Abstract
Plasmodium falciparum is the most deadly human malarial parasite, responsible for an estimated 207 million cases of disease and 627,000 deaths in 2012. Recent studies reveal that the parasite actively regulates a large fraction of its genes throughout its replicative cycle inside human red blood cells and that epigenetics plays an important role in this precise gene regulation. Here we discuss recent advances in our understanding of three aspects of epigenetic regulation in P. falciparum: changes in histone modifications, nucleosome occupancy and the three-dimensional genome structure. We compare these three aspects of the P. falciparum epigenome to those of other eukaryotes, and show that large-scale compartmentalization is particularly important in determining histone decomposition and gene regulation in P. falciparum. We conclude by presenting a gene regulation model for P. falciparum that combines the described epigenetic factors, and by discussing the implications of this model for the future of malaria research.
Keywords: malaria, nucleosome occupancy, histone modifications, three-dimensional genome organization, epigenetics, gene regulation, virulence genes
Introduction
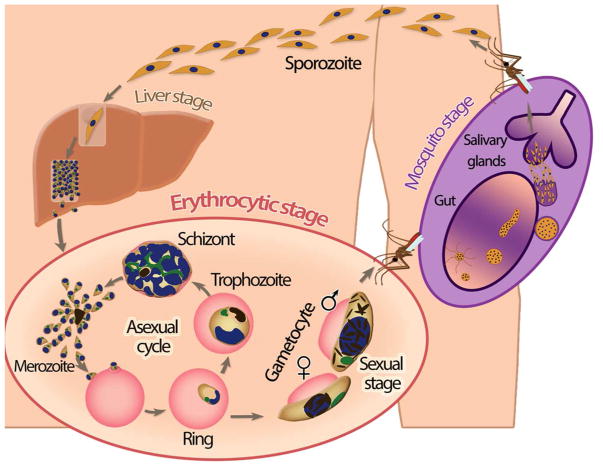
The complex life cycle of Plasmodium falciparum includes multiple stages in both the human host and the mosquito vector (reviewed in [1]) (Fig. 1). Human infection starts with the bite of an infected female Anopheles mosquito, resulting in the transfer of sporozoites that quickly migrate to the liver. Inside liver cells (hepatocytes), these sporozoites multiply extensively over a period of approximately two weeks and are then released into the bloodstream in the form of thousands of merozoites (Fig. 1 - liver stage). During the next stage of its life cycle, the parasite replicates in red blood cells (erythrocytes) by means of an unusual process of cell division called schizogony. While the parasite progresses through three distinct developmental stages (ring, trophozoite and schizont), it undergoes multiple rounds of nuclear replication followed by division of the multinucleated parasite into 16 to 32 daughter merozoites (Fig. 1 - asexual cycle). Upon bursting out of the host cell, these merozoites are released into the bloodstream and will invade new erythrocytes. During the asexual cycle, the parasite can commit to sexual development (reviewed in [2]), resulting in differentiation into a male or female gametocyte (Fig. 1 - sexual stage). The uptake of mature gametocytes by a feeding mosquito followed by the further development of the parasite in the mosquito midgut completes the P. falciparum life cycle (Fig. 1 - mosquito stage).
Figure 1.
Overview of the P. falciparum life cycle.
The asexual replication cycle is responsible for symptomatic disease and for the complications that are associated with severe malaria, such as anemia due to rupturing of red blood cells. In addition, severe disease can result from cytoadherence, the attachment of P. falciparum-infected erythrocytes to the smallest blood vessels, preventing clearance by the spleen and causing organ dysfunction. This cytoadherence is mediated by a family of parasite virulence proteins that are expressed on the erythrocyte surface: Plasmodium falciparum Erythrocyte Membrane Protein 1 (PfEMP1) [3–5]. Each P. falciparum parasite has approximately 60 different PfEMP1 variants encoded by var genes, only one of which is expressed at any time. Switching var gene expression enables the parasite to escape from host immune responses [6,7]. This process of antigenic variation is one example of the excellent adaptation of the parasite to survive in the human host.
The development of P. falciparum through the different stages of its life cycle is thought to be driven by coordinated changes in gene expression. Over the last decade, it has become clear that the parasite relies on an unusual combination of regulatory mechanisms for gene expression, and that these mechanisms are largely dependent on epigenetic processes (reviewed in [8–13]). In multicellular eukaryotes, gene expression is often mediated by transcription factors that bind to cell- or tissue-specific promoters and give rise to the expression of a subset of genes specific to that cell type or tissue [14]. However, despite extensive computational searches, relatively few transcription factors have been identified in P. falciparum [15,16], only a handful of which are known to be specific to a certain stage [17]. A notable example is PfAP2-G -- a member of the ApiAP2 transcription factor family -- which drives expression of gametocyte-specific genes and is crucial for the development of gametocytes [18,19]. On the other hand, a relatively large number of genes are predicted to encode proteins involved in chromatin structure, mRNA decay and translation rates [16], suggesting that alternative mechanisms of gene regulation, at the epigenetic as well as post-translational levels, may be more important for gene regulation in P. falciparum.
Here we focus on three important aspects of epigenetic gene regulation in P. falciparum, all of which are related to how DNA is packed in the nucleus (see [20–24] for articles discussing post-transcriptional regulation and see [25] for a discussion on DNA methylation, which is not well-characterized in P. falciparum). Similar to other eukaryotes, P. falciparum packages its DNA in the form of a condensed DNA-protein complex called chromatin. The basic packaging unit is a nucleosome, a stretch of approximately 147 bp of DNA wrapped around a core of eight histone proteins. Several layers of higher-order compaction of these strings of nucleosomes together create a highly structured nucleus. The organization of chromatin at both local and global levels is known to be involved in transcriptional regulation [26–29]. Local chromatin structure encompasses two main regulatory processes: the post-translational modification (PTM) of histone proteins that form nucleosomes, and nucleosome occupancy, which comprises the location, frequency, binding strength and protein composition (i.e., variant versus canonical histones) of nucleosomes on DNA.
At the global level, the organization of chromatin has been studied extensively, initially using gene-by-gene approaches such as immunofluorescent microscopy and fluorescent in situ hybridization (FISH) and, more recently, with chromatin conformation capture (3C)-based next-generation sequencing assays. 3C-based assays have enabled genome-wide profiling of chromatin contacts for various organisms including human, mouse, fruit fly, budding yeast and P. falciparum [30–35]. These profiles have yielded significant insights into the relationship between chromatin organization and transcription, revealing for example the compartmentalization of the genome into regions of transcriptionally active euchromatin and transcriptionally silent heterochromatin. Furthermore, for the haploid P. falciparum genome, the 3D models inferred from these contact profiles allowed the tracking of changes in nuclear organization throughout different stages of the parasite life cycle [30].
In the following sections, we provide an overview of our current understanding of chromatin organization and its role in transcriptional regulation in P. falciparum. We first describe various characteristics of local chromatin structure and subsequently focus on three-dimensional genome architecture. Finally, we combine these local and global views of chromatin to provide a model that explains our current understanding of the overall nuclear organization in P. falciparum and the role of the epigenome in regulating gene expression.
Histone modification landscape of the P. falciparum genome favors euchromatin
Post-translational modification of histone proteins
Histone proteins consist of a globular core structure and an N-terminal tail that protrudes from this core domain. Many amino acid residues in the core domain and in particular in the N-terminal tail can be chemically modified, producing various effects on chromatin organization (Fig. 2). In general, the addition of an acetyl group neutralizes the positive charge of histone proteins and thereby disrupts the stability of the DNA-histone interaction. This destabilization results in a more open chromatin structure and promotes a transcriptionally permissive state. On the other hand, methylations are uncharged and do not directly interfere with the interaction between histones and DNA. Rather, methylations mostly function by recruiting other effector molecules to the locus, resulting in further modifications of the chromatin.
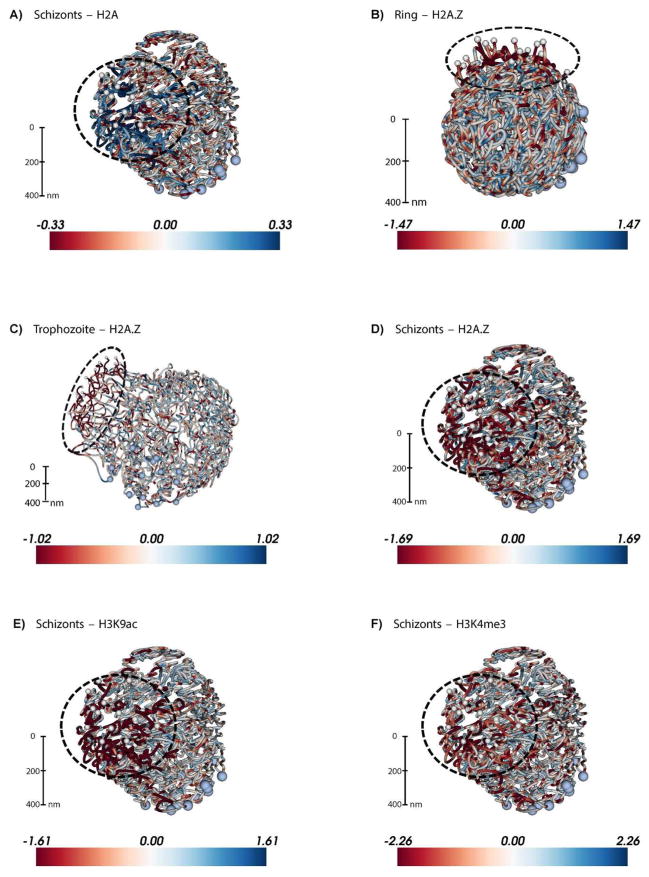
Figure 2. Large-scale depletion of the transcriptionally permissive histone variant H2A.Z and activating histone marks in the telomeric cluster visualized on the 3D P. falciparum genome.
ChIP-seq data from Bartfai et al. [45] for 4 histone variants or marks were downloaded from GEO (accession number: GSE23787) and mapped to the P. falciparum genome (PlasmoDB v9.0) using the short read alignment mode of BWA (v0.5.9) [117] with default parameter settings. Reads were post-processed, and only the reads that map uniquely with a quality score above 30 and with at most two mismatches were retained for further analysis. Retained reads were subjected to PCR duplicate elimination and then were aggregated for each non-overlapping 5 kb bin across the P. falciparum genome. The number of reads for each 5 kb bin was normalized using the overall sequencing depth of the corresponding ChIP-seq library. Plotted are the log2 ratios of sequence-depth normalized number of reads from the ChIP-seq library versus the corresponding input library (red: depletion, blue: enrichment) for A: H2A at 40 hours post invasion (hpi), B: H2A.Z at 10 hpi, C: H2A.Z at 30 hpi, D: H2A.Z at 40 hpi, E: H3K9ac at 40 hpi, and F: H3K4me3 at 40 hpi. 3D models for the ring, trophozoite and schizont stages were generated in Ay et al. [30] and were colored with ChIP-seq enrichment/depletion from 10, 20 and 40 hpi, respectively. Light blue and white spheres indicate centromeres and telomeres, respectively. The black dashed circle denotes the telomeric cluster for each stage. See http://noble.gs.washington.edu/proj/plasmo-epigenetics for the rotating 3D figure of each available ChIP-seq library.
Activating histone marks are abundant and broadly distributed
In P. falciparum, mass spectrometry experiments have identified at least 50 different histone post-translational modifications (PTMs), including methylation, acetylation, phosphorylation, ubiquitylation, and sumoylation [36–39]. Subsequent chromatin immunoprecipitation (ChIP) studies have given us insight into the genome-wide distribution of these histone marks in the asexual cycle (Table 1). In contrast to multicellular eukaryotes, a large proportion of the genome in P. falciparum is constitutively acetylated [37,40]. An abundance of activating marks has also been observed for other unicellular organisms, such as Saccharomyces cerevisiae and Tetrahymena thermophila [41]. Inhibition of histone acetyltransferase and deacetylase activity influences the expression levels of the majority of genes and interferes with parasite growth [42–44], indicative of the importance of acetylation for regulating transcription levels. Activating marks H3K9ac and H3K4me3 are mainly located in intergenic regions [45–47]. Highly transcribed genes carry more H3K9ac marks in their promoter [45], and this marking extends into the 5′ coding region [47].
Table 1.
Overview of most-studied histone modifications and variants in P. falciparum and comparison of their genome-wide distribution or function in other eukaryotes.
| Histone PTM/variant | Other eukaryotes | P. falciparum |
|---|---|---|
| H3K4me3 | Promoters of active genes [105–108] | Widely distributed in intergenic regions [45,47] |
| H3K9ac | Promoters of active genes [107,109] | Widely distributed in intergenic regions [45,47] |
| H3K9me3 | Silent genes [107,108] | Repressed var genes [40,48,49] |
| H3K27me3 | Promoters of silent/poised genes [107,108,110], absent in yeast [111] | Not detected [39] |
| H3K36me3 | Enriched in pericentromeric heterochromatin [112]; Transcribed regions of active genes [107,108] | TSS of repressed var genes [46]; 3′ end coding region active genes [46] |
| H4K20me3 | Silencing of telomeres, transposons and long terminal repeats [108,110]; inactive promoters [107] | Repressed var genes [46] and broad distribution across additional loci [40] |
| H2A.Z | Enriched in nucleosomes bordering active promoter (reviewed in [113,114]) | Widely distributed in intergenic regions [66,67] |
| H2B.Z | Lineage-specific variants with specialized functions, for example enriched at TSS in Trypanosoma brucei [68] | Widely distributed in intergenic regions [66,67] |
Repressive histone marks are scarce, and localized to specific regions
Typical repressive marks, in particular H3K9me3, are almost exclusively found in repressive clusters containing genes belonging to the virulence families, such as var, rifin, stevor, and pfmc-2tm [40,46,48,49]. Interestingly, H3K9me3 is also present at several additional loci, including the gene encoding the gametocyte-specific transcription factor PfAP2-G [40] that is tightly repressed during the asexual cycle. Transcription start sites of silent var genes are also enriched for H3K36me3 [46], while this modification is found at equal levels inside coding regions of active and repressed var genes [46,50]. H3K36me3 is present at lower levels in the rest of the genome and is enriched at the 3′ end of coding regions of active P. falciparum genes [46], in agreement with its role in transcriptional elongation in other eukaryotes. The repressive mark H4K20me3 is also mainly present in var gene clusters, although its enrichment is not as strong as for H3K9me3 and H3K36me3 [46]. On the other hand, the single active var gene, out of ~60 family members, is enriched in active histone marks, such as H3K9ac, H3K4me3, and H4 acetylations [46,49]. Finally, the repressive mark H3K27me3 has not been detected in the parasite [39], similar to yeast. The P. falciparum genome organization thus seems unusual in that a large fraction of its chromatin is continuously in a transcriptionally permissive state, while the formation of heterochromatin seems to be limited to virulence and specific sexual genes.
Histone variants and nucleosome occupancy are associated with gene expression
Relative to other eukaryotes, Plasmodium exhibits a distinctive nucleosome landscape around coding regions
Nucleosome occupancy plays an important role in regulating gene expression by allowing or restricting access of the transcription machinery to the DNA. Nucleosomes are not placed uniformly along the genome, but show a distinct distribution around coding regions [51–55]. In yeast, human and mouse, the promoter is characterized by a nucleosome-depleted region, bordered on either side by strongly positioned −1 and +1 nucleosomes, respectively, both of which are enriched for the variant histone H2A.Z [56–58]. The +1 nucleosome is located at a fixed distance relative to the transcription start site (TSS), although this distance varies between organisms [55]. The +2, +3 and subsequent nucleosomes form an array of nucleosomes with increasingly more fuzzy positioning towards the 3′ end of the gene. Finally, the transcription stop site is again demarcated by a strongly positioned nucleosome, followed by another nucleosome-depleted region.
Nucleosome organization in P. falciparum is similar to that in other eukaryotes in several respects. First, the promoter region is depleted of nucleosomes [59–61], the level of which correlates with transcriptional activity. Second, highly expressed genes have a more open chromatin organization at their core promoter than silent genes [59,60]. However, the P. falciparum nucleosome landscape also exhibits a number of unusual features. Notably, the TSS is not marked by a strongly positioned +1 nucleosome; instead, the strongest nucleosomes are the first and last nucleosomes within the coding region [59,60]. Furthermore, telomeric repeats and subtelomeric regions that contain the virulence gene families (var, rifin, etc) have higher nucleosome occupancy levels than the bulk of the genome [59,61,62]. Intergenic regions, on the other hand, contain lower nucleosome levels than coding regions [59,61–63], which is likely to be related to their extremely high AT-content (90–95%). AT-rich DNA is inherently inflexible, hampering the winding of DNA around the histone core [64,65]. Finally, intergenic regions in P. falciparum are exclusively occupied by nucleosomes composed of histone variants H2A.Z and H2B.Z [66,67], which are thought to have adopted a specialized function in P. falciparum to allow nucleosome assembly in these highly AT-rich regions. These histone variants are thus not restricted to promoter flanking nucleosomes but have a much broader distribution.
Nucleosome dynamics change in concordance with transcriptional activity during the asexual cycle
Another unconventional feature of nucleosome organization in P. falciparum is that nucleosome levels vary considerably during the asexual replication cycle, in parallel with changes in transcriptional activity [59,63]. At the transcriptionally most active trophozoite stage, histone levels decrease by approximately two-fold [59,63]. This nucleosome depletion occurs in a genome-wide fashion and is not restricted to genes that are expressed in the trophozoite stage. As the asexual cycle progresses into the schizont stage, nucleosomes are re-assembled, resulting in condensation of DNA as the parasites prepare for egress and invasion of a new red blood cell. Given the correlation between nucleosome density in promoter regions and gene expression levels, the dynamic nucleosome landscape in P. falciparum may have evolved to compensate for a paucity of specific transcription factors. Interestingly, Trypanosoma brucei, a parasite causing sleeping sickness in humans, has also developed an unusual nucleosome landscape, where certain combinations of canonical and variant histones mark the transcription initiation and termination sites in its genome [68]. Reminiscent of the lack of transcription factors in P. falciparum, transcription factors have remained elusive in T. brucei, indicating that these parasites may have followed parallel evolutionary pathways towards the use of the nucleosome landscape as a mechanism to regulate gene expression.
Three-dimensional conformation of the P. falciparum genome
Principles of nuclear organization in P. falciparum
It has long been known that the eukaryotic nucleus is a highly structured entity. In addition to the three-dimensional conformation of the chromatin-packaged DNA, key structural landmarks include the nuclear envelope, nuclear pores and nucleoli. For decades, various microscopic imaging techniques have been the “go-to” tools for understanding nuclear organization and chromatin architecture in many different organisms [69–71]. In P. falciparum, FISH applications have been instrumental in demonstrating important characteristics of genome organization in the parasite. In particular, silent var genes were shown to colocalize with each other near the nuclear periphery, while the single active var gene is located elsewhere [40,72,73]. Together with the other epigenetic mechanisms outlined above — histone modifications, histone variants and nucleosome occupancy — the non-random organization of DNA into repressive centers is believed to play a crucial role in the one-at-a-time expression of 60 genes in the var family. Another intriguing discovery from FISH experiments was that the ribosomal DNA loci that are distributed in a seemingly random fashion on different P. falciparum chromosomes show non-random colocalization in 3D [74]. A more recent study employed several ultrastructural microscopy techniques to study the distribution of nuclear pore complexes and chromatin throughout the P. falciparum asexual cycle [75], demonstrating a striking increase in pore density during the transcriptionally active trophozoite stage, as well as chromatin decomposition near the nuclear envelope. These changes parallel previously observed changes in transcriptional activity and nucleosome occupancy that have been discussed above [63].
Profiling of eukaryotic genome architecture using next-generation sequencing applications
Within the last decade, the field of genome architecture has been revolutionized by breakthroughs in combining next generation sequencing with molecular assays that measure proximities of DNA regions to certain nuclear landmarks (e.g., lamina, nucleolus) or to other regions in cis or trans (e.g., 4C, Hi-C, ChIA-PET) [32,34,76–80] (see [81] for review). Applications of these techniques to multiple genomes including human and mouse have revealed the organizational hallmarks of genome architecture. These include localization of gene-rich regions near the nuclear center and heterochromatin near the nuclear lamina [77], colocalization of ribosomal DNA loci near nucleoli [78], and megabase-scale open/closed chromatin compartments [34]. In addition, these two mammalian genomes are partitioned into megabase-sized topologically associated domains (TADs) that are enriched for interactions within but not across domains, and are separated from each other by insulator proteins [31,82,83] (see [28] for review). Finally, these studies have provided us with examples of cell type-specific chromatin loops bringing distal regulatory elements into close 3D proximity. Long-range chromatin loops that play regulatory roles in gene expression include Hox cluster silencing [84,85], control of SHH gene by an enhancer that is located 1 Mb away in human [86] and a validated set of cell type-specific enhancers in mouse [87].
Profiling of P. falciparum genome architecture during the asexual cycle
As is the case for many other next generation sequencing-based assays, application of these genome architecture assays has been challenging for the AT-rich genome of P. falciparum. However, within the last year, two groups have published their results using Hi-C, one profiling the genome architecture of different P. falciparum strains [33] and the other modeling the 3D structure of P. falciparum-3D7 at three key stages during its asexual replication cycle within human red blood cells [30]. These studies revealed key characteristics of P. falciparum genome structure (Table 2), including colocalization of centromeres, colocalization of telomeres near the nuclear periphery, colocalization of both internal and subtelomeric virulence gene clusters near the telomeres, colocalization of rDNA loci that are active in ring stage parasites and maintenance of chromosomes territories (see Ay et al. [30] for details). Furthermore, Hi-C profiles from Ay et al. exhibit different polymer behavior in the most transcriptionally active trophozoite stage compared to the other two stages, suggesting a link between overall chromatin compaction and transcriptional activity. The degree of telomere colocalization and the repressive effect of the telomeric compartment is also most pronounced in this trophozoite stage, suggesting a strict compartmentalization to segregate genes that need to be repressed from the rest. Finally, both the Hi-C contact maps and the 3D models inferred from them suggest a tight correlation between the 3D location of a gene and its expression. Gene pairs located nearby in 3D have significantly higher expression correlation compared to other pairs, even after discarding intra-chromosomal pairs that would be biased by their genomic distance in 1D [30]. Overall, these observations suggest that P. falciparum chromatin is highly structured at the large scale and that this structure provides a potential epigenetic mechanism to regulate gene expression.
Table 2.
Summary of organizational features of P. falciparum nucleus and genome at three distinct stages during asexual parasite replication in human red blood cells (asexual cycle).
| Feature | Ring | Trophozoite | Schizont |
|---|---|---|---|
| Nuclear size | Small (~700 nm diameter) [75,115] | Large (~1,700 nm diameter) [75,115] | Small (~850 nm diameter) [75,115] |
| Nuclear pores | Few (3–7), clustered together [75] | Many (12–58), uniformly distributed [75] | Few per daughter nucleus (2–6), clustered together [75] |
| Nucleosome occupancy | High [59,63] | Low [59,63] | High [59,63] |
| Chromatin compaction | Compact [30,59,63,75] | Open [30,59,63,75] | Compact [30,59,63,75] |
| Chromosome territories | Conflicting reports (absent [33] vs present [30]) | Partially lost [30] | Present [30] |
| Centromere locations | Conflicting reports (colocalized [30] vs dispersed [33,116]) | Colocalized [30,116] | Colocalized [30,116] |
| Telomere locations | Colocalized near periphery [30,72] | Colocalized near periphery [30,72] | Colocalized near periphery [30,72] |
| Virulence gene locations | Colocalized [33] near periphery [30,40,72] | Colocalized near periphery [30,40,72] | Colocalized near periphery [30,40,72] |
| rDNA gene locations | Conflicting reports (all loci clustered [74] vs strong clustering of only active loci [30,33]) | Conflicting reports (dispersed [74] vs weak clustering of only active loci [30]) | Conflicting reports (dispersed [74] vs weak clustering of only active loci [30]) |
The folded chromosome structure seen in P. falciparum is similar to what has been observed in budding and fission yeast [32,88]. However, chromosome looping to achieve localization of var genes in repressive perinuclear compartments results in a more complex three-dimensional organization of the P. falciparum genome compared to yeast, even though these organisms have similarly sized genomes [30]. Interestingly, the clonal var gene expression and clustering of all remaining var genes in repressive heterochromatin is strikingly similar to the epigenetic signature of the ~1,400 olfactory receptor genes in the mouse, all except one of which are located in heterochromatic foci enriched for H3K9me3 and H4K20me3, resulting in monogenic and monoallelic expression [89,90]. In comparison to human, mouse and fly genomes, the P. falciparum genome organization is relatively simple and does not display TADs. The nuclear architecture in P. falciparum thus exploits features from both unicellular and multicellular organisms. Even though the parasite is a unicellular organism throughout its entire life cycle, it adopts drastically different forms during the various life cycle stages to be able to survive in a variety of cells in both the human and the mosquito host. The regulation of life cycle progression and cell differentiation is therefore likely to be more constrained and complex than for most unicellular organisms. From an evolutionary standpoint, this may explain why the parasite has developed such complex nuclear architecture relative to the size of its genome.
A combined model of epigenetic gene regulation in P. falciparum
Nuclear organization and gene regulation
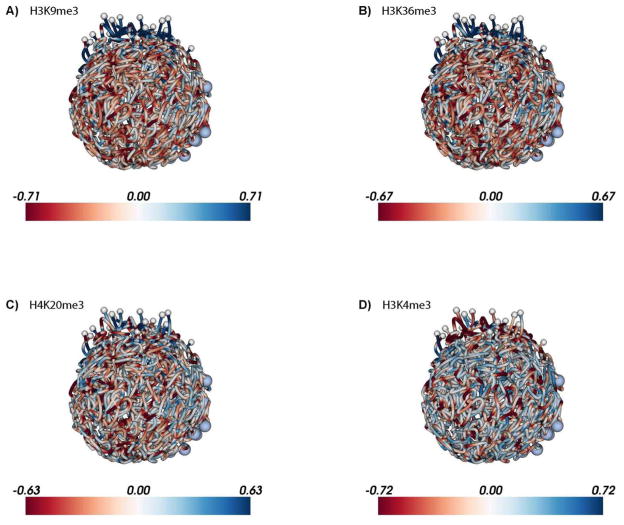
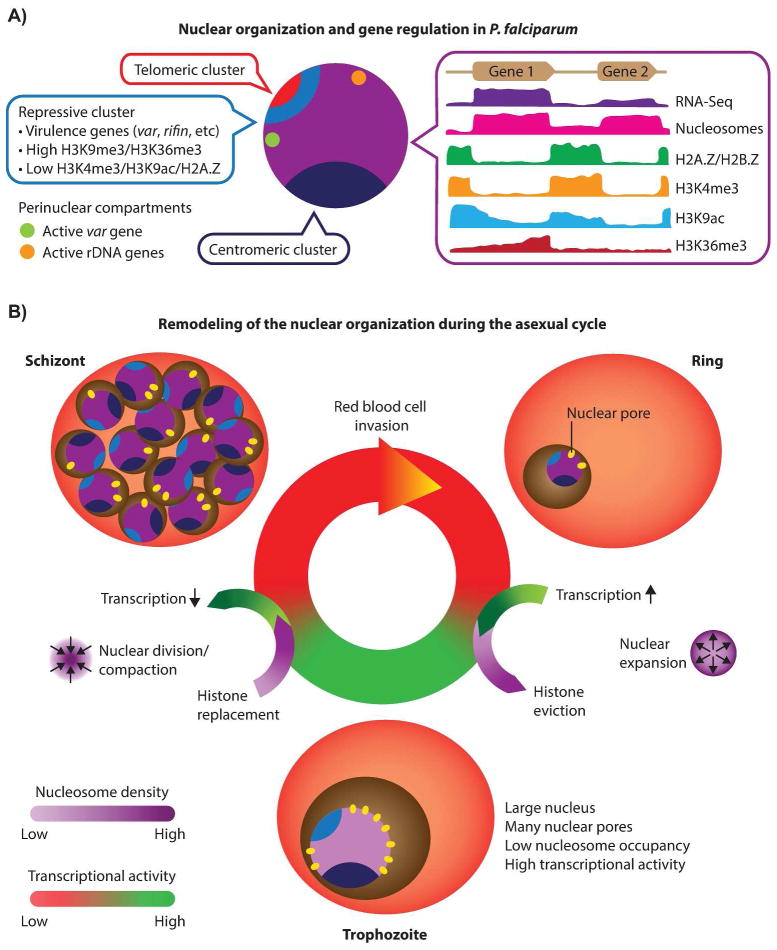
The epigenetic makeup of the P. falciparum genome, as outlined above, points towards a binary nuclear organization, with the majority of the genome present in the form of euchromatin, while a limited number of genes are organized into strongly repressed heterochromatin. This heterochromatin is localized at the nuclear periphery and is characterized by high nucleosome density (Fig. 2A), the presence of repressive histone marks H3K9me3, H3K36me3 and H4K20me3 (Fig. 3A–C), and the absence of the transcription-associated histone variant H2A.Z (Fig. 2B–D) and histone marks H3K9ac (Fig. 2E) and H3K4me3 (Fig. 2F and 3D). It was recently demonstrated that heterochromatin protein 1 (HP1) and P. falciparum histone deacetylase 2 (PfHda2) are both essential for maintaining heterochromatic regions [91,92]. Depletion of either HP1 or PfHda2 resulted in an arrest of parasite development at the trophozoite stage and a loss of var gene repression. In addition, an increase in the number of parasites differentiating into gametocytes was observed, indicating that the gametocyte transcription factor locus pfap2-g is also under strict epigenetic control. The remaining euchromatic fraction of the genome has several notable features, including perinuclear compartments containing the active var gene or active rDNA genes (Fig. 4A). In addition, clustering of silent genes that are specific to other stages of the parasite’s life cycle [30] suggests the presence of small heterochromatic islands, as observed at the trophozoite stage by advanced transmission and scanning electron microscopy [75].
Figure 3. Enrichment of repressive histone marks in the telomeric cluster visualized on the 3D P. falciparum genome at the ring stage.
ChIP-seq data from Jiang et al. [46] for 5 histone marks were downloaded from SRA (accession number: SRP022761) and processed as described in the caption of Figure 2. Due to lack of input libraries from this publication, the input libraries from Bartfai et al. at different time points were pooled into one aggregated input library which was then used for normalization of each Jiang et al. ChIP-seq library. Similar to Figure 2, log2 ratios of ChIP-seq versus input were plotted for A: H3K9me3, B: H3K36me3, C: H4K20me3, and D: H3K4me3 at 18 hpi. The 3D model for the ring stage from Ay et al. [30] was used to visualize enrichment/depletion of each histone mark. See http://noble.gs.washington.edu/proj/plasmo-epigenetics for the rotating 3D figure of each available ChIP-seq library.
Figure 4. Model for P. falciparum epigenetic gene regulation.
A: Nuclear organization and gene regulation in P. falciparum. Centromeric (dark blue) and telomeric (red) clusters are localized at the nuclear periphery. Subtelomeric virulence genes (blue) are anchored to the nuclear perimeter and cluster with internally located var genes in repressive center(s), characterized by repressive histone marks H3K9me3 and H3K36me3. The single active var gene (green) is located in a perinuclear compartment away from the repressive center(s). In addition, active rDNA genes (orange) also cluster at the nuclear periphery. The remaining genome (purple) is largely present in an open, euchromatic state with a number of notable features. (i) Nucleosome levels are high in genic and lower in intergenic regions, while gene expression correlates with nucleosome density at the transcription start site. (ii) Intergenic regions are bound by nucleosomes containing histone variants H2A.Z and H2B.Z. (iii) Intergenic regions contain H3K4me3, the level of which does not influence transcriptional activity. (iv) H3K9ac is mainly found in intergenic regions and extends into 5′ ends of coding regions, with highly expressed genes showing higher levels of H3K9ac. (v) Active genes are marked with H3K36me3 towards their 3′ end. B: Remodeling of the nuclear organization during the asexual cycle. Extensive remodeling of the nucleus takes place as the parasite progresses through the ring, trophozoite and schizont stages. In the transition from the relatively inert ring stage to the transcriptionally active trophozoite stage, the size of the nucleus and the number of nuclear pores increase, accompanied by a decrease in genome-wide nucleosome levels, resulting in an open chromatin structure that allows high transcription rates. In the schizont stage, the nucleus divides and recompacts, histones are re-assembled and transcription is shutdown, to facilitate egress of the parasites’ daughter cells and invasion of new red blood cells.
Remodeling of the nuclear organization during the asexual cycle
Microarray and RNA-seq studies have shown that 70–80% of all genes are expressed in the asexual replication cycle, in particular during the trophozoite stage [24,93,94]. During the 48-hour cycle, the nucleus and chromatin are drastically remodeled to facilitate this high transcriptional activity (Fig. 4B and Table 2). First, the nucleus expands in size [75], which can also be readily observed in microscopy images of Giemsa stained parasites [30]. Second, the number of nuclear pores increases greatly, from 3–7 clustered pores in the ring stage to 12–58 pores that are uniformly distributed around the nucleus in the trophozoite stage [75]. Third, in line with the increased nuclear volume, the chromatin opens up [30,75], accompanied by removal of nucleosomes [59,63] and increased intermingling of chromosomes [30]. Despite these large-scale nuclear dynamics, the centromeres, telomeres and repressed var genes remain clustered. The correlation of nucleosome density of gene promoters with transcriptional activity of individual genes suggests that local chromatin organization may play an important role in regulating the level of gene expression [59]. The transitioning of the parasite from the trophozoite stage to the schizont stage is characterized by a reversion of nuclear changes, including reassembly of nucleosomes and re-establishment of chromosomal territories, which results in recompaction of the genome. Finally, during DNA replication, the nucleus divides into multiple small daughter nuclei, each with a small number of the nuclear pores, i.e., those that were present in the original nucleus [75].
Outstanding questions
Does the nucleus harbor one or more repressive centers near the periphery?
Whether heterochromatin containing silent var genes is organized into a single large repressive center or is divided over a small number of perinuclear foci remains a topic of debate. FISH images visualizing the location of telomere-associated repeat elements or var gene promoters typically show 2–6 foci distributed around the nucleus [40,72,73,95]. On the other hand, single foci were observed by immunofluorescence microscopy for H3K9me3, H3K36me3, and heterochromatin protein 1 [50,96], all of which are strongly associated with the repressed var genes. In addition, the Hi-C-derived three-dimensional models of the P. falciparum genome showed strong clustering of centromeres and telomeres [30] (Fig. 4A), a chromosome configuration that has been observed in other organisms [32,88,97]. These models suggested the organization of subtelomeric var genes into a single cluster at the nuclear perimeter. Such organization, even though seemingly contradicting the FISH data, may be due to aggregation across a large population of cells for Hi-C experiments. If each var gene cluster is randomly located in one of multiple repressive clusters in each cell, then the aggregate signal would suggest colocalization of all var genes. However, it may conceivably be beneficial to locate all repressed genes in close proximity of each other to regulate the expression of a single var gene and the tight repression of all remaining family members. Additional experiments will be necessary to unravel the precise mechanisms by which var gene expression is controlled, by further dissecting the effect of gene localization, nuclear architecture, and gene-to-gene communication on this process. In particular, HiC experiments on single cells would likely provide significant insight into the localization of active and repressed var genes, as well as the extent of cell-to-cell variability.
Could the mediators of epigenetic control and nuclear remodeling be the next drug targets for malaria?
Drastic remodeling of the nucleus and chromatin are likely to be driving forces behind the wave of transcriptional activity during the trophozoite stage. Components involved in these dynamic processes may thus be promising targets for antimalarial drugs. Future research should therefore focus on understanding the molecular mechanisms involved in chromatin and nuclear remodeling. For example, very little is known about proteins and enzymes that regulate the formation of heterochromatin and the global nuclear architecture, with the exception of the role of HP1 in maintaining repressive perinuclear chromatin containing the var genes and the pfap2-g locus. A multitude of such proteins has been identified in other organisms, most notably RNA polymerase III-associated factor (TFIIIC), cohesin and CCCTC binding factor (CTCF) (reviewed in [98]), that are likely to have homologues in P. falciparum. Other potential drug targets include key components involved in expansion of the nuclear membrane and chromatin remodeling enzymes that regulate the global nucleosome eviction and re-assembly during the trophozoite and schizont stages. Analysis of chromatin-associated proteins by proteomics-based approaches will likely identify many candidates that may be involved in these processes. In addition, the application of novel genetic engineering tools in P. falciparum, such as the CRISPR/Cas9 system [99–101], may enable us to study the effect of gene deletion or translocation on genome structure to better understand the determinants of nuclear architecture.
Are the epigenetic control mechanisms similar in other stages of P. falciparum life cycle and in other parasites?
The epigenetic regulation model we present here is based on profiles taken during the asexual replication cycle. During this phase of the parasite’s life cycle, the genome seems to be largely shaped by the strict one-at-a-time expression of the var genes. The absence of var gene expression in all other parasite stages may have a large impact on chromatin organization. In addition, while some genes may be constitutively expressed during the parasite’s life cycle, others may be silenced or activated in these alternative and highly variable stages, ranging from the male and female gametocyte, via the diploid zygote in the mosquito midgut, to the haploid sporozoite. Therefore, we expect generating genome-wide profiles of histone modifications, nucleosome landscape and three-dimensional architecture during these other parasite stages to be of great interest to further explore the epigenetic regulatory mechanisms in P. falciparum. Furthermore, we know very little about the role of epigenetic control in transcriptional regulation in other Plasmodium species. P. vivax, for example, has a much lower AT-content (on average 57%), which is likely to influence the binding kinetics and preferences of nucleosomes. In addition, P. vivax expresses a large proportion of its gene family encoding for variant surface proteins (vir) during the blood stage, whereas P. falciparum expresses only one out of its 60 var genes in this stage [102,103]. In P. vivax the absence of clonal expression, as seen for the var family in P. falciparum, may reduce the requirements for strictly repressive heterochromatin. Therefore, determining the nucleosome landscape, the location of histone modifications and the three-dimensional structure of the P. vivax genome will be extremely informative for our understanding of commonalities and differences between epigenetic gene regulation in these two parasites, especially pertaining to regulation of virulence genes.
Conclusions and prospects
An increasing amount of data highlights the importance of epigenetic mechanisms in regulating gene expression in P. falciparum and other eukaryotes, including human and mouse [30–35]. Here we have discussed multiple layers of epigenetic control, including histone modifications, nucleosome occupancy, histone variants and genome architecture, which are involved in the precise gene regulation during the asexual replication cycle of the malaria parasite, P. falciparum. We summarized the current understanding of the interplay among these different layers and how these layers shape the overall nuclear organization and connect with overall transcriptional activity and to the one-at-a-time expression of var genes.
Better characterization of epigenetic regulation in P. falciparum will stimulate interest in several exciting directions in malaria research. Further studies into the establishment and maintenance of strong repressive compartments in the nucleus may reveal the underlying regulatory mechanisms and lead to the identification of proteins involved in this process. Disrupting the function of proteins responsible for maintaining heterochromatin, such as HP1 [91], could be an effective strategy to block parasite replication during the asexual cycle. Another important event in the malaria life cycle is gametocytogenesis, which was recently shown to be driven by the transcription factor PfAP2-G [18,19]. It would be interesting to fully characterize the epigenetic factors, such as genome architecture, that help PfAP2-G target and regulate gametocyte-specific genes. In addition to layers of epigenetic regulation that we have focused on here, post-transcriptional and translational controls are likely to be involved in the timing of protein expression [22–24,104]. Increased insight into these regulatory processes would significantly advance our understanding of parasite biology and could facilitate major breakthroughs in our fight against malaria.
Acknowledgments
This study was financially supported by a Computing Research Association CIFellows award (NSF award CIF 1136996 to FA), the Human Frontier Science Program (grant LT000507/2011-L to EMB), the National Institutes of Health (grants R01 AI85077-04 to KLR and R01 AI106775-02 to WSN/KLR), the European Research Council (grant SMAC-ERC-280032 to JPV), the European Commission (grant HEALTH-F5-2012-305626 to JPV/NV) and the French National Research Agency (grant ANR-11-BINF-0001 to JPV/NV).
Abbreviations
- PfEMP1
Plasmodium falciparum Erythrocyte Membrane Protein 1
- var
family of genes that encode PfEMP1 proteins
- ApiAP2
a family of transcription factors in Plasmodium
- mRNA
messenger RNA
- FISH
fluorescent in situ hybridization
- 3C
chromatin conformation capture
- 4C
circularized chromatin conformation capture
- Hi-C
chromatin conformation capture coupled to next-generation sequencing
- ChIA-PET
Chromatin Interaction Analysis by Paired-End Tag Sequencing
- PTM
post-translational modification
- TSS
transcription start site
- ChIP
chromatin immunoprecipitation
- TAD
topologically associated domain
- H4K20me3
histone H4 lysine 20 trimethylation
- H3K9ac
histone H3 lysine 9 acetylation
- H3K{N}me3
histone H3 lysine {N} trimethylation
- H2A
histone H2A
- H2A.Z
H2B.Z, variants of histone H2A and H2B
- SHH
sonic hedgehog gene
- Hox
a group of homeobox genes
- OR
olfactory receptors
- hpi
hours post invasion
Footnotes
The authors have declared no conflict of interest.
References
- 1.Greenwood BM, Fidock DA, Kyle DE, Kappe SH, et al. Malaria: progress, perils, and prospects for eradication. The Journal of clinical investigation. 2008;118:1266–76. doi: 10.1172/JCI33996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker DA. Malaria gametocytogenesis. Mol Biochem Parasitol. 2010;172:57–65. doi: 10.1016/j.molbiopara.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baruch DI, Pasloske BL, Singh HB, Bi X, et al. Cloning the P. falciparum gene encoding PfEMP1, a malarial variant antigen and adherence receptor on the surface of parasitized human erythrocytes. Cell. 1995;82:77–87. doi: 10.1016/0092-8674(95)90054-3. [DOI] [PubMed] [Google Scholar]
- 4.Smith JD, Chitnis CE, Craig AG, Roberts DJ, et al. Switches in expression of Plasmodium falciparum var genes correlate with changes in antigenic and cytoadherent phenotypes of infected erythrocytes. Cell. 1995;82:101–10. doi: 10.1016/0092-8674(95)90056-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Su XZ, Heatwole VM, Wertheimer SP, Guinet F, et al. The large diverse gene family var encodes proteins involved in cytoadherence and antigenic variation of Plasmodium falciparum-infected erythrocytes. Cell. 1995;82:89–100. doi: 10.1016/0092-8674(95)90055-1. [DOI] [PubMed] [Google Scholar]
- 6.Bull PC, Lowe BS, Kortok M, Molyneux CS, et al. Parasite antigens on the infected red cell surface are targets for naturally acquired immunity to malaria. Nat Med. 1998;4:358–60. doi: 10.1038/nm0398-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roberts DJ, Craig AG, Berendt AR, Pinches R, et al. Rapid switching to multiple antigenic and adhesive phenotypes in malaria. Nature. 1992;357:689–92. doi: 10.1038/357689a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cui L, Miao J. Chromatin-mediated epigenetic regulation in the malaria parasite Plasmodium falciparum. Eukaryot Cell. 2010;9:1138–49. doi: 10.1128/EC.00036-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duffy MF, Selvarajah SA, Josling GA, Petter M. The role of chromatin in Plasmodium gene expression. Cell Microbiol. 2012;14:819–28. doi: 10.1111/j.1462-5822.2012.01777.x. [DOI] [PubMed] [Google Scholar]
- 10.Hoeijmakers WA, Stunnenberg HG, Bartfai R. Placing the Plasmodium falciparum epigenome on the map. Trends Parasitol. 2012;28:486–95. doi: 10.1016/j.pt.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 11.Horrocks P, Wong E, Russell K, Emes RD. Control of gene expression in Plasmodium falciparum - ten years on. Mol Biochem Parasitol. 2009;164:9–25. doi: 10.1016/j.molbiopara.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 12.Deitsch K, Duraisingh M, Dzikowski R, Gunasekera A, et al. Mechanisms of gene regulation in Plasmodium. The American journal of tropical medicine and hygiene. 2007;77:201–8. [PubMed] [Google Scholar]
- 13.Voss TS, Bozdech Z, Bartfai R. Epigenetic memory takes center stage in the survival strategy of malaria parasites. Curr Opin Microbiol. 2014;20:88–95. doi: 10.1016/j.mib.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 14.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Balaji S, Babu MM, Iyer LM, Aravind L. Discovery of the principal specific transcription factors of Apicomplexa and their implication for the evolution of the AP2-integrase DNA binding domains. Nucleic Acids Res. 2005;33:3994–4006. doi: 10.1093/nar/gki709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coulson RM, Hall N, Ouzounis CA. Comparative genomics of transcriptional control in the human malaria parasite Plasmodium falciparum. Genome Res. 2004;14:1548–54. doi: 10.1101/gr.2218604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campbell TL, De Silva EK, Olszewski KL, Elemento O, et al. Identification and genome-wide prediction of DNA binding specificities for the ApiAP2 family of regulators from the malaria parasite. PLoS Pathog. 2010;6:e1001165. doi: 10.1371/journal.ppat.1001165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kafsack BF, Rovira-Graells N, Clark TG, Bancells C, et al. A transcriptional switch underlies commitment to sexual development in malaria parasites. Nature. 2014;507:248–52. doi: 10.1038/nature12920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sinha A, Hughes KR, Modrzynska KK, Otto TD, et al. A cascade of DNA-binding proteins for sexual commitment and development in Plasmodium. Nature. 2014;507:253–7. doi: 10.1038/nature12970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chung DW, Ponts N, Cervantes S, Le Roch KG. Post-translational modifications in Plasmodium: more than you think! Mol Biochem Parasitol. 2009;168:123–34. doi: 10.1016/j.molbiopara.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 21.Le Roch KG, Chung DW, Ponts N. Genomics and integrated systems biology in Plasmodium falciparum: a path to malaria control and eradication. Parasite Immunol. 2012;34:50–60. doi: 10.1111/j.1365-3024.2011.01340.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suvorova ES, White MW. Transcript maturation in apicomplexan parasites. Curr Opin Microbiol. 2014;20C:82–7. doi: 10.1016/j.mib.2014.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kramer S. RNA in development: how ribonucleoprotein granules regulate the life cycles of pathogenic protozoa. Wiley interdisciplinary reviews. RNA. 2014;5:263–84. doi: 10.1002/wrna.1207. [DOI] [PubMed] [Google Scholar]
- 24.Bunnik EM, Chung DW, Hamilton M, Ponts N, et al. Polysome profiling reveals translational control of gene expression in the human malaria parasite Plasmodium falciparum. Genome Biol. 2013;14:R128. doi: 10.1186/gb-2013-14-11-r128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ponts N, Fu L, Harris EY, Zhang J, et al. Genome-wide mapping of DNA methylation in the human malaria parasite Plasmodium falciparum. Cell host & microbe. 2013;14:696–706. doi: 10.1016/j.chom.2013.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 27.Zentner GE, Henikoff S. Regulation of nucleosome dynamics by histone modifications. Nat Struct Mol Biol. 2013;20:259–66. doi: 10.1038/nsmb.2470. [DOI] [PubMed] [Google Scholar]
- 28.Nora EP, Dekker J, Heard E. Segmental folding of chromosomes: a basis for structural and regulatory chromosomal neighborhoods? Bioessays. 2013;35:818–28. doi: 10.1002/bies.201300040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Belmont AS. Large-scale chromatin organization: the good, the surprising, and the still perplexing. Curr Opin Cell Biol. 2014;26:69–78. doi: 10.1016/j.ceb.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ay F, Bunnik EM, Varoquaux N, Bol SM, et al. Three-dimensional modeling of the P. falciparum genome during the erythrocytic cycle reveals a strong connection between genome architecture and gene expression. Genome Res. 2014;24:974–88. doi: 10.1101/gr.169417.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dixon JR, Selvaraj S, Yue F, Kim A, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–80. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duan Z, Andronescu M, Schutz K, McIlwain S, et al. A three-dimensional model of the yeast genome. Nature. 2010;465:363–7. doi: 10.1038/nature08973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lemieux JE, Kyes SA, Otto TD, Feller AI, et al. Genome-wide profiling of chromosome interactions in Plasmodium falciparum characterizes nuclear architecture and reconfigurations associated with antigenic variation. Mol Microbiol. 2013 doi: 10.1111/mmi.12381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–93. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sexton T, Yaffe E, Kenigsberg E, Bantignies F, et al. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–72. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 36.Lasonder E, Treeck M, Alam M, Tobin AB. Insights into the Plasmodium falciparum schizont phospho-proteome. Microbes and infection / Institut Pasteur. 2012;14:811–9. doi: 10.1016/j.micinf.2012.04.008. [DOI] [PubMed] [Google Scholar]
- 37.Miao J, Fan Q, Cui L, Li J. The malaria parasite Plasmodium falciparum histones: organization, expression, and acetylation. Gene. 2006;369:53–65. doi: 10.1016/j.gene.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 38.Treeck M, Sanders JL, Elias JE, Boothroyd JC. The phosphoproteomes of Plasmodium falciparum and Toxoplasma gondii reveal unusual adaptations within and beyond the parasites’ boundaries. Cell host & microbe. 2011;10:410–9. doi: 10.1016/j.chom.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trelle MB, Salcedo-Amaya AM, Cohen AM, Stunnenberg HG, et al. Global histone analysis by mass spectrometry reveals a high content of acetylated lysine residues in the malaria parasite Plasmodium falciparum. J Proteome Res. 2009;8:3439–50. doi: 10.1021/pr9000898. [DOI] [PubMed] [Google Scholar]
- 40.Lopez-Rubio JJ, Mancio-Silva L, Scherf A. Genome-wide analysis of heterochromatin associates clonally variant gene regulation with perinuclear repressive centers in malaria parasites. Cell host & microbe. 2009;5:179–90. doi: 10.1016/j.chom.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 41.Garcia BA, Hake SB, Diaz RL, Kauer M, et al. Organismal differences in post-translational modifications in histones H3 and H4. J Biol Chem. 2007;282:7641–55. doi: 10.1074/jbc.M607900200. [DOI] [PubMed] [Google Scholar]
- 42.Cui L, Miao J, Furuya T, Fan Q, et al. Histone acetyltransferase inhibitor anacardic acid causes changes in global gene expression during in vitro Plasmodium falciparum development. Eukaryot Cell. 2008;7:1200–10. doi: 10.1128/EC.00063-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cui L, Miao J, Cui L. Cytotoxic effect of curcumin on malaria parasite Plasmodium falciparum: inhibition of histone acetylation and generation of reactive oxygen species. Antimicrobial agents and chemotherapy. 2007;51:488–94. doi: 10.1128/AAC.01238-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chaal BK, Gupta AP, Wastuwidyaningtyas BD, Luah YH, et al. Histone deacetylases play a major role in the transcriptional regulation of the Plasmodium falciparum life cycle. PLoS Pathog. 2010;6:e1000737. doi: 10.1371/journal.ppat.1000737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bartfai R, Hoeijmakers WA, Salcedo-Amaya AM, Smits AH, et al. H2A.Z demarcates intergenic regions of the plasmodium falciparum epigenome that are dynamically marked by H3K9ac and H3K4me3. PLoS Path. 2010;6:e1001223. doi: 10.1371/journal.ppat.1001223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang L, Mu J, Zhang Q, Ni T, et al. PfSETvs methylation of histone H3K36 represses virulence genes in Plasmodium falciparum. Nature. 2013;499:223–7. doi: 10.1038/nature12361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Salcedo-Amaya AM, van Driel MA, Alako BT, Trelle MB, et al. Dynamic histone H3 epigenome marking during the intraerythrocytic cycle of Plasmodium falciparum. Proc Natl Acad Sci U S A. 2009;106:9655–60. doi: 10.1073/pnas.0902515106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chookajorn T, Dzikowski R, Frank M, Li F, et al. Epigenetic memory at malaria virulence genes. Proc Natl Acad Sci U S A. 2007;104:899–902. doi: 10.1073/pnas.0609084103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lopez-Rubio JJ, Gontijo AM, Nunes MC, Issar N, et al. 5′ flanking region of var genes nucleate histone modification patterns linked to phenotypic inheritance of virulence traits in malaria parasites. Mol Microbiol. 2007;66:1296–305. doi: 10.1111/j.1365-2958.2007.06009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ukaegbu UE, Kishore SP, Kwiatkowski DL, Pandarinath C, et al. Recruitment of PfSET2 by RNA polymerase II to variant antigen encoding loci contributes to antigenic variation in P. falciparum. PLoS Pathog. 2014;10:e1003854. doi: 10.1371/journal.ppat.1003854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brogaard K, Xi L, Wang JP, Widom J. A map of nucleosome positions in yeast at base-pair resolution. Nature. 2012;486:496–501. doi: 10.1038/nature11142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Buenrostro JD, Araya CL, Chircus LM, Layton CJ, et al. Quantitative analysis of RNA-protein interactions on a massively parallel array reveals biophysical and evolutionary landscapes. Nat Biotechnol. 2014;32:562–8. doi: 10.1038/nbt.2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jansen A, Verstrepen KJ. Nucleosome positioning in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2011;75:301–20. doi: 10.1128/MMBR.00046-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee W, Tillo D, Bray N, Morse RH, et al. A high-resolution atlas of nucleosome occupancy in yeast. Nat Genet. 2007;39:1235–44. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- 55.Mavrich TN, Jiang C, Ioshikhes IP, Li X, et al. Nucleosome organization in the Drosophila genome. Nature. 2008;453:358–62. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Raisner RM, Hartley PD, Meneghini MD, Bao MZ, et al. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell. 2005;123:233–48. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guillemette B, Bataille AR, Gevry N, Adam M, et al. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol. 2005;3:e384. doi: 10.1371/journal.pbio.0030384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tolstorukov MY, Kharchenko PV, Goldman JA, Kingston RE, et al. Comparative analysis of H2A.Z nucleosome organization in the human and yeast genomes. Genome Res. 2009;19:967–77. doi: 10.1101/gr.084830.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bunnik EM, Polishko A, Prudhomme J, Ponts N, et al. DNA-encoded nucleosome occupancy is associated with transcription levels in the human malaria parasite Plasmodium falciparum. BMC Genomics. 2014;15:347. doi: 10.1186/1471-2164-15-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ponts N, Harris EY, Lonardi S, Le Roch KG. Nucleosome occupancy at transcription start sites in the human malaria parasite: a hard-wired evolution of virulence? Infection, genetics and evolution : journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2011;11:716–24. doi: 10.1016/j.meegid.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Westenberger SJ, Cui L, Dharia N, Winzeler E. Genome-wide nucleosome mapping of Plasmodium falciparum reveals histone-rich coding and histone-poor intergenic regions and chromatin remodeling of core and subtelomeric genes. BMC Genomics. 2009;10:610. doi: 10.1186/1471-2164-10-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Segal E, Fondufe-Mittendorf Y, Chen L, Thastrom A, et al. A genomic code for nucleosome positioning. Nature. 2006;442:772–8. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ponts N, Harris EY, Prudhomme J, Wick I, et al. Nucleosome landscape and control of transcription in the human malaria parasite. Genome Res. 2010;20:228–38. doi: 10.1101/gr.101063.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tillo D, Hughes TR. G+C content dominates intrinsic nucleosome occupancy. BMC Bioinformatics. 2009;10:442. doi: 10.1186/1471-2105-10-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Segal E, Widom J. Poly(dA:dT) tracts: major determinants of nucleosome organization. Curr Opin Struct Biol. 2009;19:65–71. doi: 10.1016/j.sbi.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hoeijmakers WA, Salcedo-Amaya AM, Smits AH, Francoijs KJ, et al. H2A.Z/H2B.Z double-variant nucleosomes inhabit the AT-rich promoter regions of the Plasmodium falciparum genome. Mol Microbiol. 2013;87:1061–73. doi: 10.1111/mmi.12151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Petter M, Selvarajah SA, Lee CC, Chin WH, et al. H2A.Z and H2B.Z double-variant nucleosomes define intergenic regions and dynamically occupy var gene promoters in the malaria parasite Plasmodium falciparum. Mol Microbiol. 2013;87:1167–82. doi: 10.1111/mmi.12154. [DOI] [PubMed] [Google Scholar]
- 68.Siegel TN, Hekstra DR, Kemp LE, Figueiredo LM, et al. Four histone variants mark the boundaries of polycistronic transcription units in Trypanosoma brucei. Genes Dev. 2009;23:1063–76. doi: 10.1101/gad.1790409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cremer T, Kreth G, Koester H, Fink RH, et al. Chromosome territories, interchromatin domain compartment, and nuclear matrix: an integrated view of the functional nuclear architecture. Crit Rev Eukaryot Gene Expr. 2000;10:179–212. [PubMed] [Google Scholar]
- 70.Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 71.Takizawa T, Meaburn KJ, Misteli T. The meaning of gene positioning. Cell. 2008;135:9–13. doi: 10.1016/j.cell.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Freitas-Junior LH, Bottius E, Pirrit LA, Deitsch KW, et al. Frequent ectopic recombination of virulence factor genes in telomeric chromosome clusters of P. falciparum. Nature. 2000;407:1018–22. doi: 10.1038/35039531. [DOI] [PubMed] [Google Scholar]
- 73.Ralph SA, Scheidig-Benatar C, Scherf A. Antigenic variation in Plasmodium falciparum is associated with movement of var loci between subnuclear locations. Proc Natl Acad Sci U S A. 2005;102:5414–9. doi: 10.1073/pnas.0408883102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mancio-Silva L, Zhang Q, Scheidig-Benatar C, Scherf A. Clustering of dispersed ribosomal DNA and its role in gene regulation and chromosome-end associations in malaria parasites. Proc Natl Acad Sci. 2010;107:15117–22. doi: 10.1073/pnas.1001045107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Weiner A, Dahan-Pasternak N, Shimoni E, Shinder V, et al. 3D nuclear architecture reveals coupled cell cycle dynamics of chromatin and nuclear pores in the malaria parasite Plasmodium falciparum. Cell Microbiol. 2011;13:967–77. doi: 10.1111/j.1462-5822.2011.01592.x. [DOI] [PubMed] [Google Scholar]
- 76.Fullwood MJ, Liu MH, Pan YF, Liu J, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Guelen L, Pagie L, Brasset E, Meuleman W, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–51. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 78.van Koningsbruggen S, Gierlinski M, Schofield P, Martin D, et al. High-resolution whole-genome sequencing reveals that specific chromatin domains from most human chromosomes associate with nucleoli. Molecular biology of the cell. 2010;21:3735–48. doi: 10.1091/mbc.E10-06-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vogel MJ, Peric-Hupkes D, van Steensel B. Detection of in vivo protein-DNA interactions using DamID in mammalian cells. Nature protocols. 2007;2:1467–78. doi: 10.1038/nprot.2007.148. [DOI] [PubMed] [Google Scholar]
- 80.Zhao Z, Tavoosidana G, Sjolinder M, Gondor A, et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet. 2006;38:1341–7. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- 81.van Steensel B, Dekker J. Genomics tools for unraveling chromosome architecture. Nat Biotechnol. 2010;28:1089–95. doi: 10.1038/nbt.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nora EP, Lajoie BR, Schulz EG, Giorgetti L, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–5. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sofueva S, Yaffe E, Chan WC, Georgopoulou D, et al. Cohesin-mediated interactions organize chromosomal domain architecture. The EMBO journal. 2013;32:3119–29. doi: 10.1038/emboj.2013.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ferraiuolo MA, Rousseau M, Miyamoto C, Shenker S, et al. The three-dimensional architecture of Hox cluster silencing. Nucleic Acids Res. 2010;38:7472–84. doi: 10.1093/nar/gkq644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rousseau M, Crutchley JL, Miura H, Suderman M, et al. Hox in motion: tracking HoxA cluster conformation during differentiation. Nucleic Acids Res. 2014;42:1524–40. doi: 10.1093/nar/gkt998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li G, Ruan X, Auerbach RK, Sandhu KS, et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148:84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shen Y, Yue F, McCleary DF, Ye Z, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–20. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tanizawa H, Iwasaki O, Tanaka A, Capizzi JR, et al. Mapping of long-range associations throughout the fission yeast genome reveals global genome organization linked to transcriptional regulation. Nucleic Acids Res. 2010;38:8164–77. doi: 10.1093/nar/gkq955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Magklara A, Yen A, Colquitt BM, Clowney EJ, et al. An epigenetic signature for monoallelic olfactory receptor expression. Cell. 2011;145:555–70. doi: 10.1016/j.cell.2011.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lyons DB, Allen WE, Goh T, Tsai L, et al. An epigenetic trap stabilizes singular olfactory receptor expression. Cell. 2013;154:325–36. doi: 10.1016/j.cell.2013.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Brancucci NM, Bertschi NL, Zhu L, Niederwieser I, et al. Heterochromatin protein 1 secures survival and transmission of malaria parasites. Cell host & microbe. 2014;16:165–76. doi: 10.1016/j.chom.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 92.Coleman BI, Skillman KM, Jiang RH, Childs LM, et al. A Plasmodium falciparum histone deacetylase regulates antigenic variation and gametocyte conversion. Cell host & microbe. 2014;16:177–86. doi: 10.1016/j.chom.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Le Roch KG, Zhou Y, Blair PL, Grainger M, et al. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science. 2003;301:1503–8. doi: 10.1126/science.1087025. [DOI] [PubMed] [Google Scholar]
- 94.Otto TD, Wilinski D, Assefa S, Keane TM, et al. New insights into the blood-stage transcriptome of Plasmodium falciparum using RNA-Seq. Mol Microbiol. 2010;76:12–24. doi: 10.1111/j.1365-2958.2009.07026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Voss TS, Healer J, Marty AJ, Duffy MF, et al. A var gene promoter controls allelic exclusion of virulence genes in Plasmodium falciparum malaria. Nature. 2006;439:1004–8. doi: 10.1038/nature04407. [DOI] [PubMed] [Google Scholar]
- 96.Dahan-Pasternak N, Nasereddin A, Kolevzon N, Pe’er M, et al. PfSec13 is an unusual chromatin-associated nucleoporin of Plasmodium falciparum that is essential for parasite proliferation in human erythrocytes. J Cell Sci. 2013;126:3055–69. doi: 10.1242/jcs.122119. [DOI] [PubMed] [Google Scholar]
- 97.Umbarger MA, Toro E, Wright MA, Porreca GJ, et al. The three-dimensional architecture of a bacterial genome and its alteration by genetic perturbation. Mol Cell. 2011;44:252–64. doi: 10.1016/j.molcel.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gomez-Diaz E, Corces VG. Architectural proteins: regulators of 3D genome organization in cell fate. Trends Cell Biol. 2014 doi: 10.1016/j.tcb.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ghorbal M, Gorman M, Macpherson CR, Martins RM, et al. Genome editing in the human malaria parasite Plasmodium falciparum using the CRISPR-Cas9 system. Nat Biotechnol. 2014;32:819–21. doi: 10.1038/nbt.2925. [DOI] [PubMed] [Google Scholar]
- 100.Zhang C, Xiao B, Jiang Y, Zhao Y, et al. Efficient Editing of Malaria Parasite Genome Using the CRISPR/Cas9 System. mBio. 2014:5. doi: 10.1128/mBio.01414-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wagner JC, Platt RJ, Goldfless SJ, Zhang F, et al. Efficient CRISPR-Cas9-mediated genome editing in Plasmodium falciparum. Nat Methods. 2014 doi: 10.1038/nmeth.3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bozdech Z, Mok S, Hu G, Imwong M, et al. The transcriptome of Plasmodium vivax reveals divergence and diversity of transcriptional regulation in malaria parasites. Proc Natl Acad Sci U S A. 2008;105:16290–5. doi: 10.1073/pnas.0807404105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fernandez-Becerra C, Pein O, de Oliveira TR, Yamamoto MM, et al. Variant proteins of Plasmodium vivax are not clonally expressed in natural infections. Mol Microbiol. 2005;58:648–58. doi: 10.1111/j.1365-2958.2005.04850.x. [DOI] [PubMed] [Google Scholar]
- 104.Le Roch KG, Johnson JR, Florens L, Zhou Y, et al. Global analysis of transcript and protein levels across the Plasmodium falciparum life cycle. Genome Res. 2004;14:2308–18. doi: 10.1101/gr.2523904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–81. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 106.Kim TH, Barrera LO, Zheng M, Qu C, et al. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–80. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang Z, Zang C, Rosenfeld JA, Schones DE, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Barski A, Cuddapah S, Cui K, Roh TY, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 109.Nishida H, Suzuki T, Kondo S, Miura H, et al. Histone H3 acetylated at lysine 9 in promoter is associated with low nucleosome density in the vicinity of transcription start site in human cell. Chromosome research : an international journal on the molecular, supramolecular and evolutionary aspects of chromosome biology. 2006;14:203–11. doi: 10.1007/s10577-006-1036-7. [DOI] [PubMed] [Google Scholar]
- 110.Mikkelsen TS, Ku M, Jaffe DB, Issac B, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–60. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lachner M, Sengupta R, Schotta G, Jenuwein T. Trilogies of histone lysine methylation as epigenetic landmarks of the eukaryotic genome. Cold Spring Harb Symp Quant Biol. 2004;69:209–18. doi: 10.1101/sqb.2004.69.209. [DOI] [PubMed] [Google Scholar]
- 112.Chantalat S, Depaux A, Hery P, Barral S, et al. Histone H3 trimethylation at lysine 36 is associated with constitutive and facultative heterochromatin. Genome Res. 2011;21:1426–37. doi: 10.1101/gr.118091.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zlatanova J, Thakar A. H2A.Z: view from the top. Structure. 2008;16:166–79. doi: 10.1016/j.str.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 114.Talbert PB, Henikoff S. Histone variants--ancient wrap artists of the epigenome. Nat Rev Mol Cell Biol. 2010;11:264–75. doi: 10.1038/nrm2861. [DOI] [PubMed] [Google Scholar]
- 115.Bannister LH, Margos G, Hopkins JM. Making a home for Plasmodium post-genomics: ultrastructural organization of the blood stages. In: Sherman IW, editor. Molecular Approaches to Malaria. Washington DC: ASM Press; 2005. pp. 24–49. [Google Scholar]
- 116.Hoeijmakers WA, Flueck C, Francoijs KJ, Smits AH, et al. Plasmodium falciparum centromeres display a unique epigenetic makeup and cluster prior to and during schizogony. Cell Microbiol. 2012;14:1391–401. doi: 10.1111/j.1462-5822.2012.01803.x. [DOI] [PubMed] [Google Scholar]
- 117.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–95. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]