Summary
Over the last few years, the technology to create targeted knockout and knockin zebrafish animals has exploded. We have gained the ability to create targeted knockouts through the use of zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs) and clustered regularly interspaced short palindromic repeats/CRISPR associated system (CRISPR/Cas). Furthermore, using the high-efficiency TALEN system, we were able to create knockin zebrafish using a single-stranded DNA (ssDNA) protocol described here. Through the use of these technologies, the zebrafish has become a valuable vertebrate model and an excellent bridge between the invertebrate and mammalian model systems for the study of human disease.
Keywords: TALEN, Genome Engineering, Zebrafish, HDR
1. Introduction
Over the last 40 years, the zebrafish (Danio rerio) has gained notable momentum as a valuable non-mammalian vertebrate model system, particularly in understanding developmental mechanisms. Its exogenously fertilized embryos allow real-time, in vivo observation of development from the single-cell stage. Furthermore, as a vertebrate the zebrafish have many similarities with humans, including the nervous system, skin, cartilage and bone, blood and vasculature, kidney, liver, pancreas, gut, and innate and adaptive immune systems. This combination of features makes the zebrafish an excellent model for studying development, human disease and for high-throughput drug studies. The zebrafish life cycle and housing requirements tend to make experiments conducted in this system faster and cheaper than contemporary mammalian animals, making the zebrafish an excellent bridge model system between invertebrates and mammalian systems.
Advancements in genetic manipulation technologies have helped the zebrafish to approach its full potential as a vertebrate development and disease model system. The first steps were transient over-and under-expression experiments. Simple overexpression experiments involved adding exogenous DNA and messenger RNA through microinjection (1). Next, the application of morpholino antisense technology enabled targeted, transient decrease in gene expression, often called a ‘knockdown’, of most genes during embryonic development (2). Random chemical and retroviral mutagenesis followed by DNA analyses were the first well-used methods to screen for mutations in desired loci (TILLING (3); retrovirus (4)). However, true targeted knockout and knock-in experiments, allowing for long-term, heritable genomic changes remained out of reach in this animal system for many years.
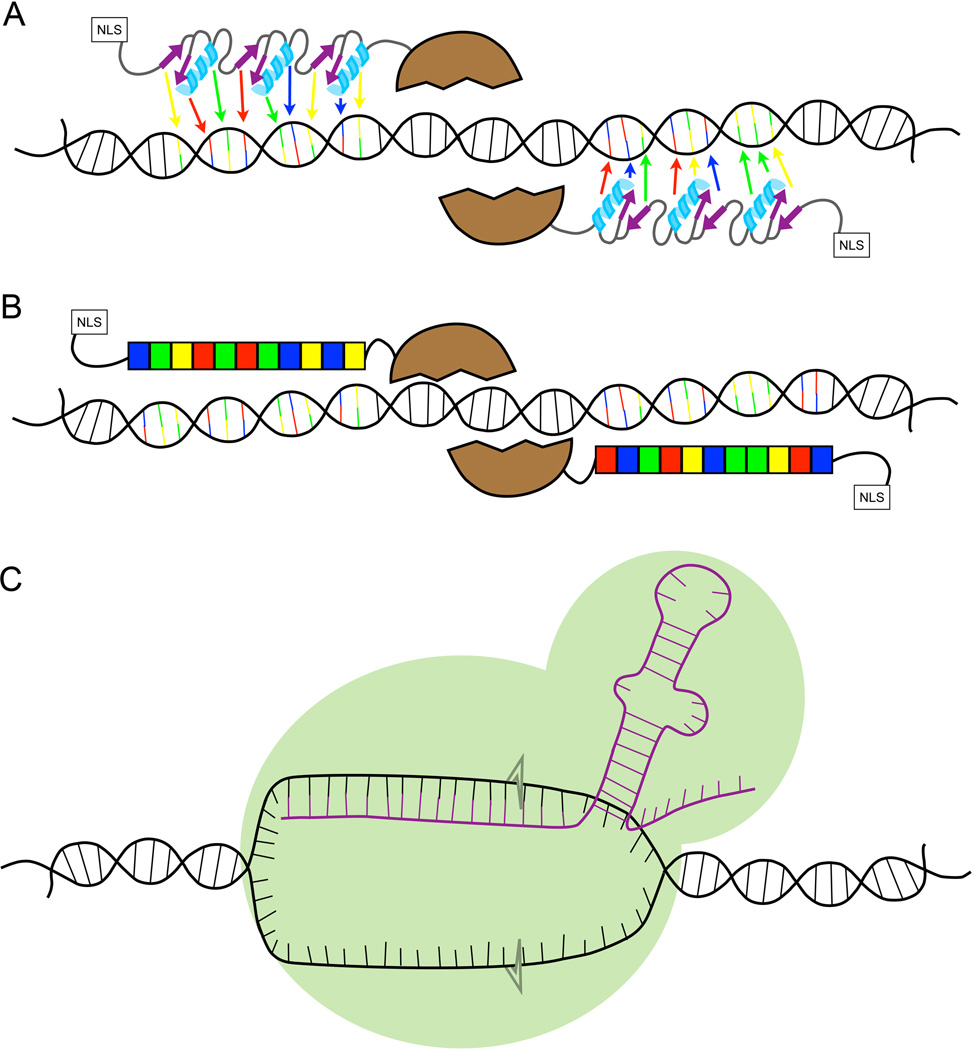
Three major molecular technologies altered the zebrafish genome engineering landscape: zinc finger nucleases (ZFNs; Fig 1A; (5, 6)), transcription activator-like effector nucleases (TALENs; Fig 1B; (7, 8); for review see (9)) and clustered regularly interspaced short palindromic repeats/CRISPR associated system (CRISPR/Cas; Fig 1C; (10, 11). All three systems are active in zebrafish and can be used to create targeted lesions in the genome. ZFNs and TALENs employ custom-designed, locus-specific proteins fused to a sequence-independent nuclease domain to generate targeted mutations, while CRISPRs (Fig 1C) use a different, RNA-guided mechanism.
Figure 1. Diagrams of each of the double strand break technologies.
A) ZFNs are made from two different arms that each typically comprise of three binding proteins recognizing three bases each, depicted by the arrows pointing to the bases to which the individual motifs would bind. Attached to the N-terminus is the nuclease domain from the FokI restriction enzyme. This dimeric protein creates double strand breaks. On the opposite end is the nuclear localization signal (NLS) B) TALENs are made of repeats that have two amino acids in the center required for sequence-specific DNA binding. Each repeat binds a single DNA base, represented by the boxes of the same color as the base to which it binds. C) The CRISPR/Cas9 system requires an RNA (shown in purple) that hybridizes to the unwound DNA. The Cas 9 protein (the green surrounding the RNA) creates the double strand breaks at the DNA/RNA hybrid.
The protein-based ZFN (Fig 1A) and TALEN (Fig 1B) systems use sequence-specific binding motifs to create targeted double-stranded breaks (DSBs; for review see (12)). The core DNA binding motif is attached to a nuclease, usually Fok I, which must homodimerize to catalyze a DSB (13, 14). Therefore, for nuclease homodimerization to occur, ZFNs and TALENs are designed as a pair of nuclease-guiding proteins that bind to both sides of the intended DSB site. The localized dimerization requirement increases nuclease specificity in this approach.
Targeted DSBs facilitate the engineering of site-specific knockouts because changes may occur through cellular repair processes. The most common mechanism that introduces genomic alterations is the non-homologous end joining (NHEJ) pathway, that typically deletes or adds random sequences to the DNA around the DSB. If the custom restriction enzymes target an open reading frame in a gene, this often creates out-of-frame proteins (15, 16).
ZFNs were the first site-specific knockout system whose in vivo use was pioneered in zebrafish (Fig 1A; (17, 18)). Each lab-made ZFN typically consists of three fingers (6). Each ZF motif is ~30 amino acids long and recognizes three bases (Fig 1A; (19)). A single ZFN consists of three motifs recognizing 9 bases. Constructing a highly functional ZFN is a technically complex endeavor because each finger can influence the binding of its neighboring finger. These interactions make it difficult to predict how well an entire ZFN will bind. Therefore, both bacterial (18) and in vitro (20) systems were created to screen multiple zinc fingers to identify which would best bind a desired DNA sequence. This relatively long and technically challenging process has made ZFNs not very accessible for high-throughput knockout projects or for smaller laboratories. Despite their limitations, including often-modest efficacy at introducing germline mutations in zebrafish, ZFNs established the modern paradigm for genome engineering techniques using custom restriction enzymes in living animals, and the more than two decades’ worth of ZFN research has been heavily used as the basis for subsequent custom restriction enzyme work using other systems (21).
Transcription activator-like effector nucleases (TALENs) were subsequently developed just a few years ago and rapidly used for in vivo targeting in zebrafish (Fig 1B; (7, 22)). TALEs were first discovered in the plant pathogen Xanthomonas (23). Each TALE domain consists of a ~30 amino acid repetitive motif with two specific amino acids that bind to each DNA base (Fig 1B; (23, 24)).These repeats are modular and therefore do not influence the binding of the next (24, 25). This feature makes designing TALENs against a particular sequence relatively simple, with very high success (95% or higher) in the latest TALEN designs (for review see (9)). To further increase the utility of the system, Golden Gate modular assembly systems were deployed using a system of unique restriction nucleases to create TALENs in two steps (26). Golden Gate assembly has since been expanded and simplified ((27, 28)) for high-throughput assembly (29), making multi-gene targeted knockout projects considerably more feasible and affordable, even for smaller laboratories. Furthermore, the TALEN system has been optimized for rapid, specific cutting that is efficiently transmitted to the germline (7, 30). To date, TALENs remain the most versatile custom restriction enzyme system capable of targeting nearly any sequence-specific genomic location and with the lowest off-targeting rates in zebrafish (see more, below).
A third site-specific mutagenesis system based on the type II CRISPR/Cas9 system has been very effective for in vivo gene targeting in zebrafish (Fig 1C; (10, 11, 31, 32)). CRISPRs were discovered in prokaryotes as an adaptive defensive system against viruses (33, 34), and their mechanism for creating DSB is different than ZFN and TALENs. The current artificial system uses a single synthetic guide RNA (sgRNA) that, with the help of the Cas9 protein, hybridizes with the DNA sequence of interest through Watson-Crick base pairing. The sgRNA recognition sequence is downstream of a 3 base protospacer adjacent motif (35). The Cas9-RNA-DNA complex creates a double-strand break in the corresponding genomic DNA (Fig 1C). This technology is relatively simple to use, and the target system easier to create than TALENs. One complication of the CRISPR system is its reduced target specificity relative to ZFNs and TALENs.
Using the Cas9 protein, the system will still bind and create DSB despite several mismatched bases. This creates many non-specific mutations within the genome; in some cases, off-target cutting was at a higher rate than on-target nuclease activity (36). Recently, this complication has been addressed using in vitro systems by mutating Cas9 to create a protein that only cuts on a single strand, a version of Cas9 called a nickase (37). This nickase-based Cas9 protein system reduces off-target effects by over 1000x, thanks to the increased specificity from the use of two guide RNAs. In these cell-based systems, the Cas9 nickase-based off-target rates are, however, still higher than what have been noted for TALENs (37), and their efficacy in zebrafish has yet to be described.
In conjunction with these targeted knockout technologies, zebrafish researchers now have the ability to insert exogenous sequences at DSB target sites. This was first accomplished by co-delivering a high-efficiency TALEN with either single-stranded DNA (ssDNA) (30) or double-stranded DNA (dsDNA) (38). The ssDNA serves as a template to induce homology directed repair (HDR), enabling changes designed into the ssDNA template to ‘knock-in’ specific sequences at the target site. To date, small changes (such as a new restriction enzyme sequence), as well as longer sequence tags such as a loxP site have been engineered into the zebrafish genome (30). Using dsDNA, longer sequences, such as enhanced green fluorescent protein has been added to the genome (38). Germline transmission of exogenous sequences through HDR has been demonstrated, though at a lower frequency than germline transmission of NHEJ-created mutations.
The ability to easily create site-specific mutations in vivo and pioneered in zebrafish has opened the world of targeted genome engineering to a large number of cellular and animal model systems (39, 40). What was once a costly enterprise has now become routine. The ability to genome engineer has been particularly transformative for the zebrafish community. Prior to these technologies, only transient knockdown and overexpression experiments were readily accessible, and generating knockouts was a tedious and challenging exercise. Now, zebrafish researchers can create designer mutations using site-specific genome engineering. This opens up the model to be used for more targeted, and human disease-specific, experimentation (41). For example, it is now possible to design a TALEN-ssDNA experiment that specifically changes the zebrafish genome to model known human sequence variations, including those associated with disease and those of unknown significance. These fish can be bred to homozygosity, and the phenotype can be defined in this rich in vivo vertebrate. These engineered animal models can consequently be used to elucidate the mechanism of the human disease mutation. Furthermore, these fish can be subsequently used to screen for drugs that would stabilize or attenuate the phenotype and be used for human drug trials. Therefore, the ability to engineer model vertebrate genomes will enhance our abilities to understand basic developmental and disease mechanisms. Furthermore, the combination of a vertebrate model with strong genome engineering capabilities and easily obtaining large numbers of mutated embryos will enable faster and more efficient drug screening.
While these genome editing technologies themselves are interesting and unique, it is the new, functionally limitless opportunities for hypothesis-driven, human disease-focused experimentation that truly excites our imagination. Therefore, with these new tools, we believe the zebrafish has become a terrific bridge between the high-throughput but invertebrate genetic models such as Drosophila and the nematode, and the classic human disease-relevant models such as the mouse. With the now highly accessible zebrafish genome and ease of the generation of large numbers of animals makes this aquatic species an outstanding vertebrate biology model with increasing impact on human health science for years to come.
The following protocol is a method using TALENs and ssDNA for creating knockout and knockin zebrafish. We used TALENs because of their high efficiency and low off-target rates. In principle, any of the systems described above can be used to create double-strand breaks and be used with ssDNA to create knockin animals. The most difficult and important part of this protocol is designing the TALENs and ssDNA. The implementation is straightforward and uses largely standard tools in the field such as microinjection and PCR-based genotyping.
2. Materials
2.1 Kits used in this protocol
RNAeasy MinElute Cleanup Kit (Ambion).
TALEN Golden Gate kit (Addgene; from Dan Voytas).
pC-GoldyTALEN (Addgene plasmid 38143; from Daniel Carlson).
T3 mMessage Kit (Ambion).
QIAprep Spin Miniprep kit (Qiagen).
PCR purification kit (Qiagen).
Nucleotide removal kit (Qiagen).
2.2 Tricaine (Ethyl 3-aminobenzoate methanesulfonate) at 0.4g/100ml (Sigma)
0.8 g Tricaine, 199 ml of embryo water (or fish water from the zebrafish facility), 1.6 g of sodium bicarbonate to bring the pH to 7.0.
Stir 1–3 together and check pH.
2.3 TAE Buffer
Make 50X stock TAE buffer; 242 g Tris base, 57.1 ml 100% acetic acid, 100 ml 0.5 M sodium EDTA, bring the volume up to 1 liter nanopure water.
Make 1X TAE by diluting 20 ml of 50X TAE with 980 ml of nanopure water.
2.4 5X Cresol Red loading dye (0.4 mg/ml)
Dissolve 4mg of cresol red into 1 ml of nanopure water. Mix until completely dissolved.
Dissolve 60g of sucrose in 80 ml of nanopure water.
Add the 4 mg/ml cresol red with the sucrose water and bring the volume up to 100 ml.
3. A Method for Genome Editing
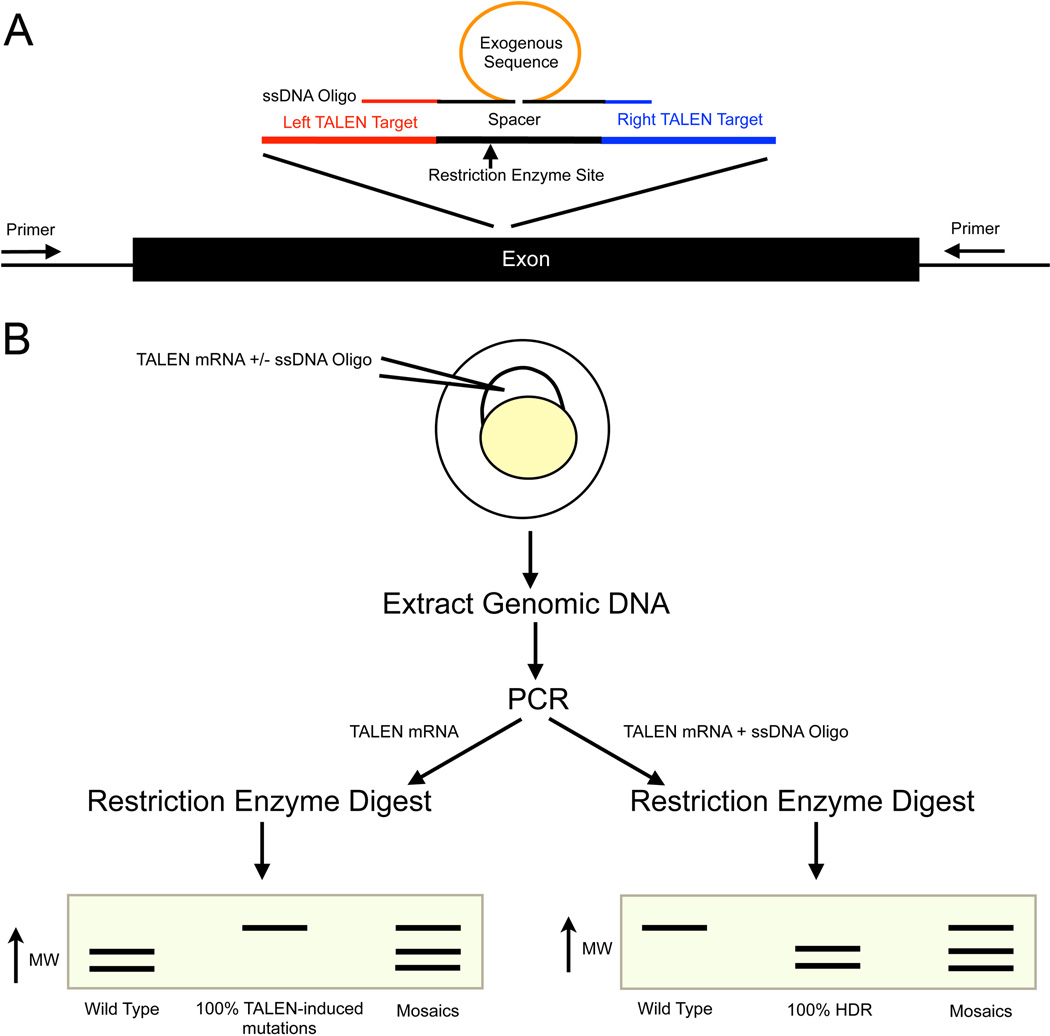
3.1 TALEN Design (Fig 2A)
Figure 2. Experimental design of zebrafish knockin and knockout experiments.
A) A diagram of a genetic locus to be targeted. To create a knockout mutation, TALENs should target a conserved exon early exon. Exon one is often an isoform-specific exon, so it is often better to target a downstream exon. Two TALENs (left and right) bind to the genomic sequence on opposite strands and create a double-strand break. Within the spacer region, we typically design around a local unique restriction enzyme for simplified downstream screening. For knockin experiments, a ssDNA oligo is designed as shown. The homology arms do not normally include the entire TALEN binding site. An exogenous sequence can be added to the center of the spacer region. B) Experimental design for creating knockout or knockin zebrafish. Inject TALEN mRNA to create a knockout or inject TALEN mRNA plus ssDNA oligo for HDR. Extract the genomic DNA from the embryos, perform a PCR and subsequently digest the PCR product. For knockout experiments (injected only with TALEN mRNA), a mutation will result in loss of the restriction enzyme binding site and a larger band on the enzyme analysis on the PCR product. For the knockin experiments such as when adding a new restriction enzyme site to the spacer region, successful HDR results in a digested band and unmodified (wild type) is therefore uncut in the enzymatic assay.
3.2 TALEN Constructs
TALEN assembly of the repeat-variable di-residues (RVD)-containing repeats was conducted using the Golden Gate kit (26). This a five-day restriction digest/ligation-based protocol that can be performed in any molecular biology laboratory.
Confirm appropriate RVD assembly by sequencing.
Clone the RVDs into pC-GoldyTALEN backbone (Addgene) using BsmB1 restriction enzyme.
Transform bacteria, using the standard laboratory procedure, with the completed TALEN.
Using a single colony, grow 1–5 ml of the bacteria containing the TALEN.
Purify the plasmid using the QIAprep spin miniprep kit.
Digest the plasmid using Sac1 endonuclease at 37°C for 2–3 hours. Use at least 5–10 mg of plasmid for restriction enzyme digest.
Purify the digested plasmid using the PCR purification kit.
Use the T3 mMessage Machine kit (Ambion) to create mRNA (see Note 4).
Purify the mRNA using either the phenol/chloroform extraction (T3 mMessage Machine kit user manual protocol) or RNeasy MinElute clean up kit (Ambion) for injection.
3.3 ssDNA Oligonucleotide (Oligo) Design for Genome Editing
Design the oligo to target the spacer sequence between the TALEN cut sites (see Note 3 and 5).
Add desired insertion sequences to the center of the oligo (see Note 6, 7 and 8).
The oligos were ordered from a company (such as Integrated DNA Technologies).
Purify the oligos using the Nucleotide Removal Kit (Qiagen) (see Note 9).
Use RNAse-free water to remove the oligo from the column.
3.4 Creating knockout or knockin zebrafish embryos (Fig 2B)
Dilute TALEN pair mRNA and/or ssDNA oligo to multiple different concentrations using RNAse-free water. Inject the embryos (1, 42) and create a lethal dose 50 (LD50) curve (see Note 11 and 12).
Choose the highest concentration at which less than 50% of the injected embryos are dead.
Inject TALEN mRNA alone for knockout experiments using the concentration determined above.
Inject TALEN mRNA + ssDNA oligo for knockin experiments (see Note 13 and 14). Any experiments with greater than 50% dead should be disregarded.
Isolate genomic DNA to assess for somatic knockout or knockin from 2–5 day-old injected embryos (see Method 3.5).
Raise the remainder of the injected embryos for germline screening. These fish can be raised, mated and the resulting embryos can be screened for germline mutation (see Method 3.7; Fig 2B).
3.5 Individual embryo genomic DNA isolation (see Note 15)
Place a single zebrafish larvae (2–5 days old) into 0.2 ml strip tubes using a defined amount of water, we used 10 microliters.
Add 10 microliters of 100 mM NaOH, for a final concentration of 50mM NaOH.
Vortex.
Incubate at 95°C for approximately 10 minutes or until the embryos have dissolved (see Note 16).
Cool to 4°C in the −20 degrees centigrade freezer.
Add 1/10 volume 1M Tris-HCL pH 8.0 to neutralize the solution.
Centrifuge to pellet the debris and use the supernatant for PCR (see Note 17).
3.6 Isolate genomic DNA from fin clips of zebrafish at least 2 months old
Anesthetize the fish using 200 µg/ml of Tricaine diluted in water from the fish room.
Trim the most caudal 2–3 mm of fin using a fresh razor blade for each fish to prevent contamination.
Place the tissue in a 0.2 ml tube, which has been placed on ice, until all fin biopsies were collected.
Add 150 µl of 50mM NaOH to the fin clips.
Incubate at 95°C for approximately 10 minutes or until the tissue has dissolved (see Note 16).
Cool to 4°C in the −20 degrees centigrade freezer.
Adding 1/10 volume 1M Tris-HCL pH 8.0 to neutralize the solution.
Centrifuge to pellet the debris and use the supernatant for PCR.
3.7 Screening for somatic and germline knockout zebrafish (see Note 18)
Use the same protocol for screening both somatic and germline mutations.
Design primers to the genomic sequence so that the PCR product is visible when digested with the restriction enzyme (see Note 19 and 3).
Use 1–2 microliters of genomic DNA (see Method 3.5 or Method 3.6) for the PCR (see Note 17).
Digest 1–5 microliters of the PCR, depending upon the PCR efficiency, using the unique restriction enzyme (see Note 20).
Run digested production on an agarose gel. The percentage agarose gel depends upon the size of the digested product (see Method 3.10; see Note 21).
If the TALENs were active in creating mutations, the restriction enzyme site should be lost and the PCR product should not be digested (Fig 2B).
Positive injections should be raised for germline screening (see Method 3.9; see Note 22).
3.8 Screening for somatic and germline knockin zebrafish
Screening for sequence knockins depends upon the sequence that is added. In this example a new restriction enzyme is added to the locus (see Note 23).
Design primers to the genomic sequence so that the PCR product is visible when digested with the restriction enzyme (see Note 15 and 3).
Use 1–2 microliters of genomic DNA (see Method 3.5 or Method 3.6) for the PCR.
Digest 1–5 microliters of the PCR, depending upon the PCR efficiency, using the unique restriction enzyme (see Note 20).
Run digested production on an agarose gel. The percentage agarose gel depends upon the size of the digested product (see Method 3.10; see Note 21).
If there is HDR the restriction enzyme site should digest the sample creating two unique bands. The wild type sequence will not digest and a single, uncut band is seen (Fig 2B).
Positive injections should be raised for germline screening (see Method 3.9).
To increase the yield of germline screening, the F0-injected fish can be screened at 2 months of age for somatic insertion of the sequence (see Method 3.6; see Note 24).
3.9 Germline Screening
3.10 1% Agarose Gel
Measure 1g of agarose (increase or decrease the amount of agarose depending on the percentage desired) and pour the powder into a flask
Add 100mL of 1x TAE.
Microwave the solution until the agarose is dissolved. Stop every 30 seconds to 1 minute and swirl the solution.
Let agarose solution cool down until it is around 60 degrees Celsius.
Add 2–3 µl of ethidium bromide (10mg/ml) (see Note 27).
Pour the agarose into a gel tray with the well comb in place.
Let the gel sit at room temperature until it is solid.
Fill the gel box with 1X TAE until the gel is completely covered
Remove the comb.
Add 5X cresol red loading dye to each samples so that the final concentration is 1x cresol red and load the samples into the well. A ladder should be placed either first or last well for a size comparison.
Run the gel at 80–150V until the dye line is over 2/3 of the way down the gel.
Visualize the gel using a UV lightbox and filter (see Note 28).
Acknowledgements
Grants: State of Minnesota grant H001274506; NIH GM63904; NIH grant P30DK084567; NIH grant DK083219; Mayo Foundation.
Footnotes
It is possible to screen for mutations without the restriction enzyme through use of random sequencing or through point-mutation screening methods not discussed here.
To simplify the TALEN design process, use an open access software such as Mojo Hand (www.talendesign.org; (43)).
The zebrafish genome contains a great deal of sequence diversity within standard laboratory strains, especially within non-exonic regions. Genomic sequences of fish within even an individual laboratory’s facility frequently differ from the published genomic sequence. TALENs and primers designed against a published genomic sequence will not bind if an investigator’s sequence of interest in the experimental animals differs from the consensus. Confirming the targeted genomic sequence in advance and in the selected specific zebrafish strain can avoid false negative TALEN efficacy due to natural polymorphisms.
The enzyme mix was modified from the mMessage kit by using 1.5 µliters of enzyme mix added to 0.5 µliters of RNAse inhibitor.
The tails of the oligo are usually relatively short and do not normally contain the whole TALEN binding site. To date, any sequences tested that contained the whole TALEN binding site has not resulted in HDR.
To date, only small insertion sequences have been published, such as EcoRV and LoxP sites.
For single base-pair changes, it is possible to use the wobble bases around the desired change to create a restriction enzyme site while leaving the protein sequence unchanged.
ssDNA oligos are regularly less than 100. Larger sizes have been shown to have markedly reduced efficiency due to an unknown mechanism.
Oligos must be purified prior to injection into the zebrafish. The standard oligo purification leaves chemicals that kill the embryos.
(42) is an excellent review for injection set up, calibration and loading embryos.
Usually, the TALENs concentration ranged from 25 pg to 400 pg.
The ssDNA oligo ranged from 25 pg to 100 pg.
Do not inject more than 9 nl total of pure water into the zebrafish embryo. Greater than that causes developmental problems.
Inject the TALEN mRNA is injected separately from the ssDNA oligo to ensure that the mRNA is not degraded.
DNA can also be isolated from pools of 10 larveal zebrafish using the the DNAeasy Blood and Tissue kit (Qiagen).
Using a vortexing incubator increases the DNA yield and ensures the entire embryo is dissolved.
When using the supernatant, make sure no cellular debris is pipetted because it can inhibit the PCR reaction.
How screening is conducted for knockout efficacy depends upon the TALEN site chosen. If the spacer region contained a unique restriction enzyme site within the nearby genomic locus, restriction enzyme analysis on a locus-specific PCR product can be easily used to screen for mutations. www.talendesign.org (43) is the support software we use to help with TALEN design.
It is ideal that the restriction enzyme sequence is asymmetrically located within the PCR product so two bands can be visualized after digest.
Most restriction enzymes do not require purification of the PCR product to allow for digestion. However, it is best to test your enzyme in the PCR buffer. Also, have a positive control to ensure the enzyme is active.
Most sequences can be resolved using a 1–2% agarose gel. However, this depends upon the size of the bands needing to be resolved using the specified genotyping assay.
Screening embryos somatic mutations is ideal. This ensures that the injection was successful. However, given the efficiency of the GoldyTALENs, it is feasible to skip the somatic screening step and raise the injections.
If larger sequences are added, for example a LoxP site, a PCR primer can be made to bind the LoxP site. This can, then, be used to screen for the insertion of the sequence. However, this screening method is significantly more sensitive than PCR and restriction enzyme digestion.
If stable somatic HDR activity is detected, there can be an increased likelihood that the insertion is also in the germline, and these fish can be prioritized for subsequent analyses. Through fin clip screening, the number of fish that need to be mated and screened for germline mutations is significantly decreased.
Genomic DNA isolation for germline screening can be done either as individual embryos or as groups of 10 embryos.
For knockout GoldyTALEN injections, the efficiency is high enough that screening 10–20 embryos is sufficient. However, knockin experiments are significantly less efficient. Therefore, screening up to 100 embryos increases the chances of finding the germline mutation.
Ethidium bromide binds to DNA, therefore, it is a carcinogen. Gloves should be worn whenever it is used.
There are many gel imaging systems. Currently, the system used in the laboratory is a Fotodyne gel imaging system.
References
- 1.Hyatt TM, Ekker SC. Vectors and techniques for ectopic gene expression in zebrafish. Methods Cell Biol. 1999;59:117–126. doi: 10.1016/s0091-679x(08)61823-3. [DOI] [PubMed] [Google Scholar]
- 2.Nasevicius A, Ekker SC. Effective targeted gene "knockdown" in zebrafish. Nature Genetics. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- 3.Sood R, English MA, Jones M, et al. Methods for reverse genetic screening in zebrafish by resequencing and TILLING. Methods. 2006;39:220–227. doi: 10.1016/j.ymeth.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 4.Amsterdam A, Hopkins N. Retrovirus-mediated insertional mutagenesis in zebrafish. Methods Cell Biol. 1999;60:87–98. doi: 10.1016/s0091-679x(08)61895-6. [DOI] [PubMed] [Google Scholar]
- 5.Zhu C, Smith T, McNulty J, et al. Evaluation and application of modularly assembled zinc-finger nucleases in zebrafish. Development. 2011;138:4555–4564. doi: 10.1242/dev.066779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Urnov FD, Rebar EJ, Holmes MC, et al. Genome editing with engineered zinc finger nucleases. Nature Reviews Genetics. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 7.Huang P, Xiao A, Zhou M, et al. Heritable gene targeting in zebrafish using customized TALENs. Nature Biotechnology. 2011;29:699–700. doi: 10.1038/nbt.1939. [DOI] [PubMed] [Google Scholar]
- 8.Sander JD, Cade L, Khayter C, et al. Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nature Biotechnology. 2011;29:697–698. doi: 10.1038/nbt.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campbell JM, Hartjes KA, Nelson TJ, et al. New and TALENted genome engineering toolbox. Circulation research. 2013;113:571–587. doi: 10.1161/CIRCRESAHA.113.301765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hwang WY, Fu Y, Reyon D, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nature Biotechnology. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blackburn PR, Campbell JM, Clark KJ, et al. The CRISPR system--keeping zebrafish gene targeting fresh. Zebrafish. 2013;10:116–118. doi: 10.1089/zeb.2013.9999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaj T, Gersbach CA, Barbas CF., III ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends in Biotechnology. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith J, Bibikova M, Whitby FG, et al. Requirements for double-strand cleavage by chimeric restriction enzymes with zinc finger DNA-recognition domains. Nucleic Acids Research. 2000;28:3361–3369. doi: 10.1093/nar/28.17.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vanamee ES, Santagata S, Aggarwal AK. FokI requires two specific DNA sites for cleavage. Journal of Molecular Biology. 2001;309:69–78. doi: 10.1006/jmbi.2001.4635. [DOI] [PubMed] [Google Scholar]
- 15.Porteus MH, Carroll D. Gene targeting using zinc finger nucleases. Nature Biotechnology. 2005;23:967–973. doi: 10.1038/nbt1125. [DOI] [PubMed] [Google Scholar]
- 16.Wyman C, Kanaar R. DNA double-strand break repair: all's well that ends well. Annu Rev Genet. 2006;40:363–383. doi: 10.1146/annurev.genet.40.110405.090451. [DOI] [PubMed] [Google Scholar]
- 17.Doyon Y, McCammon JM, Miller JC, et al. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nature Biotechnology. 2008;26:702–708. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meng X, Noyes MB, Zhu LJ, et al. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nature Biotechnology. 2008 doi: 10.1038/nbt1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Segal DJ, Crotty JW, Bhakta MS, et al. Structure of Aart, a designed six-finger zinc finger peptide, bound to DNA. Journal of Molecular Biology. 2006;363:405–421. doi: 10.1016/j.jmb.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 20.Foley JE, Yeh JR, Maeder ML, et al. Rapid mutation of endogenous zebrafish genes using zinc finger nucleases made by Oligomerized Pool ENgineering (OPEN) PLoS ONE. 2009;4:e4348. doi: 10.1371/journal.pone.0004348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lawson ND, Wolfe SA. Forward and reverse genetic approaches for the analysis of vertebrate development in the zebrafish. Dev Cell. 2011;21:48–64. doi: 10.1016/j.devcel.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 22.Liu Y, Luo D, Zhao H, et al. Inheritable and precise large genomic deletions of non-coding RNA genes in zebrafish using TALENs. PLoS ONE. 2013;8:e76387. doi: 10.1371/journal.pone.0076387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boch J, Scholze H, Schornack S, et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 24.Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- 25.Mahfouz MM, Li L, Shamimuzzaman M, et al. De novo-engineered transcription activator-like effector (TALE) hybrid nuclease with novel DNA binding specificity creates double-strand breaks. Proc Natl Acad Sci U S A. 2011;108:2623–2628. doi: 10.1073/pnas.1019533108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cermak T, Doyle EL, Christian M, et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Research. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liang J, Chao R, Abil Z, et al. FairyTALE: A High-Throughput TAL Effector Synthesis Platform. ACS synthetic biology. 2013 doi: 10.1021/sb400109p. [DOI] [PubMed] [Google Scholar]
- 28.Ma AC, Lee HB, Clark KJ, et al. High efficiency In Vivo genome engineering with a simplified 15-RVD GoldyTALEN design. PLoS ONE. 2013;8:e65259. doi: 10.1371/journal.pone.0065259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Z, Zhang S, Huang X, et al. Rapid Assembly of Customized TALENs into Multiple Delivery Systems. PLoS ONE. 2013;8:e80281. doi: 10.1371/journal.pone.0080281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bedell VM, Wang Y, Campbell JM, et al. In vivo genome editing using a high-efficiency TALEN system. Nature. 2012;491:114–118. doi: 10.1038/nature11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang N, Sun C, Gao L, et al. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell research. 2013;23:465–472. doi: 10.1038/cr.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jao L-E, Wente SR, Chen W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc Natl Acad Sci U S A. 2013;110:13904–13909. doi: 10.1073/pnas.1308335110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barrangou R, Fremaux C, Deveau H, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 34.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 35.Jinek M, Chylinski K, Fonfara I, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cho SW, Kim S, Kim Y, et al. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2014;24:132–141. doi: 10.1101/gr.162339.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ran FA, Hsu PD, Lin C-Y, et al. Double Nicking by RNA-Guided CRISPR Cas9 for Enhanced Genome Editing Specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zu Y, Tong X, Wang Z, et al. TALEN-mediated precise genome modification by homologous recombination in zebrafish. Nature Methods. 2013;10:329–331. doi: 10.1038/nmeth.2374. [DOI] [PubMed] [Google Scholar]
- 39.Carlson DF, Tan W, Hackett PB, et al. Editing livestock genomes with site-specific nucleases. Reproduction, Fertility and Development. 2013;26:74–82. doi: 10.1071/RD13260. [DOI] [PubMed] [Google Scholar]
- 40.Xu L, Zhao P, Mariano A, et al. Targeted Myostatin Gene Editing in Multiple Mammalian Species Directed by a Single Pair of TALE Nucleases, Molecular therapy. Nucleic acids. 2013;2:e112. doi: 10.1038/mtna.2013.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmid B, Haass C. Genomic editing opens new avenues for zebrafish as a model for neurodegeneration. Journal of neurochemistry. 2013;127:461–470. doi: 10.1111/jnc.12460. [DOI] [PubMed] [Google Scholar]
- 42.Bill BR, Petzold AM, Clark KJ, et al. A primer for morpholino use in zebrafish. Zebrafish. 2009;6:69–77. doi: 10.1089/zeb.2008.0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neff KL, Argue DP, Ma AC, et al. Mojo Hand, a TALEN design tool for genome editing applications. BMC Bioinformatics. 2013;14:1. doi: 10.1186/1471-2105-14-1. [DOI] [PMC free article] [PubMed] [Google Scholar]