Abstract
Song-learning birds and humans share independently evolved similarities in brain pathways for vocal learning that are essential for song and speech and are not found in most other species. Comparisons of brain transcriptomes of song-learning birds and humans relative to vocal nonlearners identified convergent gene expression specializations in specific song and speech brain regions of avian vocal learners and humans. The strongest shared profiles relate bird motor and striatal song-learning nuclei, respectively, with human laryngeal motor cortex and parts of the striatum that control speech production and learning. Most of the associated genes function in motor control and brain connectivity. Thus, convergent behavior and neural connectivity for a complex trait are associated with convergent specialized expression of multiple genes.
Vocal learning is the ability to learn to produce vocalizations by imitating a model. This complex trait convergently evolved in a few lineages of mammals and birds (fig. S1). These include humans, cetaceans (whales and dolphins), pinnepeds (seals and sea lions), bats, and elephants among mammals, and songbirds, parrots, and hummingbirds among birds (1). Although nonhuman primates have a limited ability to modify their innate vocalizations, no strong evidence exists that they learn novel vocalizations (1–3). Vocal-learning species also share the presence of babbling, deafness-induced deterioration of learned vocalizations, dialects, and forebrain circuits that control production and learning of vocalizations (1, 4–6). These circuits include a corticostriatal loop and a unique direct connection from motor cortical areas [human laryngeal motor cortex (LMC) and songbird robust nucleus of the arcopallium (RA)] to brainstem vocal motor and respiratory neurons for phonation (Fig. 1).
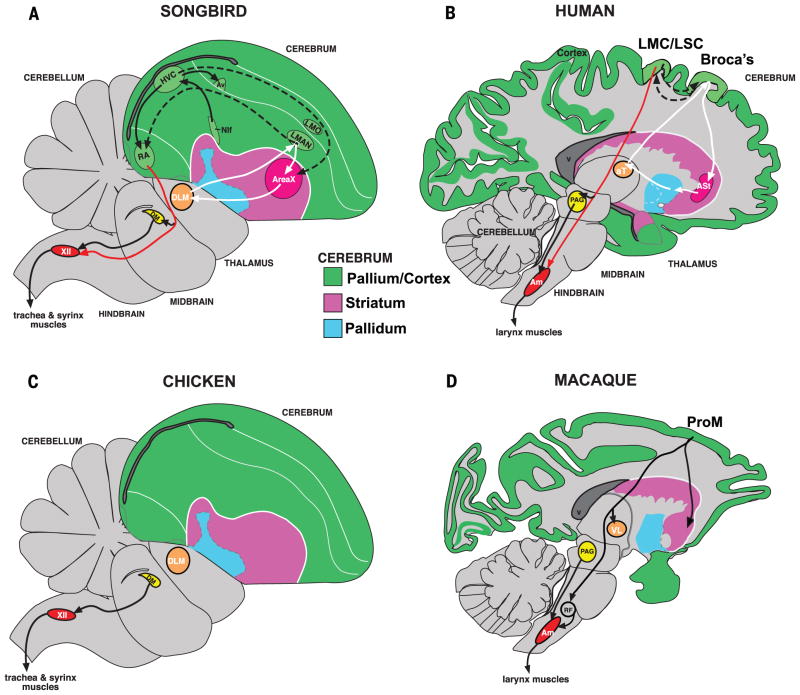
Fig. 1. Comparative brain relationships, connectivity, and cell types among vocal learners and nonlearners.
(A) Drawing of a zebra finch male brain section showing profiled song nuclei: Area X, HVC, LMAN, RA, and the 12th motor nucleus (XII). (B) Drawing of a human brain section showing proposed vocal pathway connectivity including LMC/LSC in the precentral gyrus. Black arrows, connections and regions of the posterior vocal motor pathway; white arrows, connections and regions of the anterior vocal pathway; dashed arrows, connections between the two pathways. The thick blue arrows show the analogous brain regions predicted by this study across finch and human. Red arrows show the direct projections found only in vocal learners, from vocal motor cortex regions to brainstem vocal motor neurons. (C) Known connectivity of a vocal-nonlearning bird showing absence of forebrain song nuclei. (D) Known connectivity of vocal-nonlearning primates (i.e., macaque) showing presence of forebrain regions that have an indirect projection to nucleus ambiguus (Amb) but have no known role in production of vocalizations.
A potential mechanism for convergent evolution of brain systems is one in which a larger brain structure (superregion) specializes into multiple subregions, each of which performs a distinct function (7–9). For vocal learning, it has been hypothesized that the superregions of a motor learning or auditory perception pathway have specialized into subregions that control the production of learned vocalizations (9–11). These functional brain region specializations in song-learning birds (12–17) and in humans (18) are associated with some molecular specializations. One gene, encoding parvalbumin, was found to share specialized expression in brainstem vocal-oral nuclei between song-learning birds and humans (16). Some glutamate receptors were found to have specialized expression in forebrain song and speech areas in birds (19) and humans (18), but a direct comparison between species was not made. The FOXP2 transcription factor is required for vocal learning in both humans (20) and songbirds (21–24), which suggests that neural circuits for vocal learning across clades may have evolved overlapping molecular mechanisms.
We reasoned that one or more genes underlying a complex trait could show convergent evolution across species, even those separated by tens to hundreds of millions of years from a common ancestor. For example, convergent identical amino acid substitutions have been identified in the opsin gene for light absorption across different vertebrate lineages (25); in the melanocorticotropin receptor in mammals and birds for skin and feather color, respectively (26); in a gene for yellow wing spots used for courtship in flies (27); in Pitx transcription factors for gain and loss of limbs in fish and marine mammals (28); in a sodium channel for electric organs in independent lineages of fish (29); and in the prestin gene for echolocation in bats and cetaceans (30). In addition, for echolocating mammals, genome-scale convergence in amino acid sequence was recently found in many genes involved in hearing (31). Likewise, convergent changes in amino acid sequence among vocal-learning birds and mammals have recently been reported (32, 33). However, we are not aware of examples of widespread convergence in gene expression patterns in brain regions involved in convergently evolved behavioral traits.
We examined whether complex behavioral and anatomical convergence is associated with molecular convergence in song-learning birds and humans, species separated from a common ancestor by more than 68 million years (among birds) or 310 million years (between birds and humans) (34–36). We leveraged the expression profiles of thousands of genes that we (supplementary materials section SM1) and others collected from the brains of vocal-learning and vocal-nonlearning birds (15, 16, 37), humans (http://human.brain-map.org, June 2013) (38), and nonhuman primates (39). We developed a hierarchical computational framework to compare brain region specializations across distantly related species.
Framework for finding anatomical molecular similarities across species
To test hypotheses regarding shared molecular specializations between songbird and human brain regions for song and speech control, we collected data from six experiments: (i) 24 total samples from laser-microdissected song control nuclei—Area X, HVC (a letter-based name), LMAN (lateral magnocellular nucleus of the nidopallium), and RA (Fig. 1A)—of three to six adult male zebra finches (37), the most commonly studied vocal-learning songbird; (ii) 17 total paired samples from punch biopsy–dissected Area X and adjacent ventral striatum (VS) of male zebra finches hybridized to microarrays of our design (15); (iii) 12 total paired samples from laser-microdissected RA analogs and the adjacent motor arcopallium (mAC) in three individuals of each species representing all three vocal-learning avian lineages (songbird, parrot, and hummingbird) and two non-vocal mAC regions (9) of two vocal-nonlearning lineages (dove and quail) (16), generated for this project (SM1); (iv) from these same animals, laser-microdissected brainstem vocal (nXIIts; 12th tracheosynringeal nucleus) and neck (SSp; supraspinal nucleus) motor neurons, which are found in all vertebrate species; (v) 3702 samples dissected from 231 subregions (table S1A) of the much larger postmortem human brains of six subjects from the Allen Human Brain Atlas (38); and (vi) 258 laser-captured samples from different cortical layers of 12 regions of four rhesus macaques, including major motor and sensory cortices (39).
The songbird RA and HVC are part of a vocal motor pathway that controls the production of learned vocalizations, whereas Area X and LMAN are part of a pathway that controls imitation and exploration of vocalizations (Fig. 1A) (40, 41). We used only animals that did not sing after an overnight period of silence alone, because neural activity associated with singing causes changes in expression of thousands of genes in the song nuclei (37, 42). We do not know the premortem vocalizing state of the humans, but did not expect this to prevent us from identifying convergent gene expression specializations as long as one species was silent (the birds), the genes are not sensitive to immediate vocalizing behavior, and the human samples were collected 10 to 30 hours postmortem, when many of the vocalizing-regulated gene expression changes have subsided. The avian samples were profiled on our custom songbird oligonucleotide microarray [zebra finch Agilent oligoarray v2.2 (37)]; human samples were profiled on a human microarray (Agilent custom 8×60K array) containing 7473 orthologous genes we identified between human and finch (table S2 and SM2 to SM4).
Because brain regions of different species may show convergent specializations for different sets of genes relative to homologous surrounding brain regions (or for the same genes within more divergent homologous surrounding brain regions), we developed a computational framework to accommodate these and other hypotheses (SM5). First, an anatomical gene expression specialization tree was created for each species to hierarchically organize the expression profiles of all genes from more global to specific regions (e.g., forebrain > cortex > motor cortex > subregion of motor cortex). Although the hierarchy is based on anatomy and not gene expression, the two correspond relatively well, especially at higher levels (Fig. 2A and fig. S2). A node in the tree is a vector of expression levels of all genes for each brain sample or higher grouping of samples (regions); a branch is the difference (specialization) in expression of all genes between two nodes. Second, we calculated the pairwise distances and associated Pearson correlation of all nodes and branches between two species. Third, these distance values were given as input to a dynamic programming algorithm we developed that finds the optimal gene expression specialization alignment of one species’ anatomical expression tree (e.g., avian) with another’s (e.g., human) while preserving the hierarchical structure of each tree (SM6).
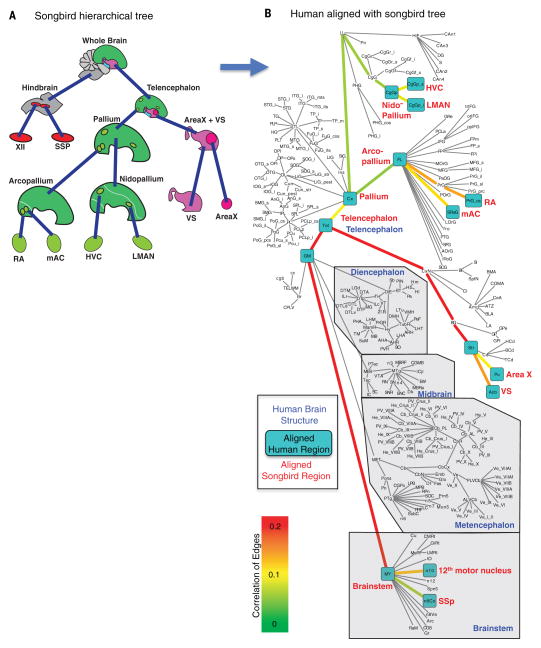
Fig. 2. Optimal alignment of human and zebra finch brain hierarchies.
(A) Tree representing zebra finch brain regions based on hierarchical expression of gene expression profiles. (B) Tree representing the hierarchy of the human brain based on the current knowledge of mammalian brain organization (http://human.brain-map.org). Each node (name) in the tree is a brain region. The daughters of a node are the subregions found within a brain region. Each edge (line) is a specialization of a subregion relative from the brain region that encompasses it. Turquoise boxes are human regions (black text) where the zebra finch brain regions (red text) optimally aligned. The blue font highlights larger human brain structures. Edges are colored by the correlation value (0 to 0.2) of the aligned avian and human specializations. Human region abbreviations are annotated (table S1A).
Similarities in gene expression specializations may be due to homology (common origin) or convergence, or may result from shared expression profiles of one or a few cell types within brain regions between species. We found that the Pearson correlations between specializations were low, even in biological replicates, because of noise and large numbers of genes that are not specialized in expression. For example, the mean correlation between biological replicates of zebra finch RA relative to the adjacent mAC specialization is 0.22. Values above 0.09 were typically statistically significant (see below).
Molecular specializations shared between songbird and human brains
We first compared the gene expression profiles between songbird (zebra finch) and human, for which we had the most samples. The dynamic programming algorithm found an optimal alignment between the brain gene expression specialization trees of songbird (Fig. 2A) and human (Fig. 2B; see also Table 1). At high levels of the alignment, the songbird telencephalic regions corresponded most closely to the human telencephalon (Tel) and the songbird brainstem nuclei corresponded to human brainstem nuclei. Within the telencephalon, the songbird pallial regions corresponded most closely to the human cortex (Cx), and the songbird striatal regions corresponded to the human striatum. The songbird pallial regions sampled did not have a strong similarity to the human claustrum (Cl) and had an inverse gene expression relationship with the amygdala (Amg), meaning that the specialization was in the opposite direction (Table 1).
Table 1. Support and rejection for hypothesized relationships between human and zebra finch brain regions.
We allowed Wernicke’s area to be any subregion in the superior temporal gyrus; Broca’s area to be either the opercular part or the triangular part of the inferior frontal gyrus; the supplementary motor area to be any part of the encompassing superior frontal gyrus. The specialization of the finch forebrain to Area X + VS and finch forebrain to pallium was compared to the human specializations of the amygdala, claustrum, cortex, globus pallidus, and striatum. The finch striatum (Area X + VS) to Area X was compared to caudate, putamen, and nucleus accumbens. The finch specialization of the pallium to RA, HVC, and LMAN was compared to all subregions of the human cortex relative to the entire cortex, as well as each subregion/cortical layer combination in macaque. Significantly supported hypotheses are in bold.
| Finch specialization | Human region/macaque cortex layer | Reference | Correlation | Permutation
|
Abbreviation | |
|---|---|---|---|---|---|---|
| P | FWER | |||||
| Forebrain to pallium | Telencephalon to cortex | (4, 41) | 0.105 | 0.068 | 0.218 | Cx |
| Telencephalon to claustrum | (45) | 0.007 | 0.537 | 0.595 | Cl | |
| Telencephalon to amygdala | (45) | −0.083 | 0.840 | 1.000 | Amg | |
| Forebrain to Area X + VS | Telencephalon to striatum | (41, 44) | 0.207 | 0.001 | 0.001 | Str |
| Telencephalon to globus pallidus | (44, 46, 48) | −0.068 | 0.800 | 1.000 | GP | |
| Striatum to Area X | Striatum to putamen | (41, 44) | 0.108 | 0.031 | 0.082 | Pu |
| Striatum to caudate | (41, 44) | 0.037 | 0.001 | 0.393 | Cd (BCd) | |
| Striatum to nucleus accumbens | (48) | −0.141 | 0.995 | 1.000 | Acb | |
| Pallium to RA | Precentral gyrus, central sulcus | (4, 41, 49, 84) | 0.139 | 0.003 | 0.017 | PrG_cs |
| Layer 5, motor cortex | (44) | 0.132 | 0.000 | 0.011 | PrGxL5 | |
| Cingulate cortex | (4) | 0.089 | 0.000 | 0.051 | CgGf (CgGf_i) | |
| Pallium to HVC | Wernicke’s area | (4) | 0.045 | 0.302 | 0.999 | STG |
| Broca’s area | (4, 84) | 0.006 | 0.505 | 1.000 | opIFG or trIFG | |
| Supplementary motor area | (4) | −0.004 | 0.491 | 1.000 | SFG | |
| Layers 2/3 | (85) | 0.046 | 0.208 | 0.908 | PoGxL2 | |
| Layer 4 | (68) | 0.026 | 0.469 | 0.998 | TAxL4 | |
| Pallium to LMAN | Broca’s area | (41) | 0.036 | 0.148 | 0.997 | opIFG or trIFG |
| Pre-SMA | (41) | −0.010 | 1.000 | 0.556 | SFG | |
| Anterior cingulate | (84) | −0.085 | 1.000 | 0.849 | CgGf | |
| Layer 4 | (68) | 0.089 | 0.070 | 0.249 | PoGxL4 | |
| Brainstem to XII | Myencephalon to hypoglossal nucleus | (44) | 0.009 | 0.440 | 1.000 | n12 |
| Myencephalon to vagus motor nucleus | No proposal | 0.125 | 0.040 | 0.040 | n10 | |
| Myencephalon to nucleus ambiguus | (54) | NA | NA | NA | NA | |
These results support the cortex hypothesis (33, 43, 44) and contradict the claustrum-amygdala hypothesis of homologies between avian and mammalian pallial brain regions (33, 45). Although some avian striatal regions (Area X and VS) are known to have a sparse population of pallidum-like neurons (46–48), we did not find a strong correlation with the human pallidum (Table 1), supporting the conclusion that the avian regions are predominantly striatal (33, 44). These findings show that our approach works, in that it recapitulates the consensus comparative anatomy view of avian and mammalian brains; in addition, it provides evidence of competing hypotheses concerning more specific homologies.
At lower levels of the tree alignment (individual brain regions), songbird Area X best aligned to the human putamen (Pu) (Fig. 2B and Table 1), supporting a prediction made on the basis of other evidence (41, 44, 48). The avian ventral striatum (VS) adjacent to Area X that does not participate in vocal learning best aligned to the human ventral striatum [i.e., nucleus accumbens (Acb) ventral to the putamen] (Fig. 2B). Songbird RA best aligned to the human central sulcus (PrG_cs) within the primary motor cortex [i.e., within the precentral gyrus (PrG)] (Fig. 2B), supporting a prediction of the cortex hypothesis (4, 33, 41, 49); the human central sulcus and songbird RA have some of the most robust direct projections to brainstem motor neurons known (Fig. 1) (41, 50). The mAC lateral to RA, which does not participate in song production (9, 51) and does not make direct projections (52, 53), aligned weakly to the human superior rostral gyrus (SRoG) of the frontal lobe (Fig. 2B). Both HVC and LMAN aligned best to the parietal part of the cingulate cortex, but the correlations were low (Fig. 2B). None of the regions hypothesized to be similar to HVC or LMAN had a strong secondary match (correlations > 0.09), but of the hypothesized regions, the highest correlation values were HVC with Wernicke’s auditory speech processing region and LMAN with Broca’s speech production and processing region (Table 1). Within the brainstem, the songbird vocal (nXIIts) and neck (SSp) motor neurons could not be aligned to their proposed human homologs—the nucleus ambiguus and supraspinal nucleus, respectively (16, 44, 49, 54)—because they were not isolated as part of the Allen Human Brain Atlas data set. However, the closest match for nXIIts in the available human data was to the vagus motor neurons (n10) (Fig. 2B and Table 1). These findings support a number of predictions and enable new predictions about molecular similarities between avian and human brain regions.
Confirming brain region specializations between human and songbird
The dynamic programming algorithm provided an unbiased global alignment between human and songbird brain regions, and the input tree distance measures provided correlation values. However, the statistical significance obtained from the Pearson correlation calculations might be artificially inflated if large numbers of genes have correlated expression. Thus, to estimate the significance of the similarities and perform an independent statistical test, we (i) computed a more stringent permutation P value (0.05 threshold) and associated familywise error rate (FWER, 0.2 threshold) for each proposed relationship on the basis of the Pearson correlations between species (SM7), and (ii) performed a linear regression on the nodes of the tree to determine the number of significantly specialized genes in common (SM8), relative to what would be expected by chance (SM9).
Both the permutation and gene number methods gave moderate (P ≈ 0.05) to strong (P < 0.001) statistical support for most zebra finch and human regions aligned by the dynamic programming algorithm at Pearson correlations of ~0.09 and above (Fig. 2B, Table 1, and fig. S3). In particular, songbird striatal regions (Area X and VS) were confirmed by both methods to have the most similar specialized gene expression to the human striatum (Fig. 3A and fig. S3A; Table 1 and table S3A). The avian pallial regions were confirmed to be most similar to the human cortex, with a lower significance by the permutation analyses (P = 0.068) and corresponding FWER analyses (0.218) but with the highest number of specialized genes in common of all regions compared (Fig. 3B and fig. S3B; Table 1 and table S3B). The lower significance could possibly reflect greater differences between the avian pallium and human cortex than between the avian and human striatum (33, 47).
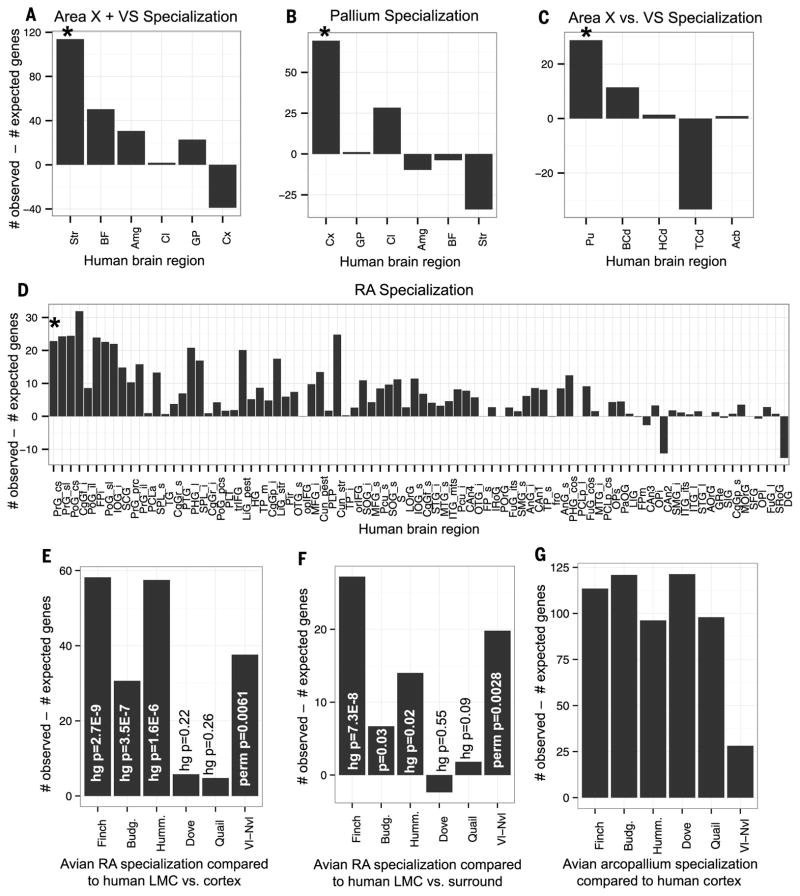
Fig. 3. Relative number of genes with significantly shared specialized expression between avian and human brain regions.
Each panel shows a plot of the number of genes significantly specialized (P < 0.05; hypergeometric test) in common between the avian and human samples relative to the number of genes expected to be specialized by chance. (A) Finch Area X + VS specialization compared to all subregions of the human telencephalon. (B) Finch pallial region (RA, neighboring arcopallium, HVC, and LMAN combined) specialization compared to all subregions of the human telencephalon. (C) Finch Area X specialization compared to all subregions of the human striatum. (D) Finch RA specialization compared to the specialization of every subregion from the human cortex, which optimally aligned to the zebra finch pallium. (E) Avian RA analogs (vocal learners) and mAC (nonlearners) relative to the adjacent arcopallium compared to human LMC/dLSC relative to cortex. (F) Avian RA analogs (vocal learners) and mAC (nonlearners) relative to the arcopallium compared to human LMC/dLSC relative to PrG/PoG. (G) Avian arcopallium versus whole brain specialized genes compared to human cortex versus whole brain specialized genes. In (A) to (D), asterisks denote the human specializations determined to be similar to the avian specialization on the basis of the optimal alignment and correlation. In (E) to (G), P values less than 0.05 indicate that the number of specialized genes is greater than chance according to a hypergeometric test.
Among the striatal regions, both tests confirmed Area X to have the strongest similarity with the putamen (Fig. 3C and fig. S3C; Table 1 and table S3C) but a significant secondary relationship with the adjacent body of the caudate, consistent with the hypothesis that Area X is similar to adjacent parts of the caudate and putamen (41). Among the pallial regions, both tests confirmed RA to have the strongest similarity to the central sulcus part of the human primary motor cortex (PrG_cs) but also to the adjacent primary somatosensory part of the central sulcus (PoG_cs) and to a region where the ventral LMC is located (PrG_sl) (Fig. 3D and fig. S3D; Table 1 and table S3D). The similarity to the somatosensory cortex is not too surprising, considering the underappreciated overlap of cells with motor and somatosensory functions in both walls (PrG_cs and PoG_cs) of the central sulcus (55). This is supported by a strong similarity in gene expression between PrG and PoG samples (fig. S2C). Another region proposed to be functionally analogous to RA, the cingulate gyrus (4), also had strong similarity from both tests (Fig. 3D and fig. S3D; Table 1 and table S3D). For HVC and LMAN, of the hypothesized human brain regions, again the highest similarities were to Wernicke’s area (plus Heschl’s and transverse auditory gyri; HG, TG) and Broca’s area, respectively, but the relationships were not significant according to the permutation and FWER tests (Table 1, fig. S3F, and table S3F). These results reveal that similarities identified by the dynamic programming algorithm are significant, and further reveal more refined similarities with other brain regions according to the anatomical annotations of the Allen Human Brain Atlas.
Zebra finch Area X and RA are most similar to human speech brain regions
The above analyses gave equal weight to samples with the same anatomical annotation, ignoring possible variation that might exist across samples or samples annotated differently according to other hypotheses of mammalian or avian brain organization. To address these concerns, we examined the correlation between the specialization of a finch brain region and each of the individual human brain samples within a matching region of interest, overlaid with the coordinates of brain regions activated during speech production (SM10) (56–62).
We found that the zebra finch Area X specialization best matched specific human putamen and caudate samples that overlapped and extended with the coordinates of brain regions activated during speech production (Fig. 4A) (56–58). Similarly, the zebra finch RA specialization best matched specific samples at or near coordinates of a recently hypothesized dorsal LMC (dLMC) and adjacent somatosensory laryngeal cortex (dLSC) (62, 63) within the PrG and PoG, respectively, as well as to vLMC samples within PrG, all regions that have the highest functional activation during speech production (Fig. 4B) (59). Moreover, several of the human samples with the highest molecular scores shared with songbird RA, which controls the syrinx and not the tongue, beak, or jaw (41), are in proximity to the center of a region activated during the production of vowels (generated by the LMC connections to the larynx), as opposed to the production of tongue, lip, or jaw movements (Fig. 4B) (59). The results suggest that the match of RA to LMC/dLSC contributed to the relationship of RA with the broader human primary motor cortex.
Fig. 4. Overlap of regions with convergent specialized expression and speech activation.
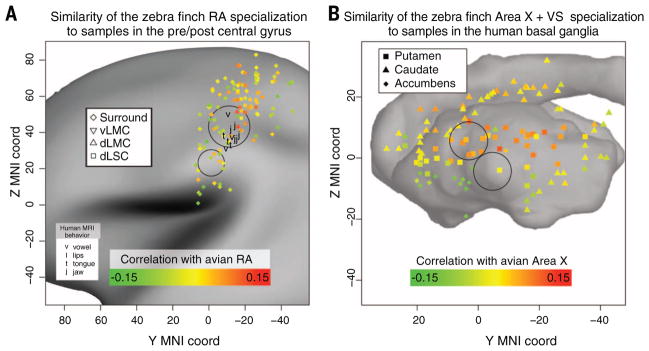
Shown are the coordinates in MNI (Montreal Neurological Institute) space of each human brain microarray sample from the striatum (A) and the precentral/postcentral gyrus (B). The different subregions within the striatum and precentral/postcentral gyrus are labeled by differently shaped symbols. These points are placed on top of a representative image of the cortical surface map from the Allen Human Brain Atlas (38). Only the left hemisphere, which has higher sample density, is shown. In (A), each point is colored on the basis of the correlation between the specialization of that sample relative to the entire human striatum, and of Area X relative to Area X and VS together. Circles represent high-confidence regions of speech activation from multiple coordinates in multiple studies (56–62). In (B), each point is colored on the basis of the correlation between the specialization of that sample relative to the entire human PrG/PoG, and of zebra finch’s RA specialization. A similar plot with the average of all vocal learners is shown in fig. S4C.
All vocal-learning birds and humans show convergent gene expression specializations
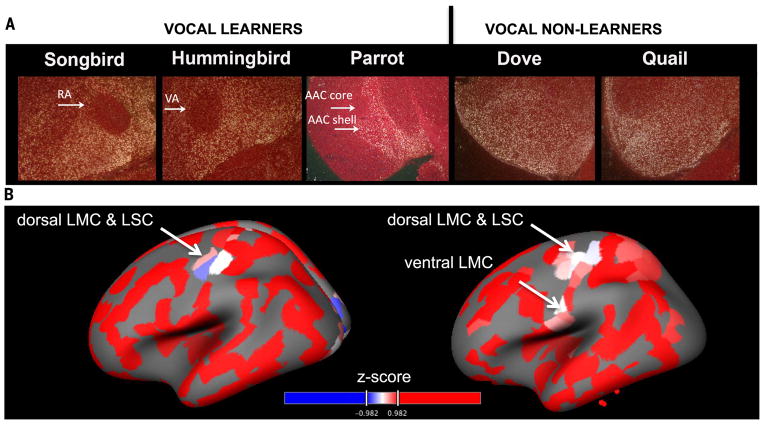
A case for convergent specialized gene regulation in vocal-learning brain regions as a general principle would be supported if other avian vocal-learning lineages showed similar convergent specializations with humans, and negative control vocal-nonlearning birds and vocal-nonlearning primates did not. We first looked for possible specializations in the gene expression profiles of the RA analog from species representing the two other vocal-learning lineages (parrots and hummingbirds) compared to the neighboring arcopallium and comparably located regions of the arcopallium of species representing two experimentally determined vocal-nonlearning lineages (dove and quail) (41, 64, 65). Because the avian samples were run on the zebra finch Agilent microarray, we had to develop a strict filtering pipeline to include only oligonucleotides that mapped to and hybridized to avian genomic DNA across species, yielding a total of 3044 genes each represented by at least one oligonucleotide also present on the human Agilent microarray (SM3).
This reduced set of genes applied to our dynamic programming algorithm recapitulated the strong relationship between gene expression specializations in zebra finch RA and human PrG_cs (fig. S4A). This relationship was stronger for the RA analog of all three vocal learners (fig. S4, A to C) relative to the similarly located central arcopallium of both vocal nonlearners (fig. S4, D and E). The correlations increased for the vocal-learning avian species, but not for the vocal-nonlearning avian species, when comparisons were made to the specific human putative LMC (dLMC and vLMC) and dLSC samples within the PrG_cs and PoG_cs relative to the cortex (fig. S4C, triangles and squares); the correlations were still present when the comparison was relative to only the remaining PrG and PoG (table S4, A to C). Likewise, the RA analogs of all three vocal learners had many more genes specialized in common with the human LMC/dLSC than did the arcopallium of vocal nonlearners, and the number of specialized genes in common was significant only for the vocal learners (Fig. 3, E and F, and table S4, B and C). For a control analysis, we identified the common specialized genes between all the arcopallium samples of each avian species and the entire human frontal lobe, and found them to be similar across all avian species (Fig. 3G).
To test whether the same set of genes led to these independent patterns of convergent evolution, we extended our linear regression model to identify genes that were differentially specialized with humans in all three vocal-learning avian species relative to two vocal-nonlearning avian species (SM9). This vector of genes and expression levels in all vocal-learning birds had a positive correlation with genes specialized in LMC/dLSC relative to the surrounding PrG and PoG (0.14, permutation P = 0.016). A significant number of genes overlapped between the human LMC/dLSC and the vocal-learning minus vocal-nonlearning birds (Fig. 3, E and F, permutation test; Table 1). Overall, these findings indicate that the avian RA analogs of each vocal-learning lineage have mostly the same convergent molecular specializations with human LMC/dLSC.
Songbird RA shows greater similarity to human LMC than to nonhuman primate motor cortex
We conducted the macaque comparisons separately because the macaque microarray data were from more limited regions (mainly in the cortex), contained different cell layers of each cortical region (as opposed to all cortical layers combined in each human sample), and had fewer orthologous genes (4702) with zebra finch than with human (7473). All of our approaches (correlation, gene number, and permutation) showed that songbird RA had the highest match to layer 5 cells of macaque primary motor cortex (with a secondary match to layer 5 of premotor cortex) relative to all other layers within the primary motor cortex and to all layers of all other cortical regions sampled (cingulate, auditory, visual, and other cortices; Table 1, fig. S5, A and B, and tables S3G and S4D). HVC had the best match, although weaker, to macaque layer 2 of the PoG and other cortical regions (Table 1, fig. S5, C and D, and table S3H). Both findings are consistent with the cortex hypothesis of homologies (41, 66, 67). However, LMAN, also part of the nidopallium, had the best match to layer 4 of the PoG and other cortical regions, contradicting previous hypotheses (41, 67) (Table 1, fig. S5, E and F, and table S3I) but consistent with a recent study comparing chicken nidopallium to mouse cortical layers (68).
Comparing the arcopallium samples from each of the avian species with the specializations of human LMC/dLSC versus macaque layer 5, we found a weak, nonsignificant difference in the correlation between the arcopallium regions of avian vocal learners versus nonlearners and macaque layer 5 cells (table S4D), but a stronger significant correlation between the RA analogs of avian vocal learners versus nonlearners and human LMC/dLSC versus macaque layer 5 cells (table S4E). These findings demonstrate the similarity of avian RA and arcopallium to mammalian layer 5 cells, but a greater similarity of each vocal-learning species’ RA analog to human laryngeal cortex regions (which also contain layer 5 cells) relative to nonhuman primate motor cortex.
Function of genes specialized between avian and human brains
We conducted a linear regression analysis to identify the specific genes with statistically significant differences for each species (SM8) and performed functional enrichment analyses relative to annotated pathways (SM11). We found that 288 genes contributed significantly to the shared specialization between the avian and human striatum (table S3A). These included those with positive control in situ hybridization data showing higher expression that distinguishes the striatum from other regions in both birds and mammals (fig. S6, A and B, and table S3A) (43). Functional enrichment analysis of these genes found movement disorders (68 genes) and disorders of the basal ganglia (55 genes) as two of the top categories (table S5A), consistent with known functions of the striatum. Similarly, 342 genes contributed to the shared specialization between avian pallium and human cortex, and also included those validated by in situ hybridization (fig. S6, C and D, and table S3B) (43). The top enrichment categories were behavior (64 genes) and neurotransmission (38 genes; table S5B). These findings show that our linear regression approach correctly identifies genes contributing to shared specializations between avian and mammalian brains.
We identified 78 genes that contributed significantly to the shared specialized expression between songbird Area X and human putamen (fig. S7 and table S3C). In situ hybridization analyses of 13 randomly selected genes revealed that those with a permutation P < 0.015 had a true positive rate of ~90% for differential expression in Area X (fig. S7, highlighted genes, fig. S8, and table S3C). The top enrichment categories were movement disorders (21 genes) and ataxia (8 genes; table S5C), consistent with motor-driven neural activity and gene regulation in Area X (4, 42), and with functional magnetic resonance activation during speech production in the matching human striatal regions (57).
We identified 40 genes that contributed to the shared specialized expression between songbird RA and human PrG_cs (table S3D); a subset of these (10 genes) contributed ~50% to the shared specialization between songbird RA and macaque layer 5 of motor cortex (table S3D). In contrast, we found a mostly nonoverlapping set of 55 genes (only 3 overlapping) that contributed to the convergent shared specialized expression between the RA analogs of vocal learners and human LMC/dLSC (Fig. 5 and table S6). These findings indicate that the similarity between RA and the broader primary motor cortex in humans is due in part to similarity with its layer 5 neurons, and is separate from the similarities to LMC/dLSC within primary motor cortex.
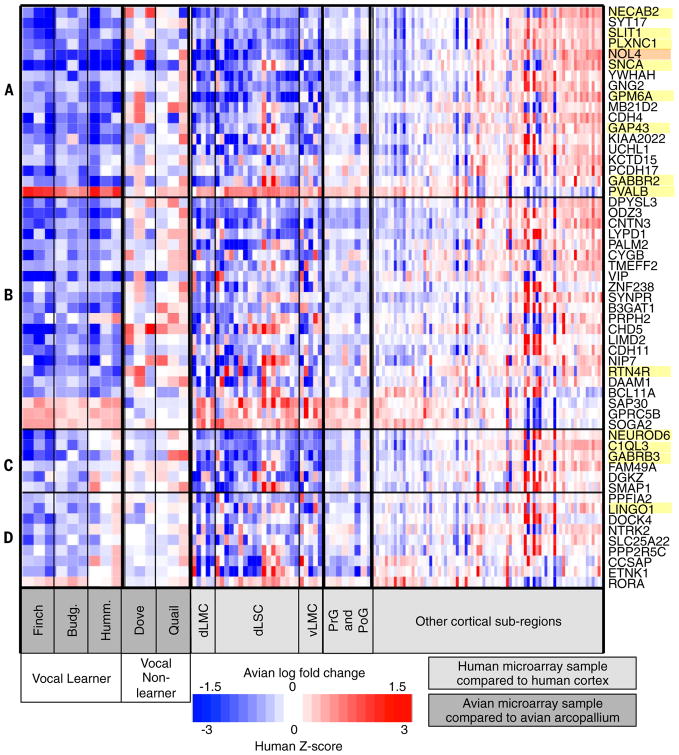
Fig. 5. Heat map of gene expression specialization in avian RA analogs versus the arcopallium and human LMC/dLSC regions versus other cortical regions.
(A to D) Each row represents a gene (table S4), sorted according to whether or not they are significant in hummingbird RA analog [(A) and (B)], significant in individual human LMC/LSC regions relative to the entire PrG and PoG [(A) and (C)], or all LMC/dLSC regions (dLMC, vLMC, LSC) combined (D). Samples within each section are ordered on the basis of estimated log fold difference in LMC/LSC versus PrG and PoG surround. Each column is a microarray sample from an avian species (dark gray) or human (light gray) as listed at the bottom. For the avian species samples, each entry in the heat map shows the log fold change between each microarray sample and median gene expression value for the entire finch arcopallium (needed one species and microarray platform to normalize). For the human samples, each entry is the Z-score specialization relative to the entire cortex for the human sample or brain region. Red, higher expression; blue, lower expression; white, no difference between the compared regions. In the hummingbird microarray data set, one animal was an outlier for some genes [(C) and (D); third column], which we believe is due to an error in the laser capture dissection for a subset of sections (fig. S17); in situ hybridization data validated the hummingbird profiles of one of these genes as an example (fig. S11). Yellow highlights show validated genes; orange highlights were not able to be validated.
In situ hybridization of 15 of the 55 genes (selected because of their axon guidance functions or previously published profiles) revealed a ~95% true positive rate for differential expression in zebra finch RA relative to one or more parts of the adjacent arcopallium (Fig. 6, figs. S9 and S10, and table S5). The majority of identified genes (50 genes; ~91%) had decreased expression in the RA analogs and human LMC/dLSC relative to the surrounding regions, as opposed to only four that had increased levels (Fig. 6). This is in contrast to Area X and the analogous putamen/caudate regions, where more than 70% of the genes had increased expression (fig. S7). The top enrichment categories among the 40 genes shared with motor cortex cells were movement disorders and abnormal morphology of neurons (table S5D), whereas the 55 genes shared more specifically with LMC/dLSC were related to cognitive behaviors (cognition, learning, and memory) and development of neural connections (neuritogenesis, axon and dendrite growth) for different gene subsets (5 to 15 genes each; table S5E). The latter finding suggests that the specialized genes could play a role in mediating the differential connectivity of avian RA and of human LMC.
Fig. 6. Convergent differential regulation of SLIT1 in the RA analog and human LMC.
(A) In situ hybridization of SLIT1 in the RA analog and arcopallium of vocal-learning and vocal-nonlearning avian species. Shown are frontal sections; dorsal is up, right is midline. White, SLIT1 mRNA detected by 35S riboprobe in dark-field view; red, cresyl violet stain of brain cells. (B) Cortical surface map of SLIT1 relative gene expression levels (Z-score) in the human brain measured by microarrays (http://human.brain-map.org/static/brainexplorer). Red, higher expression; blue, lower expression. Two example persons are shown (left hemisphere), one with both dLMC/LSC and vLMC. Dorsal is up, front is left.
In situ patterns of commonly specialized avian RA and human LMC genes
To achieve even better resolution, we analyzed all 55 candidate human LMC/dLSC genes with the use of microarray expression cortical surface profiles from the Allen Human Brain Atlas (http://human.brain-map.org/static/brainexplorer) and analyzed the 15 candidate avian genes by means of in situ hybridization on zebra finch brains (SM12). The anatomical profiles of the genes fell into three categories (Fig. 6 and figs. S9 and S10): (i) those with expression specialization in the RA analog and human dLMC/dLSC cortex; (ii) those with expression specialization in the RA plus immediately adjacent intermediate arcopallium and LMC plus the adjacent human primary motor cortex; and (iii) those with a more varied pattern of expression within the avian or human brain, but with local differences in avian RA and human motor cortex. On the basis of these results, we analyzed a subset of 10 of the 15 genes by in situ hybridization in our predicted dLMC/dLSC region of the other hemisphere (right) of two human brain specimens used for the microarrays; although we noted a possible stronger relationship of RA with the speech-dominant left LMC/dLSC hemisphere, the right hemisphere was also correlated (SM8).
A candidate with one of the most distinct specializations was the axon guidance ligand SLIT1, which showed verified down-regulation in the RA analog of all vocal-learning bird species, but no down-regulation in the arcopallium of the vocal-nonlearning species (Fig. 6, A and B); it was also partially down-regulated in the adjacent mAC of zebra finches. Likewise, macaques have recently been shown to have lower SLIT1 in primary motor cortex (69), which we also found in marmosets (fig. S13). However, like songbird RA, our putative human dLMC in the central sulcus showed further down-regulation, particularly in the upper cortical layers. The adjacent dLSC showed striking down-regulation throughout the region (Fig. 7). In nonhuman primates, a ventral premotor region (area 6v) is hypothesized to be a precursor of the human primary LMC and makes an indirect connection to vocal motor neurons in the brainstem, However, unlike vocal-learning birds and humans, this region is not required for vocalization (61) and did not show differential expression of SLIT1 (fig. S13). In parrots, the RA analog has a core portion that makes a direct projection to brainstem vocal motor neurons, like songbird and hummingbird RA analogs and the human LMC, as well as a more ventral shell portion unique to parrots that projects to other song nuclei (70). Only the core portion of the parrot RA analog showed SLIT1 down-regulation (Fig. 6A) (70). SLIT1 promoter is a target of the FOXP2 transcription factor (71), and FOXP2 showed lower expression in the dLMC region (fig. S12).
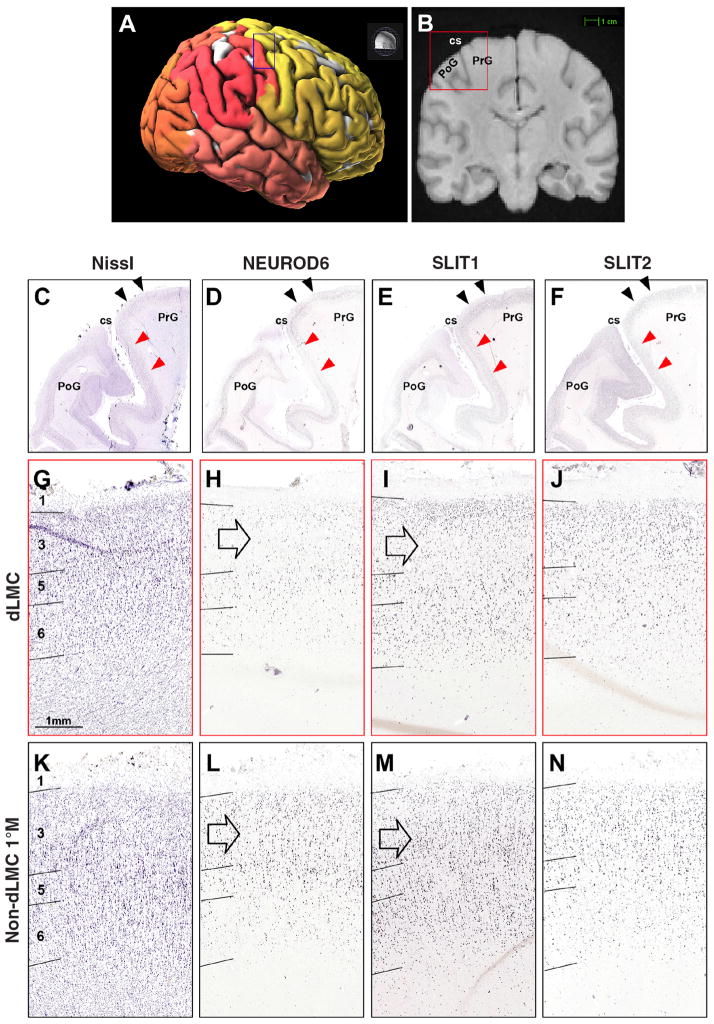
Fig. 7. In situ hybridization localization of the putative dLMC/dLSC in the human brain.
(A) Surface image of a human brain showing the different cortical lobes (colors) and the region dissected for the in situ hybridization analyses (box). (B) Magnetic resonance image showing the location of the region dissected for in situ hybridization analyses in the right hemisphere (box). (C) Nissl stain of the examined region. (D) NEUROD6 down-regulation in a distinct region of the PrG, and in the upper layers of the adjacent PoG. (E) SLIT1 down-regulation in the same PrG dLMC region (see I versus M), as well as in the adjacent PoG. (F) SLIT2 control showing no noticeable difference. (G to N) Red arrows correspond to the boundaries of the regions represented in the higher-power images of (G) to (J); black arrows correspond to (K) to (N). Down-regulation in dLMC is strongest in layer 3 (open arrow), but also in layer 5 for NEUROD6.
Another candidate gene was the NEUROD6 transcription factor, involved in brain development and differentiation. Although weakly down-regulated in zebra finch RA, the pattern of down-regulation in the putative human dLMC was distinct such that it defined the boundaries of the region (Fig. 7). We examined negative control genes, some belonging to the same gene families (such as SLIT2), and did not find differential expression in RA analogs (70) and human LMC (Fig. 7). We also examined RNA expression patterns for 10 of the candidate genes available in the Allen Mouse Brain Atlas (http://mouse.brain-map.org) in a putative rudimentary LMC region of the mouse brain that makes a very sparse direct projection to vocal motor neurons (72), and did not identify examples of differential expression (fig. S14), except for PVALB but with reduced expression (fig. S14); FOXP2 was also in the opposite direction, with up-regulation in layer 5 cells of primary motor cortex (M1; fig. S14) (73). A comparison of expression levels in the microarray data reveals that some of these genes have some of the largest log fold expression differences among all 7473 orthologs in songbird RA and human LMC regions, with SLIT1, NEUROD6, C1QL3, and PVALB being among the most differentially expressed genes in the dLMC and RA of both species (fig. S15). A further characterization of the expression profiles of these and other genes can be found in the legends of figs. S9 to S14.
Proteomics supports difference between zebra finch RA and surrounding arcopallium
To determine whether the gene expression specializations we found in RA were reflected in the corresponding protein products, we performed nanoscale capillary tandem mass spectrometry experiments on zebra finch RA and the adjacent mAC with a microproteomic method developed for laser capture–microdissected cells (74). We identified 81 proteins as differentially expressed in RA (fig. S16A) and detected corresponding mRNA gene expression differences in the microarray experiment (table S7). Three of the specialized down-regulated proteins, GAP43, SNCA, and UCHL1, were also identified in the mRNA analyses for avian RA and human LMC. Overall, individual mRNA and protein level differences in RA and the surrounding arcopallium had a modest but significant correlation (R2 = 0.18, P = 9.4 × 10−5). Notably, the genes that had lower mRNA expression in RA showed a stronger correlation with protein levels (t test, P = 0.006; fig. S16B). Concordant with the mRNA expression analysis, the top disease categories for the protein analyses included movement disorders (P = 8.7 × 10−14, 29 genes), and other categories included neural connectivity and brain development (table S5F). The top biological pathway gene ontology category overall was endocytosis of vesicles involved in cellular assembly and organization (P = 6.6 × 10−11, 7 genes), consistent with a role in neural connectivity.
Discussion
Our study indicates that behavioral and neuroanatomical convergence for the vocal-learning trait is associated with molecular convergence of gene expression changes in the circuits that control the behavior. The findings identify specific molecularly analogous brain regions for song and speech between birds and humans, and further support broader homologous brain regions in which these specialized song and speech regions are located. Although the correlations we find at first glance may not seem strong, they are highly significant, are predominantly driven by the specialized expression of tens to hundreds of genes (depending on the level in the anatomical tree) out of thousands of genes, and for vocal-learning regions are supported by lack of such findings in vocal-nonlearning birds and primates. We believe that the success of identifying these genes arises from a combination of the number of tissue sources from both birds and primates, the selection of brain regions we profiled, and the computational approach we developed, based on an anatomical gene expression specialization tree.
We find the convergent gene expression for RA/LMC and Area X/putamen remarkable in that they involve species separated from a common ancestor by 68 million to 310 million years (34–36), different vertebrate lineages, and many genes. Consistent with the function of the genes in neural connectivity and motor behavior, the avian RA and human primary motor cortex make more direct contacts with vocal motor neurons than do motor cortex neurons in any other vertebrates tested to date (6, 61). Direct cortical projections to lower motor neurons are correlated with finer motor control and more complex motor learning (75). In contrast, the avian arcopallium, like the motor cortex in many mammals, is thought to make mostly indirect contact with lower motor neurons (6). We caution that our analyses do not show that the specializations we discovered are functionally related to the projections of these neurons or to vocal learning, because the difference in gene expression could be a consequence, rather than a cause, of the differential connectivity within and across species. Nonetheless, even if only a consequence, the genes and their expression differences can serve as candidates that can lead us to the cause.
Our findings on SLIT1 add to a growing body of studies implicating an important role for this gene in speech and vocal learning. SLIT1 was found to be more differentially up-regulated by the human FOXP2 transcription factor than by the chimpanzee FOXP2 (71). SLIT1 is a ligand for the ROBO1 axon guidance receptor, and ROBO1 mutations cause dyslexia and speech disorders in humans (76). ROBO1 is one of five candidate genes with convergent amino acid substitutions in vocal-learning mammals (32). ROBO1 itself was also found to be differentially expressed in fetal human speech brain regions (77) and in the RA analog of avian vocal learners (70). Thus, because binding of SLIT1 to ROBO1 causes repulsion of the axonal processes from cell bodies (78), we hypothesize that the down-regulation of SLIT1 and other neural connectivity genes in RA and LMC may represent a permissive mechanism to allow certain neural connections to form, such as the unique direct projection to vocal motor neurons.
Among the very few genes with convergent up-regulation in RA and LMC, the calcium-buffering protein parvalbumin has been noted (16) to be present at unusually high levels in the RA neurons that project to vocal motor neurons in songbirds and in the analogous layer 5 neurons of oral motor cortex (containing vLMC) of humans and chimpanzees, but not in the projection neurons of the adjacent arcopallium or in the oral motor cortex of other primates or mammals (79, 80). Normally, parvalbumin is expressed in fast-spiking inhibitory interneurons of the cortex. It has been hypothesized that the increased expression in RA projection neurons (16), and we propose here also in LMC, is to protect these brain regions from Ca2+ toxicity due to highly active neurons that control some of the fastest muscles in the body (syrinx in birds and larynx in humans). This is supported by the fact that the RA neurons have a higher spontaneous firing rate than the surrounding arcopallium (81).
The enriched neurotransmission functions of the convergently specialized genes in songbird Area X and parts of the human putamen/caudate are consistent with findings that Area X does not appear to have large differences in neural connectivity relative to the surrounding striatum (82), but like in RA, the neural activity in Area X is commonly known to differ. The match of Area X to the human striatum and of RA analogs to the cortex enables specific hypotheses about which brain regions are analogous in song-learning birds and humans, as well as which regions in humans may be specialized for speech. The role of the human striatum in speech acquisition and production has begun to be appreciated (57, 83), but specific loci have not been as well defined as for the cortex. Similarly, the dLMC was also only recently described (63), and our results add to the growing body of work (62) that shows this region could viably be a specialized part of the speech control circuit.
Our lack of finding significant convergent specializations of the songbird HVC or LMAN with humans does not mean that shared specializations of more specific regions within the cortex do not exist. Identifying these may require analyses of the transcriptomes of the adjacent non-song neurons in the avian nidopallium in which HVC and LMAN are located; for RA and Area X, in contrast, the immediate adjacent areas were profiled. We attempted to analyze an available data set of HVC and the surrounding auditory nidopallium shelf, generated with cDNA microarrays (12). However, the overlap between our Agilent microarray was not high enough to incorporate these data into our framework.
At higher levels of brain organization, our cortical results are in contrast with findings of a recent study that used brain transcriptome profiles to argue that the avian pallium is equally and only weakly similar to the mouse cortex, claustrum, or amygdala (68). We believe our ability to identify a stronger relationship to the cortex was because we organized the expression profiles as specialization trees, developed a dynamic programming algorithm requiring hierarchical anatomical alignment of brain similarities, and performed avian brain dissections using more precise laser capture microdissections. It is also possible that even though the pallial song nuclei (which made up 75% of the avian pallial samples) have more convergent similarities with the entire human cortex, the avian pallial subdivisions in which the song nuclei are housed have similarities with the claustrum and amygdala. However, our results across species suggest that this is not the case for the arcopallium, the region most often suggested to be similar to the mammalian amygdala (34, 44).
Our study has identified molecularly analogous brain regions that function in song and speech in vocal-learning birds and humans. The gene expression similarities we find across species, combined with circuit and functional similarities (40, 41), suggest that avian vocal learners can be used as models for speech production at a molecular level. Our experiments provide a candidate set of genes involved in neural connectivity and cell communication functions. Future studies will be necessary to decipher the specific function of these genes in vocal communication and the mechanisms through which they are differentially expressed and selected. Insight into the latter question is beginning to be revealed, as in our companion study we found convergent mutations in the genomic regulatory regions of some of the genes we find here with specialized expression in the RA analog across vocal-learning lineages (33). Our computational approach can also be applied to other anatomical molecular profiling experiments across species. The fact that convergent neural circuits for vocal learning are accompanied by convergent molecular changes of multiple genes in species separated by millions of years from a common ancestor indicates that brain circuits for complex traits may have limited ways in which they could have evolved from a common ancestor.
Supplementary Material
Acknowledgments
We thank H. Dressman and L.-L. Rowlette for microarray hybridizations, the Allen Institute for Brain Science for early access to human microarray data and human brain in situ hybridizations (and the Allen Institute founders, P. G. Allen and J. Allen, for their encouragement of that project), K. Simonyan for her coordinates of meta-analyses for speech regions, and S. Jarvis for help with imaging. Supported by NIH Directors Pioneer Award DP1 OD000448, National Institute on Deafness and Other Communication Disorders grant R01DC007218, and the Howard Hughes Medical Institute (E.D.J.). The brain gene expression atlas (ZEBrA) is supported by National Institute of General Medical Sciences grant R24GM092842 (C.V.M.). Marmoset experiments were supported by the Funding Program for World-leading Innovative R&D on Science and Technology (A.I.). Microarray data are available in the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo) under accession numbers GSE33365 and GSE34819 (for songbird Area X), GSE28395 (for avian arcopallium regions), GSE33667 (for avian brainstem regions), and GSE31613 (for macaque brain regions); human microarray data are available from the Allen Human Brain Atlas site (http://human.brain-map.org/static/download). E.D.J., A.R.P., E.H., M.R., and P.R. conceived and designed the project; E.D.J., A.R.P., and A.J.H. wrote the paper; E.D.J., A.J.H., M.A.M., A.I., E.L., and C.V.M. co-supervised parts of the project; A.R.P. conducted most computational analyses; G.G., T.B., E.J.S., and A.J.H. conducted other computational analyses and supervision; E.H., M.R., O.W., and J.T.H. collected avian samples and performed microarray experiments; J.M., M.W., P.V.L., E.H., R.W., M.K., A.Bo., A.Be., C.V.M., and E.D.J. performed in situ hybridizations and analyses across species; G.Z., M.T.P.G., and E.D.J. provided genomes; and P.R., M.A.M., J.W.T., and E.J.S. performed proteomic experiments.
Footnotes
REFERENCES AND NOTES
- 1.Petkov CI, Jarvis ED. Birds, primates, and spoken language origins: Behavioral phenotypes and neurobiological substrates. Front Evol Neurosci. 2012;4:12. doi: 10.3389/fnevo.2012.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jürgens U. The neural control of vocalization in mammals: A review. J Voice. 2009;23:1–10. doi: 10.1016/j.jvoice.2007.07.005;. [DOI] [PubMed] [Google Scholar]
- 3.Egnor SE, Hauser MD. A paradox in the evolution of primate vocal learning. Trends Neurosci. 2004;27:649–654. doi: 10.1016/j.tins.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 4.Doupe AJ, Kuhl PK. Birdsong and human speech: Common themes and mechanisms. Annu Rev Neurosci. 1999;22:567–631. doi: 10.1146/annurev.neuro.22.1.567. [DOI] [PubMed] [Google Scholar]
- 5.Bolhuis JJ, Okanoya K, Scharff C. Twitter evolution: Converging mechanisms in birdsong and human speech. Nat Rev Neurosci. 2010;11:747–759. doi: 10.1038/nrn2931. [DOI] [PubMed] [Google Scholar]
- 6.Arriaga G, Jarvis ED. Mouse vocal communication system: Are ultrasounds learned or innate? Brain Lang. 2013;124:96–116. doi: 10.1016/j.bandl.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oakley TH, Rivera AS. Genomics and the evolutionary origins of nervous system complexity. Curr Opin Genet Dev. 2008;18:479–492. doi: 10.1016/j.gde.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 8.Barrett HC. A hierarchical model of the evolution of human brain specializations. Proc Natl Acad Sci USA. 2012;109(suppl 1):10733–10740. doi: 10.1073/pnas.1201898109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feenders G, et al. Molecular mapping of movement-associated areas in the avian brain: A motor theory for vocal learning origin. PLOS ONE. 2008;3:e1768. doi: 10.1371/journal.pone.0001768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Margoliash D, et al. Distributed representation in the song system of oscines: Evolutionary implications and functional consequences. Brain Behav Evol. 1994;44:247–264. doi: 10.1159/000113580. [DOI] [PubMed] [Google Scholar]
- 11.Mello CV, Vates GE, Okuhata S, Nottebohm F. Descending auditory pathways in the adult male zebra finch (Taeniopygia guttata) J Comp Neurol. 1998;395:137–160. doi: 10.1002/(SICI)1096-9861(19980601)395:2<137::AID-CNE1>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 12.Lovell PV, Clayton DF, Replogle KL, Mello CV. Birdsong “transcriptomics”: Neurochemical specializations of the oscine song system. PLOS ONE. 2008;3:e3440. doi: 10.1371/journal.pone.0003440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lovell PV, Carleton JB, Mello CV. Genomics analysis of potassium channel genes in songbirds reveals molecular specializations of brain circuits for the maintenance and production of learned vocalizations. BMC Genomics. 2013;14:470. doi: 10.1186/1471-2164-14-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsunaga E, Suzuki K, Kobayashi T, Okanoya K. Comparative analysis of mineralocorticoid receptor expression among vocal learners (Bengalese finch and budgerigar) and non-vocal learners (quail and ring dove) has implications for the evolution of avian vocal learning. Dev Growth Differ. 2011;53:961–970. doi: 10.1111/j.1440-169X.2011.01302.x. [DOI] [PubMed] [Google Scholar]
- 15.Hilliard AT, Miller JE, Horvath S, White SA. Distinct neurogenomic states in basal ganglia subregions relate differently to singing behavior in songbirds. PLOS Comput Biol. 2012;8:e1002773. doi: 10.1371/journal.pcbi.1002773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hara E, Rivas MV, Ward JM, Okanoya K, Jarvis ED. Convergent differential regulation of parvalbumin in the brains of vocal learners. PLOS ONE. 2012;7:e29457. doi: 10.1371/journal.pone.0029457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horita H, et al. Specialized motor-driven dusp1 expression in the song systems of multiple lineages of vocal learning birds. PLOS ONE. 2012;7:e42173. doi: 10.1371/journal.pone.0042173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amunts K, et al. Broca’s region: Novel organizational principles and multiple receptor mapping. PLOS Biol. 2010;8:e1000489. doi: 10.1371/journal.pbio.1000489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wada K, Sakaguchi H, Jarvis ED, Hagiwara M. Differential expression of glutamate receptors in avian neural pathways for learned vocalization. J Comp Neurol. 2004;476:44–64. doi: 10.1002/cne.20201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lai CS, Fisher SE, Hurst JA, Vargha-Khadem F, Monaco AP. A forkhead-domain gene is mutated in a severe speech and language disorder. Nature. 2001;413:519–523. doi: 10.1038/35097076. [DOI] [PubMed] [Google Scholar]
- 21.Haesler S, et al. FoxP2 expression in avian vocal learners and non-learners. J Neurosci. 2004;24:3164–3175. doi: 10.1523/JNEUROSCI.4369-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haesler S, et al. Incomplete and inaccurate vocal imitation after knockdown of FoxP2 in songbird basal ganglia nucleus Area X. PLOS Biol. 2007;5:e321. doi: 10.1371/journal.pbio.0050321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scharff C, Petri J. Evo-devo, deep homology and FoxP2: Implications for the evolution of speech and language. Philos Trans R Soc London Ser B. 2011;366:2124–2140. doi: 10.1098/rstb.2011.0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Teramitsu I, Kudo LC, London SE, Geschwind DH, White SA. Parallel FoxP1 and FoxP2 expression in songbird and human brain predicts functional interaction. J Neurosci. 2004;24:3152–3163. doi: 10.1523/JNEUROSCI.5589-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yokoyama S, Radlwimmer FB. The molecular genetics and evolution of red and green color vision in vertebrates. Genetics. 2001;158:1697–1710. doi: 10.1093/genetics/158.4.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arendt J, Reznick D. Convergence and parallelism reconsidered: What have we learned about the genetics of adaptation? Trends Ecol Evol. 2008;23:26–32. doi: 10.1016/j.tree.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 27.Wittkopp PJ, Williams BL, Selegue JE, Carroll SB. Drosophila pigmentation evolution: Divergent genotypes underlying convergent phenotypes. Proc Natl Acad Sci USA. 2003;100:1808–1813. doi: 10.1073/pnas.0336368100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shapiro MD, et al. Genetic and developmental basis of evolutionary pelvic reduction in threespine sticklebacks. Nature. 2004;428:717–723. doi: 10.1038/nature02415. [DOI] [PubMed] [Google Scholar]
- 29.Zakon HH, Lu Y, Zwickl DJ, Hillis DM. Sodium channel genes and the evolution of diversity in communication signals of electric fishes: Convergent molecular evolution. Proc Natl Acad Sci USA. 2006;103:3675–3680. doi: 10.1073/pnas.0600160103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y, et al. Convergent sequence evolution between echolocating bats and dolphins. Curr Biol. 2010;20:R53–R54. doi: 10.1016/j.cub.2009.11.058. [DOI] [PubMed] [Google Scholar]
- 31.Parker J, et al. Genome-wide signatures of convergent evolution in echolocating mammals. Nature. 2013;502:228–231. doi: 10.1038/nature12511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang R. thesis. Duke University; 2011. http://dukespace.lib.duke.edu/dspace/bitstream/handle/10161/5630/Wang_duke_0066D_10911.pdf?sequence=1. [Google Scholar]
- 33.Zhang G, et al. Comparative genomics reveals insights into avian genome evolution and adaptation. Science. 2014;346:1311–1320. doi: 10.1126/science.1251385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jarvis ED, et al. Avian brains and a new understanding of vertebrate brain evolution. Nat Rev Neurosci. 2005;6:151–159. doi: 10.1038/nrn1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar S, Hedges SB. A molecular timescale for vertebrate evolution. Nature. 1998;392:917–920. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- 36.Jarvis ED, et al. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science. 2014;346:1320–1331. doi: 10.1126/science.1253451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whitney O, et al. Science. 2014;346:1256780. doi: 10.1126/science.1256780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hawrylycz MJ, et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489:391–399. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bernard A, et al. Transcriptional architecture of the primate neocortex. Neuron. 2012;73:1083–1099. doi: 10.1016/j.neuron.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brainard MS, Doupe AJ. Translating birdsong: Songbirds as a model for basic and applied medical research. Annu Rev Neurosci. 2013;36:489–517. doi: 10.1146/annurev-neuro-060909-152826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jarvis ED. Learned birdsong and the neurobiology of human language. Ann NY Acad Sci. 2004;1016:749–777. doi: 10.1196/annals.1298.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jarvis ED, Nottebohm F. Motor-driven gene expression. Proc Natl Acad Sci USA. 1997;94:4097–4102. doi: 10.1073/pnas.94.8.4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jarvis ED, et al. Global view of the functional molecular organization of the avian cerebrum: Mirror images and functional columns. J Comp Neurol. 2013;521:3614–3665. doi: 10.1002/cne.23404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reiner A, et al. The Avian Brain Nomenclature Forum: Terminology for a new century in comparative neuroanatomy. J Comp Neurol. 2004;473:E1–E6. doi: 10.1002/cne.20119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Puelles L, et al. Pallial and subpallial derivatives in the embryonic chick and mouse telencephalon, traced by the expression of the genes Dlx-2, Emx-1, Nkx-2.1, Pax-6, and Tbr-1. J Comp Neurol. 2000;424:409–438. doi: 10.1002/1096-9861(20000828)424:3<409::AID-CNE3>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 46.Carrillo GD, Doupe AJ. Is the songbird Area X striatal, pallidal, or both? An anatomical study. J Comp Neurol. 2004;473:415–437. doi: 10.1002/cne.20099. [DOI] [PubMed] [Google Scholar]
- 47.Reiner A, Laverghetta AV, Meade CA, Cuthbertson SL, Bottjer SW. An immunohistochemical and pathway tracing study of the striatopallidal organization of area X in the male zebra finch. J Comp Neurol. 2004;469:239–261. doi: 10.1002/cne.11012. [DOI] [PubMed] [Google Scholar]
- 48.Farries MA, Perkel DJ. A telencephalic nucleus essential for song learning contains neurons with physiological characteristics of both striatum and globus pallidus. J Neurosci. 2002;22:3776–3787. doi: 10.1523/JNEUROSCI.22-09-03776.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nottebohm F, Stokes TM, Leonard CM. Central control of song in the canary, Serinus canarius. J Comp Neurol. 1976;165:457–486. doi: 10.1002/cne.901650405. [DOI] [PubMed] [Google Scholar]
- 50.Rathelot JA, Strick PL. Subdivisions of primary motor cortex based on cortico-motoneuronal cells. Proc Natl Acad Sci USA. 2009;106:918–923. doi: 10.1073/pnas.0808362106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mandelblat-Cerf Y, Las L, Denisenko N, Fee MS. A role for descending auditory cortical projections in songbird vocal learning. eLife. 2014;3:e04371. doi: 10.7554/eLife.04371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dubbeldam JL. The neural substrate for ‘learned’ and ‘nonlearned’ activities in birds: A discussion of the organization of bulbar reticular premotor systems with side-lights on the mammalian situation. Acta Anat. 1998;163:157–172. doi: 10.1159/000046494. [DOI] [PubMed] [Google Scholar]
- 53.Bottjer SW, Brady JD, Cribbs B. Connections of a motor cortical region in zebra finches: Relation to pathways for vocal learning. J Comp Neurol. 2000;420:244–260. doi: 10.1002/(SICI)1096-9861(20000501)420:2<244::AID-CNE7>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 54.Wild JM, Li D, Eagleton C. Projections of the dorsomedial nucleus of the intercollicular complex (DM) in relation to respiratory-vocal nuclei in the brainstem of pigeon (Columba livia) and zebra finch (Taeniopygia guttata) J Comp Neurol. 1997;377:392–413. doi: 10.1002/(SICI)1096-9861(19970120)377:3<392::AID-CNE7>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 55.Nii Y, Uematsu S, Lesser RP, Gordon B. Does the central sulcus divide motor and sensory functions? Cortical mapping of human hand areas as revealed by electrical stimulation through subdural grid electrodes. Neurology. 1996;46:360–367. doi: 10.1212/WNL.46.2.360. [DOI] [PubMed] [Google Scholar]
- 56.Sörös P, et al. Clustered functional MRI of overt speech production. Neuroimage. 2006;32:376–387. doi: 10.1016/j.neuroimage.2006.02.046. [DOI] [PubMed] [Google Scholar]
- 57.Brown S, et al. The somatotopy of speech: Phonation and articulation in the human motor cortex. Brain Cognit. 2009;70:31–41. doi: 10.1016/j.bandc.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bohland JW, Guenther FH. An fMRI investigation of syllable sequence production. Neuroimage. 2006;32:821–841. doi: 10.1016/j.neuroimage.2006.04.173. [DOI] [PubMed] [Google Scholar]
- 59.Grabski K, et al. Functional MRI assessment of orofacial articulators: Neural correlates of lip, jaw, larynx, and tongue movements. Hum Brain Mapp. 2012;33:2306–2321. doi: 10.1002/hbm.21363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Simonyan K, Ostuni J, Ludlow CL, Horwitz B. Functional but not structural networks of the human laryngeal motor cortex show left hemispheric lateralization during syllable but not breathing production. J Neurosci. 2009;29:14912–14923. doi: 10.1523/JNEUROSCI.4897-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Simonyan K, Horwitz B. Laryngeal motor cortex and control of speech in humans. Neuroscientist. 2011;17:197–208. doi: 10.1177/1073858410386727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Simonyan K. The laryngeal motor cortex: Its organization and connectivity. Curr Opin Neurobiol. 2014;28:15–21. doi: 10.1016/j.conb.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brown S, Ngan E, Liotti M. A larynx area in the human motor cortex. Cereb Cortex. 2008;18:837–845. doi: 10.1093/cercor/bhm131. [DOI] [PubMed] [Google Scholar]
- 64.Nottebohm F, Nottebohm ME. Vocalizations and breeding behaviour of surgically deafened ring doves (Streptopelia risoria) Anim Behav. 1971;19:313–327. doi: 10.1016/S0003-3472(71)80012-X. [DOI] [PubMed] [Google Scholar]
- 65.Derégnaucourt S, Saar S, Gahr M. Dynamics of crowing development in the domestic Japanese quail (Coturnix coturnix japonica) Proc Biol Sci. 2009;276:2153–2162. doi: 10.1098/rspb.2009.0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jarvis ED, et al. A framework for integrating the songbird brain. J Comp Physiol A. 2002;188:961–980. doi: 10.1007/s00359-002-0358-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Karten HJ. Homology and evolutionary origins of the ‘neocortex’. Brain Behav Evol. 1991;38:264–272. doi: 10.1159/000114393. [DOI] [PubMed] [Google Scholar]
- 68.Belgard TG, et al. Adult pallium transcriptomes surprise in not reflecting predicted homologies across diverse chicken and mouse pallial sectors. Proc Natl Acad Sci USA. 2013;110:13150–13155. doi: 10.1073/pnas.1307444110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sasaki T, Komatsu Y, Watakabe A, Sawada K, Yamamori T. Prefrontal-enriched SLIT1 expression in Old World monkey cortex established during the postnatal development. Cereb Cortex. 2010;20:2496–2510. doi: 10.1093/cercor/bhp319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang R, et al. Convergent differential regulation of ROBO-SLIT axon guidance pathway genes in the brains of vocal learners. J Comp Neurol. 2014 doi: 10.1002/cne.23719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Konopka G, et al. Human-specific transcriptional regulation of CNS development genes by FOXP2. Nature. 2009;462:213–217. doi: 10.1038/nature08549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Arriaga G, Zhou EP, Jarvis ED. Of mice, birds, and men: The mouse ultrasonic song system has some features similar to humans and song-learning birds. PLOS ONE. 2012;7:e46610. doi: 10.1371/journal.pone.0046610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hisaoka T, Nakamura Y, Senba E, Morikawa Y. The forkhead transcription factors, Foxp1 and Foxp2, identify different subpopulations of projection neurons in the mouse cerebral cortex. Neuroscience. 2010;166:551–563. doi: 10.1016/j.neuroscience.2009.12.055. [DOI] [PubMed] [Google Scholar]
- 74.Roulhac PL, Ward JM, Thompson JW, Soderblom EJ, Silva M, Mosely MA, III, Jarvis ED. Cold Spring Harb Protoc. 2011 doi: 10.1101/pdb.prot5573. pdb.prot5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lemon RN. Descending pathways in motor control. Annu Rev Neurosci. 2008;31:195–218. doi: 10.1146/annurev.neuro.31.060407.125547. [DOI] [PubMed] [Google Scholar]
- 76.Fisher SE, Francks C. Genes, cognition and dyslexia: Learning to read the genome. Trends Cognit Sci. 2006;10:250–257. doi: 10.1016/j.tics.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 77.Johnson MB, et al. Functional and evolutionary insights into human brain development through global transcriptome analysis. Neuron. 2009;62:494–509. doi: 10.1016/j.neuron.2009.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cornide-Petronio ME, Barreiro-Iglesias A. Role of Slit and Robo proteins in the development of dopaminergic neurons. Dev Neurosci. 2013;35:285–292. doi: 10.1159/000351023. [DOI] [PubMed] [Google Scholar]
- 79.Wild JM, Williams MN, Suthers RA. Parvalbumin-positive projection neurons characterise the vocal premotor pathway in male, but not female, zebra finches. Brain Res. 2001;917:235–252. doi: 10.1016/S0006-8993(01)02938-9. [DOI] [PubMed] [Google Scholar]
- 80.Sherwood CC, Holloway RL, Erwin JM, Hof PR. Cortical orofacial motor representation in Old World monkeys, great apes, and humans. II. Stereologic analysis of chemoarchitecture. Brain Behav Evol. 2004;63:82–106. doi: 10.1159/000075673. [DOI] [PubMed] [Google Scholar]
- 81.Adret P, Margoliash D. Metabolic and neural activity in the song system nucleus robustus archistriatalis: Effect of age and gender. J Comp Neurol. 2002;454:409–423. doi: 10.1002/cne.10459. [DOI] [PubMed] [Google Scholar]
- 82.Doupe AJ, Perkel DJ, Reiner A, Stern EA. Birdbrains could teach basal ganglia research a new song. Trends Neurosci. 2005;28:353–363. doi: 10.1016/j.tins.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 83.Simonyan K, Horwitz B, Jarvis ED. Dopamine regulation of human speech and bird song: A critical review. Brain Lang. 2012;122:142–150. doi: 10.1016/j.bandl.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Okanoya K, Merker B. In: Emergence of Communication and Language. Lyon C, Nehaniv CL, Cangelosi A, editors. Springer; London: 2007. pp. 421–434. [Google Scholar]
- 85.Karten HJ, Shimizu T. The origins of neocortex: Connections and lamination as distinct events in evolution. J Cognit Neurosci. 1989;1:291–301. doi: 10.1162/jocn.1989.1.4.291. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.