Abstract
In cyanobacteria many compounds, including chlorophylls, carotenoids, and hopanoids, are synthesized from the isoprenoid precursors isopentenyl diphosphate (IPP) and dimethylallyl diphosphate. Isoprenoid biosynthesis in extracts of the cyanobacterium Synechocystis strain PCC 6803 grown under photosynthetic conditions, stimulated by pentose phosphate cycle substrates, does not appear to require methylerythritol phosphate pathway intermediates. The sll1556 gene, distantly related to type 2 IPP isomerase genes, was disrupted by insertion of a Kanr cassette. The mutant was fully viable under photosynthetic conditions although impaired in the utilization of pentose phosphate cycle substrates. Compared to the parental strain the Δsll1556 mutant (i) is deficient in isoprenoid biosynthesis in vitro with substrates including glyceraldehyde-3-phosphate, fructose-6-phosphate, and glucose-6-phosphate; (ii) has smaller cells (diameter ca. 13% less); (iii) has fewer thylakoids (ca. 30% less); and (iv) has a more extensive fibrous outer wall layer. Isoprenoid biosynthesis is restored with pentose phosphate cycle substrates plus the recombinant Sll1556 protein in the Δsll1556 supernatant fraction. IPP isomerase activity could not be demonstrated for the purified Sll1556 protein under our in vitro conditions. The reduction of thylakoid area and the effect on outer wall layer components are consistent with an impairment of isoprenoid biosynthesis in the mutant, possibly via hopanoid biosynthesis. Our findings are consistent with an alternate metabolic shunt for biosynthesis of isoprenoids.
Isoprenoids are required for many cell processes including photosynthesis, membrane stability, electron transport, and the cellular production of carotenoids, rubber, and fragrances. More than 30,000 isoprenoid compounds have been described. Synthesis of the essential 5-carbon building blocks of isoprenoids, isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP), can be attributed to one of two pathways. The mevalonic acid pathway, the sole pathway in animals and in many bacteria, also occurs in the cytoplasm of plant cells, where it is responsible for synthesis of sterols and ubiquinones (18, 20). The 2-C-methyl-d-erythritol-4-phosphate (MEP) pathway occurs in Escherichia coli and many other bacteria as well as in cyanobacteria. It is also present in chloroplasts, where it provides substrates for the synthesis of carotenoids, chlorophylls, and quinones. This pathway is believed important for cyanobacterial photosynthetic pigment biosynthesis including that of carotenoids and the phytolation of chlorophyll and in hopanoid synthesis of bacterial and cyanobacterial cell walls and membranes (8, 20, 24). Most of the MEP pathway genes have been functionally verified in the gram-negative heterotrophic bacterium E. coli (reviewed in references 4, 14, 18, and 20), and this bacterium has thus become the standard for defining the MEP pathway in other organisms. The MEP pathway is typically represented as a linear sequence of reactions commencing with pyruvate (PYR) and glyceraldehyde-3-phosphate (GA3P) as substrates with a condensation to 1-deoxy-d-xylulose-5-phosphate (DXP), leading to the synthesis of IPP and DMAPP, and is usually assumed to involve an IPP isomerase for IPP and DMAPP interconversion (4, 14, 18, 20).
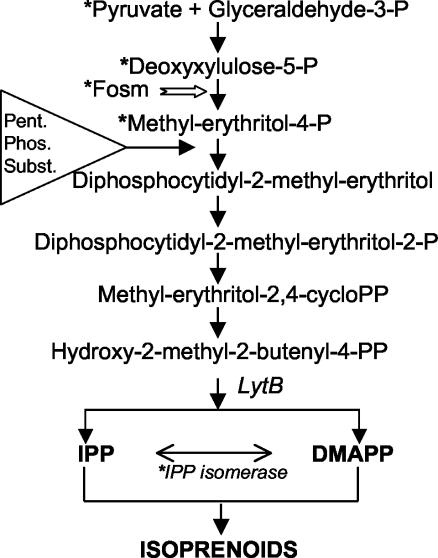
Our studies have concentrated on the photosynthetic prokaryote Synechocystis strain PCC 6803, which possesses homologs of all the essential genes for the MEP pathway (10). These recent studies (5, 6) provided evidence that isoprenoid biosynthesis in cells grown photoautotrophically exhibits some major differences from the pathway predicted from E. coli as summarized in Fig. 1. This cyanobacterium does not utilize the predicted MEP pathway substrates PYR and DXP in vitro. Instead it utilizes products of photosynthesis as substrates. In addition it was shown that isoprenoid biosynthesis in cultures of Synechocystis strain PCC 6803 was not affected in vivo or in vitro by fosmidomycin (6), the inhibitor of the key MEP enzyme deoxyxylulose phosphate reductoisomerase. Collectively, these results strongly suggest that alternate substrate paths are used in isoprenoid biosynthesis. In accordance with our data, we proposed that in this cyanobacterium the linear MEP pathway as defined for E. coli is not the sole pathway by which isoprenoids are synthesized under photosynthetic conditions but rather that products of the pentose phosphate cycle serve as substrates, and it was hypothesized that they could subsequently enter the MEP pathway downstream of MEP (Fig. 1; also Fig. 6 of reference 6). It was previously found that LytB, which is at the branch point of IPP and DMAPP synthesis, is essential for the survival of Synechocystis strain PCC 6803 (3) and therefore is likely required for the formation of IPP and/or DMAPP. It is generally expected that an IPP isomerase is involved in DMAPP production from IPP (13, 14, 16). However, in our previous work with Synechocystis strain PCC 6803 we were unable to demonstrate IPP isomerase activity consonant with a lack of the type 1 IPP isomerase gene (10), and it was suggested that DMAPP and IPP are separately generated (5, 6), which had been independently also suggested for plant cell cultures (1). It should be noted that a possible homolog of a type 2 IPP isomerase in Synechocystis, similar to the Streptomyces type 2 IPP isomerase (9), was suggested from sequence identity (32%) from the sll1556 open reading frame (ORF). To test this possibility, we created the knockout mutant Δsll1556 and examined the recombinant protein from this ORF for IPP isomerase activity.
FIG. 1.
The isoprenoid biosynthetic MEP pathway as determined for E. coli beginning with PYR plus GA3P leading to the formation of MEP from DXP and through subsequent steps via the LytB enzyme to IPP and DMAPP (3, 14, 17, 18, 19, 20, 30). Fosmidomycin (white arrow) inhibits the synthesis of MEP and blocks the growth of the bacterium. Asterisks denote differences in Synechocystis strain PCC 6803 where PYR, DXP, and MEP do not serve as substrates in vitro; where IPP isomerase activity has not been observed; and where fosmidomycin does not inhibit isoprenoid biosynthesis or growth in the cyanobacterium (6), and the triangle indicates where pentose phosphate cycle substrates may possibly enter downstream of MEP.
FIG. 6.
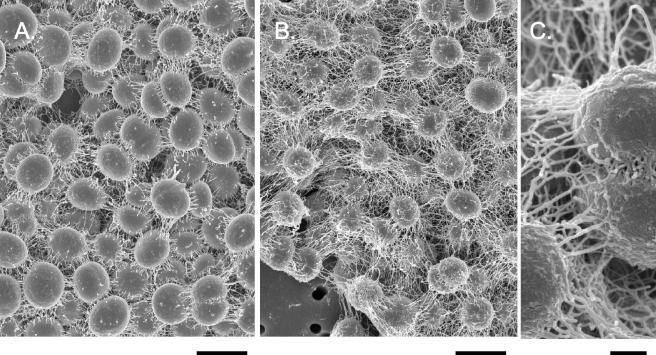
Scanning electron micrographs of Synechocystis strain PCC 6803 WT (A) and Δsll1556 mutant (B) cells from log-phase cultures (as is evident from several new daughter cells that are kidney shaped). The WT cells have a relatively smoother outer surface than do the mutant cells, and the mutant cell surfaces have greater fibrous extensions, shown at greater magnification in panel C, than do the WT cells. Bars, 2 (A and B) and 0.15 (C) μm.
In the present work we report that Sll1556 is not essential but that the nonlethal Synechocystis strain PCC 6803 mutant was impaired in the utilization of pentose phosphate cycle substrates for isoprenoid biosynthesis in vitro. The impairment is correlated with a significant reduction of thylakoid membranes in the mutant and an increase in outer wall components. We suggest that DMAPP and IPP biosynthesis may occur by more than one metabolic path leading to isoprenoid biosynthesis.
MATERIALS AND METHODS
Cell culture and fractionation.
Liquid cultures of the glucose-tolerant Synechocystis strain PCC 6803 (obtained from Wim Vermaas, Arizona State University) were routinely grown at ca. 30°C in continuous light (15 to 20 μmol/m2/s) with continuous shaking and slow bubbling of 5% CO2 in air. The parental strain (wild type [WT]) of Synechocystis, referred to throughout the paper as WT, is the widely used glucose-tolerant strain (29). The culture medium BG-11 was supplemented with 5 mM potassium-TES [N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid, pH 8.3].
For in vitro assays Synechocystis strain PCC 6803 cells were harvested in the log phase of growth. Cells pelleted by centrifugation were quickly rinsed in 100 mM HEPES-KOH (pH 7.7) and 1 mM dithiothreitol (DTT), treated with lysozyme (10 mg/ml, 60 min, 37°C), rinsed in the same buffer, and broken in a French pressure cell (20,000 lb/in2, 4°C). The supernatant fraction (3 to 5.5 mg of protein per ml), after centrifugation at 60,000 × g for 1 h, was stored at −80°C for use within less than 3 weeks. An alternate improvement in the procedure was to prepare the 60,000 × g supernatant directly from pelleted frozen cells (−80°C; 0.7 ml), previously rinsed in 100 mM HEPES-1 mM DTT (pH 7.7) and broken (for 30 s with intermittent cooling, four times) in buffer with a Mini-Bead beater (Biospec Products Inc., Bartlesville, Okla.). Also, the reaction mixture was preincubated with unlabeled IPP for 20 min (37°C) before addition of [14C]IPP and further incubation and assayed from 0 to 60 min. The preincubation served to deplete any residual DMAPP that might have been present in the cells extracts. It should be noted that the results were essentially the same as with the previous procedure (6), except that variation in activity with time of stored supernatant was eliminated, as was a small stimulation from residual MEP previously observed (as in Fig. 3 of reference 6).
FIG. 3.
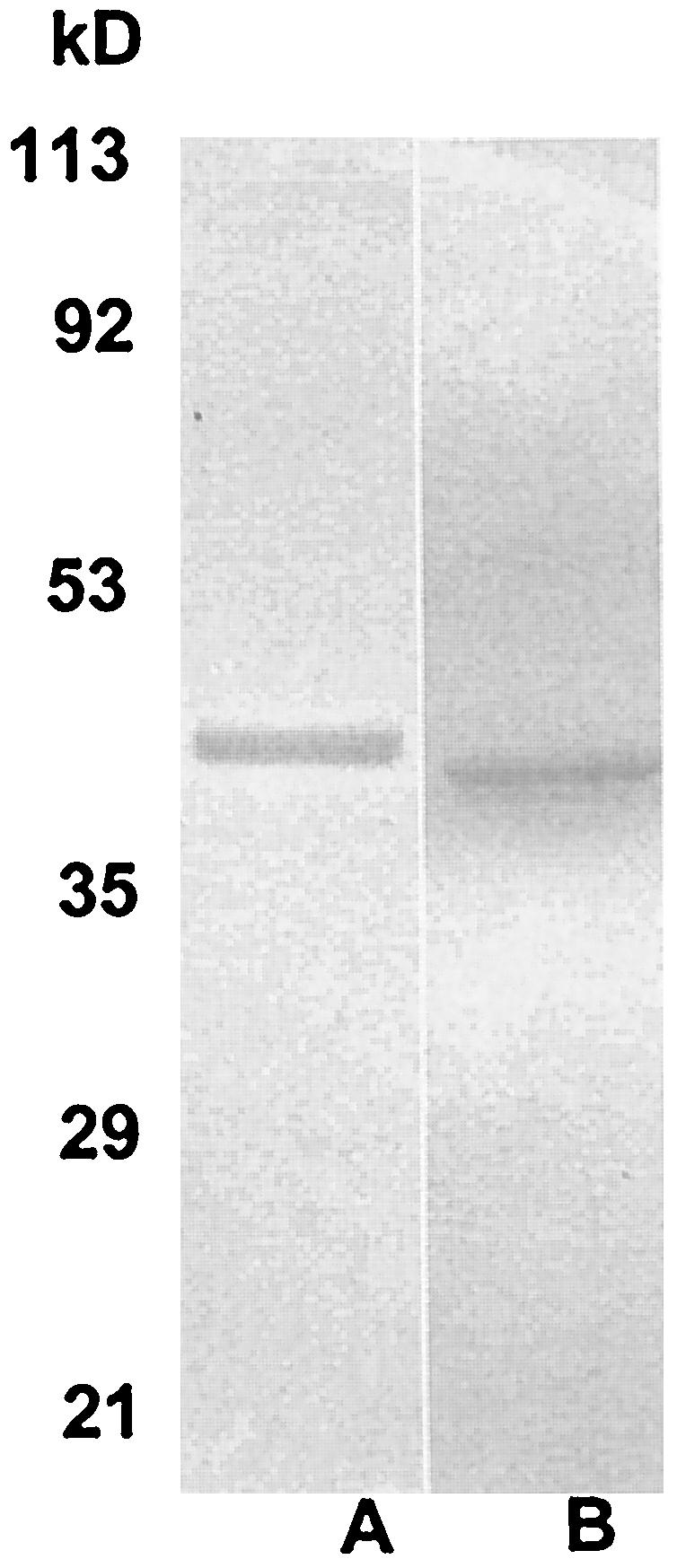
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of His-tagged recombinant proteins purified on Ni-NTA columns and stained with Coomassie blue. (A) Sll1556 protein (ca. 41.6 kDa) of Synechocystis strain PCC 6803; (B) type 2 IPP isomerase (37 kDa) of Streptomyces sp. strain CL190. The molecular mass range is as indicated on the left.
Isoprenoid biosynthesis with radiolabeled IPP incorporation.
The incorporation of [14C]IPP into the acid-labile fraction of a petroleum ether extract (6) was used as an indirect assay for DMAPP synthesis in extracts of Synechocystis strain PCC 6803 cells. The reaction mixture was composed of the cell-free supernatant fraction (60,000 × g) with 100 mM HEPES-KOH (pH 7.7), 5 mM glutathione, 5 mM MgCl2, 2.5 mM MnCl2, 500 μM ATP, 250 μM CTP, 100 μM thiamine-PP, 10 μM coenzyme B12 (5′-deoxyadenosyl-cobalamine), 1 mM NADPH, 500 μM NADP, and 1 mM flavin adenine dinucleotide (FAD). Each incubation was carried out in a total volume of 1 ml with a final concentration of 8.5 μM [1-14C]IPP (Amersham) (and 8.25 × 105 dpm/ml) at 37°C. Following a 20-min preincubation with 3.0 μM IPP, [14C]IPP and compounds tested were added individually or in combination: DMAPP (2 μM), DXP (500 μM) (Echelon Research Labs Inc., Salt Lake City, Utah), GA3P (1 mM), fructose-6-phosphate (FR6P; 500 μM), glucose-6 phosphate (GL6P; 500 μM), MEP (500 μM) (Echelon Research Labs. Inc.), and PYR (500 μM). Aliquots of 0.2 ml (0.2 to 0.46 mg of protein) were assayed for radioactivity.
The incorporation of [14C]IPP into allylic prenols was verified by extraction into petroleum ether (boiling point, 55 to 110°C) after hydrolysis (0.5 N HCl, 37°C, 20 min) as in the work of Ershov et al. (5, 6), and each extracted sample was counted in 10 ml of ScintiSafe Econo 2 cocktail (Fisher Scientific). Verification that [14C]IPP was incorporated into isoprenoids had been previously established by reversed-phase column chromatography (5). Each 0.5-ml sample of the petroleum ether extract was applied to a silica gel 6, RP-18 (EM Industries) column (24 by 1 cm), previously equilibrated in and then eluted with 100% acetonitrile. The following alcohols served as calibration standards: isopentenyl alcohol (C5), geraniol (C10), linalool (C10), farnesol (C15), nerolidol (C15), and geranyl geraniol (C20) (Aldrich). Controls for phosphatase activity showed that the petroleum ether extract had very few counts without acid hydrolysis, indicating little or no phosphatase activity. Furthermore, the 14C incorporation into isoprenoids was time course dependent, where longer chains, i.e., >C10, became predominant with increased time of incubation (data not shown).
Cloning and disruption of Synechocystis strain PCC 6803 gene sll1556.
The nucleotide sequence of Synechocystis strain PCC 6803 ORF sll1556 (10) was obtained at Cyanobase (http://www.kazusa.or.jp/cyanobase/). Oligonucleotide primers sll1556N (GAGAGGATCCATGGATAGCACCCCCCACCGTAAG; the initiation codon is underlined) and sll1556C (TCGTCAACCAGAGCAAAATGTC) were designed, with the assistance of the program Primer3 (21), to amplify the entire ORF and introduce NcoI and BamHI sites at the N terminus. Genomic DNA was extracted from a Synechocystis strain as earlier described (29). The PCR was performed using an MJ Research (Waltham, Mass.) PTC-150-25 MiniCycler with a heated lid. The Advantage-HF2 PCR kit (Clontech Laboratories, Inc.) was used in a reaction volume of 50 μl in 100-μl thin-walled tubes. After an initial denaturation for 1 min at 94°C, amplification was for 30 cycles at 94, 60, and 68°C for 10, 60, and 150 s, respectively, with a final extension for 10 min at 68°C. The amplified sll1556 PCR product was purified by extraction with phenol-chloroform, precipitated with sodium acetate and ethanol, washed with 70% ethanol, dried, and resuspended in Tris-EDTA (TE) buffer (23). After digestion with BamHI, a 1.1-kb fragment containing sll1556 was purified by gel electrophoresis (1% agarose), recovered using the Geneclean kit (Bio 101, Inc., Carlsbad, Calif.), and cloned in frame in the BglII and PvuII sites of plasmid vector pBAD/His-B (Invitrogen) to give plasmid pBAD/His-sll1556. The sll1556 insert was sequenced to confirm the reading frame and the absence of mutations introduced by PCR.
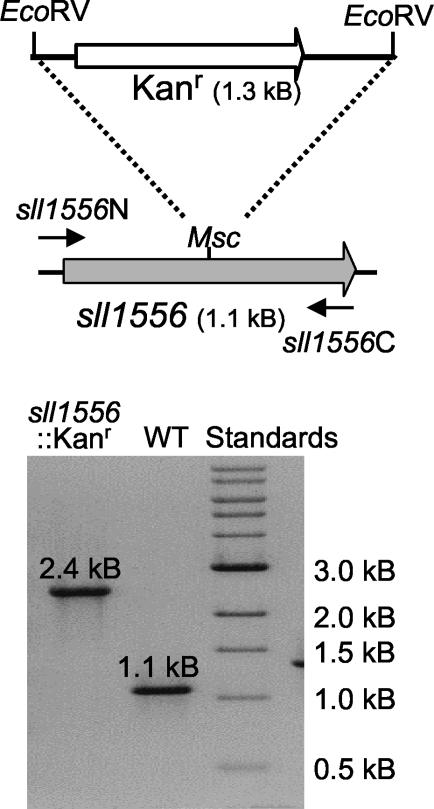
A kanamycin resistance gene, originally obtained from Tn903, was excised as a 1.3-kb EcoRV-EcoRV fragment and cloned into the MscI site within sll1556 (Fig. 2) to give plasmid pBAD/His-sll1556Kan. This plasmid was linearized by digestion with EcoRI (a site present in the multiple cloning site of the vector) and then extracted with phenol-chloroform, precipitated, and resuspended in TE as described above. This linearized plasmid preparation was used to transform Synechocystis strain PCC 6803 essentially as described by Williams (29). Segregation of the sll1556::Kanr (Δsll1556) mutant was confirmed by PCR (Fig. 2) with the primers and reaction conditions described above.
FIG. 2.
(Top) Schematic illustration of Synechocystis strain PCC 6803 genomic DNA encompassing gene sll1556 (gray arrow). A kanamycin-resistance gene (Kanr; white arrow) was inserted in the MscI site to inactivate the gene. (Bottom) Agarose gel electrophoresis of PCR products obtained using genomic DNA from the WT strain (WT) and mutant sll1556::Kanr indicates complete segregation of the mutant (i.e., all gene copies have been inactivated).
Expression and purification of proteins.
E. coli strain Rosetta (Novagen) containing plasmid pBAD/His-sll1556 (see above) was grown in Luria-Bertani medium supplemented with ampicillin (150 μg/ml) at 30°C at 300 cycles of shaking/min until the culture reached an A600 of approximately 0.5. At this point 20% arabinose was added to give a final concentration of 0.2%. After 16 h of further growth at 20°C, the culture flask was chilled in ice water and cells were harvested at 500 × g for 10 min at 4°C. Well-drained cell pellets were frozen at −80°C. The recombinant protein was also produced using growth at 30°C with 3 h of induction in Top10 cells (Invitrogen).
For production of recombinant Streptomyces type 2 IPP isomerase, conditions were the same except that isopropyl-β-d-thiogalactopyranoside (IPTG; 2 mM final concentration) was used as the inducer, with cells being grown at 20°C for 16 h before harvest. Cells contained the plasmid pQCLD41 (9).
Recombinant proteins were purified after cell breakage by sonication and eluted from a Ni-nitrilotriacetic acid (NTA) column with 100 mM imidazole-Tris-HCl, pH 8.0. Alternatively, cells were extracted with the B-PER bacterial protein extraction reagent (Pierce Biotechnology Co.) and the recombinant protein was purified on Ni-NTA minicolumns (Qiagen, Chatsworth, Calif.) by elution with buffered 125 mM imidazole (50 mM phosphate, 300 mM NaCl, 1 mM DTT, pH 8.0). In Fig. 3 are shown the purified proteins on sodium dodecyl sulfate-polyacrylamide gels after Coomassie blue staining. The size estimates of 41.6 kDa for Sll1556 of Synechocystis sp. strain PCC 6803 and of 37 kDa for the type 2 IPP isomerase of Streptomyces sp. strain CL190, with the use of molecular mass markers (Low Range) from Bio-Rad Laboratories, are consistent with the expected molecular masses (including the His tag).
Enzymatic activity tests. (i) IPP isomerase assay.
For the type 2 IPP isomerase assay we used the procedure developed by Kaneda et al. (9). Assays were conducted with FAD with and without NADPH or with flavin mononucleotide (FMN) with and without NADPH. We thus confirmed the requirement for FAD (or FMN) and NADPH. Briefly, the activity was measured by [14C]IPP incorporation into the petroleum ether layer, dependent on the acid lability of DMAPP as described above. Each reaction mixture at 37°C (0.5-ml final volume) contained 8.5 μM [14C]IPP, 100 mM HEPES (pH 7.7), 2 mM DTT, 5 mM MgCl2, 5 mM MnCl2, 1 mM NADPH, 1 mM FAD (or FMN), and recombinant protein (over a range of 0.1 to 50 μg/0.5 ml). We also used the standard assay developed for the type 1 IPP isomerase according to the work of Spurgeon et al. (25) to ascertain activity of the recombinant proteins with and without cell supernatants.
(ii) Glycolate oxidase.
The Sll1556 protein was tested for glycolate oxidase activity by two separate methods. The first was according to the work of Zelitch (33), involving the loss of color at 620 nm of 2,6-dichlorophenylindophenol (DCPIP). The oxidase assay in 1.0 ml (final volume) contained 30 mM potassium phosphate buffer (pH 8.0), 0.003% DCPIP, with or without 5 mM FMN, 42 mM imidazole, 30 mM potassium cyanide, and Sll1556 protein (ca. 3 μg). After a 20-min incubation the validity of the glycolate oxidase assay was verified by addition of spinach glycolate oxidase (EC 1.1.2.15) (ca. 5 μg). Phenylhydrazone formation was used for the second assay by measuring the increased optical density at 324 nm. Reaction conditions were as described above, except that 3 mM phenylhydrazine replaced DCPIP and cyanide, and the assay validity was verified by addition of glyoxylic acid.
(iii) Lactate dehydrogenase.
The Sll1556 protein was tested for lactate dehydrogenase activity by the decrease of optical density at 340 nm by modifying the procedure in reference 12. A 1.0-ml incubation mixture contained 30 mM Tris-HCl, 5 mM MgCl2, with or without 5 mM FMN, 50 mM pyruvic acid, 1 mM NADH (or NADPH), 1 mM DTT, and Sll1556 protein (ca. 3 μg). Lactate dehydrogenase in NADH (rabbit muscle EC 1.1.1.27) was added to validate the incubation conditions.
Protein concentrations were determined as in the work of Ershov et al. (5) by the bicinchoninic acid assay and the Micro BCA protein assay (Pierce Biotechnology Co.) with bovine serum albumin as the standard. Except where otherwise indicated, the chemicals used in this study were purchased from Sigma Chemical Co.
Electron microscopy.
Cells from the log phase of growth, rinsed in 0.25 M phosphate buffer (pH 7.0), were fixed in 2.0% phosphate-buffered glutaraldehyde (2 h) and rinsed several times in buffer. The secondary fixation in 1% OsO4 (2 h) was also followed by several phosphate buffer rinses. For transmission electron microscopy cells were dehydrated in ethanol and propylene oxide before embedment in Epon resin. Sections were stained with 1% uranyl acetate and lead citrate and examined in a Zeiss EM10 CA microscope.
For scanning electron microscopy cells were fixed in glutaraldehyde as described above and were then collected on a Nuclepore (0.5-μm-pore-size) filter, dehydrated in ethanol (75 to 100%) for critical point drying, and shadowed with gold-palladium in a Denton Vacuum DCP-1 apparatus for examination with a Hitachi S-4700 scanning electron microscope.
Multiple sequence alignment and construction of neighbor-joining tree.
The predicted amino acid sequences of bacterial type 2 IPP isomerases and polypeptides of undetermined function related to these enzymes were aligned using the program Clustal X (27) with the Blosum series weight matrix. All parameters were set at their default values. A neighbor-joining tree (22) was constructed from the alignment, with gaps excluded from the analysis and correction made for multiple substitutions (11). One thousand bootstrap trials were carried out with a random number generator seed of 111. The tree was displayed using the program NJplot (15).
RESULTS
Sll1556 and IPP isomerase activity in vitro.
The gene sll1556 of Synechocystis strain PCC 6803 was identified as a probable type 2 IPP isomerase based on 32% amino acid sequence identity with a homologous gene of Streptomyces (9). In fact, Kaneda et al. (9) were the first to identify the FMN- or FAD-plus-NADPH-requiring novel type 2 IPP isomerase, which lacks homology with the well-characterized IPP isomerase (type 1) (7, 16, 25). The type 2 IPP isomerase activity has also been shown in Bacillus subtilis (26) and in Staphylococcus aureus (9) (Table 1). In several protein databases the ORF sll1556 is annotated as IPP isomerase type 2 or as FMN-dependent lactate dehydrogenase. Genes related to sll1556 of Synechocystis strain PCC 6803 are also found in a number of other cyanobacteria. However, notably absent are predicted homologs in three strains of Prochlorococcus marinus (SS 120, MED 4, and MIT 9313) as well as in Synechococcus sp. strain WH 8102 and “Gloeobacter violaceus” PCC 742 (Table 1). To our knowledge, activity for neither IPP isomerase (type 2) nor lactate dehydrogenase has been functionally demonstrated for Sll1556 nor for a related protein in any of the cyanobacteria listed in Table 1. This prompted us to first assay the IPP isomerase activity of the expressed Sll1556 recombinant protein of Synechocystis and to examine the consequences of inactivating this gene.
TABLE 1.
Demonstrated IPP isomerase function in bacteria and genes (by gene bank identification numbers) related to IPP isomerase in cyanobacteria
| Organism | Type 1 | Type 2 | Source reference or databasea |
|---|---|---|---|
| Organisms with IPP isomerase function demonstrated: | |||
| Escherichia coli | + | 7 | |
| Bacillus subtilis | + | 26 | |
| Streptomyces sp. strain CL190 | + | 9 | |
| Staphylococcus aureus | + | 9 | |
| Cyanobacteria with genes related to type 2: | |||
| Anabaena (Nostoc) strain PCC 7120 | gi:17133727 (all4519) | Kazusa DNA Research Institute, Japan | |
| “Nostoc punctiforme” PCC 73102 | gi:23128293 | DOE Joint Genome Institute, USA | |
| Synechocystis strain PCC 6803 | gi:2829616 (sll1556) | Kazusa DNA Research Institute, Japan | |
| “Synechococcus elongatus” PCC 7942 | Draft genome prediction | DOE Joint Genome Institute, USA | |
| “Thermosynechococcus elongatus” BP-1 | gi:22295127 (tll1403) | Kazusa DNA Research Institute, Japan | |
| “Trichodesmium erythraeum” BP-1 | gi:23041526 | DOE Joint Genome Institute, USA | |
| Cyanobacteria lacking type 2-related genes: | |||
| “Gloeobacter violaceus” PCC 742 | Kazusa DNA Research Institute, Japan | ||
| Prochlorococcus marinus SS 120 | Genoscope, France | ||
| Prochlorococcus marinus MED 4 | DOE Joint Genome Institute, USA | ||
| Prochlorococcus marinus MIT 9313 | DOE Joint Genome Institute, USA | ||
| Synechococcus sp. strain WH 8102 | DOE Joint Genome Institute, USA |
Kazusa DNA Research Institute, Kazusa, Japan; DOE Joint Genome Institute, Walnut Creek, Calif.; Genoscope, Evry, France. USA, United States of America.
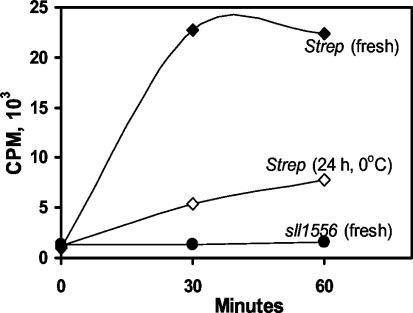
IPP isomerase activity could not be demonstrated (Fig. 4) with the recombinant Sll1556 protein purified on Ni-NTA columns (Fig. 3) under the conditions used for the homologous Streptomyces gene product. Yet, as seen in Fig. 4, IPP isomerase activity was obtained with the recombinant protein of Streptomyces, consistent with the previous report (9). Our results confirm the initial observation that the type 2 isomerase requires FAD (or FMN) and NADPH (data not shown). The recombinant enzyme of Streptomyces was very active initially but declined over time at 0°C (Fig. 4), and its activity was totally abolished by freezing with or without 50% glycerol. Numerous alternate purification conditions were also tried in isolating the protein including breakage of E. coli cells by sonication, decreasing or omitting DTT, Tris-HCl (pH 7.9) buffer substitution, FMN presence throughout, and varying the protein concentration, but definitive IPP isomerase activity could not be demonstrated for the sll1556 gene product. It was concluded that if this gene product is an IPP isomerase it must require conditions as yet unknown or that the protein has a different function.
FIG. 4.
[14C]IPP conversion to DMAPP (as described in Materials and Methods) by IPP isomerase type 2 (of Streptomyces sp. strain CL190) but not by the Sll1556 protein (of Synechocystis strain PCC 6803). Symbols: •, freshly purified recombinant Sll1556 protein of Synechocystis strain PCC 6803 (1.0 μg/500 μl); ⧫, Streptomyces sp. freshly purified enzyme (0.57 μg/500 μl); ⋄, Streptomyces sp. freshly purified enzyme stored for 24 h at 0°C (1.14 μg/500 μl). The Sll1556 protein lacks IPP isomerase activity, whereas freshly purified Streptomyces sp. strain CL190 protein is clearly active, and this activity declined upon storage. The incubation mixture consisted of 8.5 μM [14C]IPP, 100 mM HEPES, 2 mM DTT, 5 mM MgCl2, 5 mM MnCl2, 1 mM NADPH, and 1 mM FAD, pH 7.7 (37°C).
Pentose phosphate cycle substrate stimulation of isoprenoid biosynthesis is impaired in the Δsll1556 mutant.
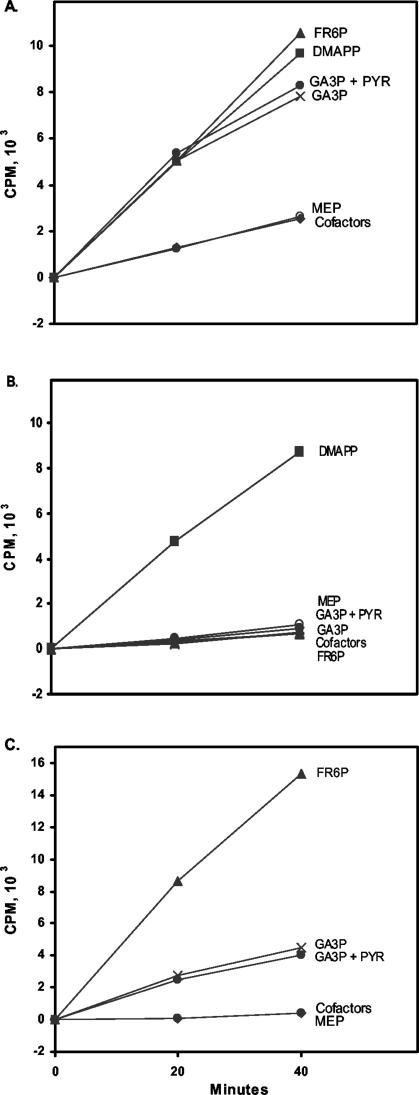
If the sll1556 gene product is involved in isoprenoid biosynthesis, then this should be reflected in the mutant where the gene has been inactivated (as in Fig. 2). Isoprenoid biosynthesis, requiring both DMAPP and IPP for formation of compounds of C10 and greater, was measured in vitro for WT and Δsll1556 as in the work of Ershov et al. (6). Basically, the [14C]IPP incorporated in the cell supernatant extract (60,000 × g) is retrieved in the petroleum ether fraction (after acid hydrolysis). Analysis by reverse-phase chromatography verified that the labeled isoprenoids were compounds of C10 and greater (5) (data not shown). This incorporation into isoprenoids larger than C5 indicates DMAPP production. Furthermore, stimulation of isoprenoid biosynthesis was obtained by the addition of exogenous DMAPP with both WT and Δsll1556 mutant supernatant (Fig. 5A and B, respectively). Hence, we equate [14C]IPP uptake with DMAPP production under our in vitro conditions. Also, in WT supernatant [14C]IPP stimulation was observed with pentose phosphate cycle intermediates (Fig. 5A), in addition to stimulation by DMAPP. Typical MEP pathway intermediates such as MEP, PYR, and DXP (data not shown) did not stimulate [14C]IPP incorporation above that obtained with the incubation cofactor mixture alone in WT. Pentose phosphate cycle intermediates (such as GL6P and FR6P) which may feed into the MEP pathway downstream (or possibly at alternate sites) were again active as had been previously shown for the WT (6).
FIG. 5.
(A) [14C]IPP incorporation into the isoprenoid fraction of the supernatant of WT is stimulated by FR6P and GA3P, indicating DMAPP production. (B) In the supernatant of the Δsll1556 mutant the same substrates failed to stimulate [14C]IPP incorporation. (C) Reconstitution of activity by addition of Sll1556 protein at an 0.15-μg/ml final concentration in the reaction mixture. Symbols: ▴, FR6P (500 μM); ▪, DMAPP (2 μM); ×, GA3P (1 mM); •, GA3P (1 mM) plus PYR (500 μM); ○, MEP (500 μM); ⧫, incubation cofactor mixture (500 μM ATP, 250 μM CTP, 100 μM thiamine PP, 1 mM NADPH, 500 μM NADP, 1 mM FAD, 5 mM glutathione, 5 mM MgCl2, 2.5 mM MnCl2, 10 μM coenzyme B12, 100 mM HEPES-KOH, pH 7.7) incubated at 37°C. Incorporation with radiolabeled [14C]IPP (8.5 μM) followed preincubation with IPP (3 μM) (Materials and Methods).
However, with the Δsll1556 mutant there was no stimulation of isoprenoid synthesis with FR6P, GA3P, MEP, or GL6P (data not shown) nor with the cofactor mixture alone (Fig. 5B), indicating a lack of DMAPP production with these substrates. These results clearly imply that the missing factor, probably an enzyme, is required for incorporation of these pentose phosphate cycle components into the formation of a pool of isoprenoids.
The stimulation of pentose phosphate cycle substrates could be restored upon addition of the Sll1556 protein to the mutant Δsll1556 supernatant as seen in Fig. 5C. The inferred production of DMAPP, as indicated by [14C]IPP incorporation, occurred with GA3P, FR6P, and GL6P (data not shown) but not with MEP or the Sll1556 protein alone. In fact, the restoration of activity with FR6P under the same experimental conditions was equivalent to, or greater than, that obtained with the WT (with the addition of 2.0 μM DMAPP-0.5 mM FR6P). The Sll1556 protein is likely an enzyme as judged by the increased activity upon higher Sll1556 concentrations (data not shown) and because heating of the protein (2 min, 100°C) destroyed the activity. The nature of this apparent enzyme was subsequently further investigated.
Other suggested Sll1556 enzyme functions.
There is little doubt that the Sll1556 protein has activity as evident from the reconstitution experiments (Fig. 5C), which suggests that the protein influences another route to DMAPP production and isoprenoid biosynthesis. A BlastP search suggests that the sll1556 gene product is functionally similar to FMN-dependent alpha-hydroxy acid dehydrogenases, as well as IPP isomerase (type 2). Glycolate oxidase and lactate dehydrogenase are suggested albeit with rather low similarities. Hence, their substrate utilization was assayed by standard procedures (see Materials and Methods) with enzymes of known function to verify the assay conditions. We could not confirm glycolate oxidase activity nor activity for lactate dehydrogenase.
The Δsll1556 mutation affects membrane development.
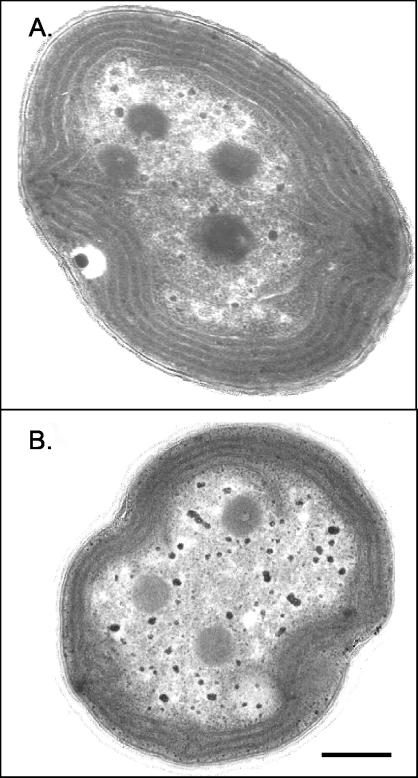
A comparison of the cell morphology of WT Synechocystis strain PCC 6803 and the Δsll1556 mutant was made using cultures in the log phase of growth and of similar cell densities (Table 2). Cells of the WT, when grown under moderate-light conditions (20 μmol/m2/s) are typically coccoid in shape except during division, when the daughter cells are oblong (Fig. 6). In scanning electron micrographs the cell diameter is 1.53 ± 0.12 μm for mutant cells and 1.75 ± 0.11 μm for WT cells. The outer wall layer of the mutant is considerably more fibrous than that of the WT. These fibrous extensions may be elongated stretches of outer wall layers. They are more coarse than fimbriae (28) but could be related to some of the coarser pili observed in Synechocystis strain PCC 6803 (2, 32). Negatively stained images of whole mounts of fixed cells did not reveal pili on either WT or mutants, which does not rule out their presence. Thylakoid membranes in sectioned cells (Fig. 7) tended to be peripherally arranged but could also fill the cell center. Also common were discontinuations of thylakoid membranes near the cell wall, suggestive of recent cell divisions. In the mutant there were fewer thylakoids (ca. 6.8) per central cell section than in WT (ca. 9.4). Growth over a range of light intensities (ca. 4, 20, and 200 μmol/m2/s) varied (data not shown), but the mutant and WT cultures had about the same growth rate at the respective light intensities, suggesting that the lesser thylakoid content in the mutant was not a limiting factor.
TABLE 2.
Comparison of Synechocystis strain PCC 6803 WT and Δsll1556 mutant
| Characteristic | WT | Δsll1556 mutant |
|---|---|---|
| Cell diam (μm) | 1.75 ± 0.1 (SD) | 1.53 ± 0.1 (SD) |
| No. of thylakoids/section | 9-11 | 5-8 |
| Outer wall | Smooth | More fibrous |
| IPP isomerase | Deficient | Deficient |
| Isoprenoid production in vitro | Pentose phosphate cycle substrate stimulation | No pentose phosphate cycle substrate stimulation |
FIG. 7.
Transmission electron micrographs of a Synechocystis strain PCC 6803 WT cell (A) and a Δsll1556 mutant cell (B). Thylakoid density in Δsll1556 mutant cells was typically less than that in WT cells. Bar, 0.3 μm.
DISCUSSION
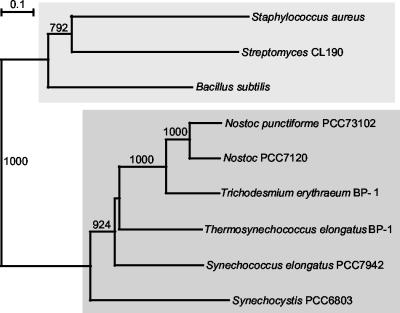
That the Sll1556 protein is an IPP isomerase (type 2) was not demonstrated under our in vitro conditions, although we were able to confirm the activity of the Streptomyces homolog (Fig. 4). With an amino acid identity of only 32% we must consider that the function of Sll1556 may not be that of an IPP isomerase. A BlastP analysis together with searches of databases of cyanobacterial genomes revealed a number of gene sequences related to sll1556 as a putative IPP isomerase type 2 (Table 1), but as noted above a functional confirmation for purified gene products is lacking. Activities suggested from gene sequence annotations, such as glycolate oxidase and FMN-dependent lactate dehydrogenase, were also not obtained. Sequence comparisons with Sll1556 indicate that homologous proteins similar to that in Synechocystis strain PCC 6803 are present in five other cyanobacteria: Anabaena (Nostoc) strain PCC, “Nostoc punctiforme” PCC 73102, “Synechococcus elongatus” PCC 7942, “Thermosynechococcus elongatus” BP-1, and “Trichodesmium erythraeum” BP-1. For the species listed in Table 1 and shown in the tree (Fig. 8) the sll1556-related gene similarities are ca. 30% between bacteria and cyanobacteria and ca. 50% among cyanobacteria. Many of these were designated as IPP isomerases in databases, but the functional annotation can be considered valid only for the three bacterial species for which type 2 IPP isomerase has been functionally verified (9, 26). Even though a meaningful relatedness is suggested from the tree (Fig. 8), any functional designation for the Sll1556 protein and its probable cyanobacterial homologs (ca. 50% similarity) is at this time premature.
FIG. 8.
Neighbor-joining tree of the known bacterial type 2 IPP isomerases and related polypeptides encoded in cyanobacterial genomes. Type 2 IPP isomerase has been functionally determined for only the top group. Bootstrap values greater than 50% (for 1,000 trials) are indicated. See Table 1 for accession numbers of the polypeptides.
A requirement for an IPP isomerase would apply only if an organism had only one pathway by which IPP and DMAPP are produced, as implied by a linear pathway as assumed for E. coli (Fig. 1). Yet, even for E. coli it has been shown that an IPP isomerase is not essential (7). In addition, Kuzuyama and Seto (14) further reported (cf. reference 14 and Table 1) that a number of bacteria do not possess genes for either type of IPP isomerase, thus indicating considerable diversity.
There is little doubt that the sll1556 gene product affects isoprenoid biosynthesis in vitro. Since the Δsll1556 mutant was impaired in inferred DMAPP production by lack of stimulation by FR6P and other pentose phosphate cycle substrates (Fig. 5B), it can be assumed that it was deficient in an alternate path of isoprenoid production. The rather significant membrane reduction in the mutant implies that this alternate path could be very significant under certain conditions. These results require further exploration of pentose phosphate cycle substrates under varied in vivo and in vitro conditions to clarify the nature and role of the Sll1556 protein.
The analysis of the WT Synechocystis strain PCC 6803 and the Δsll1556 mutant, as summarized in Table 2, reveals some notable physiological and morphological phenotypical characteristics. It is well known that hopanoids are present in the membranes (thylakoids, cell membranes, and wall layers) of cyanobacteria (8, 24). Hence, a reduction in thylakoid synthesis in the mutant (Fig. 7) is readily attributable to reduced isoprenoid biosynthesis, since isoprenoid precursors are essential building blocks of carotenoids, chlorophylls, and certain quinones required for construction and assembly of the photosynthetic apparatus. Modifications in normal isoprenoid biosynthesis are also consistent with alterations of the outer wall layers (glycocalyx or pili?), as suggested by the greater fibrous appearance of the outer surface in the mutant (Fig. 6B and C). It is likely that these combined effects result from a deficiency, but not absence, of isoprenoid biosynthesis.
Under normal photosynthetic conditions the Sll1556 protein is dispensable: the growth of the mutant culture in mineral medium did not differ significantly from that of the WT (at the same light intensity). It is likely that the cells rely more heavily on an alternate metabolic path. Differences in metabolite utilization can depend on the organism's environment. For example, recently Yang et al. (31) analyzed the fluxes of central metabolites by monitoring the products from [13C]glucose in Synechocystis strain PCC 6803 under different conditions. They found significant differences of the flow through the pentose phosphate pathway and through the glycolytic path between cells grown heterotrophically and those grown photomixotrophically.
This brings us back to the presumption of a linear isoprenoid production pathway via MEP in Synechocystis strain PCC 6803. For E. coli there is clear support for the proposed MEP pathway (14, 17, 18, 19, 20, 30) (Fig. 1) for isoprenoid biosynthesis as mentioned in the introduction. Both organisms have essential genes for the MEP pathway. However, as noted above for Synechocystis, the MEP pathway is not the only pathway of isoprenoid biosynthesis under photosynthetic growth conditions. It was previously shown that pentose phosphate cycle substrates lead to isoprenoid biosynthesis (6), which we confirmed here. Furthermore, our present results with the Δsll1556 mutant show that in addition to the pentose phosphate cycle substrates there may yet be another path to DMAPP biosynthesis, and perhaps also to IPP synthesis. Although we fully recognize the essential nature of the MEP pathway for the survival of Synechocystis strain PCC 6803, our previous and present results lead us to suggest alternate paths.
These studies of the sll1556 mutant have provided further evidence to suggest that isoprenoid biosynthesis in this photosynthetic cyanobacterium is more complex than predicted from E. coli. It appears that biosynthesis of isoprenoids in this organism is not linear but involves more than one pool of substrates and probably at least one alternate path to DMAPP biosynthesis. An alternate route to DMAPP production from pentose phosphate cycle substrates can be metabolically advantageous for a photosynthetic organism at optimal growth conditions where an increased supply of isoprenoids would enhance thylakoid and cell wall synthesis.
Acknowledgments
The help of T. Maugul, director of the electron microscope facility, is greatly appreciated. We are grateful to T. Kuzuyama for providing the pQCLD41 expression plasmid.
Research was supported by DOE grant no. DE-FG02-98ER20302.
REFERENCES
- 1.Arigoni, D., W. Eisenreich, C. Latzel, S. Sagner, T. Radykewicz, M. H. Zenk, and A. Bacher. 1999. Dimethylallyl pyrophosphate is not the committed precursor of isopentenyl pyrophosphate during terpenoid biosynthesis from 1-deoxyxylulose in higher plants. Proc. Natl. Acad. Sci. USA 96:1309-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhaya, D., N. R. Bianco, D. Bryant, and A. Grossman. 2000. Type IV pilus biogenesis and motility in the cyanobacterium Synechocystis sp. PCC6803. Mol. Microbiol. 37:1365-2958. [DOI] [PubMed] [Google Scholar]
- 3.Cunningham, F. X., T. P. Lafond, and E. Gantt. 2000. Evidence of a role for LytB in the nonmevalonate pathway of isoprenoid biosynthesis. J. Bacteriol. 182:5841-5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eisenreich, W., F. Rohdich, and A. Bacher. 2001. Deoxyxylulose phosphate pathway to terpenoids. Trends Plant Sci. 6:78-84. [DOI] [PubMed] [Google Scholar]
- 5.Ershov, Y., R. R. Gantt, F. X. Cunningham, and E. Gantt. 2000. Isopentenyl diphosphate isomerase deficiency in Synechocystis sp. strain PCC6803. FEBS Lett. 473:337-340. [DOI] [PubMed] [Google Scholar]
- 6.Ershov, Y., R. R. Gantt, F. X. Cunningham, and E. Gantt. 2002. Isoprenoid biosynthesis in Synechocystis sp. strain PCC6803 is stimulated by compounds of the pentose phosphate cycle but not by pyruvate or deoxyxylulose-5-phosphate. J. Bacteriol. 184:5045-5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hahn, F. M., A. P. Hurlburt, and C. D. Poulter. 1999. Escherichia coli open reading frame 696 is idi, a nonessential gene encoding isopentenyl diphosphate isomerase. J. Bacteriol. 181:4499-4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jürgens, U. J., P. Simonin, and M. Rohmer. 1992. Localization and distribution of hopanoids in membrane systems of the cyanobacterium Synechocystis PCC 6714. FEMS Microbiol. Lett. 92:285-288. [DOI] [PubMed] [Google Scholar]
- 9.Kaneda, K., T. Kuzuyama, M. Takagi, Y. Hayakawa, and H. Seto. 2001. An unusual isopentenyl diphosphate isomerase found in the mevalonate pathway gene cluster from Streptomyces sp. strain CL190. Proc. Natl. Acad. Sci. USA 98:932-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaneko, T., S. Sato, H. Kotani, A. Tanaka, E. Asamizu, Y. Nakamura, N. Miyajima, M. Hirosawa, N. Sugiura, S. Sasamoto, T. Kimura, T. Hosouchi, A. Matsuno, A. Muraki, N. Nakazaki, K. Naruo, S. Okumura, S. Shimpo, C. Takeuchi, T. Wada, A. Watanabe, M. Yamada, M. Yasuda, and S. Tabata. 1996. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 3:185-209. [DOI] [PubMed] [Google Scholar]
- 11.Kimura, M. 1980. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 12.Kornberg, A. 1955. Lactic dehydrogenase of muscle. Methods Enzymol. 1:441-449. [Google Scholar]
- 13.Koyama, T., and K. Ogura. 1999. Isopentenyl diphosphate isomerase and prenyltransferases, p. 69-96. In D. Barton and K. Nakanishi (ed.), Comprehensive natural products chemistry. Elsevier Science Ltd., Oxford, United Kingdom.
- 14.Kuzuyama, T., and H. Seto. 2003. Diversity of the biosynthesis of the isoprene units. Nat. Prod. Rep. 20:171-183. [DOI] [PubMed] [Google Scholar]
- 15.Perrière, G., and M. Gouy. 1996. WWW-Query: an on-line retrieval system for biological sequence banks. Biochimie 78:364-369. [DOI] [PubMed] [Google Scholar]
- 16.Ramos-Valdivia, A. C., R. van der Heijden, and R. Verpoorte. 1997. Isopentenyl diphosphate isomerase: a core enzyme in isoprenoid biosynthesis. A review of its biochemistry and function. Nat. Prod. Rep. 14:591-603. [DOI] [PubMed] [Google Scholar]
- 17.Rodriguez-Concepción, M., N. Campos, L. M. Lois, C. Maldonado, J. F. Hoeffler, C. Grosdemange-Billiard, M. Rohmer, and A. Boronat. 2000. Genetic evidence of branching in the isoprenoid pathway for the production of isopentenyl diphosphate and dimethylallyl diphosphate in Escherichia coli. FEBS Lett. 473:328-332. [DOI] [PubMed] [Google Scholar]
- 18.Rodriguez-Concepción, M., and A. Boronat. 2002. Elucidation of the methylerythritol phosphate pathway for isoprenoid biosynthesis in bacteria and plastids. A metabolic milestone achieved through genomics. Plant Physiol. 130:1079-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rohdich, F., F. Zepeck, P. Adam, S. Hecht, J. Kaiser, R. Laupitz, T. Gräwert, S. Amslinger, W. Eisenreich, A. Bacher, and D. Arigoni. 2003. The deoxyxylulose phosphate pathway of isoprenoid biosynthesis: studies on the mechanisms of the reactions catalyzed by IspG and IspH protein. Proc. Natl. Acad. Sci. USA 100:1586-1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rohmer, M. 1999. The discovery of a mevalonate-independent pathway for isoprenoid biosynthesis in bacteria, algae and higher plants. Nat. Prod. Rep. 16:565-574. [DOI] [PubMed] [Google Scholar]
- 21.Rozen, S., and H. J. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers, p. 365-386 In S. Krawetz and S. Misener (ed.), Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, N.J. [DOI] [PubMed]
- 22.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Simonin, P., U. J. Jürgens, and M. Rohmer. 1996. Bacterial triterpenoids of the hopanes series from the prochlorophytes Prochlorothrix hollandica and their intercellular localization. Eur. J. Biochem. 241:865-871. [DOI] [PubMed] [Google Scholar]
- 25.Spurgeon, S. L., N. Sathyamoorthy, and J. W. Porter. 1984. Isopentenyl pyrophosphate and prenyltransferases from tomato fruit and plastids. Arch. Biochem. Biophys. 230:446-454. [DOI] [PubMed] [Google Scholar]
- 26.Steinbacher, S., J. Kaiser, S. Gerhardt, W. Eisenreich, R. Huber, A. Bacher, and F. Rohdich. 2003. Crystal structure of the type II isopentenyl diphosphate:dimethylallyl diphosphate isomerase from Bacillus subtilis. J. Mol. Biol. 329:973-982. [DOI] [PubMed] [Google Scholar]
- 27.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vaara, T., and M. Vaara. 1988. Cyanobacterial fimbriae. Methods Enzymol. 167:189-195. [Google Scholar]
- 29.Williams, J. G. K. 1988. Construction of specific mutations in photosystem II photosynthetic reaction center by genetic engineering methods in Synechocystis 6803. Methods Enzymol. 167:766-778. [Google Scholar]
- 30.Wolff, M., M. Seemann, B. Tse Sum Bui, Y. Frapart, D. Tritsch, A. G. Estrabot, M. Rodriguez-Concepción, A. Boronat, A. Marquet, and M. Rohmer. 2003. Isoprenoid biosynthesis via the methylerythritol phosphate pathway: the (E)-4-hydroxy-3-methylbut-2-enyl diphosphate reductase (LytB/IspH) from Escherichia coli is a [4Fe-4S] protein. FEBS Lett. 541:115-120. [DOI] [PubMed] [Google Scholar]
- 31.Yang, C., Q. Hua, and K. Shimizu. 2002. Metabolic flux analysis in Synechocystis using isotope distribution from 13C-labeled glucose. Metab. Eng. 4:202-216. [DOI] [PubMed] [Google Scholar]
- 32.Yoshimura, H., S. Yoshihara, S. Okamoto, M. Ikeuchi, and M. Ohmori. 2002. A camp receptor protein, SYCRP1, is responsible for the cell motility of Synechocystis sp. PCC6803. Plant Cell Physiol. 43:460-463. [DOI] [PubMed] [Google Scholar]
- 33.Zelitch, I. 1955. Glycolic acid oxidase and glyoxylic acid reductase. Methods Enzymol. 1:528-535. [Google Scholar]