14, 32), and BtuB (7, 8) reveal an identical basic design consisting of a β-barrel composed of 22 antiparallel β-strands into which a globular domain is incorporated that closes the channel of the β-barrel (2). In the FecA crystal structure, loop 7 moves 11 Å and loop 8 moves 15 Å upon binding of dinuclear ferric citrate (14, 32), resulting in closure of the external pocket through which ferric citrate enters the high-affinity binding site. Closure of the binding pocket prevents escape of ferric citrate into the medium and may facilitate its unidirectional diffusion into the periplasm after opening of the channel, which presumably occurs through input of energy of the cytoplasmic membrane potential mediated by the proteins TonB, ExbB, and ExbD (3, 28). In BtuB, loops 2, 3, and 4 are disordered. Upon binding of Ca2+, loops 2 and 3 become ordered and loop 4 becomes partially ordered. With additional binding of cyanocobalamin, all three loops are ordered (7, 8). In the crystal structures of FhuA, no large loop movements upon binding of ferrichrome (15, 25), albomycin (13), and rifamycin CGP 4832 (16) are apparent. These results raise the question of whether these transporters function through distinct mechanisms despite having very similar structures or whether crystal forces prevent the movement of loops. It is also possible that the high osmolality of the solutions used for crystallization hinders loop movements, as has spectroscopically been demonstrated for BtuB, where the substrate-induced order-disorder transition of the N-terminal TonB box has not been observed in the crystallization buffer (11).
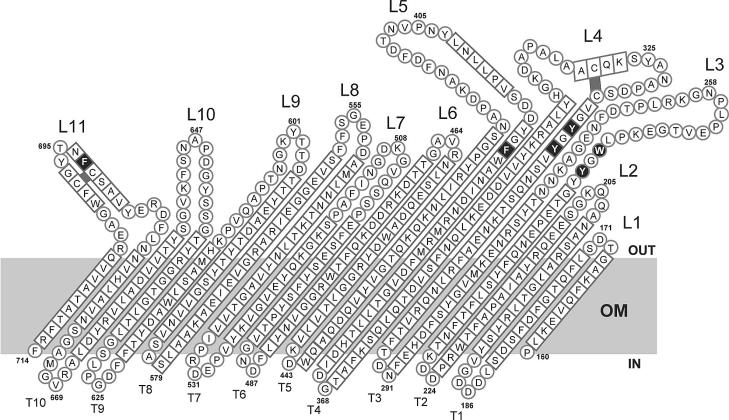
To determine whether loops 7 and 8 of FhuA are essential for substrate transport, as they apparently are in FecA, each of these loops was deleted (Fig. 1). In addition, loops 3, 4, 5, 9, 10, and 11 were each deleted to examine the FhuA transport activities for ferrichrome, albomycin, and microcin J25 and also the FhuA receptor functions for the phages T1, T5, and φ80 and for colicin M. Loops 1, 2, and 6 are very short; therefore, corresponding deletion mutants were not constructed. Loop 11 was deleted such that the highly conserved NLFD motif of the FhuA class of outer membrane transporters (7) was retained. To prevent structural restrictions in β-barrel formation, we inserted peptide NSEG, which forms part of loop 2 of OmpF (9), or NSEGS (Table 1), the latter in cases where insertion of NSEG would have changed the sequence of the flanking region. The deletion mutants were constructed by PCR. The sequences of the primers used will be provided upon request. The mutations were verified by nucleotide sequencing of the entire fhuA genes. Wild-type and mutant fhuA genes were each cloned into plasmid pT7-6. Binding of ferrichrome to fhuA transformants of E. coli MB1859 ΔfhuACDB tonB aroB, transport of ferrichrome into fhuA transformants of E. coli MB98 ΔfhuA aroB, and sensitivity to the FhuA-specific ligands listed in Table 2 were determined as previously described (10, 22). Both mutants lack the entire chromosomal fhuA gene.
FIG. 1.
Unfolded FhuA β-barrel as inserted into the outer membrane (OM). Loops L1 to L11 and turns T1 to T11 connect the 22 antiparallel β-strands. The amino acid residues represented as circles are those present in loops and turns, and those represented as squares are residues contained in β-strands. Black squares indicate the ferrichrome binding sites in the β-barrel. The two disulfide bridges are also marked in black. The numbers indicate the amino acid residues of mature FhuA. Modified from a figure in reference 4 with permission of the publisher.
TABLE 1.
E. coli strains and plasmids used in this study
| Strain or plasmid | Genotype and/or phenotype | Reference or source |
|---|---|---|
| Strains | ||
| MB98 | aroB tsx malT thi ΔfhuA | 10 |
| MB99 | aroB tsx malT thi ΔfhuA tonB | 2 |
| MB1859 | aroB tsx malT thi ΔfhuACDB tonB | Michael Braun |
| Plasmids | ||
| pHK763 | pT7-6 fhuA wild type | 23 |
| pFE46 | pT7-6 fhuA (Δ502-515 NSEG)a | This study |
| pFE47 | pT7-6 fhuA (Δ394-419 NSEG) | This study |
| pFE48 | pT7-6 fhuA (Δ598-611 NSEGS) | This study |
| pFE61 | pT7-6 fhuA (Δ243-273 NSEGS) | This study |
| pFE62 | pT7-6 fhuA (Δ640-654 NSEGS) | This study |
| pFE69 | pT7-6 fhuA (Δ318-339 NSEGS) | This study |
| pFE73 | pT7-6 fhuA (Δ552-558 NSEG) | This study |
| pFE74 | pT7-6 fhuA (Δ689-701 NSEG A702S) | This study |
| pFE75 | pHSG576 fhuA (Δ243-273 NSEGS) | This study |
| pFE76 | pHSG576 fhuA (Δ318-339 NSEGS) | This study |
| pFE77 | pHSG576 fhuA (Δ394-419 NSEG) | This study |
| pFE78 | pHSG576 fhuA (Δ502-515 NSEG) | This study |
| pFE79 | pHSG576 fhuA (Δ552-558 NSEG) | This study |
| pFE80 | pHSG576 fhuA (Δ598-611 NSEGS) | This study |
| pFE81 | pHSG576 fhuA (Δ640-654 NSEGS) | This study |
| pFE82 | pHSG576 fhuA (Δ689-701 NSEG A702S) | This study |
| pHK570 | pHSG576 fhuA wild type | 20 |
| pT7-6 | ColE1; Ampr; T7 gene 10 promoter | 30 |
| pHSG576 | Cmr; pSC101 origin | 31 |
Numbers in parentheses correspond to encoded amino acid residues deleted by the mutation. The indicated peptide was inserted to prevent structural restrictions in β-barrel formation. Plasmids pFE74 and pFE82 carry amino acid change A702S.
TABLE 2.
Phenotypes of FhuA loop deletion mutants
| Strain or mutationa | Deleted residues | Fc transport rates (%)b | Level of Fc binding (%)c | Sensitivities tod:
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| Colicin M | Albomycin | Microcin J25 | φ80 | T1 | T5 | ||||
| Wild type | 100, 100 | 100 | 2 (3), 2 (3) | 6 (7, 8), 6 (7, 8) | 2 (3), 2 (3) | 5 (6, 7), 5 (6, 7) | 6, 6 | 4 (5), 4 (5) | |
| ΔLoop 3 | 243-273 | 2, 0 | 0 | −, − | −, − | 2 (3), 2 (3) | 3 (4, 5), 3 (4, 5) | 6, 6 | 4 (5, 6), 4 (5) |
| ΔLoop 4 | 318-339 | 117, 109 | 76 | 2 (3), 2 (3) | 6 (7, 8), 6 (7, 8) | 2 (3), 2 (3) | (1-3), (1, 2) | −, − | 4 (5), 4 (5) |
| ΔLoop 5 | 394-419 | 73, 49 | 4 | 2 (3), 2 (3) | 5 (6), 3 (4, 5) | −, − | (1), − | 3, 3 | 4 (5), 4 (5) |
| ΔLoop 7 | 502-515 | 46, 15 | 82 | −, − | 4 (5), 3 (4, 5) | −, − | 5 (6), 5 (6) | 6, 6 | 4 (5), 4 (5) |
| ΔLoop 8 | 552-558 | 73, 37 | 123 | −, − | 4 (5, 6), 3 (4, 5) | (0, 1), (0, 1) | −, − | (1-3), − | (1-3), (1-3) |
| ΔLoop 9 | 598-611 | 88, 102 | 53 | 2 (3), 2 (3) | 6 (7, 8), 6 (7, 8) | 3 (4), 3 (4) | 5 (6, 7), 5 (6, 7) | 6, 6 | 4 (5), 4 (5) |
| ΔLoop 10 | 640-654 | 92, 115 | 134 | 2 (3), 2 (3) | 8, 7 (8) | 4, 3 (4) | 5 (6), 5 (6) | 6, 6 | 3 (4), 3 (4) |
| ΔLoop 11 | 689-701 | 0, 0 | 0 | 1 (2), (1, 2) | −, − | (1-4), (1-4) | 3 (4), (1-3) | 5, 6 | 4 (5), 4 (5) |
Wild-type and mutant fhuA alleles were cloned into plasmid pT7-6 or pHSG576 and examined in E. coli MB98 ΔfhuA or E. coli MB1859 ΔfhuACDB tonB.
The ferrichrome (Fc) transport rates (numbers of iron ions per cell per minute) were determined with pT7-6 and pHSG576 transformants of MB98s, respectively, calculated in the nearly linear region between minutes 5 and 13, and compared to the ferrichrome transport rates for wild-type FhuA (100%).
The levels of ferrichrome (Fc) binding were determined with pT7-6 transformants of MB1859 and listed as percentages of the wild-type level.
Sensitivities to the ligands were tested using MB98 freshly transformed with the plasmids listed in Table 1 expressing the indicated FhuA derivatives. Sensitivities were tested by spotting 4 μl of 10-fold (colicin M and phages T1, T5, and φ80) or three fold (microcin J25 and albomycin) dilutions onto tryptone-yeast extract (TY) agar plates overlaid with TY soft agar containing the strain to be tested. The results are given as the last of a dilution series that resulted in a clear zone of growth inhibition; e.g., 4 means that the phage stock suspension could be diluted 104-fold and the albomycin solution could be diluted 34-fold to yield a clear zone of cell lysis. Numbers in parentheses indicate turbid zones of growth inhibition. Tabulated values from the sensitivity tests were derived from a single experiment, but little variation was seen in repeated trials. −, no growth inhibition. Results were obtained with pT7-6 and pHSG576 transformants, respectively.
Deletion of loop 7 reduced the ferrichrome transport rate to 46% of the FhuA wild-type rate (Table 2) but had little effect on ferrichrome binding (82% of the wild-type level). Deletion of loop 8 reduced the transport rate somewhat, although the level of binding was higher than that of binding to wild-type FhuA. No ferrichrome binding site has been observed in loops 7 and 8 (15, 25). The crystal structures predict residues R81, G99, Q100, and Y116 in the globular domain and residues Y244, W246, Y313, Y315, F391, and F693 in the β-barrel to bind ferrichrome. Deletion of loop 3 abolished ferrichrome binding and transport, which is consistent with the removal of the two ferrichrome binding sites Y244 and W246. Interestingly, uptake of microcin J25 was unchanged, although microcin J25 uptake is usually abolished in most FhuA mutants which still display activities for other ligands (2, 10). Apparently, these ferrichrome binding sites are not involved in microcin J25 binding. The microcin J25 binding site may be contained in loop 5, 7, 8, or 11; deletion of any one of these loops led to complete or almost complete microcin J25 resistance. However, the altered ferrichrome transport rates of the loop 7 and 11 deletion mutants indicated general transport defects that were not specific for microcin J25. In contrast, deletion of loop 9 or 10 rendered cells more sensitive to microcin J25. Even though the deletion of loop 4 reduced ferrichrome binding to 76% of the wild-type level, transport of ferrichrome was higher than that into FhuA wild-type cells (117%). Removal of loop 5 abolished ferrichrome binding even though no binding site is found in this loop, but transport activity was retained at a relatively high level. Low binding but high transport activity has also been observed with the R81A mutant (F. Endriß and V. Braun, unpublished results). Binding was determined by washing radiolabeled cells on filters with 0.1 M LiCl. After this procedure, only tightly bound [55Fe3+]ferrichrome remains bound to the cells. If bound [55Fe3+]ferrichrome is separated from unbound [55Fe3+]ferrichrome by centrifugation of radiolabeled cells through an oil layer, mutants which reveal no binding by the filter technique show low-affinity binding (19). It is likely that the loop 5 mutant retains a low level of binding. Deletion of loop 9 or 10 reduced transport slightly, although binding to the loop 10 deletion mutant was enhanced. Neither loop contains a ferrichrome contact site. Deletion of loop 11 completely abolished ferrichrome binding and transport, although this loop contains only a single binding site, F693. Sensitivity to albomycin, which uses the same binding sites as ferrichrome, was in approximate agreement with the ferrichrome transport data. Deletion of the residues that bind the antibiotic moiety of albomycin but do not serve as ferrichrome binding sites—Q505, part of the loop 7 deletion, and F557 and F558, part of the loop 8 deletion—may reduce albomycin sensitivity slightly more than the ferrichrome transport rate.
Previously, binding sites in loop 4 of FhuA have been mapped by using synthetic hexapeptides identical in sequence to defined segments of loop 4 that cause temperature-dependent release of DNA of the TonB-independent phage T5 and strongly reduced infection by the other phages (21). The inhibitory hexapeptides displayed high sequence specificity. Deletion of loop 4 conferred phage T1 and φ80 resistance, thereby confirming the previous results for these phages (Table 2). However, the loop 4 deletion mutant was as sensitive to phage T5 as were FhuA wild-type cells, suggesting that loop 8 is sufficient for T5 infection. The T5 data can be reconciled by the assumption that loop 4 contributes to the T5 binding site but that loop 8 is sufficient when loop 4 is absent. When loop 4 is present, T5 must for steric reasons bind to it. It is also possible that removal of loop 4 alters the access to loop 8 or the conformation of loop 8 such that it serves as a single T5 binding site. Loop 4 is not close to loop 8, but the tail of phage T5 is much larger than FhuA (1) and may contact several loops at the same time. Deletion of loop 5 also resulted in resistance to phage φ80, strongly reduced sensitivity to phage T1, and full sensitivity to phage T5. The loop 3 deletion strongly affected only phage φ80 infection.
Cells became resistant to colicin M when loop 3, 7, or 8 was deleted, and cells displayed reduced sensitivity when they synthesized the FhuA loop 11 deletion protein. Loop 7 apparently serves specifically as a binding site for colicin M since phage sensitivity was not affected by this deletion whereas deletion of loop 3 or 11 reduced sensitivity to phage φ80 also.
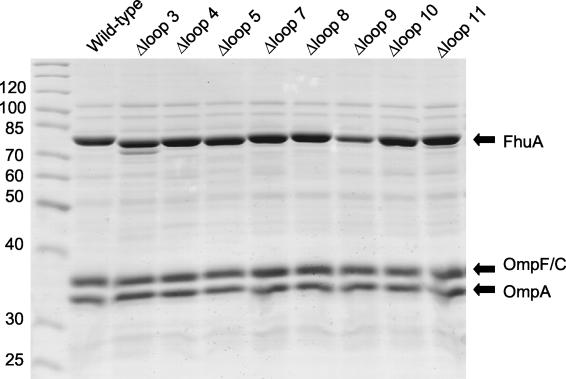
The described experiments were performed with fhuA deletion derivatives cloned into the medium-copy-number vector pT7-6 and compared with wild-type fhuA cloned into the same vector. The fhuA genes were under the control of the fhuA promoter and transcribed by the E. coli RNA polymerase. Ferrichrome transport rates and sensitivities of cells to albomycin, the phages, colicin M, and microcin J25 may be influenced by the amount of synthesized FhuA protein. Therefore, we determined the content of the FhuA mutant proteins relative to the content of the FhuA wild-type protein under the same conditions used for measuring the transport and receptor activities. The proteins of outer membrane preparations were separated by polyacrylamide gel electrophoresis in the presence of sodium dodecyl sulfate (10). The amounts of the mutant FhuA proteins were similar to the amount of wild-type FhuA and comparable to the amounts of the major outer membrane proteins OmpF/OmpC and OmpA (Fig. 2). The different phenotypes among the FhuA mutants and wild-type FhuA were therefore not caused by different amounts of protein. The proteins were also not sensitive to cellular proteases, except for the FhuA loop 3 deletion protein, which was partially degraded to a smaller product. Proteolytic cleavage is supported by the generation of a similar product upon addition of trypsin to isolated FhuA (17).
FIG. 2.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of outer membrane proteins (FhuA, OmpF/OmpC, and OmpA) of E. coli MB98 transformed with the fhuA loop deletion mutants cloned into plasmid pT7-6 as listed in Table 1. The gel was stained with Serva blue. Molecular masses in kilodaltons of the markers (left lane) are indicated on the left.
Figure 2 reveals rather high expression of the FhuA proteins, higher than what is observed for chromosomally encoded FhuA under natural, iron-limiting growth conditions. Although the data obtained with the FhuA loop deletion mutants were not given as absolute values but related to data for wild-type FhuA, the possibility was not excluded that some of the mutants showed higher relative activities than they would under the haploid state. Therefore, the fhuA wild-type and mutant genes were cloned into the low-copy-number plasmid pHSG576 (31) and their activities were determined. The transport-inactive loop 3 and 11 mutants remained inactive, and the highly active loop 4, 9, and 10 mutants remained highly active (Table 2). The loop 5, 7, and 8 mutants with intermediate activity with the fhuA genes in the medium-copy-number plasmid showed lower activity with the fhuA genes in the low-copy-number plasmid. Lower ferrichrome transport activity was also revealed by low albomycin sensitivity (Table 2). These results support the participation of loops 5, 7, and 8 in ferrichrome and albomycin transport, as suggested by the results obtained with the fhuA genes in the medium-copy-number plasmid. Low expression did not alter, or only slightly altered, colicin M and phage sensitivities of the mutants relative to those of the wild-type (Table 2).
Previously it was noted that sensitivity to microcin J25 is abolished in mutants which still display other FhuA activities (2). Reduction of the fhuA wild-type and mutant genes abolished microcin sensitivity but clearly revealed the increased sensitivity of the loop 9 and 10 mutants (Table 2).
Deletion of a loop may cause long-range alterations in FhuA structure so that a channel in FhuA is opened. Therefore, we tested whether E. coli MB99 ΔfhuA tonB transformants with plasmid-carried FhuA loop deletion mutations could grow on ferrichrome as a sole iron source. Since the tonB mutation prevents active transport of ferrichrome across the outer membrane, ferrichrome can pass through FhuA only by diffusion if the loop deletions open a channel. Growth promotion was tested on agar plates containing nutrient broth (Difco Laboratories) to which 0.2 mM dipyridyl was added to reduce the available iron. The transformants carrying wild-type fhuA or one of the mutant fhuA genes were seeded into nutrient broth top agar and plated. Various concentrations of ferrichrome (1, 0.3, 0.1, 0.03, and 0.01 mM) were spotted onto paper disks, and formation of growth zones was scored after incubation overnight (10). Of the FhuA deletion mutations, only the FhuA loop 3 deletion mutation allowed growth of MB99 around a paper disk spotted with 1 mM ferrichrome. In comparison, growth of the E. coli MB98 ΔfhuA transformants carrying wild-type fhuA or any one of the other fhuA mutant genes was supported by ferrichrome down to concentrations of 0.01 mM, except for the loop 11 mutant, which did not grow at any ferrichrome concentration. Since the FhuA loop 3 deletion transformant of E. coli MB98 ΔfhuA did not actively transport ferrichrome (Table 2) and the test was done with the MB99 tonB mutant, growth promotion by ferrichrome could result only from diffusion.
The enhanced outer membrane permeability of cells synthesizing the FhuA loop 3 deletion mutant was studied further. Sensitivity to antibiotics to which wild-type E. coli shows no or only low sensitivity owing to the outer membrane permeability barrier (27) was determined. Compared to the other FhuA deletion derivatives, the loop 3 and loop 11 deletion mutants displayed increased sensitivities to novobiocin, erythromycin, and rifamycin (Table 3). The loop 11 deletion mutant was also sensitive to bacitracin. All of the FhuA deletion mutants were more sensitive to the antibiotics than wild-type FhuA cells, but loop 3 and loop 11 deletion mutants were the most sensitive.
TABLE 3.
Sensitivities of E. coli FhuA loop deletion mutants to antibiotics
| Strain or mutation | Deleted residues | Sensitivity toa:
|
|||
|---|---|---|---|---|---|
| Novobiocin (30 μg; 634 Da) | Erythromycin (15 μg; 734 Da) | Rifamycin (5 μg; 823 Da) | Bacitracin (30 μg; 1,421 Da) | ||
| Vector | no FhuA | 10 | 7 | − | − |
| Wild type | 10 | 7 | − | − | |
| ΔLoop 3 | 243-273 | 14 | 11 | 14 | − |
| ΔLoop 4 | 318-339 | 12 | 8 | 7 | − |
| ΔLoop 5 | 394-419 | 12 | 8 | 10 | − |
| ΔLoop 7 | 502-515 | 12 | 8 | 10 | − |
| ΔLoop 8 | 552-558 | 11 | 7 | 6 | − |
| ΔLoop 9 | 598-611 | 12 | 11 | 8 | − |
| ΔLoop 10 | 640-654 | 12 | 9 | 9 | − |
| ΔLoop 11 | 689-701 | 15 | 11 | 13 | 8 |
Sensitivities to the ligands were tested using E. coli MB98 ΔfhuA freshly transformed with the plasmids listed in Table 1 expressing the indicated FhuA derivatives cloned into pT7-6. Sensitivities were tested by placing filter disks containing the various inhibitors onto TY agar plates overlaid with TY soft agar containing the strain to be tested. The size of the inhibition zone (in millimeters) is given without subtraction of the size of the paper disk (6 mm) used to supply the inhibitors at the concentration given. The molecular masses of the inhibitors are indicated. Numbers in parentheses indicate turbid zones of growth inhibition. −, no sensitivity.
FhuA displays many activities that can advantageously be used to uncover differences in the phenotypes of mutants. All the FhuA loop deletion mutants had properties distinct from cells that synthesized wild-type FhuA. However, specific properties of the different deletions were encountered. Deletion of loop 7 or 8 reduced, but did not abolish, FhuA transport activity. If these loops move upon binding of ferrichrome to the high-affinity binding site, movement is not essential for ferrichrome uptake. However, reduction of the transport rate to 46 or 73%, respectively, with the mutated fhuA genes in the medium-copy-number plasmid and to 15 or 37% with mutant fhuA in the low-copy-number plasmid may indicate that closure of the binding cavity contributes to the diffusion of ferrichrome into the periplasm. In the absence of loop 7 or 8, a portion of the ferrichrome may escape into the medium and thus reduce the transport rate. Interaction with TonB presumably changes the geometry of the FhuA binding residues such that ferrichrome is released. In addition, the globular domain must move to open the channel of the β-barrel.
In contrast to deletion of loop 7 or 8, deletion of loop 3 or 11 completely abolished FhuA transport activity. Since both FhuA derivatives still functioned as phage binding sites, which for phage T1 and φ80 infection required a response to TonB, the FhuA derivatives must have been properly integrated into the outer membrane and were not altered much in their structure. However, the loop 3 deletion FhuA mutant supported diffusion of ferrichrome and three of the four tested antibiotics, and the loop 11 deletion FhuA mutant supported diffusion of all four antibiotics. Removal of one of these two loops may alter the structure of FhuA such that the globular domain no longer fits perfectly into the β-barrel channel.
Loop deletions specifically affected receptor activities: removal of loop 3, 7, or 8 abolished sensitivity to colicin M; removal of loop 4, 5, or 8 abolished sensitivity to phage φ80; removal of loop 4 or 8 abolished sensitivity to phage T1; and removal of loop 8 abolished sensitivity to phage T5. The involvement of these loops, except loop 4, in receptor activities has not been determined previously. Deletions constructed prior to the determination of the FhuA crystal structures were located outside loops or removed portions of loops and adjacent β-strands (6, 18). Inserted peptides with 4 and 16 residues were not placed in loops, except at positions 321, 338, 511, and 646 (24, 26). Insertions after residue 321 strongly reduce sensitivity to phages T1 (24, 26) and φ80 and T5 (24); insertions after residue 338 have no effects; insertions after residue 511 strongly reduce colicin M sensitivity with little effect on phage sensitivity (24); and insertions after residue 646 moderately reduce sensitivity to colicin M and φ80 (24). The phenotypes of the mutants with peptide insertions after residues 321 and 511 agree with the phenotypes of the loop 3 and 7 deletion mutants. The insertion after residue 338 is positioned at the end of loop 4 and has no effect; the insertion after residue 646 has a smaller effect than deletion of the entire loop.
FhuA loops contribute six residues to ferrichrome binding. During transport, ferrichrome is released from the high-affinity binding site (Kd [binding constant], 0.6 nM [29]), for which process structural changes must occur. The loops are mobile and interact with one another and with loops of the globular domain. Ten-nanosecond-duration molecular dynamics simulations of the ferrichrome-free and ferrichrome-loaded states of FhuA show that the conformation of extracellular loops is sensitive to the presence of ferrichrome at its binding site (12). They suggest that the loops are internally stable but move with respect to the β-barrel. Loops 4 to 7, in particular loop 7 and especially loop 8, show structural drifts indicating a swinging motion. The simulations also show an extensive solvation of the interface between the globular domain and the β-barrel, which may facilitate movement of the globular domain within or out of the β-barrel. A network of salt bridges, hydrogen bonds, and van der Waals contacts spans the entire molecule. Therefore, mutations can result not only in local structural changes but also in long-distance alterations. The differential effects of the mutations on ferrichrome binding, ferrichrome transport, albomycin and microcin J25 transport, and sensitivity to the phages T1, T5, and φ80 and colicin M allowed certain structural changes to be related to specific functions. Loops 3, 5, 7, 8, and 11 contribute to ferrichrome transport; loops 3, 5, and 11 are essential for ferrichrome binding; loop 4 is essential for T1 and φ80 infection; loop 5 is essential for φ80 infection; loop 8 is essential for T1, T5, and φ80 infection; and loops 3, 7, and 8 are essential for colicin M sensitivity. No loop deletion prevented all FhuA functions or insertion into the outer membrane. FhuA does not contain a single active site. Its entire exposed surface serves as a contact site for the various ligands. Presumably FhuA evolved as a transporter for ferric siderophores of the ferrichrome type and later was used as a receptor for the phages and colicin M, which selected their appropriate binding sites.
Acknowledgments
We thank Karen A. Brune for critical reading of the manuscript and Michael Braun for providing E. coli strain MB1859.
This work was supported by the Deutsche Forschungsgemeinschaft (Forschergruppe “Bakterielle Zellhülle: Synthese, Funktion und Wirkort”) and by the Fonds der Chemischen Industrie.
REFERENCES
- 1.Böhm, J., O. Lambert, A. S. Frangakis, L. Letellier, W. Baumeister, and J. L. Rigau. 2001. FhuA-meditated phage genome transfer into liposomes: a cryo-electron tomography study. Curr. Biol. 11:1168-1175. [DOI] [PubMed] [Google Scholar]
- 2.Braun, M., F. Endriß, H. Killmann, and V. Braun. 2003. In vivo reconstitution of the FhuA transport protein of Escherichia coli K-12. J. Bacteriol. 185:5508-5518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braun, V. 2003. Iron uptake by Escherichia coli. Front. Biosci. 8:1409-1421. [DOI] [PubMed] [Google Scholar]
- 4.Braun, V., and M. Braun. 2002. Active transport of iron and siderophore antibiotics. Curr. Opin. Microbiol. 5:194-201. [DOI] [PubMed] [Google Scholar]
- 5.Buchanan, S. K., B. S. Smith, L. Venkatramani, D. Xia, L. Esser, M. Palnitkar, R. Chakraborty, D. van der Helm, and J. Deisenhofer. 1999. Crystal structure of the outer membrane active transporter FepA from Escherichia coli. Nat. Struct. Biol. 6:56-63. [DOI] [PubMed] [Google Scholar]
- 6.Carmel, G., and J. W. Coulton. 1991. Internal deletions in the FhuA receptor of Escherichia coli K-12 define domains of ligand interactions. J. Bacteriol. 173:4394-4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chimento, D. P., R. Kadner, and M. Wiener. 2003. The Escherichia coli outer membrane transporter BtuB: structural analysis of the calcium and substrate binding, and identification of orthologous transporters by sequence/structure conservation. J. Mol. Biol. 332:999-1014. [DOI] [PubMed] [Google Scholar]
- 8.Chimento, D. P., A. K. Mohanty, R. J. Kadner, and M. C. Wiener. 2003. Substrate-induced transmembrane signaling in the cobalamin transporter BtuB. Nat. Struct. Biol. 10:394-401. [DOI] [PubMed] [Google Scholar]
- 9.Cowan, S. W., T. Schirmer, G. Rummel, M. Steiert, R. Ghosh, R. A. Pauptit, J. N. Jansonius, and J. P. Rosenbusch. 1992. Crystal structures explain functional properties of two E. coli porins. Nature 358:727-733. [DOI] [PubMed] [Google Scholar]
- 10.Endriß, F., M. Braun, H. Killmann, and V. Braun. 2003. Mutant analysis of the Escherichia coli FhuA protein reveals sites of FhuA activity. J. Bacteriol. 185:4683-4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fanucci, G. E., J. Y. Lee, and D. S. Cafiso. 2003. Spectroscopic evidence that osmolytes used in crystallization buffers inhibit a conformational change in a membrane protein. Biochemistry 42:13106-13112. [DOI] [PubMed] [Google Scholar]
- 12.Faraldo-Gómez, J. D., G. S. Smith, and M. S. P. Sansom. 2003. Molecular dynamics simulations of the bacterial outer membrane protein FhuA: a comparative study of the ferrichrome-free and bound states. Biophys. J. 85:1406-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferguson, A. D., V. Braun, H. P. Fiedler, J. W. Coulton, K. Diederichs, and W. Welte. 2000. Crystal structure of the antibiotic albomycin in complex with the outer membrane transporter FhuA. Protein Sci. 9:956-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferguson, A. D., R. Chakraborty, B. S. Smith, L. Esser, D. van der Helm, and J. Deisenhofer. 2002. Structural basis of gating by the outer membrane transporter FecA. Science 295:1715-1719. [DOI] [PubMed] [Google Scholar]
- 15.Ferguson, A. D., E. Hofmann, J. W. Coulton, K. Diederichs, and W. Welte. 1998. Siderophore-mediated iron transport: crystal structure of FhuA with bound lipopolysaccharide. Science 282:2215-2220. [DOI] [PubMed] [Google Scholar]
- 16.Ferguson, A. D., J. Ködding, G. Walker, C. Bös, J. W. Coulton, K. Diederichs, V. Braun, and W. Welte. 2001. Active transport of an antibiotic rifamycin derivative by the outer-membrane protein FhuA. Structure 9:707-716. [DOI] [PubMed] [Google Scholar]
- 17.Hoffmann, H., E. Fischer, H. Schwarz, and V. Braun. 1986. Overproduction of the proFhuA outer membrane receptor protein of Escherichia coli K-12: isolation, properties, and immunocytochemical localization at the inner side of the cytoplasmic membrane. Arch. Microbiol. 145:334-341. [DOI] [PubMed] [Google Scholar]
- 18.Killmann, H., and V. Braun. 1992. An aspartate deletion mutation defines a binding site of the multifunctional FhuA outer membrane receptor of Escherichia coli K-12. J. Bacteriol. 174:3479-3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Killmann, H., and G. Gestwa. 2002. Determination of ferrichrome binding to the FhuA outer membrane transport protein, periplasmic accumulation of ferrichrome, or transport of ferrichrome into cells using a three-layer oil technique. Anal. Biochem. 130:55-60. [DOI] [PubMed] [Google Scholar]
- 20.Killmann, H., C. Herrmann, A. Torun, G. Jung, and V. Braun. 2002. TonB of Escherichia coli activates FhuA through interaction with the beta-barrel. Microbiology 148:3497-3509. [DOI] [PubMed] [Google Scholar]
- 21.Killmann, H., G. Videnov, G. Jung, H. Schwarz, and V. Braun. 1995. Identification of receptor binding sites by competitive peptide mapping: phages T1, T5, and φ80 and colicin M bind to the gating loop of FhuA. J. Bacteriol. 177:694-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Killmann, H., M. Braun, C. Herrmann, and V. Braun. 2001. FhuA barrel-cork hybrids are active transporters and receptors. J. Bacteriol. 183:3476-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Killmann, H., R. Benz, and V. Braun. 1993. Conversion of the FhuA transport protein into a diffusion channel through the outer membrane of Escherichia coli. EMBO J. 12:3007-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koebnik, R., and V. Braun. 1993. Insertion derivatives containing segments of up to 16 amino acids identify surface- and periplasm-exposed regions of the FhuA outer membrane receptor of Escherichia coli K-12. J. Bacteriol. 175:826-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Locher, K. P., B. Rees, R. Koebnik, A. Mitschler, L. Moulinier, J. P. Rosenbusch, and D. Moras. 1998. Transmembrane signaling across the ligand-gated FhuA receptor: crystal structures of free and ferrichrome-bound states reveal allosteric changes. Cell 95:771-778. [DOI] [PubMed] [Google Scholar]
- 26.Moeck, G. S., B. S. F. Bazzaz, M. F. Gras, T. S. Ravi, M. J. Ratcliffe, and J. W. Coulton. 1994. Genetic insertion and exposure of a reporter epitope in the ferrichrome-iron receptor of Escherichia coli K-12. J. Bacteriol. 176:4250-4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nikaido, H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67:593-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Postle, K., and R. J. Kadner. 2003. Touch and go: tying TonB to transport. Mol. Microbiol. 49:869-882. [DOI] [PubMed] [Google Scholar]
- 29.Scott, D. C., Z. Cao, Z. Qi, M. Bauler, J. D. Igo, S. M. C. Newton, and P. E. Klebba. 2001. Exchangeability of N termini in the ligand-gated porins of Escherichia coli. J. Biol. Chem. 276:13025-13033. [DOI] [PubMed] [Google Scholar]
- 30.Tabor, S., and C. C. Richardson. 1985. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc. Natl. Acad. Sci. USA 82:1074-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takeshita, S., M. Sato, M. Toba, M. Masahashi, and T. Hashimoto-Gotoh. 1987. High-copy-number and low-copy-number plasmid vectors for lacZ alpha complementation and chloramphenicol- or kanamycin-resistance selection. Gene 61:63-74. [DOI] [PubMed] [Google Scholar]
- 32.Yue, W. W., S. Grizot, and S. K. Buchanan. 2003. Structural evidence for iron-free citrate and ferric citrate binding to the TonB-dependent outer membrane transporter FecA. J. Mol. Biol. 332:353-368. [DOI] [PubMed] [Google Scholar]