Abstract
Enterococcus faecalis and Enterococcus faecium infections are increasingly difficult to treat due to high levels of resistance to antibiotics. PlyV12, a bacteriophage lytic enzyme, was isolated and shown to effectively kill both E. faecalis and E. faecium (including vancomycin-resistant strains), as well as other human pathogens. We propose its development and use as an alternative therapeutic tool.
Enterococcus faecalis and Enterococcus faecium are gram-positive bacteria that commensally colonize the lower intestinal tract, oral cavity, and vaginal tract of humans. In healthy individuals, E. faecalis and E. faecium colonization normally has no adverse effect on the host; however, the acquisition of virulence factors and high-level antibiotic resistance by enterococci are causing these organisms to emerge as a leading source of nosocomial infections, particularly in immunocompromised patients (3, 8, 9, 16). Common diseases caused by enterococcal infections include endocarditis, abdominal abscesses, bacteremia, and urinary tract infections.
We are currently developing a novel approach to the control of pathogenic microorganisms through the action of purified bacteriophage lytic enzymes, termed lysins, produced during the natural life cycle of the bacteriophage. Lysins have evolved to rapidly break down the bacterial cell wall in order to release progeny phage (23). Structurally, lysins are commonly found as modular proteins with an amino-terminal domain that confers the enzymatic activity for a peptidoglycan bond and a carboxy-terminal domain that confers binding specificity to a carbohydrate epitope in the bacterial cell wall (13-15, 20). These highly evolved enzymes are normally very specific to the bacterial host of the phage from which they are derived (5, 6). When lysins are purified and applied extrinsically, their binding efficiency and catalytic activity can be harnessed to achieve targeted killing of select pathogenic bacteria with minimal effects on other commensal bacteria; this capacity is an advantage over conventional antibiotics. The efficacy of various lysins in killing Bacillus anthracis (19), Streptococcus pyogenes (a group A streptococcus) (17), and Streptococcus pneumoniae (10, 11) has been demonstrated both in vitro and in animal models of colonization and/or infection with these pathogens.
In this report, we describe a lysin, PlyV12, from the enterococcal bacteriophage Φ1 which infects the host, E. faecalis strain V12. Φ1 (obtained from the d'Herelle collection, Laval University, Quebec, Canada) was originally isolated from sewage in 1975 and belongs to the Myoviridae family, whose members are characterized by contractile tails and icosahedral heads (2, 7). PlyV12 is a proposed amidase that exhibits a substantial lytic effect on multiple E. faecalis strains. Significantly, PlyV12 also lyses vancomycin-resistant strains of E. faecalis and strains of the closely related enterococcal pathogen E. faecium. Distinct from other reported lysins, PlyV12 was also found to be active against several disease-causing streptococcal and staphylococcal strains, making it one of the first lysins to demonstrate a spectrum of activity outside that of the host and closely related bacterial strains. This broad activity spectrum suggests the presence of a unique cell wall carbohydrate epitope common to these diverse human pathogens.
PlyV12 was identified based on a previously described genetic screening process (12) whereby a Φ1 genomic library was screened for lytic agents active against E. faecalis strain V12. A 945-bp open reading frame was identified after DNA sequencing of a lytic clone. The open reading frame translates into a protein of 314 amino acids with a molecular mass of approximately 34 kDa. The plyV12 gene was initially cloned into several Escherichia coli expression systems; however, the enzyme yields were very low in all cases. PlyV12 (as other lytic enzymes) is translated without a leader sequence; thus, it remains in the cytoplasm, where it cannot exert an effect on its peptidoglycan substrate, therefore making it possible to be expressed in a wide range of systems. Subsequently, the plyV12 gene and approximately 100 bp of the flanking sequence were amplified by PCR and cloned into the E. coli-Bacillus shuttle vector pDG148 (21), yielding pPY1. plyV12 was then expressed in Bacillus megaterium strain WH320 (MoBiTec, Marco Island, Fla.) by induction with isopropyl-β-d-thiogalactopyranoside for 1 h, at which time the culture was centrifuged. The pelleted bacteria were suspended in 100 ml of a lysis reagent, BugBuster (diluted from a 10× concentration to a 1× concentration in phosphate-buffered saline [PBS] containing 100 μg of lysozyme/ml) (Novagen, San Diego, Calif.). This cell lysate was allowed to remain at 37°C for 1 h, followed by centrifugation at 4,000 rpm in an Eppendorf centrifuge to pellet the cell debris. The supernatant containing the crude lysin was diluted 1:3 in 50 mM morpholineethanesulfonic acid at pH 6.7 prior to being loaded on three 5-ml HiTrap SP HP columns (Amersham Biosciences, Piscataway, N.J.) connected in a series. The columns were washed with the 50 mM morpholineethanesulfonic acid buffer until the optical density at a wavelength of 280 nm (OD280) reached the baseline. PlyV12 was eluted with a 20-column-volume linear gradient to a concentration of 1 M sodium chloride (NaCl), and a peak at ∼330 mM NaCl was found to contain the lytic activity. Furthermore, sodium dodecyl sulfate-polyacrylamide gel electrophoresis revealed a 34-kDa band corresponding to this peak, which matched the predicted size of PlyV12. Fractions constituting the peak were pooled and concentrated to 25 U/ml (see below) in an Amicon Ultra unit with a cutoff of 10 kDa (Millipore, Billerica, Mass.). The final PlyV12 sample was purified >85% based on spot densitometry (AlphaImager; Alpha Innotech, San Leandro, Calif.) of the gel images of crude and purified PlyV12 that were obtained by sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
PlyV12 lytic activity was expressed in units per milliliter, in which units represent the reciprocal of the highest dilution of enzyme resulting in a 50% reduction in bacterial OD600 in 15 min. Subsequent bacterial killing experiments were carried out with a PlyV12 preparation of 25 U/ml.
For the lysin killing assays, the bacterial strains to be assayed were grown to mid-logarithmic phase in brain heart infusion broth, pelleted, and resuspended in PBS at pH 7.4 to an OD600 of 2.0. The pH profile experiments for PlyV12 activity were performed after the bacterial cells were resuspended in 100 mM acetate buffer at pH 5.2, 100 mM morpholinepropanesulfonic acid at pH 6.5, or 100 mM Tris at pH 7.5 or 8.5 instead. A 100-μl volume of bacterial suspension was mixed with 100 μl of PlyV12 at 25 U/ml. For the OD assays, the OD600 of the mixture was monitored every 15 s with shaking over a 15-min period on an automated 96-well plate reader (SpectraMax Plus384; Molecular Devices, Sunnyvale, Calif.). For viability assays, the mixture of a bacteria and PlyV12 was incubated at room temperature with gentle shaking for specified amounts of time, and at designated time points, aliquots were removed, serially diluted, and plated on agar plates for viability counts.
An alignment of the PlyV12 sequence from E. faecalis bacteriophage Φ1 with lysins from bacteriophages of Streptococcus agalactiae (a group B streptococcus), S. pyogenes, S. pneumoniae, Streptococcus mitis, and Streptococcus thermophilus revealed similarities primarily limited to the amino termini of these molecules, which correspond to the catalytic domains of most lysins (Fig. 1). Since several of these lysins are known to confer N-acetylmuramyl-l-alanine amidase activity, PlyV12 may also be an amidase. The diminished sequence similarity among the carboxy-terminal portions of all these lysins suggests that they have distinct cell wall binding epitopes and hence different bacterial specificities.
FIG. 1.
Alignment of the amino acid sequence of PlyV12 from E. faecalis bacteriophage Φ1 (Ef) with lysins from five other gram-positive bacteriophages. Those phages include λSa2 from S. agalactiae, a group B streptococcus, (Sa); 315.3 from S. pyogenes, a group A streptococcus (Spy); Dp-1 from S. pneumoniae (Spn); SM1 from S. mitis (Sm); and Sfi19 from S. thermophilus (St). Identical residues are highlighted by black boxes, while conserved residues are highlighted with gray boxes.
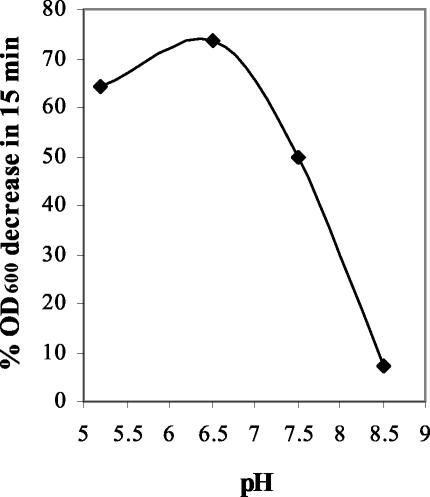
PlyV12 was found to have maximal activity at about pH 6.0 while retaining partial yet significant activity at pH values of 5.0 and 7.5 (Fig. 2). The activity decreased rapidly at higher pHs, resulting in near inactivation at pH 8.5, a profile common among bacteriophage lysins (11). Although not optimum, all subsequent assays were carried out in PBS at pH 7.4 to determine the PlyV12 activity under physiological conditions. While PlyV12 functioned well at physiological salt concentrations, sodium chloride at concentrations upwards of 250 mM deteriorates its activity (results not shown).
FIG. 2.
pH profile of PlyV12 activity. PlyV12 activity against E. faecalis strain V12 was tested at pHs 5.2, 6.5, 7.5, and 8.5. PlyV12 activity was measured by an OD assay and plotted as the percent decrease in OD600 in 15 min.
In addition to killing its host bacterial strain, E. faecalis strain V12, PlyV12 also had lytic activity against 14 clinical and laboratory E. faecalis and E. faecium strains tested, including two vancomycin-resistant E. faecalis strains and three vancomycin-resistant E. faecium strains (Table 1 and Fig. 3). Interestingly, PlyV12 also exhibited a significant killing effect on strains of pathogenic streptococci, including S. pyogenes (a group A streptococcus), which are responsible for pharyngitis, toxic shock syndrome, and rheumatic fever (4); group B streptococci, which are responsible for neonatal meningitis (1); and group C streptococci, which constitute an emerging pathogen group causing diseases similar to those caused by S. pyogenes (18). While the sequence alignment of PlyV12 from the E. faecalis bacteriophage Φ1 with lysins of group A and group B streptococcal bacteriophages (from S. pyogenes and S. agalactiae, respectively) (Fig. 1) suggests that these lysins target different cell wall epitopes, the ability of PlyV12 to lyse strains of E. faecalis, S. pyogenes, and group B streptococci shows that PlyV12 recognizes a receptor common among these different pathogens. Additionally, PlyV12 had some lytic effect against Staphylococcus aureus, another high-level, antibiotic-resistant nosocomial pathogen that is responsible for sepsis, necrotizing pneumonia, and toxic shock syndrome (22).
TABLE 1.
Bacterial strains tested for lysin activity
| Bacterial straina | Sourceb |
|---|---|
| Enterococci | |
| Enterococcus faecalis V12 | 1 |
| Enterococcus faecalis A220 | 1 |
| Enterococcus faecalis A936 | 1 |
| Enterococcus faecalis B722 | 1 |
| Enterococcus faecalis D76 | 1 |
| Enterococcus faecalis JH2-2 | 2 |
| Enterococcus faecalis EF-24 | 2 |
| Enterococcus faecalis EF-25 | 2 |
| Enterococcus faecalis HER1044 | 1 |
| Enterococcus faecalis HER1323 | 1 |
| Enterococcus faecalis (VRE) EF-1 | 2 |
| Enterococcus faecalis (VRE) EF-17 | 2 |
| Enterococcus faecium (VRE) EFSK-2 | 2 |
| Enterococcus faecium (VRE) EFSK-16 | 2 |
| Enterococcus faecium (VRE) EFSK-33 | 2 |
| Lancefield group streptococci | |
| Group A streptococcus strain D471 | 1 |
| Group A streptococcus strain CS24 | 1 |
| Group A streptococcus strain CS112 | 1 |
| Group A streptococcus strain A486 variant | 1 |
| Group B streptococcus strain NC11237 | 1 |
| Group B streptococcus strain A349 | 1 |
| Group B streptococcus strain A909 | 1 |
| Group C streptococcus strain 2GRP66 | 1 |
| Group E streptococcus strain K131 | 1 |
| Group F streptococcus strain F68C | 1 |
| Group G streptococcus strain D166B | 1 |
| Group L streptococcus strain D167A | 1 |
| Group N streptococcus strain C559 | 1 |
| Other streptococci | |
| Streptococcus pneumoniae GB2162 | 1 |
| Streptococcus pneumoniae PK1850 | 1 |
| Streptococcus uberis 45 | 1 |
| Streptococcus uberis ATCC27958 | 1 |
| Streptococcus gordonii FS12 | 1 |
| Streptococcus gordonii PK488 | 1 |
| Streptococcus mutans ATCC25175 | 1 |
| Streptococcus mutans 10449 | 1 |
| Streptococcus oralis PK34 | 1 |
| Streptococcus salivarius ATCC9222 | 1 |
| Streptococcus rattus BHT | 1 |
| Streptococcus sobrinus 6715 | 1 |
| Streptococcus mitis J22 | 1 |
| Streptococcus intermedius PK2821 | 1 |
| Streptococcus parasanguis PK2564 | 1 |
| Streptococcus crista PK1408 | 1 |
| Staphylococci | |
| Staphylococcus aureus RN4220 | 1 |
| Staphylococcus aureus RN6390 | 1 |
| Staphylococcus aureus HER1283 | 1 |
| Staphylococcus epidermidis BJ0018 | 1 |
VRE, vancomycin-resistant enterococcus.
1, The Rockefeller University Bacteria Collection; 2, Alexander Tomasz, The Rockefeller University, New York, N. Y.
FIG. 3.
Range of activity of PlyV12. PlyV12 activity against enterococcal, streptococcal, and staphylococcal strains was measured as the rate of decrease in OD per minute, expressed in milliunits of OD600 per minute. This initial velocity measurement allowed direct comparison of PlyV12 activities among bacterial species. VRE, vancomycin-resistant enterococci. (Inset) Viability assay of E. faecalis strain V12 treated with PlyV12. Viability was measured in CFU per milliliter. E. faecium strains EFSK-2, EFSK-16, and EFSK-33 as well as E. faecalis strains JH2-2, EF-24, EF-25, EF-1, and EF-17 were obtained from A. Tomasz. All other strains are part of the Rockefeller University Bacteria Collection.
The assays described above were carried out using a measurement of OD, which corresponds with bacterial viability. However, to better quantitate the extent of bacterial killing, a direct viability assay was performed with E. faecalis strain V12 incubated with PlyV12. The results revealed a >4-log reduction in viability when the mixture was exposed to 2.5 U of PlyV12 for 15 min, which corresponds to a drop of 200 mOD600/min in the OD assay (Fig. 3, inset). The viability of this strain continued to decrease upon further incubation with PlyV12.
PlyV12 has bactericidal activity against all strains of E. faecalis and E. faecium tested, which is very significant for clinical strains and strains resistant to the antibiotic vancomycin. Vancomycin is considered the last line of defense against many bacterial pathogens that are already resistant to other available antibiotics (3). Enterococci are a leading cause of nosocomial infections, and treating these infections with conventional antibiotics has become increasingly difficult in light of the acquisition of antibiotic resistance genes by these organisms. It is anticipated that soon enterococci will be untreatable by current antibiotics, and alternative means of combating infections caused by these organisms will be urgently needed. We have shown that PlyV12 is a viable candidate for such an antienterococcal therapeutic agent.
PlyV12 exhibited significant lethal activity against other pathogens, such as S. pyogenes and group B, C, E, and G streptococci, with minimal effects against commensal bacteria (with the exception of Streptococcus gordonii). This finding suggests a broader application for this lysin whereby PlyV12 is used as a therapeutic agent specific for multiple pathogens that are responsible for serious infections. While the organization of its primary structure and the nature of its enzymatic activity make PlyV12 consistent with other bacteriophage lysins previously studied, PlyV12 is believed to be the first bacteriophage lysin described to have activity against Enterococcus species. In addition, previously studied bacteriophage lysins generally have a narrow spectrum of activity specific to the host bacterial species from which the phage was isolated. PlyV12's comparatively broad spectrum of activity against numerous bacterial strains indicates that this is a truly unique enzyme. Furthermore, the PlyV12 binding activity determined in this study suggests a previously undiscovered surface structure common to several different pathogens, which could serve as a target in the development of new antibiotics.
Nucleotide sequence accession number.
The nucleotide sequence of PlyV12 has been deposited into GenBank under accession number AY581208.
Acknowledgments
This work was supported by a grant from the Defense Advance Research Project Agency (DARPA) to V.A.F.
We thank Alexander Tomasz for providing the bacterial strains and Vasant Kumar for plasmid pDG148.
REFERENCES
- 1.Balter, S., C. G. Whitney, and A. Schuchat. 2000. Epidemiology of group B streptococcal infections, p. 154-162. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens. American Society for Microbiology, Washington, D.C.
- 2.Caprioli, T., F. Zaccour, and S. S. Kasatiya. 1975. Phage typing scheme for group D streptococci isolated from human urogenital tract. J. Clin. Microbiol. 2:311-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cetinkaya, Y., P. Falk, and C. G. Mayhall. 2000. Vancomycin-resistant enterococci. Clin. Microbiol. Rev. 13:686-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cunningham, M. W. 2000. Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 13:470-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fischetti, V. A. 2003. Novel method to control pathogenic bacteria on human mucous membranes. Ann. N. Y. Acad. Sci. 987:207-214. [DOI] [PubMed] [Google Scholar]
- 6.Fischetti, V. A. 2001. Phage antibacterials make a comeback. Nat. Biotechnol. 19:734-735. [DOI] [PubMed] [Google Scholar]
- 7.Jarvis, A. W., L. J. Collins, and H.-W. Ackermann. 1993. A study of five bacteriophages of the Myoviridae family which replicate on different gram-positive bacteria. Arch. Virol. 133:75-84. [DOI] [PubMed] [Google Scholar]
- 8.Jett, B. D., M. M. Huycke, and M. S. Gilmore. 1994. Virulence of enterococci. Clin. Microbiol. Rev. 7:462-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson, A. P. 1994. The pathogenicity of enterococci. J. Antimicrob. Chemother. 33:1083-1089. [DOI] [PubMed] [Google Scholar]
- 10.Loeffler, J. M., D. Nelson, and V. A. Fischetti. 2001. Rapid killing of Streptococcus pneumoniae with a bacteriophage cell wall hydrolase. Science 294:2170-2172. [DOI] [PubMed] [Google Scholar]
- 11.Loeffler, J. M., S. Djurkovic, and V. A. Fischetti. 2003. Phage lytic enzyme Cpl-1 as a novel antimicrobial for pneumococcal bacteremia. Infect. Immun. 71:6199-6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loessner, M., G. Wendlinger, and S. Scherer. 1995. Heterogeneous endolysins in Listeria monocytogenes bacteriophages: a new class of enzymes and evidence for conserved holin genes within the siphoviral lysis cassettes. Mol. Microbiol. 16:1231-1241. [DOI] [PubMed] [Google Scholar]
- 13.Loessner, M., K. Kramer, F. Ebel, and S. Scherer. 2002. C-terminal domains of Listeria monocytogenes bacteriophage murein hydrolases determine specific recognition and high-affinity binding to bacterial cell wall carbohydrates. Mol. Microbiol. 44:335-349. [DOI] [PubMed] [Google Scholar]
- 14.Lopez, R., E. Garcia, P. Garcia, and J. L. Garcia. 1997. The pneumococcal cell wall degrading enzymes: a modular design to create new lysins? Microb. Drug Resist. 3:199-211. [DOI] [PubMed] [Google Scholar]
- 15.Lopez, R., M. P. Gonzalez, E. Garcia, J. L. Garcia, and P. Garcia. 2000. Biological roles of two new murein hydrolases of Streptococcus pneumoniae representing examples of module shuffling. Res. Microbiol. 151:437-443. [DOI] [PubMed] [Google Scholar]
- 16.Murray, B. E. 1990. The life and times of the enterococcus. Clin. Microbiol. Rev. 3:46-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nelson, D., L. Loomis, and V. A. Fischetti. 2001. Prevention and elimination of upper respiratory colonization of mice by group A streptococci by using a bacteriophage lytic enzyme. Proc. Natl. Acad. Sci. USA 98:4107-4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oster, H. R., and A. L. Bisno. 2000. Group C and group G streptococcal infections: epidemiologic and clinical aspects, p. 184-190. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens. American Society for Microbiology, Washington, D.C.
- 19.Schuch, R., D. Nelson, and V. A. Fischetti. 2002. A bacteriolytic agent that detects and kills Bacillus anthracis. Nature 418:884-888. [DOI] [PubMed] [Google Scholar]
- 20.Sheehan, M. M., J. L. Garcia, R. Lopez, and P. Garcia. 1997. The lytic enzyme of the pneumococcal phage Dp-1: a chimeric enzyme of intergeneric origin. Mol. Microbiol. 25:717-725. [DOI] [PubMed] [Google Scholar]
- 21.Stragier, P., C. Bonamy, and C. Karmazyn-Campelli. 1988. Processing of a sporulation sigma factor in Bacillus subtilis: how morphological structure could control gene expression. Cell 52:697-704. [DOI] [PubMed] [Google Scholar]
- 22.Tenover, F. C., and R. P. Gaynes. 2000. The epidemiology of Staphylococcus infections, p. 414-421. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens. American Society for Microbiology, Washington, D.C.
- 23.Young, R., I.-N. Wang, and W. D. Roof. 2000. Phages will out: strategies of host cell lysis. Trends Microbiol. 8:120-128. [DOI] [PubMed] [Google Scholar]