Abstract
Understanding the effects of land-use change on zoonotic disease risk is a pressing global health concern. Here, we compare prevalence of Yersinia pestis, the etiologic agent of plague, in rodents across two land-use types—agricultural and conserved—in northern Tanzania. Estimated abundance of seropositive rodents nearly doubled in agricultural sites compared with conserved sites. This relationship between land-use type and abundance of seropositive rodents is likely mediated by changes in rodent and flea community composition, particularly via an increase in the abundance of the commensal species, Mastomys natalensis, in agricultural habitats. There was mixed support for rodent species diversity negatively impacting Y. pestis seroprevalence. Together, these results suggest that land-use change could affect the risk of local transmission of plague, and raise critical questions about transmission dynamics at the interface of conserved and agricultural habitats. These findings emphasize the importance of understanding disease ecology in the context of rapidly proceeding landscape change.
Introduction
Zoonotic pathogens, disease agents transmitted to humans from nonhuman animals, such as the Lyme disease spirochete (Borrelia burgdorferi) and the anthrax bacterium (Bacillus anthracis), are significant burdens on global public health.1 In particular, bacterial zoonoses originating from wildlife constitute a growing problem, and their emergence and transmission into human populations are often correlated with poverty and rapid rates of environmental and ecological change.1,2 Geographic hotspots of emerging and reemerging infectious diseases (EIDs) have been identified at a global scale, and are frequently located in the tropics and associated with high mammal biodiversity.1,3 East Africa is one such identified EID hotspot, and harbors several emerging or re-emerging vector-borne pathogens, for example, Rift Valley fever virus, Chikungunya virus, and the plague bacterium, Yersinia pestis.4–6 The region is also experiencing high rates of land-use change, making it a crucial and informative area to examine the effects of land-use change on zoonotic disease ecology.7–9
At a more local scale, relationships between land-use change and disease ecology have been suggested for a wide array of host–pathogen systems.10–14 The “dilution effect” hypothesis, for example, suggests that the net effect of higher species diversity will be a reduction in disease risk at local scales15; therefore, conserved areas, which often host higher levels of biodiversity, may have reduced risk for human disease. The mechanisms accounting for such an effect include increases in relative abundance of high competence (amplifying hosts) and increases in total abundance of potentially susceptible hosts (susceptible host regulation).15,16 However, relationships between biodiversity and disease risk are not uniform and appear to depend on the ecological context, the ecology of the host–pathogens involved,17–20 and the metric of disease risk used.21,22 Understanding these relationships is important; if we can understand and predict changes in host and vector species assemblages as a function of land use, and the consequent risks of rodent-borne zoonotic diseases, then we can improve our ability to enact zoonotic disease control and possibly synergize both public health and conservation initiatives.
Y. pestis, the bacterium that causes plague, is an example of a vector-borne organism that constitutes an important public health concern in many parts of Africa.4,23,24 Research on the eco-epidemiology of plague in East Africa has given relatively little direct attention to the role of land-use change in disease transmission, and has focused instead on the roles of factors such as climate, context, large mammal presence, and host abundance.1,25–32
One of the most common types of landscape change underway in East Africa, and elsewhere in the global tropics, is the conversion of wildlands to agricultural uses.33–36 Cultivation in a variety of regions of East Africa has expanded by more than 70% in the last several decades.37 Given that rodent abundance and community composition are known to be strongly impacted by land use,16 we hypothesized that conversion of land to agriculture may affect landscape-level disease dynamics. To examine how this particular form of landscape change may influence the ecology of plague, we compared rodent communities and Y. pestis activity in conserved landscapes and in nearby converted agricultural fields, using a paired sampling design. Sampling was carried out in a region of northern Tanzania where plague is endemic and is a source of public health concern.38 We compare metrics of rodent community assembly (i.e., abundance, richness, and diversity) and plague ecological risk (i.e., the presence/absence of plague activity and calculated abundance of seropositive rodents) in these two landscape types. Results from this work should contribute to our understanding of how landscape change influences wildlife communities and zoonotic disease risk on the local scale and can inform relevant intervention actions.
Methods
Sampling locations.
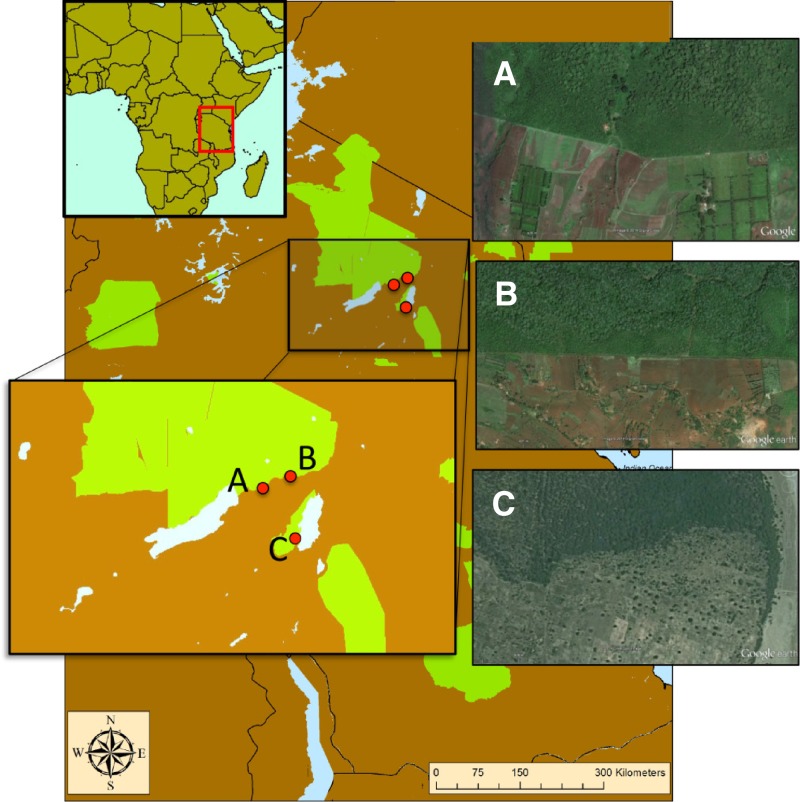
Rodent trapping and pathogen sampling were carried out in three regions of north central Tanzania: the Tloma village vicinity (03°17′S, 35°40′E); the Kambi ya Nyoka village vicinity (03°18′S, 35°36′E); and the southern Manyara region (03°42′S, 35°43′E) (Figure 1). The sampling sites in the Tloma and Kambi ya Nyoka regions are situated at approximately 1,700-m elevation and located within and along the border of the Ngorongoro Conservation Area. The Manyara sites are situated at approximately 1,000-m elevation and located within and along the border of Lake Manyara National Park. This region of Tanzania is generally characterized as having a bimodal pattern of rainfall with peaks generally occurring in March–May and November–December.39 Field sampling was carried out in this vicinity during June and July 2011, the onset of the dry season, shortly after the typical annual peak in human plague cases.23
Figure 1.
A map of the three site pairs in northern Tanzania at different scales, with nationally conserved areas designated in green. Insets of satellite images (from Google Earth) show that stark differences in land use occur directly outside the boundaries of these protected areas. These borders separate conserved and agricultural treatments used in this study at each of the three field sites: Kambi ya Nyoka (A), Tloma (B), and Manyara (C).
We sampled rodents, as well as their fleas and pathogens, at six sites for this study. All sites were placed in a matched-pair design (across three site pairs, one in each of the three regions). Each site pair included two land-use treatment types: one site with “agricultural” use and one site with “conserved” use. Agricultural sites were actively under cultivation and in private ownership; they had little or no evidence of large wildlife presence (e.g., via dung or track). The dominant crop in agricultural sites was maize, but chickpeas, vegetables, tobacco, and millet were typically interspersed at lower densities between maize plantings. Conserved sites were at least 500 m inside the boundaries of conservation areas (Ngorongoro Conservation Area or Lake Manyara National Park) in which ecosystems were fully protected and typically consisted of closed-canopy evergreen forest. Tracks and signs of large wildlife species were frequently observed in conserved sites. Agricultural sites were presumed to be formerly identical or similar to conserved sites prior to land conversion.
Rodent sampling.
At each site, we placed two sampling grids (< 1 km from each other) to encompass a wider range of environmental and agricultural (e.g., time of harvest, crops planted) variability. On each grid we placed 100 Sherman traps spaced 10 m apart. The three site pairs were separated from one another by approximately 15–50 km. Matched agricultural or conserved treatment sites within a pair were physically separated from one another by fences, fire trails, and patrolled/demarcated park boundaries. Agriculture/conserved site pairs were exposed to the same general climatic conditions and soil types. Sites within a given pair were sampled simultaneously. These a priori classifications of habitat type were confirmed by vegetation surveys at each site (see “Habitat descriptions”).
Rodent traps were baited with a mix of maize meal, oats, and peanut butter. All traps were set in the late afternoon and checked at dawn. Captured rodents were identified to species in the field, sexed, weighed, marked with ear tags, sampled for blood under full anesthesia (using halothane), and released. After recovering the rodent from the trap, it was restrained over a container of ethanol while a technician collected fleas by passing a fine-toothed flea comb repeatedly over the animal from the base of the ears to the base of the tail. We then collected these fleas from the container. All fleas collected were identified to species morphologically; to confirm identities and eliminate the possibility of cryptic species, a subset of rodents and fleas was also identified using DNA barcoding techniques with records submitted to the Barcode of Life Project and made publicly available.40 One group of morphologically and genetically distinct fleas (Ctenopthalmus sp.) could not be identified firmly to species level as all individuals from this group were either female or were damaged in sampling, making species-level identification impossible. However, based on genetic clustering we included it in diversity analyses as a separate species. For site-level abundance analyses, we used the density of total fleas present on sampled hosts per hectare (one rodent sampling grid; species pooled) as a source of further information on plague transmission risk.25
At the conclusion of trapping at a given site, a subset of rodents was collected as voucher specimens (deposited both at the Sokoine University of Agriculture and the Smithsonian's National Museum of Natural History). Identities of voucher specimens were confirmed via cranial morphology; DNA barcodes of each species were then submitted to the Barcode of Life Project. For rodents, where species-level identification was difficult to determine on live animals, the identity of all individuals in question that were not lethally sampled was subsequently determined using a DNA barcoding approach. DNA taken from blood samples of these individuals was compared with reference DNA barcodes. To further ensure that cryptic species were not missed in the field, blood spots from a random subset of the animals not killed for voucher specimens (∼10% of total catch per species) were used to confirm species identity using DNA barcodes. Our rodent identifications follow the taxonomy of Musser and Carleton,41 except where updated by more recent research.42,43
Each site was trapped for three sequential nights (which previous work has shown captures the great majority of these species44), yielding approximately 3,600 trap nights (12 grids × 100 traps × 3 nights per trap). Because of low capture and recapture rates at some sites, we assessed abundance of rodents per site using the minimum number of rodents known alive, based on the number of unique individual rodents captured per site (i.e., rather than via mark recapture estimates). We reported and analyzed patterns of abundance between land-use types for all rodents (species pooled). Rodent diversity at each trapping site was calculated using both Shannon and Simpson diversity indices as well as species richness (Supplemental Table 1). All rodent trapping and sampling was conducted in accordance with institutional animal care and use permits (Smithsonian Institution IACUC permit # 2009-04) and the guidelines of the American Society of Mammalogists for the use of wild mammals in research.45
Habitat descriptions.
To describe the habitat quantitatively at each of our sampling areas, we overlaid a vegetation sampling grid on each of our 12 rodent trapping grids, with 20 × 20 m spacing between each point. At each point we placed 5 sample pins, with 1 m spacing between each pin (thus, there were 250 pin drops per site). At each pin drop we identified the number of plants that touched the 50-cm pin and the height at which they touched.46 We recorded if the plant was an agricultural species (identifying crop type), a grass, or a forb. To describe tree and shrub cover, we recorded presence or absence of woody vegetation above 50 cm in height at each pin drop point.
Detection of Y. pestis antibodies.
Y. pestis exposure was measured using an enzyme-linked immunosorbent assay (ELISA).47,48 Blood samples collected from field-sampled rodents were immediately separated using centrifugation. The serum was then screened for the presence of antibodies against the specific antigen (fraction 1 or F1 antigen) of Y. pestis using methods previously described.49 In brief, microtitre plates were coated with Y. pestis fraction 1 capsular antigen (F1) before adding test serum samples. In this test, serum samples containing antibodies against F1 antigen (positive serum) (Centers for Disease Control and Prevention, Fort Collins, CO) blocks the F1 antigen from reacting with the enzyme labeled rabbit anti-F1 antibody (second antibody), which is not species specific, hence enabling detection of plague antibodies in wild and domestic animals and humans. In addition to positive and negative rabbit serum, positive control serum from rodents, dogs, and humans were also used. The ELISA titers in these samples (N = 106) ranged from 1:16 to 1:256.
Assessing plague activity.
Given sampling constraints (e.g., some individuals were too small to provide the blood volume needed for ELISA analyses), only a subset of the rodents captured was tested for plague (Supplemental Table 2). All species were surveyed at each land-use type where they occurred. To best assess plague activity at each site, we adapted the host community model of LoGiudice and others50 to calculate the expected density of seropositive animals at a site. The advantage of using this approach is that it can make inferences that incorporate variation in host seroconversion, without sampling all individuals at all sites, and has been broadly applied in studies of the ecology of zoonotic disease.21,50,51 In congruence with this method, the predicted number of seropositive rodents for each species (plague activity index [PAI]) was calculated by multiplying each species' abundance by its species-specific Y. pestis seroprevalence in each land-use type (i.e., conserved versus agricultural) and summing the values for all species at each site.
Statistical analysis.
To accurately calculate species richness and diversity given uneven capture success across sites, we used rarefaction curves, calculated using individual-based abundance estimates, with the analyses conducted in EstimateS 9.0. We used the Chao1 indicator for species richness, and both Simpson and Shannon metrics of diversity. For all response variables (rodent abundance, diversity, and richness; flea abundance; PAI), we used a paired t test to compare differences among land use types with site pairs as the unit of replication. We also used a linear regression approach to test for correlations among response variables, here with site rather than site pair as the unit of replication. Comparison of community composition of fleas across treatments was also done with site rather than site pair as unit of replication. Statistics were run (except as indicated above) in R.52 Results reported are mean ± SD.
Results
Habitat descriptions.
A priori descriptions of habitats as agriculture or conserved forest were clearly supported by vegetation data, which varied strongly between sites. In agricultural sites, 50.0 ± 15.1% of all vegetation sampled consisted of agricultural crop species (the remainder was largely understory grasses and forbs growing between crops and crop rows). Agricultural plants were absent (< 0.01% plant cover) from conserved plots. Woody cover, largely absent in agricultural plots (4.9 ± 4.1%), was high (80.0 ± 13%) in conserved plots. Similarly, understory stratification (defined as the mean number of contacts of vegetation per pin) was also higher in conserved sites (3.7 ± 1.1) than in agricultural sites (2.2 ± 0.8).
Rodent abundance and diversity.
A total of 240 rodents was captured, representing nine rodent species (Supplemental Table 1). The two Mus spp. < 12 g in mass (members of the cryptic Mus minutoides-musculoides taxonomic complex41) were grouped together in all analyses given the difficulties with field identification and subsequent difficulties in DNA quality in genetic sequencing.
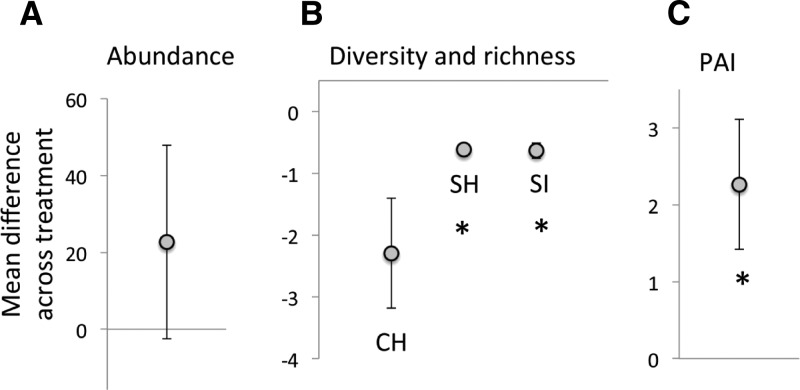
Rodent abundance (number of unique individuals; species pooled) tended to be higher in agricultural sites (156 agricultural animals versus 84 conserved animals), but the difference was not significant (t = 2.26, df = 2, P = 0.15) because of large variation among sites (Figure 2A). Both Shannon and Simpson measures of rodent diversity were significantly higher in conserved sites (Shannon t = 18.8, df = 2, P = 0.003; Simpson t = 11.0, df = 2, P = 0.008) (Figure 2B) than in paired agricultural plots. Rodent species richness was always greater in conserved sites (Supplemental Table 1), but this difference was only marginally significant (t = 2.77, df = 2, P = 0.10) (Figure 2B).
Figure 2.
Mean difference (±SD) between land-use treatments (agricultural vs. conserved) of (A) rodent abundance, (B) rodent community diversity, and (C) plague activity index (PAI); positive values indicate increases in agricultural treatments. For rodent community diversity, the following metrics were used: Chao 1 species richness (CH), Shannon diversity (SH), and Simpson diversity (SI). Asterisk (*) indicates significant differences between agricultural and conserved sites for each response variable. Rodent abundance (A) was consistently but not significantly elevated in agricultural sites as compared with adjacent conserved sites. Diversity trended significantly lower in agricultural sites for all metrics. PAI (predicted plague seropositive rodents) was roughly twice as high in agricultural vs. conserved sites.
The numerically dominant rodent species in this study was the natal multimammate mouse, Mastomys natalensis, which accounted for 56.7% of individual captures in this study. Abundance of M. natalensis was highly variable between sites, but was consistently and significantly higher in agricultural sites (44.0 ± 2.0 individuals/ha) compared with conserved sites (2.3 ± 3.2 individuals/ha; t = 47.2, df = 2, P = 0.0004). Overall, 97% of all M. natalensis captures were from agricultural sites (Supplemental Table 1). Most species in the study showed similar strong treatment preferences. Mus spp. were also much more common in agricultural plots (89% of all captures were in agriculture). All captures of Grammomys dolichurus, Graphiurus murinus, Lophuromys makundii, and Praomys taitae were in conserved plots. Of species captured more than five times, only Lemniscomys striatus was found relatively equally in both habitats (55% captures in conserved, 45% in agricultural habitats).
A total of nine flea species from five genera of rodents surveyed were identified (Table 1; host–flea associations shown in Supplemental Table 3). Prevalence of flea infestation was moderate and not significantly different across site types, with 17% of individuals in agricultural sites and 14% individuals in conserved sites having fleas. Intensity of infestation also did not differ across site types (Supplemental Table 3). Site-level flea abundance (total number of fleas on host per hectare) was consistently higher in agricultural (17.0 ± 5.7 individuals/ha) as compared with conserved (5.7 ± 9.8 individuals/ha) sites, with this pattern approaching but not achieving significance (t = 3.6, df = 2, P = 0.07). In contrast, flea diversity (estimated via Shannon index) was consistently higher in conserved (2.0 ± 0.2) versus agricultural sites (1.2 ± 0.2), although this difference did not meet significance (t = 3.9, df = 2, P = 0.06). Community composition was, however significantly different across treatments (ANOSIM R = 0.48, P = 0.03). Notably, the most common flea, Dinopsyllus lypusus (33% of fleas identified), was only found in agricultural habitats. Xenopsylla humilis was also found only in agricultural habitats, but this species was represented by only a single individual flea. There were also four flea species that were found only in conserved habitats (C. evidens, Ctenopthalmus sp., D. longifrons, and Leptopsylla aethiopica) although Leptopsylla aethiopica was also represented by only a single individual flea.
Table 1.
Flea species by habitat type (as percent of all fleas sampled). Host–flea associations are provided in Supplemental Table 3
| Species | n | Agriculture (%) | Conserved (%) |
|---|---|---|---|
| Ctenophthalmus calceatus cabirus (Jordan and Rothschild, 1913) | 7 | 6 | 8 |
| Ctenophthalmus evidens (Jordan, 1929) | 7 | 0 | 13 |
| Ctenophthalmus sp. | 5 | 0 | 10 |
| Dinopsyllus longifrons (Jordan and Rothschild, 1913) | 6 | 0 | 12 |
| Dinopsyllus lypusus (Jordan and Rothschild, 1913) | 17 | 33 | 0 |
| Leptopsylla aethiopica (Rothschild, 1909) | 1 | 0 | 2 |
| Listropsylla basilewskyi (Smit, 1960) | 2 | 2 | 2 |
| Xenopsylla brasiliensis (Baker, 1904) | 6 | 10 | 2 |
| Xenopsylla humilis (Jordan, 1925) | 1 | 2 | 0 |
Y. pestis seroprevalence.
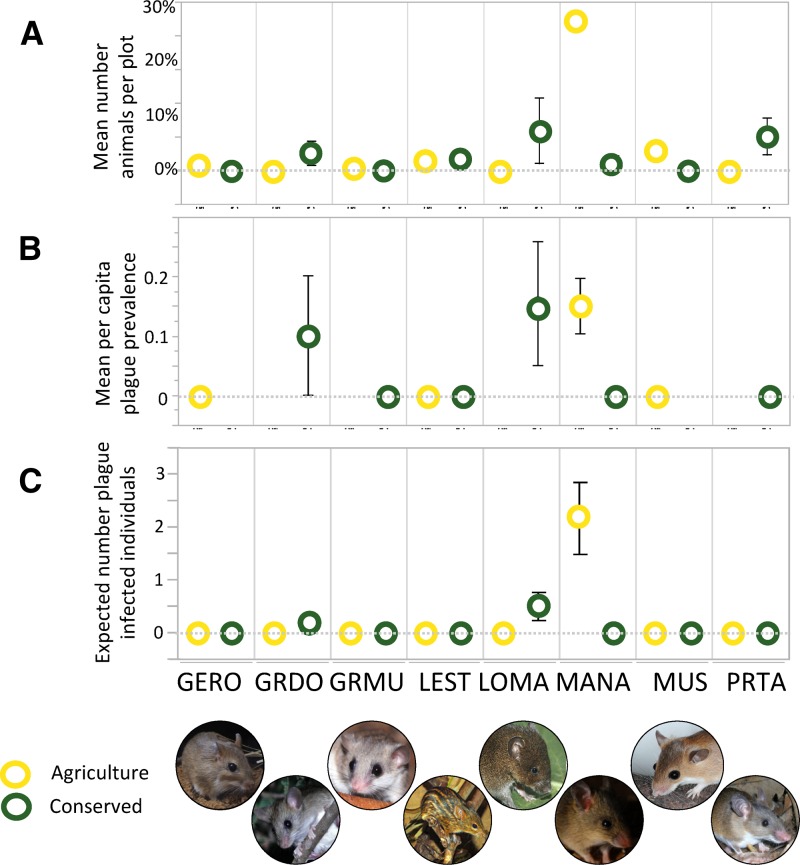
A total of 106 rodents representing eight species (43 in conserved and 63 in agricultural sites) were screened for Y. pestis antibodies (Supplemental Table 1). Three species (G. dolichurus, L. makundii, M. natalensis) tested seropositive for Y. pestis exposure. Mastomys natalensis, the most abundant species sampled and an avid human commensal, accounted for the majority of all seropositive rodents (75%, 9/12) (Supplemental Table 1; Figure 3). Seropositive M. natalensis were only found in agricultural sites, and this was the only species that was seropositive in agricultural sites.
Figure 3.
There were strong differences in the abundance (A) and plague seroprevalence (B) of rodent species and across the two land-use treatment types surveyed: agricultural (yellow) and conserved (green). Combining these two metrics provides an estimate of the likely number of plague seroprevalent rodents per species and treatment (C). Here we plot mean and SD (when appropriate). The eight species monitored in this study were Gerbilliscus robustus (GERO), Grammomys dolichurus (GRDO), Graphiurus murinus (GRMU), Lemnisomys striatus (LEST), Lophuromys makundii (LOMA), Mastomys natalensis (MANA), Mus minutoides and musculoides (MUS), and Praomys taitae (PRTA).
On the basis of the presence/absence of seropositive rodents at the site level only, plague had recently been active at five out of six of our study sites, suggesting that plague is broadly endemic across the land-use mosaic sampled in this study. The predicted abundance of seropositive rodents (PAI) in our two land-use treatments was significantly higher in agricultural sites relative to conserved sites (t = 3.6, df = 2, P = 0.04) with roughly twice as many seropositive animals predicted in agricultural sites (1.08 ± 0.03 infected animals expected per hectare, compared with 0.51 ± 0.15 in conserved sites).
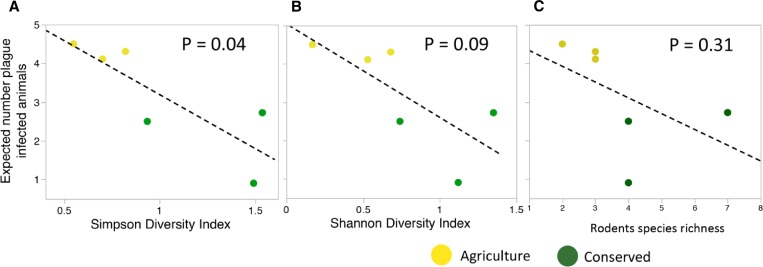
Rodent abundance (total number of individuals; species pooled) was not significantly related to calculated abundance of plague seropositives (F1,4 = 3.7, r2 = 0.48, P = 0.13) (Supplemental Figure 1), although the trend was positive. There was a strong negative relationship between species diversity (as measured using the Simpson diversity index) and PAI at our sites (F1,4 = 8.7, r2 = 0.69, P = 0.04) (Figure 4A). A similar pattern was observed using the Shannon Diversity Index, although this relationship was not quite significant (F1,4 = 4.7, r2 = 0.54, P = 0.09) (Figure 4B). There was no significant relationship between Chao1 estimates of species richness and PAI (F1,4 = 1.3, r2 = 0.24, P = 0.31) (Figure 4C). PAI was also strongly correlated with the relative abundance of the numerically dominant rodent M. natalensis (F1,4 = 7.2, r2 = 0.64, P = 0.05) and with abundance of fleas per hectare (F1,4 = 11.4, r2 = 0.74, P = 0.03).
Figure 4.
A negative slope was observed for the relationship of plague activity index (PAI) and Simpson diversity (A), Shannon diversity (B), and species richness (C), with generally higher diversity and lower PAI in conserved sites. This relationship, however, was only significant for Simpson diversity.
Discussion
In this study, land use was found to have an important influence on rodent communities. Two important measures of diversity (Shannon and Simpson) indicated that rodent communities were more diverse in conserved sites that had not been converted to agriculture (although species richness did not actually change significantly). On the whole, rodent abundance (species pooled) was not different between agricultural and conserved sites. Mastomys natalensis, however, was nearly 20 times more abundant in agricultural sites than conserved sites. These shifts in diversity and species composition are presumably the result of observed differences in vegetation structure and composition associated with land-use conversion. Such patterns are consistent with observations made in other contexts.26,53 Rodent species other than M. natalensis also exhibited strong affinities for particular land-use types. However, all five rodent species captured in agricultural sites were also detected in conserved sites.
Flea community composition was also significantly affected by land use. Agricultural land use may favor species highly competent for transmitting Y. pestis. The two most common fleas observed in agricultural habitats, D. lypusus and X. brasiliensis, are both thought to be important and efficient vectors for plague transmission.54–56 D. lypusus was completely absent from conserved sites and X. brasiliensis was five times more common in agricultural as conserved sites. Both species were specialists on M. natalensis. The only other flea species in this study known to be competent at transmitting plague, Ctenopthalmus calceatus57 was found at similarly low levels in both site types (N = 3 in agricultural sties, N = 4 in pastoral sites). However vector competency of many of the other flea species has not been studied.26 Further research on vector competence of each species, as well as their propensity to bite humans, is necessary to fully evaluate the importance of the observed changes in flea community composition on transmission risk.58
These land-use-induced shifts in rodent community diversity and abundance had a strong effect on plague activity in our study region. The predicted abundance of seropositive rodents (PAI) was roughly twice as high in agricultural sites (1.08 ± 0.03 infected rodents expected per hectare, compared with 0.51 ± 0.15 in conserved sites) relative to conserved sites. This increase appears to be largely driven by the high numbers of M. natalensis found in agricultural habitats, with the abundance of this species being very strongly correlated to plague seroprevalence. Mastomys natalensis is one of the most frequently seropositive species for Y. pestis in Africa and is a widespread commensal species38,59 (and an important host of various other human pathogens, including Lassa virus60,61). Analogous changes in community composition, with a common commensal species serving as a major reservoir and “key host” for a pathogen, have been reported elsewhere, e.g., in West Nile virus (American robin, Turdus migratorius),62 Lyme disease (white-footed mouse, Peromyscus leucopus)50 and other systems.63 Given that globally community composition change appears to be more pervasive than net biodiversity change this is an important pattern to note.64,65
Aggregate rodent abundance was not related to predicted abundance of seropositive rodents, similar to other studies in East Africa that failed to detect differences in rodent abundance between villages with or without a history of plague.26,66 On the basis of the biodiversity patterns exhibited in conserved versus agricultural lands, the dilution effect hypothesis would predict that there should be a reduced disease risk in conserved land. Our data indicate that the relationship between diversity (at the level of rodents) and plague risk (as measured using PAI) in this context depends entirely on the measure of biodiversity used: a strong negative relationship was observed using Simpson diversity index, the negative relationship approached significance but did not meet the threshold using Shannon diversity index, and no pattern observed when using Chao estimates of species richness. As these three indices respectively give increasing weight to rare species, our results suggest that differences in abundances of common species are more important in predicting the PAI. All of these metrics are routinely used in examining diversity-disease relationships, but multiple metrics are rarely reported.10,18,22,67–69
An important area for future research that will help extend the reach of these observations will be learning more about interactions between rodent communities in agricultural and conserved sites. All of the rodent species found in agricultural sites were also detected in paired conserved sites. Mastomys natalensis, the dominant agricultural species, certainly demonstrates the capacity to move long distances between habitat types such as our paired treatment sites.70 This creates the possibility for coupling of the pathogen transmission dynamics in these adjacent habitats. Mastomys natalensis populations in African agricultural fields exhibit extreme boom and bust cycles with densities of up to 1,000 rodents/ha reported during population explosions.71–73 Rodent populations in African evergreen forests also cycled, but population peaks are rarely as high as those observed in M. natalensis.74,75 The juxtaposition of agricultural sites and conserved sites could influence disease dynamics if the rodent populations interact and allow source-sink dynamics for both host and pathogen population.
We were unable to test all captured rodents for Y. pestis presence, so our conclusions need to be interpreted in light of this caveat. Nevertheless, our host community model should accommodate incomplete sampling, and the species-specific seroprevalence patterns observed in the rodents sampled in this study are compatible with other surveys of rodent seroprevalence in the same region.38 Also, although seroprevalence studies are widely adopted for plague surveillance, we note that plague seroprevalence studies offer insights into rates of Y. pestis resistance rather than infection or infectiousness, that is, animals that die of Y. pestis infection are not available to sample. Can M. natalensis act as infectious plague hosts, or do they simply exhibit resistance to infection and not play a part in infecting fleas? Furthermore, the number of seropositive rodents per hectare does not translate directly to risk or occurrence of human disease because of many other factors that can affect exposure and infection, and progression from infection to overt disease.63,76,77 Data on plague eco-epidemiology in East Africa are largely reliant on observations of disease in human populations combined with seroprevalence studies of rodents and other mammals. We currently lack a comprehensive understanding of Y. pestis reservoir competence, transmission dynamics, and plague epidemiology in East Africa.24,78
Our results suggest that agricultural conversion can cause increases in plague activity. However, they also highlight many ongoing questions about the effects of land-use change on disease ecology and on the implications for disease control efforts. Understanding how landscape conversion and the interrelationships between different land-use types affect the ecology and epidemiology of plague in East Africa better positions us to design control measures for this disease in regions where it remains a real human health risk. Lessons learned from this case are also broadly important for understanding the dynamics of other zoonotic diseases in this era of rapid landscape change.
Synthesis.
Transmission risk for vector-borne diseases at the landscape level depends on multiple factors including: 1) abundance of animal hosts and vectors, 2) pathogen dynamics among those animal hosts and vectors, and 3) probability of contact between susceptible human hosts and infectious vectors. Conversion of wildlands for agriculture almost certainly increases the rate of encounter between humans and rodent hosts and vectors through an increase in human activity in converted landscapes. Our results show that, in the study area, the conversion of conservation areas to agriculture potentially increases the risk of plague transmission to humans via multiple pathways: not only are people more present in agricultural areas but the abundance of seropositive rodents also nearly doubled. The vector community may also be changed to favor species competent at transmitting plague, although more specific research on vector competence of rarer species would be needed to confirm this finding. In the context of sub-Saharan Africa, these observations are worth particular consideration given the rapid conversion of landscapes to agriculture in this region. We suggest that future interdisciplinary work, exploring both ecological interactions between conserved to agricultural habitats, and the dynamics of realized human exposure, will be critical to understanding how land-use change affects plague risk in these areas.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ramadhani Iddi, Halid Kibwana, Yustina Kiwango, Pasha Feinberg, and Annie Adelson for assistance in the field. For intellectual and logistical support we thank the Tanzania National Parks, the Tanzania Wildlife Research Institute, the Ngorongoro Conservation Area, Lake Manyara National Park, and Lake Manyara Tree Lodge. We also thank A. Bergmann, L. Cattaneo, M. Carleton, A. Hintz, S. Miller, L. Helgen, D. Lunde, J. Ososky, A. Driskell, and H. Kafka for assistance at the Smithsonian Institution.
Footnotes
Financial support: This project was supported by the James Smithson Fund of the Smithsonian Institution, the National Geographic Society (Grants 4691-91, 8846-10, 9106-12,), the National Science Foundation (DEB-0909670), the Woods Institute for the Environment at Stanford University, the Smithsonian Barcode Network grant, and the Smithsonian Women's Committee (SWC 44).
Authors' addresses: Douglas J. McCauley and Hillary S. Young Department of Ecology, Evolution, and Marine Biology, University of California Santa Barbara, Santa Barbara, CA, E-mails: douglas.mccauley@lifesci.ucsb.edu and hillary.young@lifesci.ucsb.edu. Daniel J. Salkeld, Rodolfo Dirzo, Eric F. Lambin, Lynnee Gaffikin, Michele Barry, Woods Institute for the Environment, Department of Medicine, Stanford University, Stanford, CA, E-mails: dansalkeld@gmail.com, rdirzo@stanford.edu, elambin@stanford.edu, earthlg@gmail.com, and michele.barry@stanford. Rhodes Makundi, Sokoine University of Agriculture, Morogoro, Tanzania, E-mail: rmakundi@yahoo.com. Ralph P. Eckerlin, Natural Sciences Division, Northern Virginia Community College, Annandale, VA, E-mail: reckerlin@nvcc.edu. Kristofer M. Helgen, Division of Mammals, Smithsonian Institution, Washington, DC, E-mail: helgenk@si.edu.
References
- 1.Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, Daszak P. Global trends in emerging infectious diseases. Nature. 2008;451:990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morens DM, Folkers GK, Fauci AS. The challenge of emerging and re-emerging infectious diseases. Nature. 2004;430:242–249. doi: 10.1038/nature02759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dunn RR, Davies TJ, Harris NC, Gavin MC. Global drivers of human pathogen richness and prevalence. Proc R Soc Lond B Biol Sci. 2010;277:2587–2595. doi: 10.1098/rspb.2010.0340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neerinckx S, Bertherat E, Leirs H. Human plague occurrences in Africa: an overview from 1877 to 2008. Trans R Soc Trop Med Hyg. 2010;104:97–103. doi: 10.1016/j.trstmh.2009.07.028. [DOI] [PubMed] [Google Scholar]
- 5.Thiberville SD, Moyen N, Dupuis-Maguiraga L, Nougairede A, Gould EA, Roques P, de Lamballerie X. Chikungunya fever: epidemiology, clinical syndrome, pathogenesis and therapy. Antiviral Res. 2013;99:345–370. doi: 10.1016/j.antiviral.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sindato C, Karimuribo ED, Pfeiffer DU, Mboera LEG, Kivaria F, Dautu G, Bernard B, Paweska JT. Spatial and temporal pattern of Rift Valley fever outbreaks in Tanzania; 1930 to 2007. PLoS ONE. 2014;9:e88897. doi: 10.1371/journal.pone.0088897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fratkin E. East African pastoralism in transition: Maasai, Boran, and Rendille cases. Afr Stud Rev. 2001;44:1–25. [Google Scholar]
- 8.Homewood K, Lambin EF, Coast E, Kariuki A, Kikula I, Kivelia J, Said M, Serneels S, Thompson M. Long-term changes in Serengeti-Mara wildebeest and land cover: pastoralism, population, or policies? Proc Natl Acad Sci USA. 2001;98:12544–12549. doi: 10.1073/pnas.221053998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Homewood KM. Policy, environment and development in African rangelands. Environ Sci Policy. 2004;7:125–143. [Google Scholar]
- 10.Mills JN. Biodiversity loss and emerging infectious disease: an example from the rodent-borne hemorrhagic fevers. Biodiversity. 2006;7:9–17. [Google Scholar]
- 11.Lambin EF, Tran A, Vanwambeke SO, Linard C, Soti V. Pathogenic landscapes: interactions between land, people, disease vectors, and their animal hosts. Int J Health Geogr. 2010;9:1–13. doi: 10.1186/1476-072X-9-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vittor AY, Gilman RH, Tielsch J, Glass G, Shields T, Lozano WS, Pinedo-Cancino V, Patz JA. The effect of deforestation on the human-biting rate of Anopheles darlingi, the primary vector of falciparum malaria in the Peruvian Amazon. Am J Trop Med Hyg. 2006;74:3–11. [PubMed] [Google Scholar]
- 13.Chasar A, Loiseau C, Valkiūnas G, Iezhova T, Smith TB, Sehgal RNM. Prevalence and diversity patterns of avian blood parasites in degraded African rainforest habitats. Mol Ecol. 2009;18:4121–4133. doi: 10.1111/j.1365-294X.2009.04346.x. [DOI] [PubMed] [Google Scholar]
- 14.Pongsiri MJ, Roman J, Ezenwa VO, Goldberg TL, Koren HS, Newbold SC, Ostfeld RS, Pattanayak SK, Salkeld DJ. Biodiversity loss affects global disease ecology. Bioscience. 2009;59:945–954. [Google Scholar]
- 15.Keesing F, Holt RD, Ostfeld RS. Effects of species diversity on disease risk. Ecol Lett. 2006;9:485–498. doi: 10.1111/j.1461-0248.2006.00885.x. [DOI] [PubMed] [Google Scholar]
- 16.Young HS, Dirzo R, Helgen KM, McCauley DJ, Billeter SA, Kosoy MY, Osikowicz LM, Salkeld DJ, Young TP, Dittmar K. Declines in large wildlife increase landscape-level prevalence of rodent-borne disease in Africa. Proc Natl Acad Sci USA. 2014;111:7036–7041. doi: 10.1073/pnas.1404958111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Randolph SE, Dobson A. Pangloss revisited: a critique of the dilution effect and the biodiversity-buffers-disease paradigm. Parasitology. 2012;139:847–863. doi: 10.1017/S0031182012000200. [DOI] [PubMed] [Google Scholar]
- 18.Salkeld DJ, Padgett KA, Jones JH. A meta-analysis suggesting that the relationship between biodiversity and risk of zoonotic pathogen transmission is idiosyncratic. Ecol Lett. 2013;16:679–686. doi: 10.1111/ele.12101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Young H, Griffin RH, Wood CL, Nunn CL. Does habitat disturbance increase infectious disease risk for primates? Ecol Lett. 2013;16:656–663. doi: 10.1111/ele.12094. [DOI] [PubMed] [Google Scholar]
- 20.Wood CL, Laffert KD, DeLeo G, Young HS, Hudson PJ, Kuris AM. Does biodiversity protect humans against infectious disease? Ecology. 2014;95:817–832. doi: 10.1890/13-1041.1. [DOI] [PubMed] [Google Scholar]
- 21.Salkeld DJ, Lane RS. Community ecology and disease risk: lizards, squirrels, and the Lyme disease spirochete in California, USA. Ecology. 2010;91:293–298. doi: 10.1890/08-2106.1. [DOI] [PubMed] [Google Scholar]
- 22.Piudo L, Monteverde MJ, Walker RS, Douglass RJ. Rodent community structure and Andes virus infection in sylvan and peridomestic habitats in northwestern Patagonia, Argentina. Vector Borne Zoonotic Dis. 2011;11:315–324. doi: 10.1089/vbz.2009.0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davis S, Makundi R, Machang'u R, Leirs H. Demographic and spatio-temporal variation in human plague at a persistent focus in Tanzania. Acta Trop. 2006;100:133–141. doi: 10.1016/j.actatropica.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 24.Ogen-Odoi A, Mbidde EK, Lutwama J, Wamala J, Mucunguzi A, Mugagga M, Kagirita A, Lukwago L, Musanza MM, Talisuna A. Bubonic and pneumonic plague—Uganda, 2006. MMWR. 2009;58:778–781. [PubMed] [Google Scholar]
- 25.McCauley DJ, Keesing F, Young T, Dittmar K. Effects of the removal of large herbivores on fleas of small mammals. J Vector Ecol. 2008;33:263–268. doi: 10.3376/1081-1710-33.2.263. [DOI] [PubMed] [Google Scholar]
- 26.Laudisoit A, Leirs H, Makundi R, Krasnov BR. Seasonal and habitat dependence of fleas parasitic on small mammals in Tanzania. Integr Zool. 2009;4:196–212. doi: 10.1111/j.1749-4877.2009.00150.x. [DOI] [PubMed] [Google Scholar]
- 27.Davis S, Begon M, De Bruyn L, Ageyev VS, Klassovskiy NL, Pole SB, Viljugrein H, Stenseth NC, Leirs H. Predictive thresholds for plague in Kazakhstan. Science. 2004;304:736–738. doi: 10.1126/science.1095854. [DOI] [PubMed] [Google Scholar]
- 28.Davis S, Trapman P, Leirs H, Begon M, Heesterbeek J. The abundance threshold for plague as a critical percolation phenomenon. Nature. 2008;454:634–637. doi: 10.1038/nature07053. [DOI] [PubMed] [Google Scholar]
- 29.Stenseth NC, Samia NI, Viljugrein H, Kausrud KL, Begon M, Davis S, Leirs H, Dubyanskiy VM, Esper J, Ageyev VS. Plague dynamics are driven by climate variation. Proc Natl Acad Sci USA. 2006;103:13110–13115. doi: 10.1073/pnas.0602447103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salkeld DJ, Salathé M, Stapp P, Jones JH. Plague outbreaks in prairie dog populations explained by percolation thresholds of alternate host abundance. Proc Natl Acad Sci USA. 2010;107:14247–14250. doi: 10.1073/pnas.1002826107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moore SM, Monaghan A, Griffith KS, Apangu T, Mead PS, Eisen RJ. Improvement of disease prediction and modeling through the use of meteorological ensembles: human plague in Uganda. PLoS ONE. 2012;7:e44431. doi: 10.1371/journal.pone.0044431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andrianaivoarimanana V, Kreppel K, Elissa N, Duplantier JM, Carniel E, Rajerison M, Jambou R. Understanding the persistence of plague foci in Madagascar. PLoS Negl Trop Dis. 2013;7:e2382. doi: 10.1371/journal.pntd.0002382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Serneels S, Said M, Lambin E. Land cover changes around a major east African wildlife reserve: the Mara Ecosystem (Kenya) Int J Remote Sens. 2001;22:3397–3420. [Google Scholar]
- 34.Lambin EF, Geist HJ, Lepers E. Dynamics of land-use and land-cover change in tropical regions. Annu Rev Environ Resour. 2003;28:205–241. [Google Scholar]
- 35.Ogutu J, Owen-Smith N, Piepho H, Said M. Continuing wildlife population declines and range contraction in the Mara region of Kenya during 1977–2009. J Zool. 2011;285:99–109. [Google Scholar]
- 36.Beale CM, Rensberg SV, Bond WJ, Coughenour M, Fynn R, Gaylard A, Grant R, Harris B, Jones T, Mduma S. Ten lessons for the conservation of African savannah ecosystems. Biol Conserv. 2013;167:224–232. [Google Scholar]
- 37.Olson JM. The Spatial Patterns and Root Causes of Land Use Change in East Africa. LUCID Project. Nairobi, Kenya: International Livestock Research Institute; 2004. pp. 1–38. [Google Scholar]
- 38.Kilonzo B, Mbise T, Mwalimu D, Kindamba L. Observations on the endemicity of plague in Karatu and Ngorongoro, northern Tanzania. Tanzan J Health Res Bull. 2006;8:1–6. doi: 10.4314/thrb.v8i1.14262. [DOI] [PubMed] [Google Scholar]
- 39.Mwalyosi R. Ecological changes in Lake Manyara National Park. Afr J Ecol. 1981;19:201–204. [Google Scholar]
- 40.Ondrejicka DA, Locke SA, Morey K, Borisenko AV, Hanner RH. Status and prospects of DNA barcoding in medically important parasites and vectors. Trends Parasitol. 2014;30:582–591. doi: 10.1016/j.pt.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 41.Musser G, Carleton M. Order Rodentia. In: Wilson DE, Reeder DM, editors. Mammal Species of the World: A Taxonomic and Geographic Reference. Vol. 2. Baltimore, MD: Johns Hopkins University Press; 2005. pp. 745–1600. [Google Scholar]
- 42. Verheyen W, Hulselmans JLT, Dierckx T, Mulungu L, Liers H, Corti M, Verheyen E. 2007. The characterization of the Kilimanjaro Lophuromys aquilus True, 1892 population and the description of five new Lophuromys species (Rodentia, Muridae). Bulletin van het Koninklijk Belgisch Instituut voor Natuurwetenschappen, Biologie 77 23 75 [Google Scholar]
- 43.Carleton MD, Stanley WT. Species limits within the Praomys delectorum group (Rodentia: Muridae: Murinae) of East Africa: a morphometric reassessment and biogeographical implications. Zool J Linn Soc. 2012;165:420–469. [Google Scholar]
- 44.Young HS, McCauley DJ, Dirzo R, Goheen JR, Agwanda B, Brook C, Castillo EO, Ferguson A, Kinyua SN, McDonough MM, Palmer TM, Pringle RM, Young TP, Helgen KM. Context-dependent effects of large wildlife declines on small mammal communities in central Kenya. Ecol Appl. 2015;25:348–360. doi: 10.1890/14-0995.1. [DOI] [PubMed] [Google Scholar]
- 45.Sikes RS, Gannon WL. Guidelines of the American Society of Mammalogists for the use of wild mammals in research. J Mammal. 2011;92:235–253. doi: 10.1093/jmammal/gyw078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Young HS, McCauley DJ, Helgen KM, Goheen JR, Otárola-Castillo E, Palmer TM, Pringle RM, Young TP, Dirzo R. Effects of mammalian herbivore declines on plant communities: observations and experiments in an African savanna. J Ecol. 2013;101:1030–1041. doi: 10.1111/1365-2745.12096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Williams JE, Gentry MK, Braden CA, Leister F, Yolken RH. Use of an enzyme-linked immunosorbent assay to measure antigenaemia during acute plague. Bull World Health Organ. 1984;62:463–466. [PMC free article] [PubMed] [Google Scholar]
- 48.Esamaeili S, Azadmanesh K, Naddaf SR, Rajerison M, Carniel E, Mostafavi E. Serologic survey of plague in animals, western Iran. Emerg Infect Dis. 2013;19:1549–1551. doi: 10.3201/eid1909.121829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chu MC. Laboratory Manual of Plague Diagnostic Tests. Atlanta, GA: U.S. Department of Health and Human Services, Centers for Disease Control and Prevention and World Health Organization; 2000. pp. 61–63. [Google Scholar]
- 50.LoGiudice K, Ostfeld RS, Schmidt KA, Keesing F. The ecology of infectious disease: effects of host diversity and community composition on Lyme disease risk. Proc Natl Acad Sci USA. 2003;100:567–571. doi: 10.1073/pnas.0233733100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kilpatrick AM, Daszak P, Jones MJ, Marra PP, Kramer LD. Host heterogeneity dominates West Nile virus transmission. Proc R Soc Lond B Biol Sci. 2006;273:2327–2333. doi: 10.1098/rspb.2006.3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.R Development Core Team . R: A Language and Environment for Statistical Computing. Vienna, Austria: The R Foundation for Statistical Computing; 2014. [Google Scholar]
- 53.Neerinckx S, Peterson AT, Gulinck H, Deckers J, Kimaro D, Leirs H. Predicting potential risk areas of human plague for the Western Usambara Mountains, Lushoto District, Tanzania. Am J Trop Med Hyg. 2010;82:492–500. doi: 10.4269/ajtmh.2010.09-0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bahmanyar M, Cavanaugh DC. Plague Manual. Geneva, Switzerland: World Health Organization; 1976. pp. 1–76. [Google Scholar]
- 55.Zimba M, Loveridge J, Davies DM, Mukaratirwa S. Seasonal abundance and epidemiologial indices of potential plague vectors Dinopysyllus lypusus (Siphonaptera: Hystrichopsyllidae) and Ctenophthalmus calceatus (Siphonaptera: Ctenophthalmidae) on rodents captured from three habitat types of Hatcliffe and Dzivarasekwa suburbs of Harare, Zimbabwe. J Med Ent. 2012;49:1453–1459. doi: 10.1603/me11231. [DOI] [PubMed] [Google Scholar]
- 56.Kilonzo B, Mvena Z, Machangu R, Mbise T. Preliminary observations on factors responsible for long persistence and continued outbreaks of plague in Lushoto district, Tanzania. Acta Trop. 1997;68:215–227. doi: 10.1016/s0001-706x(97)00096-x. [DOI] [PubMed] [Google Scholar]
- 57.Gratz N. Plague manual: epidemiology, distribution, surveillance and control. World Health Organ Tech Rep Ser. 1999;99:63–96. [Google Scholar]
- 58.Eisen RJ, Borchert JN, Holmes JL, Amatre G, Van Wyk K, Enscore RE, Babi N, Atiku LA, Wilder AP, Vetter SM. Early-phase transmission of Yersinia pestis by cat fleas (Ctenocephalides felis) and their potential role as vectors in a plague-endemic region of Uganda. Am J Trop Med Hyg. 2008;78:949–956. [PubMed] [Google Scholar]
- 59.Makundi RH, Massawe AW, Mulungu LS, Katakweba A, Mbise TJ, Mgode G. Potential mammalian reservoirs in a bubonic plague outbreak focus in Mbulu District, northern Tanzania, in 2007. Mammalia. 2008;72:253–257. [Google Scholar]
- 60.Monath T, Newhouse VF, Kemp GE, Setzer HW, Cacciapuoti A. Lassa virus isolation from Mastomys natalensis rodents during an epidemic in Sierra Leone. Science. 1974;185:263–265. doi: 10.1126/science.185.4147.263. [DOI] [PubMed] [Google Scholar]
- 61.Denys C, Koulémou K, Soropogui B, Koivogui L, Doré A, Meulen JT, Akoua-Koffi C, Camara MD, Allali BK, Calvet E, Sylla O, Kouassi-Kan S, Kourouma F, Lecompte E. African Biodiversity. New York, NY: Springer US; 2005. Community analysis of Muridae (Mammalia, Rodentia) diversity in Guinea: a special emphasis on Mastomys species and Lassa fever distributions; pp. 339–350. [Google Scholar]
- 62.LaDeau SL, Kilpatrick AM, Marra PP. West Nile virus emergence and large-scale declines of North American bird populations. Nature. 2007;447:710–713. doi: 10.1038/nature05829. [DOI] [PubMed] [Google Scholar]
- 63.Streicker DG, Fenton A, Pedersen AB. Differential sources of host species heterogeneity influence the transmission and control of multihost parasites. Ecol Lett. 2013;16:975–984. doi: 10.1111/ele.12122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dornelas M, Gotelli NJ, McGill B, Shimadzu H, Moyes F, Sievers C, Magurran AE. Assemblage time series reveal biodiversity change but not systematic loss. Science. 2014;344:296–299. doi: 10.1126/science.1248484. [DOI] [PubMed] [Google Scholar]
- 65.Dirzo R, Young H, Galetti M, Ceballos G, Isaac NJB, Collen B. Defaunation in the Anthropocene. Science. 2014;345:401–406. doi: 10.1126/science.1251817. [DOI] [PubMed] [Google Scholar]
- 66.Amatre G, Babi N, Enscore RE, Ogen-Odoi A, Atiku LA, Akol A, Gage KL, Eisen RJ. Flea diversity and infestation prevalence on rodents in a plague-endemic region of Uganda. Am J Trop Med Hyg. 2009;81:718–724. doi: 10.4269/ajtmh.2009.09-0104. [DOI] [PubMed] [Google Scholar]
- 67.Schmidt KA, Ostfeld RS. Biodiversity and the dilution effect in disease ecology. Ecology. 2001;82:609–619. [Google Scholar]
- 68.Allan BF, Langerhans RB, Ryberg WA, Landesman WJ, Griffin NW, Katz RS, Oberle BJ, Schutzenhofer MR, Smyth KN, Maurice AS. Ecological correlates of risk and incidence of West Nile virus in the United States. Oecologia. 2009;158:699–708. doi: 10.1007/s00442-008-1169-9. [DOI] [PubMed] [Google Scholar]
- 69.Clay CA, Lehmer EM, Jeor SS, Dearing MD. Testing mechanisms of the dilution effect: deer mice encounter rates, Sin Nombre virus prevalence and species diversity. EcoHealth. 2009;6:250–259. doi: 10.1007/s10393-009-0240-2. [DOI] [PubMed] [Google Scholar]
- 70.Leirs H, Verheyen W, Verhagen R. Spatial patterns in Mastomys natalensis in Tanzania (Rodentia, Muridae) Mammalia. 1996;60:545–556. [Google Scholar]
- 71.Stenseth NC, Leir H, Skonhoft A, Davis SA, Pech RP, Andreassen HP, Singleton GR, Lima M, Machang'u RS, Makundi RH. Mice, rats, and people: the bio-economics of agricultural rodent pests. Front Ecol Environ. 2003;1:367–375. [Google Scholar]
- 72.Vibe-Petersen S, Leirs H, Bruyn LD. Effects of predation and dispersal on Mastomys natalensis population dynamics in Tanzanian maize fields. J Anim Ecol. 2006;75:213–220. doi: 10.1111/j.1365-2656.2006.01037.x. [DOI] [PubMed] [Google Scholar]
- 73.Leirs H. Population ecology of Mastomys natalensis (Smith, 1834) multimammate rats: possible implications for rodent control in Africa. Agricul Ed. 1995;35:1–737. [Google Scholar]
- 74.Makundi RH, Massawe AW, Mulungu LS. Breeding seasonality and population dynamics of three rodent species in the Magamba Forest Reserve, Western Usambara Mountains, north-east Tanzania. Afr J Ecol. 2007;45:17–21. [Google Scholar]
- 75.Fa JE, Purvis A. Body size, diet and population density in afrotropical forest mammals: a comparison with neotropical species. J Anim Ecol. 1997;66:98–112. [Google Scholar]
- 76.MacMillan K, Enscore RE, Ogen-Odoi A, Borchert JN, Babi N, Amatre G, Atiku LA, Mead PS, Gage KL, Eisen RJ. Landscape and residential variables associated with plague-endemic villages in the West Nile region of Uganda. Am J Trop Med Hyg. 2011;84:435–442. doi: 10.4269/ajtmh.2011.10-0571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Eisen RJ, Borchert JN, Mpanga JT, Atiku LA, MacMillian K, Boegler KA, Montenieri JA, Monaghan A, Gage KL. Flea diversity as an element for persistence of plague bacteria in an East African plague focus. PLoS ONE. 2012;7:e35598. doi: 10.1371/journal.pone.0035598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Laudisoit A, Leir H, Makundi RH, Van Dongen S, Davis S, Neerinckx S, Deckers J, Libois R. Plague and the human flea, Tanzania. Emerg Infect Dis. 2007;13:687–693. doi: 10.3201/eid1305.061084. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.