Abstract
A proteome analysis of Lactobacillus casei mutants that are affected in carbon catabolite repression revealed that a 15-kDa protein was strongly overproduced in a ptsHI47T mutant. This protein was identified as EIIA of a mannose class phosphotransferase system (PTS). A 7.1-kb DNA fragment containing the EIIA-encoding open reading frame and five other genes was sequenced. The first gene encodes a protein resembling the RpoN (σ54)-dependent Bacillus subtilis transcription activator LevR. The following pentacistronic operon is oriented in the opposite direction and encodes four proteins with strong similarity to the proteins of the B. subtilis Lev-PTS and one protein of unknown function. The genes present on the 7.1-kb DNA fragment were therefore called levR and levABCDX. The levABCDX operon was induced by fructose and mannose. No “−12, −24” promoter typical of RpoN-dependent genes precedes the L. casei lev operon, and its expression was therefore RpoN independent but required LevR. Phosphorylation of LevR by P∼His-HPr stimulates its activity, while phosphorylation by P∼EIIBLev inhibits it. Disruption of the EIIBLev-encoding levB gene therefore led to strong constitutive expression of the lev operon, which was weaker in a strain carrying a ptsI mutation preventing phosphorylation by both P∼EIIBLev and P∼His-HPr. Expression of the L. casei lev operon is also subject to P-Ser-HPr-mediated catabolite repression. The observed slow phosphoenolpyruvate- and ATP-dependent phosphorylation of HPrI47T as well as the slow phosphoryl group transfer from the mutant P∼His-HPr to EIIALev are assumed to be responsible for the elevated expression of the lev operon in the ptsHI47T mutant.
In many bacteria, the phosphoenolpyruvate (PEP):sugar phosphotransferase system (PTS) catalyzes the concomitant uptake and phosphorylation of numerous sugars, sugar alcohols, and other sugar derivatives (45). The PTS is usually composed of four soluble proteins or protein domains (EI, HPr, EIIA, and EIIB), which form a protein phosphorylation cascade, and one (EIIC) and sometimes two (EIIC and EIID) integral membrane proteins. EI and HPr are the general PTS proteins, while the EIIs are specific for one or several sugars. EI autophosphorylates at a histidine at the expense of PEP and transfers the phosphoryl group to His-15 in HPr. P∼His-HPr phosphorylates a histidyl residue in one of the various sugar-specific EIIAs usually present in bacteria. P∼EIIA subsequently transfers the phosphoryl group to a cysteyl or histidyl residue in EIIB with the same sugar specificity, which then donates the phosphoryl group to a sugar molecule bound to the corresponding membrane-integrated EIIC. The phosphorylated sugar is subsequently released into the cytoplasm (45).
In addition to its phosphocarrier activity within the PTS phosphorylation cascade, P∼His-HPr also phosphorylates several non-PTS proteins. In gram-positive bacteria, P∼His-HPr-mediated phosphorylation at a conserved histidyl residue of glycerol kinase leads to an about 10-fold increase of its enzymatic activity (5, 52). The uptake of a rapidly metabolizable PTS sugar leads to dephosphorylation of the PTS proteins and therefore to poor phosphorylation of glycerol kinase (12). As a consequence, glycerol kinase of gram-positive bacteria is less active when a rapidly metabolizable PTS sugar is taken up. This leads to reduced expression of the glpFK operon, since under these conditions, only low amounts of the inducer glycerol-3-P are formed. This regulatory mechanism was therefore called inducer exclusion (9). In a similar phosphorylation-dependent manner, P∼His-HPr also controls the activity of PTS-regulated antiterminators and transcription activators. In addition to their RNA or DNA binding domain, these proteins usually possess two PTS regulation domains (PRDs) (49). PTS-controlled transcription activators also contain EIIA and EIIB domains (14, 26). These antiterminators and transcription activators control the expression of genes encoding either sugar-specific PTS components or, in a few cases, extracellular polysaccharide-degrading enzymes (14). Their activity is regulated by P∼His-HPr- and/or P∼EIIB-mediated phosphorylation at conserved histidines in their EIIA domain and one or both PRDs (32, 34, 47).
The best-studied PRD-containing transcription activator is LevR of Bacillus subtilis, which controls the expression of the levDEFGsacC operon (35). This operon encodes the sugar-specific components for a low-capacity but high-affinity fructose or mannose-specific PTS and the extracellular enzyme levanase, which degrades the fructose polymer levan. LevR possesses an N-terminal DNA binding domain, an RpoN (σ54) interaction domain, and a short region of unknown function, which is followed by PRD1, EIIAMan, and EIIBGat domains and, finally, a truncated PRD2 (see Fig. 2B) (14, 26). LevR binds to the palindromic upstream activating sequence (UAS) located about 100 bp in front of a “−12, −24” promoter (36) and functions as enhancer binding protein by interacting with RpoN of the RNA polymerase-RpoN holoenzyme. The activity of B. subtilis LevR is controlled by two PTS-mediated phosphorylation reactions. Phosphorylation by P∼His-HPr at the phosphorylatable histidine in the EIIAMan domain of LevR stimulates its activity (14, 34). During the uptake of a rapidly metabolizable PTS sugar, the phosphoryl group of P∼His-HPr is primarily used for sugar phosphorylation, and as a consequence, LevR will be barely phosphorylated at the EIIAMan domain. The poor phosphorylation of LevR at the EIIAMan domain during the rapid uptake of a PTS sugar provides a carbon catabolite repression (CCR) mechanism. By contrast, phosphorylation of LevR by P∼EIIBLev at a histidine in PRD2 inhibits its transcription activator function (34). In the absence of a substrate for the Lev-PTS, most LevR will be phosphorylated at PRD2. By contrast, when a substrate for the Lev-PTS is present, P∼EIIBLev will donate its phosphoryl group preferentially to the sugar transported by the Lev-PTS and not to PRD2 of LevR, which will therefore be highly active. Poor phosphorylation at PRD2 of LevR due to the presence of a Lev-PTS substrate serves as an induction mechanism for the lev operon. In agreement with this concept, replacement of the phosphorylatable histidine in PRD2 of LevR or inactivation of EIIALev or EIIBLev, which are parts of the LevR/PRD2 phosphorylation cascade (6), led to constitutive expression from the lev promoter (34).
FIG. 2.
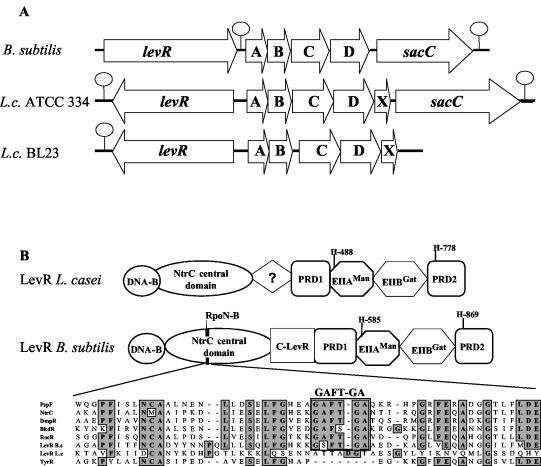
(A) Schematic representation of the lev operon and the preceding levR gene in B. subtilis and in L. casei (L.c.) strains ATCC 334 and BL23. Hairpin loops indicate transcription terminators. (B) Domain organization of the L. casei and B. subtilis transcription activator LevR. DNA-B indicates the DNA binding domain with the helix-turn-helix motif. C-LevR indicates a region located between the NtrC central domain and PRD1 in B. subtilis LevR which differs from the corresponding region in L. casei LevR. An alignment of the sequence around the RpoN binding motif GAFTGA (RpoN-B) of several proteins containing an NtrC central domain is also shown. PspF, NtrC, and TyrR are proteins from E. coli, and BkdR and RocR are from B. subtilis. The GAFTGA sequence motif is absent from TyrR and L. casei LevR, which are both RpoN independent, but is present in B. subtilis LevR, which is RpoN (SigL) dependent, and in the other transcription activators included in the alignment.
The B. subtilis lev operon is also subject to the major CCR mechanism operative in low-G+C-content gram-positive bacteria. This mechanism implies the catabolite control protein A (CcpA), a member of the LacI/GalR repressor family (27). To bind to the catabolite response elements (cre) (41), which are operator sites located in front or at the beginning of most catabolite-repressed genes and operons (14), CcpA usually requires a corepressor, which was identified as P-Ser-HPr (19). Bacilli, geobacilli, and oceanobacilli also possess an HPr-like protein, Crh, which is also implicated in CCR of a few genes and operons (14, 20, 37). The ATP-dependent phosphorylation of HPr (or Crh) at Ser-46 is catalyzed by the enzyme HPr kinase/phosphorylase (HprK/P) (21). The kinase function of HprK/P is stimulated by high amounts of glycolytic intermediates, such as fructose-1,6-bisphosphate (13, 21), the concentration of which rises in bacteria during the uptake of a rapidly metabolizable carbon source (38, 50). In the presence of high concentrations of phosphate, the bifunctional HprK/P also catalyzes the dephosphorylation of P-Ser-HPr (30), which leads to the production of pyrophosphate (phosphorolysis instead of the usual hydrolysis reaction) (38). The binding site for the P-Ser-HPr/CcpA protein complex in the B. subtilis lev operon is located upstream of the −12, −24 promoter (37). It has been proposed that the binding of P-Ser-HPr/CcpA to the lev cre would prevent the interaction of LevR fixed to the UAS with the RNA polymerase/RpoN holoenzyme.
We identified a Lactobacillus casei operon which is controlled by a LevR-like regulator and which encodes the proteins of a mannose class PTS strongly resembling the proteins of the Lev-PTS of B. subtilis. However, in contrast to the B. subtilis lev operon (10), expression of the L. casei lev operon did not require a functional RpoN (σ54). Interestingly, the replacement of Ile-47 in L. casei HPr by a threonine led to overexpression of the lev operon. The observed slow in vitro PEP- and ATP-dependent phosphorylation of the mutant HPr as well as the slow phosphoryl group transfer from histidyl-phosphorylated mutant HPr to EIIALev are assumed to be responsible for the elevated expression of the lev operon.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and plasmids.
The L. casei and Escherichia coli strains used in this study are listed in Table 1. L. casei strains were grown under static conditions in MRS fermentation medium (11) supplemented with 0.5% sugar. E. coli strains were used as hosts for cloning experiments and also to overproduce maltose binding protein (MBP)- and His-tagged proteins. They were grown in Luria-Bertani medium at 37°C under vigorous shaking. Solid MRS or Luria-Bertani medium was prepared by adding 1.5% agar. E. coli strains were transformed by electroporation with a Gene Pulser apparatus (Bio-Rad Laboratories) as recommended by the manufacturer, and L. casei strains were transformed as previously described for Lactobacillus sakei (4). E. coli and L. casei transformants were selected on solid medium containing ampicillin (100 μg/ml) or erythromycin (5 μg/ml). The plasmids used in this study are listed in Table 2.
TABLE 1.
Strains used in this study
| Strain | Genotype or characteristica | Source or reference |
|---|---|---|
| E. coli | ||
| Fit | F−ompT hsdSB(rB− mB−) gal dcm (DE3) ptsH | Takara Bio Inc. |
| DH5α | F− φ80lacZ ΔM15 Δ(lacZYA-argF)U169 deoR endA1 recA1 hsdR17(rK− mK−) supE44 thi-1 gyrA96 relA1 phoA | Invitrogen |
| NM522 | SupE thi-1 Δ(lac-proAB) Δ(mcrB-hsdSM) (rK− mK−) [F′ proAB lacIqZΔGM15] | 25 |
| L. casei | ||
| BL23 | Devoid of pLZ15 | 1 |
| BL71 | ccpA::erm | 39 |
| BL121 | ptsH1 | 51 |
| BL122 | ptsH2 | 51 |
| BL123 | ptsH3 | 51 |
| BL124 | ptsI::erm | 51 |
| LcG102 | hprK::pHKLc208(hprK208Am) | 15 |
| BL23levB | levB10(Oc) | This work |
| BL23H3levB | ptsH3 levB10(Oc) | This work |
| BL23levX | levX5(Oc) | This work |
| BL23H3levX | ptsH3 levX5(Oc) | This work |
| BL23levR | levR::pRVlevR | This work |
| BL23H3levR | ptsH3 levR::pRVlevR | This work |
| BL23sig54 | rpoN19(Oc) | This work |
| BL23H3sig54 | ptsH3 rpoN19(Oc) | This work |
Am and Oc indicate amber (UAG) and ochre (UAA) stop codons.
TABLE 2.
Plasmids used in this study
| Plasmid | Characteristic(s) | Source or reference |
|---|---|---|
| pBluescriptSK(+) | Amp | Invitrogen |
| pCDNA2.1 | Amp | Invitrogen |
| pQE30 | Amp | QIAGEN |
| pQELevA | levA cloned in pQE30 | This work |
| pQELevB | levB cloned in pQE30 | This work |
| pET-15b | Ampr | Novagen |
| pET-ptsH | ptsH cloned in pET-15b | C. D. Esteban, unpublished |
| pET-ptsH3 | ptsHI47T cloned in pET-15b | This work |
| pMal-c2X | Amp | New England BioLabs |
| pMalLevR | levR cloned in pMal-c2X | This work |
| pMalLevR-H1 | levRH1 cloned in pMal-c2X | This work |
| pMalLevR-H12 | levRH12 cloned in pMal-c2X | This work |
| pRV300 | Erm and Amp | 31 |
| pRVlevR | Internal fragment of levR cloned in pRV300 | This work |
| pRVlevB | levB allele with stop codon 10 cloned in pRV300 | This work |
| pRVlevC | levC allele with stop codon 6 cloned in pRV300 | This work |
| pRVsig54 | rpoN allele with stop codon 19 cloned in pRV300 | This work |
2-D protein gel electrophoresis.
Two independent analytical two-dimensional (2-D) gel electrophoresis experiments were carried out with L. casei BL23 or one of the five CCR-relieved mutants grown in 5 ml of MRS medium containing 0.5% glucose. When the cultures approached an optical density at 600 nm of 2, 250 μCi of a [35S]methionine-cysteine protein labeling mix (1 Ci/μmol; Perkin-Elmer Life Sciences, Boston, Mass.) was added, and the cells were kept for 24 h at 37°C. Proteins were subsequently extracted from the various strains, and the amount of radioactivity incorporated was determined by liquid scintillation counting. Equal amounts of radiolabeled proteins were loaded onto each gel, and electrophoresis was performed as previously described (24). The gels were silver stained, dried, and exposed to a storage phosphor screen for 48 h (Packard Instrument, Canberra, Australia) before radioactive spots were visualized by using a Phosphoimager (Cyclone; Packard Instrument).
To carry out preparative 2-D gel electrophoresis, L. casei cells were grown in 100 ml of MRS medium containing 0.5% glucose. The Millipore (Bedford, Mass.) Investigator 2-D electrophoresis system was used for the first dimension (pH gradient of 4.4 to 5.5). A 14% polyacrylamide gel without a stacking gel was used for the second dimension. After electrophoresis, proteins were transferred onto a polyvinylidene difluoride membrane (Immobilon; Millipore) by electroblotting (MilliBlot-Graphite Electroblotter; Millipore) according to the manufacturer's instructions. After Coomassie blue staining, selected protein spots were cut out of the membrane, and their N-terminal sequences were determined at the Institut für Biochemie, Universität Wien, Vienna, Austria.
The migration position of HPr after 2-D gel electrophoresis on polyvinylidene difluoride membranes was detected by using a rabbit polyclonal antiserum raised against HPr of Staphylococcus carnosus. The HPr-antibody complex was visualized with the ECL Western blot analysis system (Amersham International, Little Chalfond, United Kingdom).
General molecular biological methods.
For the extraction of chromosomal DNA, bacteria were grown in 80-ml cultures and harvested by centrifugation, and DNA was extracted by using the NUCLEBOND AX kit (Macherey-Nagel GmbH, Düren, Germany) according to the manufacturer's instructions. Plasmids were isolated with a QIAPrep kit (QIAGEN, Santa Clara, Calif.). Several DNA fragments from the levR gene or the lev operon were amplified by ligase-mediated PCR (16) using pBluescript SK(+) as a vector. Restriction endonucleases (EcoRI, PstI, BamHI, ClaI, HindIII, AcsI, AluI, and Sau3A), alkaline phosphatase, and T4 DNA ligase were purchased from Roche Diagnostic GmbH (Mannheim, Germany). PCRs were carried out in 25-μl assay mixtures by using PCR MasterMix (Eppendorf, Hamburg, Germany). PCR products were purified with a QIAquick kit (QIAGEN).
DNA sequencing and sequence analyses.
PCR fragments were sequenced by using the dideoxy chain termination method with the ABI Prism sequencing system (PE Biosystem). DNA sequence analyses, database searches, and sequence alignments were performed with the Mac Vector (Kodak Scientific Imaging Systems), BLAST (3), and phredPhrap (17) programs, respectively.
Northern blots.
Total RNA of L. casei BL23 or one of its mutant strains was isolated from exponentially growing cells by using the RNeasy Midi kit (QIAGEN). To carry out Northern blots, RNA was prepared from the various strains, and aliquots of 5 μg were separated by electrophoresis and subsequently transferred onto Hybond-N+ membranes (Amersham International) by using standard procedures. The size of the transcripts was estimated by comparison with an RNA ladder (0.28 to 6.6 kb) (Amersham International). The oligonucleotides EIIA3 (5′-ATGGCGTTAACCACATTGATGTCG-3′) and EIIA7 (5′-TACCGCAATAATCCCATCCA-3′) as well as LevX1 (5′-AAGAAGTTGCGTATCAGAAG-3′) and LevX2 (5′-ACTTTTTAGCTGCCTTCAAC-3′) were used to prepare PCR-amplified radiolabeled probes (7), which allowed for the detection of levA and levX mRNAs, respectively. Hybridization experiments were carried out overnight at 60°C by submerging the Hybond-N+ membranes in 0.5 M sodium phosphate buffer (pH 7.0) containing 5% sodium dodecyl sulfate (SDS) and one of the radiolabeled probes. The membranes were subsequently washed at 60°C with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) containing 0.2% SDS followed by 0.2× SSC containing 0.2% SDS and exposed to a storage phosphor screen (Packard Instrument) for 5 h.
Mapping of the transcription start site.
The 5′ ends of the mRNA of the lev operon and the levR gene were mapped by using the 5′ rapid amplification of cDNA ends (RACE) technique. The oligonucleotides used for the PCR with the 3′/5′ RACE kit (Roche), for the PCR after the polyA tailing, and for the sequencing of the PCR products are presented in Table 3.
TABLE 3.
Oligonucleotides used in this study
| Function and name | Sequencea | Used for: |
|---|---|---|
| Construction of mutants | ||
| LvRXh | ACGCGCTCGAGATCAAAGATGTGATTCTTGC | Cloning part of levR |
| LvRPs | CCAACTGCAGGAATCACAACTAATCAGGCT | |
| LvBBlunt | ACCGCAATACTGTCCGAAC | Cloning levB and inserting stop codons and restriction sites |
| LvBMut1 | CGTATTGATTAACGCATGCTTTAAGGCTTGATCACGGTTCGT | |
| LvBMut2 | ATCAAGCCTTAAAGCATGCGTTAATCAATACGTACAAACGAA | |
| LvBNh | AACCGCTTGGCTAGCCATTCATGAGTTGGAACATAAG | |
| LvXKp | GACGGCGGTACCATTAACGATATGCGT | Cloning levX and inserting stop codons and restriction sites |
| LvXMut1 | TAGACTAAGCTTTAAGAAAAAGAAGTTGCGTATCAG | |
| LvXMut2 | TAAAGCTTAGTCTATTAGTCAATTGTCATTTTGATTTT | |
| LvXEc | GCCGGAATTCCTTCACCGGATTGTC | |
| SLSp | GTTGGTACTAGTGATCCGCACATTA | Cloning part of rpoN and inserting stop codons and restriction sites |
| SLMut1 | ACGTAAGGGATCCGATAGTAAATTCAGATGTTGCAAT | |
| SLMut2 | TTTACTATCGGATCCCTTACGTTAGCACCAACTTATTG | |
| SLBlunt | TCTGCAAATACCAGCTCAG | |
| Overexpression of genes | ||
| QEABA | AGGGATGGGATCCATGAAATATTTGCTTCTTG | Cloning levA |
| QEAKP | TTTTGCTGGTACCTTAAATATCGTCGTCGTC | |
| QEBBA | AGGAGTGGGATCCATGGCAATTTCGTTTGTA | Cloning levB |
| QEBKP | TATTGTGGTACCATTAATACCCAAAGCG | |
| H3-1 | CAGATCACATATGGAAAAACGCGAAT | Cloning ptsHI47T |
| H3-2 | GCAGAGATCTCCTTCAAATGTTCAG | |
| MalRXBA | GGTGTCTAGAATGAACGCAGTAGAAAAACTTTATG | Cloning levR |
| MalRPST | CCTTTTTGTCTGCAGATCACGCCACAAAAATAGAGT | |
| MH1A | TGTGTTCGCCGGCAGCAAGCAGGATGGCATTGTA | levR His488Ala mutation |
| MH1B | ATCCTGCTTGCTGCCGGCGAACACACCGCATCGAG | |
| MH2A | TTAAGGACAAGGCCATGAAGAGATTGAGCTTTAG | levR His776Ala mutation |
| MH2B | AATCTCTTCATGGCCTTGTCCTTAATGATTGAACG | |
| Detection of the transcriptionnal start site | ||
| EIIA2 | GTTTGCTTTAAGCCACTGG | Identification of the transcription start site of the lev operon |
| EIIA5 | CATCAATGTGGTTAACGCCA | |
| EIIA6 | CCAGAACAACAAAGCTTGCA | |
| levR5 | ACGACGAATGGCTTCGTG | Identification of the transcription start site of levR |
| levR6 | TGCTTCGATTGCCAGCTG | |
| levr7 | CATCGGCAGCCTGATTAG |
Letters in boldface type indicate newly created restriction sites; newly created stop codons or mutations are underlined.
Purification of His- and MBP-tagged proteins.
Synthesis and purification of EI from B. subtilis carrying an N-terminal His6 were carried out as previously described (20).
To overproduce His-tagged LevA and LevB and MBP-tagged LevR, the genes encoding the L. casei proteins were amplified by PCR with chromosomal DNA isolated from strain BL23, Pyrobest polymerase (Takara Bio Inc., Shiga, Japan), and appropriate primers (Table 3), which added restriction sites to the 5′ and 3′ ends. The PCR fragments were cut with the restriction enzymes recognizing the sites added by the PCR and inserted into vectors cleaved with the same enzymes. We used the His tag expression vector pQE30 (QIAGEN) for inserting levA and levB, and we used the MBP tag expression vector pMAL-c2X (New England BioLabs, Herts, United Kingdom) for the cloning of levR. The resulting plasmids pQELevA, pQELevB, and pMalLevR were used to transform E. coli NM522 (25), and the correct sequence of the inserts was confirmed by DNA sequencing. LevA and LevB carrying an N-terminal His6 were synthesized and purified on Ni-nitrilotriacetic acid columns by following the standard protocol of QIAGEN. MBP-tagged LevR was also synthesized in strain NM522 and subsequently purified on an amylose column by following the protocol recommended by New England Biolabs.
To obtain the levRH1 allele, which encodes a LevR protein with a His-488-Ala replacement, a three-step PCR experiment was carried out (9). By using plasmid pMalLevR as a template and appropriate primers (Table 3), the first PCR allowed the 5′ part of levR with the desired mutation located at the 3′ end to be amplified by using primers LevRXBA and MH1B (Table 3), while the second PCR provided the 3′ part of levR and contained the mutation at the 5′ end (primers LevRPST and MH1A). These two PCR products served as a template for the third PCR, which required a special first cycle (1 min at 90°C, 10 s at 80°C, 2 min at 60°C, 10 min at 72°C, 1 min at 90°C, 2 min at 60°C, and 3 min at 72°C) before conventional PCR cycles were applied to amplify the complete levRH488A allele (with primers LevRXBA and LevRPST). The final PCR product was inserted into pMAL-c2X, thus providing plasmid pMalLevR-H1, which was used to transform NM522. To obtain the levRH12 allele encoding a LevR protein with His-488-Ala and His-776-Ala replacements, an approach identical to that used for the construction of the levRH1 allele was applied, except that plasmid pMalLevR-H1 was used as a template and the oligonucleotides MH2A and MH2B (Table 3) were used as mutagenic primers. The correct sequences of the entire inserts in plasmids pMalLevR-H1 and pMalLevR-H12 were confirmed by DNA sequencing. Overexpression of the levR alleles and purification of the MBP-tagged mutant proteins were carried out as described above for wild-type LevR.
Overproduction of L. casei HPr was carried out similarly to a previously described method (33). However, the ptsH gene was inserted into vector pET-15b instead of pET-3c (Merck KGaA, Darmstadt, Germany), which allowed us to synthesize L. casei HPr with an N-terminal His tag in the E. coli strain Fit (BL21-DE3 carrying a ptsH mutation; Takara Bio Inc.) and to purify it on a Ni-nitrilotriacetic acid affinity column (C. D. Esteban and G. Perez-Martinez, unpublished data). To overproduce HPrI47T, the corresponding ptsH allele was amplified by PCR using chromosomal DNA of the ptsHI47T strain and appropriate primers (Table 3). The PCR product was inserted into vector pET-15b. Expression of the ptsHI47T allele and purification of the His-tagged mutant HPr were carried out as described above for the wild-type protein.
Protein phosphorylation experiments with [32P]PEP.
[32P]PEP was synthesized by using the PEP-pyruvate isotope exchange method in the presence of pyruvate kinase (46). Phosphorylation of HPr and HPrI47T by EI was carried out at 37°C in 210-μl assay mixtures containing 5 μg of EI, 5 μg of wild-type or mutant HPr, 50 mM Tris-HCl (pH 7.4), 5 mM MgCl2, and 7.5 μM [32P]PEP (30 μCi/ml). Aliquots of 40 μl were withdrawn after different time intervals, and the reactions were stopped by adding 6 μl of SDS sample buffer. Proteins were separated by electrophoresis on 0.1% SDS-15% polyacrylamide gels.
To follow the phosphoryl group transfer from P∼His-HPr or P∼His-HPrI47T to EIIALev, 210-μl assay mixtures were prepared as described above and preincubated for 5 min at 37°C to allow efficient phosphorylation of HPr or HPrI47T before 50 μg of EIIALev was added. Aliquots of 40 μl were withdrawn after various time intervals, and the reactions were stopped by adding 6 μl of SDS sample buffer. Proteins were separated by electrophoresis on 0.1% SDS-15% polyacrylamide gels.
To phosphorylate the EIIAMan domain of LevR, the assay mixture (35 μl) contained 2 μg of LevR or the various mutant LevRs, 1 μg of EI, 1.5 μg of wild-type HPr, 50 mM Tris-HCl (pH 7.4), 5 mM MgCl2, and 7.5 μM [32P]PEP (30 μCi/ml); 4 μg of EIIALev and EIIBLev was additionally added for the phosphorylation of LevR or the mutant LevRs at PRD2. After incubation at 37°C for 15 min, the samples were separated on denaturing gels (0.1% SDS) containing either 8 or 15% polyacrylamide. To compare P∼His-HPr- and P∼His-HPrI47T-mediated phosphorylation at the EIIAMan domain of MBP-LevR, experiments were carried out in 210-μl assay mixtures, which were prepared as described above for the phosphorylation of EIIALev and preincubated for 5 min at 37°C before 15 μg of MBP-LevR was added. Aliquots of 40 μl were withdrawn after various time intervals, and the reactions were stopped by adding 6 μl of SDS sample buffer. Proteins were separated by electrophoresis on denaturing gels containing 0.1% SDS-15% polyacrylamide.
ATP-dependent HPr phosphorylation and P-Ser-HPr dephosphorylation.
ATP-dependent HprK/P-catalyzed phosphorylation of HPr and HPrI47T at Ser-46 was carried out at 37°C in 250-μl assay mixtures containing 300 ng of HprK/P, 25 μg of wild-type or mutant HPr, 50 mM Tris-HCl (pH 7.4), 5 mM MgCl2, 5 mM fructose-1,6-bisphosphate (FBP), and 1 mM ATP. Aliquots of 40 μl were withdrawn after various time intervals. The reactions were stopped by heating the samples for 5 min at 75°C. HPr and seryl-phosphorylated HPr were separated by electrophoresis on nondenaturing 12.5% polyacrylamide gels and visualized by staining with Coomassie blue.
For the dephosphorylation experiments, seryl-phosphorylated L. casei wild-type HPr and HPrI47T were prepared by using L. casei Val-267-Phe mutant HprK/P, which is normally active as kinase but has almost completely lost the phosphorylase function (40). About 0.3 mg of HPr or HPrI47T was incubated for 2 h at 37°C in 1.5-ml assay mixtures containing 5 μg of HprK/P(V267F), 50 mM Tris-HCl (pH 7.4), 5 mM MgCl2, 5 mM FBP, and 1 mM ATP. To inactivate HprK/P(V267F), the samples were heated for 5 min at 65°C before they were desalted on PD-10 columns (Pharmacia) to remove ATP and FBP and lyophilized. Dephosphorylation reactions with P-Ser-HPr were carried out at 37°C in 250-μl assay mixtures containing 300 ng of HprK/P, 25 μg of wild-type or mutant P-Ser-HPr, 50 mM Tris-HCl (pH 7.4), 5 mM MgCl2, and 5 mM inorganic phosphate. Aliquots of 40 μl were withdrawn after various time intervals. The reactions were stopped by heating the samples for 5 min at 65°C. HPr and seryl-phosphorylated HPr were separated on nondenaturing 12.5% polyacrylamide gels as described above.
Construction of the various L. casei mutants.
An internal DNA fragment of levR was amplified by PCR with the primers levRXh and levRPs (Table 3) and cloned in the integrative vector pRV300 (31). The resulting plasmid was used to transform the L. casei strain BL23 and the ptsHI47T mutant. One integrant of each strain resulting from a single crossover was selected, and the integrants were named BL23levR and BL23H3levR, respectively. PCR analysis and DNA sequencing confirmed the integration at the levR locus.
To construct L. casei levB, levC, levX, and rpoN mutants, DNA fragments carrying the desired mutations were amplified by the three-step PCR technique (9). In each case, specific oligonucleotides were used, which allowed the creation of stop codons located close to a newly created restriction site at the beginning of the corresponding open reading frame (ORF) (Table 3). The PCR products were cloned into pRV300 (31), and the resulting plasmids were called pRVlevB, pRVlevC, pRVlevX, and pRVsig54, respectively, and were used to transform strain BL23 and the ptsHI47T mutant. The eight different integrants obtained with the four different plasmids and the two strains were selected on erythromycin-containing solid medium. A single clone of each of the eight integrants was isolated and grown for several generations on liquid medium in the absence of the antibiotic to allow a second recombination which, depending on its location, could either restore the corresponding wild-type gene or provide the mutant allele. Erythromycin-sensitive clones were isolated, and DNA fragments containing the site of mutation were amplified by PCR. Identification of strains carrying the desired mutant allele was facilitated, since their PCR products contained newly created restriction sites. The presence of the desired mutation was confirmed by DNA sequencing of the PCR product. The levB, levC, levX, and rpoN mutants obtained with BL23 were called BL23levB, BL23levC, BL23levX, and BL23sig54, while the corresponding mutants obtained with the ptsHI47T strain were called BL23H3levB, BL23H3levC, BL23H3levX, and BL23H3sig54, respectively (Table 1).
RESULTS
Proteome analysis with wild-type L. casei and a ptsHI47T strain.
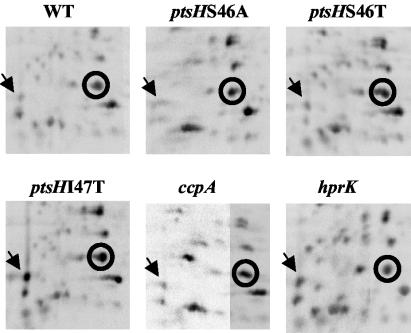
When carrying out analytical 2-D gel electrophoresis with soluble extracts from L. casei BL23 and five mutants affected in CCR, i.e., ccpA (39), ptsHS46A, ptsHS46T, ptsHI47T (51), and hprK (15), we observed numerous differences of the protein pattern between the wild-type and the mutant strains. One protein attracted our attention, as it was overproduced in the ptsHI47T strain but not in the other CCR-relieved mutants (Fig. 1; the position of this protein is indicated with an arrow). This protein was isolated by preparative 2-D gel electrophoresis, and microsequencing provided the following N-terminal amino acid sequence: MKYLLLVSHGDFSSGLKQTLGMFAGDDA. A homology search with this sequence revealed that out of its 28 amino acids, 16 were identical to the N-terminal sequence of a presumed EIIALev from Clostridium acetobutylicum (43) and 19 were identical to an EIIA of a mannose class PTS from Streptococcus mutans (2).
FIG. 1.
Analytical 2-D gel electrophoresis carried out with crude extracts of L. casei wild-type BL23 (WT) and five CCR-relieved mutants. Shown is the region around the migration position of HPr (encircled), which was determined on preparative gels by carrying out Western blots with a rabbit polyclonal antibody. The arrows indicate the position of EIIALev, which is overproduced in the ptsHI47T strain.
DNA sequence of the EIIA-encoding gene and its surrounding ORFs.
A reverse genetic approach was used to clone a DNA fragment containing a major part of the EIIA-encoding gene. For this purpose, a 20-bp oligonucleotide probe, NTP1 (5′-ATGTTTGCNGGYGAYGAYGC-3′), was designed based on the last seven amino acids of the above microsequence (MFAGDDA) and hybridized with L. casei chromosomal DNA digested with one of eight different restriction enzymes. In each case, a single radioactive hybridization band was observed, indicating that the probe was highly specific. AcsI-restricted chromosomal DNA provided a radioactive band of about 1.3 kb (data not shown). The corresponding DNA fragment was cloned into pBluescript SK(+) cut with EcoRI and subsequently sequenced. It was composed of 1,292 bp and contained almost the complete EIIA-encoding gene preceded by an incomplete ORF oriented in the opposite direction.
The entire sequence of the PTS-encoding operon and the preceding ORF was determined by cloning DNA fragments using the ligase-mediated PCR technique (see Materials and Methods) and by including information obtained from an ongoing L. casei BL23 genome sequencing project (29). In total, we determined the sequence of a 7,123-bp DNA fragment (Fig. 2A) (EMBL database accession number AJ344254). This sequence contained the EIIA-encoding gene preceded by a 2,532-bp ORF encoding a protein exhibiting a domain organization similar to that of the transcription activator LevR from B. subtilis (Fig. 2B), although characteristic differences exist, as will be explained later. The EIIA-encoding gene was followed by four ORFs oriented in the same direction (Fig. 2A). The first three ORFs encode EIIB, EIIC, and EIID of a mannose class PTS. It was therefore likely that they form an operon together with the EIIA-encoding gene and the fifth gene, although a noncoding region of 150 bp was present between the EIIB- and EIIC-encoding genes (Fig. 2A). The EII components exhibit significant sequence identity (between 25 and 60%) to the corresponding EIIs of the Lev-PTS of B. subtilis and a presumed sacC-containing Lev-PTS of C. acetobutylicum but also to the EIIs of a mannose class PTS from S. mutans with unknown sugar specificity. Based on this sequence homology, and since the newly discovered L. casei operon is preceded by a LevR-like protein, we called it lev operon. However, in contrast to B. subtilis and C. acetobutylicum, the L. casei BL23 lev operon does not contain the levanase-encoding sacC gene. Interestingly, genome sequencing of the L. casei neotype strain ATCC 334 (http://genome.jgi-psf.org/draft_microbes/lacca/lacca.home.html) revealed that this strain possesses a sacC-containing lev operon (more than 98% identical to the lev operon of strain BL23). Since genome sequencing of strain BL23 is not yet completed (about 93% of the sequence have been determined), the possibility that sacC might be present somewhere else on its genome cannot be excluded. The fifth gene of the L. casei BL23 lev operon encodes a small protein (105 amino acids) of unknown function.
Transcription analysis with wild-type L. casei and the five CCR mutants.
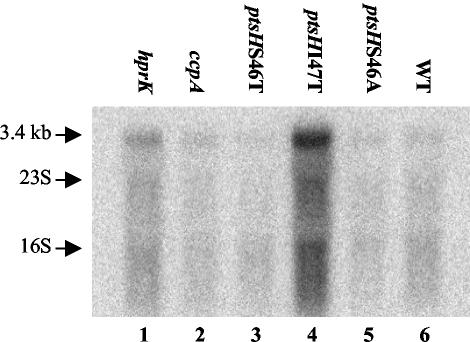
RNA was isolated from L. casei BL23 and the five CCR-relieved mutants grown in glucose-containing medium to exponential or stationary growth phase and hybridized with a probe derived from either the levA or the levX gene. Figure 3 shows the results obtained with RNA extracted from cells during exponential growth hybridized with the levA probe. A strong radioactive band of about 3.4 kb was observed with the ptsHI47T mutant (Fig. 3, lane 4). In agreement with the proteome analysis (Fig. 1), where overproduction of EIIALev was observed only in the ptsHI47T mutant, the radioactive band was much weaker in the other CCR-relieved mutants and the wild-type strain. These results suggested that a relief from CCR cannot be the only cause for the observed overexpression of the lev operon in the ptsHI47T mutant and that the transcript of the lev operon was pentacistronic (levABCDX, with a total size of 3.16 kb). This assumption was confirmed by carrying out a Northern blot with the levX-derived probe, which also provided a 3.4-kb radioactive band (data not shown). When the cells were grown to stationary phase, the lev operon transcript could no longer be detected (data not shown), not even in the ptsHI47T mutant. When the wild-type strain was grown in the presence of various carbon sources, fructose and mannose led to the strongest expression of the lev operon. A basal expression occurred during growth on glycerol or ribose, while glucose seemed to exert a repressive effect (data not shown). When a levR-derived probe was used, no transcript could be detected, indicating that under the employed experimental conditions, the levR gene was only poorly expressed (data not shown).
FIG. 3.
Northern blots carried out with 5 μg of RNA isolated from either the L. casei wild-type strain BL23 (WT) or one of the five CCR-relieved mutants grown in MRS fermentation medium complemented with glucose. A levA-specific probe was used to detect the lev operon transcript of 3.4 kb in the various samples. The positions of 16S and 23S rRNAs are indicated with arrows.
Mapping the transcription start site of levR and the lev operon.
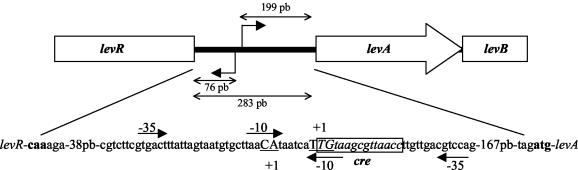
In order to locate the transcription initiation site of levR and the lev operon, RACE PCR experiments were carried out with total RNA extracted from either the wild-type strain BL23 or the ptsHI47T mutant harvested during exponential growth. In each case, a unique band was observed after electrophoresis with the 5′ RACE PCR product. The transcription start site of the lev operon was identified as G, T, or T located between 198 and 200 nucleotides upstream of the start codon, respectively. It is preceded by the putative −10 box TAACAT (Fig. 4). The transcription start site of the levR gene was found to be a G or T located 76 or 77 nucleotides upstream of the presumed start codon TTG. It is preceded by the putative −10 box TACAAT. Presumed −35 boxes are also present and are indicated in Fig. 4. A potential cre site (TGTAAGCGTTAACC) containing only one mismatch compared to the imperfect palindromic consensus sequence TGWNANCGNTNWCA (42) is overlapping the transcription start site of the lev operon and the −10 box of levR (Fig. 4).
FIG. 4.
Schematic representation of the 283-bp intergenic region between levR and levA of L. casei. This sequence contains the transcription start sites (+1, underlined capital letters), the start codons (letters in boldface type), and presumed −10 and −35 promoter regions (arrows) for both the levR gene and the levRABCDX operon as well as a potential cre site (boxed area) which overlaps the transcription start site of the lev operon and the −10 promoter sequence of levR.
L. casei lev operon expression requires LevR but not RpoN.
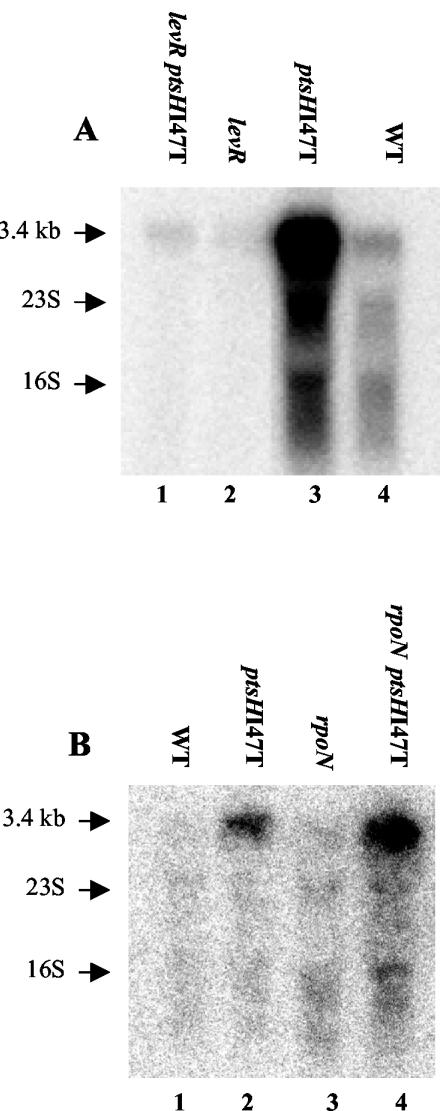
Since the lev operon of L. casei misses a −12, −24 promoter typical of RpoN-dependent transcription units, we tested whether LevR and RpoN were really necessary for the expression of the L. casei lev operon. For this purpose, the levR gene was disrupted in the wild-type strain BL23 and the ptsHI47T mutant. Northern blots revealed that the strong radioactive signal observed with the ptsHI47T mutant disappeared in the ptsHI47T levR double mutant (Fig. 5A), indicating that LevR is necessary for the expression of the lev operon. By contrast, a disruption of rpoN in the wild-type strain BL23 and the ptsHI47T mutant had no effect on lev operon expression (Fig. 5B).
FIG. 5.
Northern blots carried out with 5 μg of RNA isolated from wild-type strain BL23 (WT), the ptsHI47T or levR mutant, or the ptsHI47T levR double mutant (A) and wild-type strain BL23 (WT), the ptsHI47T or rpoN mutant, or the ptsHI47T rpoN double mutant (B). The strains were grown in MRS fermentation medium complemented with glucose. The levA-specific probe was used to detect the lev operon transcript of 3.4 kb. The positions of the 16S and 23S rRNAs are indicated with arrows.
LevR of L. casei is phosphorylated by P∼His-HPr at His-488 and by P∼His-EIIALev at His-776.
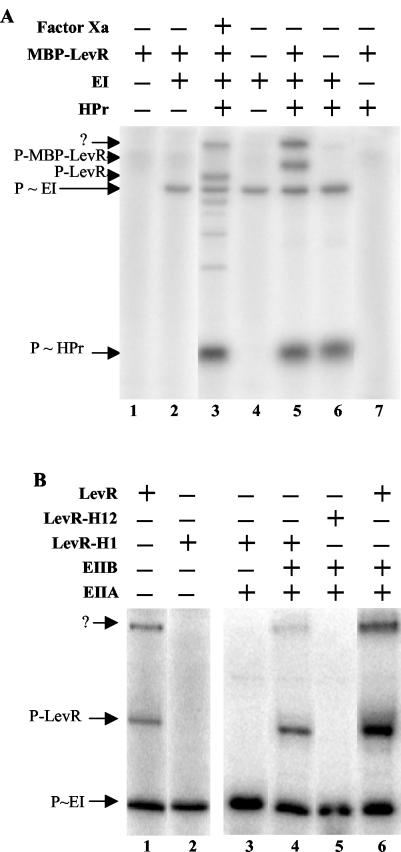
Since expression of the L. casei lev operon required LevR, we wanted to test whether, similar to LevR of B. subtilis, L. casei LevR was also phosphorylated by P∼His-HPr in the EIIA domain and by P∼His-EIIBLev in PRD2. L. casei LevR was purified as a fusion protein with the E. coli maltose binding protein (MBP-LevR). No phosphorylation of MBP-LevR could be detected when it was incubated with [32P]PEP in the absence of PTS proteins or with only EI or only HPr (Fig. 6A, lanes 1, 2, and 7, respectively). However, in the presence of both general PTS proteins, MBP-LevR was phosphorylated by [32P]PEP, as a radioactive band appeared which was absent in the phosphorylation mixture containing EI and HPr (Fig. 6A, compare lanes 5 and 6) and which migrated according to the molecular weight calculated for the MBP fusion protein. Interestingly, a second radioactive band was present which, based on its migration behavior, was assumed to correspond to MBP-LevR dimers. Since the samples could not be heated due to the instability of P∼His bonds, MBP-LevR dimers might not have been dissociated.
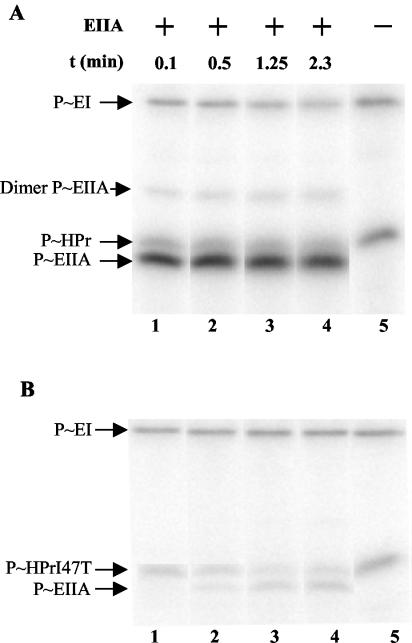
FIG. 6.
Phosphorylation of MBP-LevR by [32P]P∼His-HPr (A) and [32P]P∼EIIBLev (B). (A) Samples containing [32P]PEP and the indicated proteins were incubated at 37°C before they were separated on a 0.1% SDS-15% polyacrylamide gel which was dried and exposed to autoradiography. The sample loaded on lane 3 contained MBP-LevR, which had been treated with factor Xa before the phosphorylation reaction was carried out. The migration positions of HPr, EI, LevR, and MBP-LevR are indicated by arrows. The slowest-migrating radioactive band (arrow with the question mark) probably corresponds to MBP-LevR dimers. (B) Samples containing [32P]PEP, EI, HPr, and the indicated proteins were incubated at 37°C before they were separated on a 0.1% SDS-8% polyacrylamide gel, which was dried and exposed to autoradiography. In LevR-H1, His-488 in the EIIAMan domain was replaced with an alanine, while in LevR-H12, His-776 in PRD2 was also replaced with an alanine. The migration positions of EI and wild-type and mutant LevRs are indicated by arrows.
When MBP-LevR was treated with factor Xa, which cuts the fusion protein within the linker connecting MBP and LevR, before it was incubated with [32P]PEP, EI, and HPr, a major radioactive band migrating according to the molecular mass of LevR (93 kDa) and not of MBP (43 kDa) was detected (Fig. 6A, lane 3), confirming that the fusion protein is phosphorylated within the LevR part. The minor faster-migrating radioactive bands observed with factor Xa-treated MBP-LevR are probably due to unspecific cleavage by the protease. We also purified a mutant MBP-LevR, in which His-488, the site of phosphorylation in the EIIAMan domain, had been replaced with an alanine. When carrying out phosphorylation experiments with the MBP-LevR(H488A) mutant protein, none of the two LevR-related radioactive bands could be detected (Fig. 6B, compare lanes 1 and 2), confirming that P∼His-HPr-mediated phosphorylation occurs at His-488 (B. subtilis is phosphorylated at the equivalent His-585) (34) and further supporting the assumption that the second slowly migrating radioactive band corresponds to MBP-LevR dimers.
The MBP-LevR(H488A) mutant protein was also used to test whether LevR would be phosphorylated by P∼His-EIIBLev. When the mutant protein was incubated with [32P]PEP, EI, HPr, and EIIALev, no phosphorylation of MBP-LevR(H488A) could be observed (Fig. 6B, lane 3). However, when EIIBLev was present in addition (EIIALev and EIIBLev were purified as His-tagged proteins), a radioactive band migrating to the position of MBP-LevR and a second band, probably corresponding to MBP-LevR dimers, could be detected (Fig. 6B, lane 4). Wild-type MBP-LevR phosphorylated under identical conditions provided a stronger radioactive band (Fig. 6B, lane 6), probably because it is phosphorylated at both the EIIAMan domain and PRD2. By contrast, when an MBP-LevR fusion protein was used in which His-448 and His-776 had been replaced with alanine, no phosphorylation of LevR occurred (Fig. 6B, lane 5), confirming that His-778, which is equivalent to His-869 in B. subtilis LevR (34), represents the site of phosphorylation by EIIBLev in PRD2 of L. casei LevR (Fig. 2B).
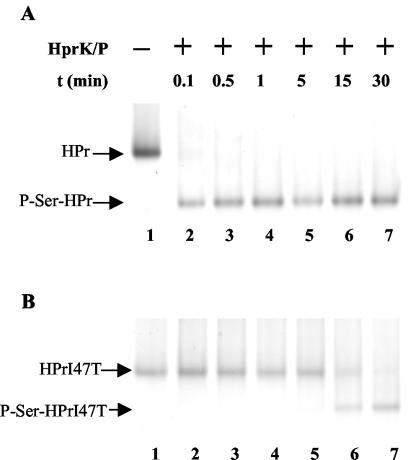
Effect of the Ile-47-Thr replacement in HPr on its ATP-dependent phosphorylation.
Since the formation of P∼His-HPr is necessary for the phosphorylation of L. casei LevR at His-488 and His-778 (Fig. 2B), and since the lev operon contains a cre sequence which is the binding site for the P-Ser-HPr/CcpA complex, we wanted to test whether the Ile-47-Thr replacement in HPr would affect its ATP- and/or PEP-dependent phosphorylation, which in turn could be responsible for the observed elevated lev operon expression in the ptsHI47T mutant. When carrying out ATP-dependent phosphorylation reactions with purified His-tagged L. casei wild-type HPr and HPrI47T, we observed that wild-type HPr was almost completely phosphorylated after a few seconds (Fig. 7A), while under identical conditions, phosphorylation of the mutant HPr was not yet completed after 30 min (Fig. 7B). Dephosphorylation experiments with wild-type and mutant P-Ser-HPr in the presence of HprK/P were also carried out. The Ile-47-Thr replacement also slowed the dephosphorylation at Ser-46 (data not shown), but the inhibitory effect was less pronounced (about 10-fold) than that observed for ATP-dependent HPr phosphorylation.
FIG. 7.
ATP-dependent HprK/P-catalyzed phosphorylation at Ser-46 of HPr (A) and HPrI47T (B). Samples were prepared as described in Materials and Methods, incubated for different time periods at 37°C, and separated on nondenaturing 12.5% polyacrylamide gels, which allowed us to separate HPr and P-Ser-HPr. The gels were stained with Coomassie blue.
Effect of the Ile-47-Thr replacement on PEP-dependent HPr phosphorylation and phosphoryl group transfer from P∼His-HPr to EIIALev.
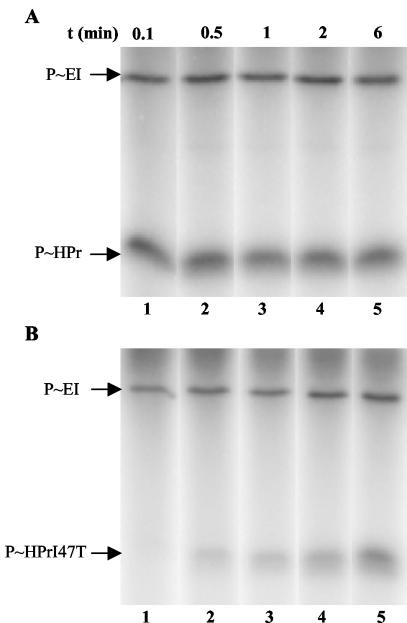
Phosphorylation experiments were carried out with [32P]PEP, EI, and wild-type HPr or HPrI47T. While in the presence of wild-type HPr, an equilibrium of the phosphoryl group transfer reaction was reached after a few seconds (Fig. 8A, lane 1); it took about 6 min with HPrI47T (Fig. 8B, lane 5).
FIG. 8.
[32P]PEP-dependent EI-catalyzed phosphorylation of HPr (A) and HPrI47T (B). Samples were prepared as described in Materials and Methods, incubated for different time periods at 37°C, and separated on 0.1% SDS-15% polyacrylamide gels which were dried and exposed to autoradiography. The migration positions of EI and HPr are indicated by arrows.
We also wanted to study whether the ptsHI47T mutation would affect the phosphoryl group transfer from P∼His-HPr to EIIALev. For this purpose, wild-type HPr and HPrI47T were preincubated with [32P]PEP and EI to allow their nearly complete phosphorylation. EIIALev was subsequently added to the reaction mixtures, and aliquots were withdrawn at the indicated time intervals. The phosphoryl group transfer from wild-type HPr to EIIALev was very fast and reached an equilibrium after a few seconds (Fig. 9A, lane 1). By contrast, under identical experimental conditions, only a small amount of EIIALev was phosphorylated after 2.5 min of incubation in the presence of histidyl-phosphorylated HPrI47T (Fig. 9B). A weak radioactive band migrating according to a molecular mass of 30 kDa was detectable in the experiments with wild-type HPr (Fig. 9A), and an equivalent faint band can also be seen in Fig. 9B (lane 4). This band most likely corresponds to EIIALev dimers, as a band migrating to this position could also be observed after electrophoresis with purified EIIALev and subsequent staining of the gel with Coomassie blue. However, when the samples containing purified EIIALev were boiled before electrophoresis, the slower-migrating band disappeared (data not shown).
FIG. 9.
Phosphoryl group transfer from [32P]P∼His-HPr (A) and [32P]P∼His-HPrI47T (B) to EIIALev. [32P]PEP, EI, and HPr or HPrI47T were preincubated to allow their exhaustive phosphorylation before EIIALev was added to the assay mixtures, which were further incubated at 37°C. Aliquots were withdrawn after different time periods and separated on 0.1% SDS-15% polyacrylamide gels which were dried and exposed to autoradiography. The migration positions of EI, HPr, HPrI47T, EIIALev, and its dimer are indicated by arrows.
We also tested whether the Ile-47-Thr replacement would affect the phosphoryl group transfer from P∼His-HPr to the EIIAMan domain in LevR of L. casei. Experiments identical to those described above for EIIALev were carried out with LevR. Phosphoryl group transfer from wild-type P∼His-HPr to LevR was very fast and was completed after 30 s, while after a 5-min incubation in the presence of P∼His-HPrI47T, only a faint radioactive band corresponding to P∼LevR could be detected (data not shown).
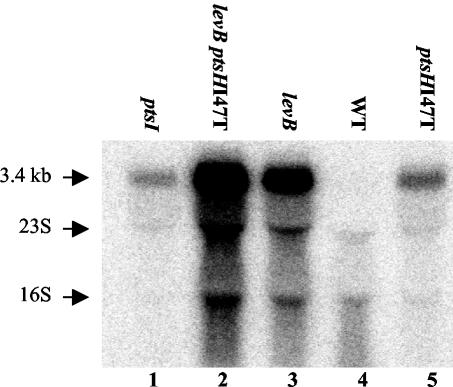
Interruption of the Lev-PTS phosphorylation cascade affects lev operon expression.
The strong inhibitory effect of the Ile-47-Thr replacement in HPr on its ATP- and PEP-dependent phosphorylation and on the phosphoryl group transfer from P∼His-HPr to EIIALev and LevR observed in in vitro experiments suggested that the elevated expression in the ptsHI47T mutant might be partly due to altered LevR phosphorylation. To test this assumption, expression of the lev operon was studied in a ptsI mutant (ptsI encodes EI) and a levB mutant, which are interrupted at the first and last steps, respectively, of the Lev-PTS phosphorylation cascade (6). A ptsI mutant, in which both the activating phosphorylation in the EIIAMan domain and the inactivating phosphorylation in PRD2 of LevR are prevented, did indeed exhibit elevated lev operon expression similar to that of the ptsHI47T strain (Fig. 10, compare lanes 1 and 5). Expression of the lev operon was even stronger in the levB mutant (Fig. 10, lane 3), in which only phosphorylation at the PRD2 domain of LevR is prevented, and was strongest in the levB ptsHI47T double mutant (Fig. 10, lane 2). These results suggest that the major factor leading to elevated lev operon expression in the ptsHI47T mutant might be the diminished phosphorylation at PRD2 of LevR. However, other factors, including altered phosphorylation of the EIIAMan domain of LevR and diminished P-Ser-HPr-mediated CCR, also seem to affect lev operon expression in the ptsHI47T mutant.
FIG. 10.
Northern blots with 5 μg of RNA isolated from either the L. casei wild-type strain BL23 (WT); the ptsHI47T, ptsI, or levB mutant; or the ptsHI47T levB double mutant, which were grown in MRS fermentation medium complemented with ribose. The levA-specific probe was used to detect the lev operon transcript of 3.4 kb. The positions of the 16S and 23S rRNAs are indicated with arrows.
DISCUSSION
Proteome analysis of several CCR-relieved mutants of L. casei allowed us to identify a small protein specifically overproduced in the ptsHI47T strain, which synthesizes an HPr in which Ile-47, a residue located next to the phosphorylatable Ser-46, was replaced with a threonine. Ile-47 (or Leu-47 in E. coli HPr) has previously been suggested to be important for the interaction of HPr with its various partner proteins EI (22), EIIAs (8), CcpA (28), and HprK/P (18). The ptsHI47T mutation was first described for Streptococcus salivarius, where it relieved several enzymes from CCR (23). This mutation did not prevent phosphorylation of HPr at His-15 or Ser-46, although it caused an increase of unphosphorylated HPr in glucose-grown cells at the expense of doubly phosphorylated HPr.
The protein overproduced in the L. casei ptsHI47T strain was identified as EIIA of a mannose class PTS, which was called Lev-PTS based on the similarity of its components to the proteins of the Lev-PTS of B. subtilis and C. acetobutylicum. However, several differences were found between the B. subtilis and the L. casei lev operons and the way their expression is regulated. The most obvious difference was the absence of the levanase-encoding sacC gene and the presence of an additional gene, levX, in L. casei. A gene encoding a similar protein is also present as a fifth cistron in the presumed lev operons of L. casei ATCC 334 and C. acetobutylicum and in operons encoding a mannose class PTS in Lactobacillus johnsonii, Lactobacillus gasseri, S. mutans, Streptococcus gordonii and Leuconostoc mesenteroides. The 12-kDa LevX protein exhibits no significant similarity to any protein of known function. It does not seem to be a transcription regulator, as disruption of the L. casei levX gene had no effect on lev operon expression either in the wild-type strain or in the ptsHI47T background (data not shown). Sequence analysis suggested that LevX contains two transmembrane helices (from amino acids 24 to 46 and 50 to 72).
Although sacC is absent from the lev operon of L. casei BL23, the L. casei neotype strain ATCC 334 possesses a sacC-containing lev operon similar to that of B. subtilis (Fig. 2A). Interestingly, compared to B. subtilis and C. acetobutylicum SacC, the presumed SacC of L. casei ATCC 334 contains a 170-amino-acid N-terminal extension. The lev genes of the two L. casei strains exhibit more than 98% sequence identity. The region of identity stops abruptly at the end of levR on one side and 15 codons before the end of levX on the other side (Fig. 2A). Even the noncoding 283-bp region containing the promoters for levR and the lev operon (Fig. 4) was completely identical. The only significant difference occurred in the intergenic region between levB and levC, where 110 bp was missing in strain ATCC 334 (Fig. 2A). It is tempting to assume that the different location of the lev operon in strains BL23 and ATCC 334 results from a gene rearrangement in BL23, during which the lev operon without sacC was translocated to a new place. The sacC gene of strain BL23 might therefore still be present at the original location of the lev operon. Due to the near identity of the Lev-PTS proteins in the two strains BL23 and ATCC 334, it is likely that they transport the same substrates. Similar to what has been observed for B. subtilis, the L. casei BL23 lev operon was induced by mannose and fructose, suggesting that these two sugars are transported by the Lev-PTS. Even if strain BL23 should not possess a functional sacC gene, its Lev-PTS might transport fructose, possibly produced by degradation of an oligo- or polysaccharide other than levan.
Differences between the B. subtilis and L. casei lev operons also exist in the promoter region. While the B. subtilis lev operon contains a RpoN (σ54)-dependent −12, −24 promoter and a UAS about 100 bp further upstream (36), no such sequences could be detected for the L. casei lev operon. In agreement with this observation, disruption of rpoN, which has been identified within the L. casei BL23 genome sequencing project (29) and is located upstream of a gene encoding another LevR-like protein (M. J. Yebra, R. Viana, and G. Pérez-Martinez, personal communication), had no effect on lev operon expression. This result suggested that, in contrast to LevR of B. subtilis, L. casei LevR does not interact with RpoN but with another sigma factor. RpoN-independent NifA/NtrC-type enhancer binding proteins have been identified before. One of them was TyrR of E. coli, which functions as an activator for two transcription units by interacting with the RNA polymerase-σ70 holoenzyme (44). The NifA/NtrC central domain of enhancer binding proteins has been shown to contain a specific sequence (GAFTGA) which is essential for the interaction with RpoN (48). Interestingly, while this sequence is present in LevR of B. subtilis, it is absent from TyrR of E. coli and LevR of L. casei (Fig. 2B). The domain separating the NifA/NtrC-like central domain and PRD1 is also different in LevR from B. subtilis and L. casei (Fig. 2B). It is not known what the function of this domain is and whether this difference is of any physiological significance. By contrast, all regulatory domains (PRD1, EIIAMan, EIIBGat, and PRD2) are also present in LevR of L. casei, and the P∼His-HPr- and P∼EIIBLev-dependent phosphorylation sites are conserved (Fig. 2B).
HPr is implicated in both LevR phosphorylation reactions, and the L. casei lev operon is preceded by a cre sequence, indicating that it is subject to CCR via the P-Ser-HPr/CcpA complex. We therefore suspected that the observed overexpression of the lev operon in the ptsHI47T mutant might be due to altered PEP- and/or ATP-dependent phosphorylation of HPr. Indeed, ATP-dependent phosphorylation by HprK/P at Ser-46 as well as PEP-dependent phosphorylation by EI at His-15 were significantly slowed for HPrI47T compared to that of wild-type HPr. In addition, transfer of the phosphoryl group from P∼His-HPr to EIIALev was also slower with the mutant protein than with wild-type HPr. It was therefore likely that in the ptsHI47T mutant strain, phosphorylation at PRD2 of LevR was drastically diminished. Since phosphorylation at PRD2 inhibits the activity of LevR, the ptsHI47T strain contains elevated amounts of active LevR, which was assumed to be partly responsible for the strong expression of the lev operon in this mutant. In agreement with this assumption, disruption of levB, which prevents phosphorylation of LevR at PRD2, also led to overexpression of the lev operon. The stronger lev operon expression in the levB mutant compared to that of the ptsHI47T strain might be due partly to the complete absence of phosphorylation of LevR at PRD2 in the levB strain. In addition, due to the slow phosphorylation of HPrI47T by PEP and EI, phosphorylation at the EIIAMan domain which, in contrast to phosphorylation at PRD2, stimulates the activity of LevR, is probably also diminished in the ptsHI47T mutant. In agreement with this assumption, a ptsI mutant in which phosphorylation at both domains EIIAMan and PRD2 is completely prevented exhibited lev operon expression similar to that of the ptsHI47T strain (Fig. 10, compare lanes 1 and 5). According to the in vitro results, the ptsHI47T mutation is also expected to diminish phosphorylation at PRD2 and the EIIAMan domain of LevR. The results with the ptsI mutant also confirm that phosphorylation of LevR by P∼His-HPr at the EIIAMan domain is not indispensable for LevR activity.
The strongest expression of the lev operon was observed in the levB ptsHI47T double mutant. The increase of LevR activity in the levB ptsHI47T strain compared to that of the ptsHI47T mutant is probably due to the complete absence of phosphorylation at PRD2 in the former strain, while the elevated lev operon expression in the levB ptsHI47T strain compared to that of the levB mutant is probably due to a relief from P-Ser-HPr/CcpA-mediated CCR in the double mutant. HPrI47T is only very slowly phosphorylated by ATP and HprK/P at Ser-46. Although P-Ser-HPrI47T is also a poor substrate for the dephosphorylation by HprK/P, accumulation of seryl-phosphorylated mutant HPr probably occurs only slowly and thus leads to diminished P-Ser-HPr/CcpA-mediated CCR. However, it cannot be excluded that the Ile-47-Thr replacement might also lower the affinity of seryl-phosphorylated HPr for CcpA. This effect has been suggested for S. salivarius, where the ptsHI47T mutation altered the amount of P-Ser-HPr only slightly (23). The lev operon of L. casei was expected to be subject to strong CCR, as the location of the presumed cre suggested that not only the expression of the lev operon but also the synthesis of its transcription activator LevR would be repressed by growth on rapidly metabolizable carbohydrates. In conclusion, the concerted effects of the ptsHI47T mutation on ATP- and PEP-dependent phosphorylation of HPr, on the phosphoryl group transfer from P∼His-HPr to EIIALev and LevR, and possibly on the interaction of P-Ser-HPr with CcpA seem to be responsible for the observed overexpression of the lev operon in the ptsHI47T mutant.
Acknowledgments
This research was supported by the Région Basse Normandie, the CNRS, the INRA, and the INA-PG.
We are thankful to G. Perez-Martinez and C. D. Esteban for providing us with strains and plasmids, to W. Hengstenberg for the gift of antibodies against S. carnosus HPr, and to M. Zagorec for valuable discussions and plasmid pRV300.
REFERENCES
- 1.Acedo-Félix, E., and G. Pérez-Martinez. 2003. Significant differences between Lactobacillus casei subsp. casei ATCC 393T and a commonly used plasmid-cured derivative revealed by a polyphasic study. Int. J. Syst. Evol. Microbiol. 53:67-75. [DOI] [PubMed] [Google Scholar]
- 2.Ajdic, D., W. M. McShan, R. E. McLaughlin, G. Savic, J. Chang, M. B. Carson, C. Primeaux, R. Tian, S. Kenton, H. Jia, S. Lin, Y. Qian, S. Li, H. Zhu, F. Najar, H. Lai, J. White, B. A. Roe, and J. J. Ferretti. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. USA 99:14434-14439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Berthier, F., M. Zagorec, M. Champonmier-Vergès, S. D. Ehrlich, and F. Morel-Deville. 1996. Efficient transformation of Lactobacillus sake by electroporation. Microbiology 142:1273-1279. [DOI] [PubMed] [Google Scholar]
- 5.Charrier, V., E. Buckley, D. Parsonage, A. Galinier, E. Darbon, M. Jaquinod, E. Forest, J. Deutscher, and A. Claiborne. 1997. Cloning and sequencing of two enterococcal glpK genes and regulation of the encoded glycerol kinases by phosphoenolpyruvate-dependent, phosphotransferase system-catalyzed phosphorylation of a single histidyl residue. J. Biol. Chem. 272:14166-14174. [DOI] [PubMed] [Google Scholar]
- 6.Charrier, V., J. Deutscher, A. Galinier, and I. Martin-Verstraete. 1997. Protein phosphorylation chain of a Bacillus subtilis fructose-specific phosphotransferase system and its participation in regulation of the expression of the lev operon. Biochemistry 36:1163-1172. [DOI] [PubMed] [Google Scholar]
- 7.Connil, N., Y. Le Breton, X. Dousset, Y. Auffray, A. Rince, and H. Prevost. 2002. Identification of the Enterococcus faecalis tyrosine decarboxylase operon involved in tyramine production. Appl. Environ. Microbiol. 68:3537-3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cornilescu, G., B. R. Lee, C. C. Cornilescu, G. Wang, A. Peterkofsky, and G. M. Clore. 2002. Solution structure of the phosphoryl transfer complex between the cytoplasmic A domain of the mannitol transporter IIMannitol and HPr of the Escherichia coli phosphotransferase system. J. Biol. Chem. 277:42289-42298. [DOI] [PubMed] [Google Scholar]
- 9.Darbon, E., P. Servant, S. Poncet, and J. Deutscher. 2002. Antitermination by GlpP, catabolite repression via CcpA, and inducer exclusion elicited by P∼GlpK dephosphorylation control Bacillus subtilis glpFK expression. Mol. Microbiol. 43:1039-1052. [DOI] [PubMed] [Google Scholar]
- 10.Débarbouillé, M., I. Martin-Verstraete, A. Klier, and G. Rapoport. 1991. The transcriptional regulator LevR of Bacillus subtilis has domains homologous to both σ54- and phosphotransferase system-dependent regulators. Proc. Natl. Acad. Sci. USA 88:2212-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Man, J. C., M. Rogosa, and M. E. Sharp. 1960. A medium for the cultivation of Lactobacilli. J. Appl. Bacteriol. 23:130-135. [Google Scholar]
- 12.Deutscher, J., B. Bauer, and H. Sauerwald. 1993. Regulation of glycerol metabolism in Enterococcus faecalis by phosphoenolpyruvate-dependent phosphorylation of glycerol kinase catalyzed by enzyme I and HPr of the phosphotransferase system. J. Bacteriol. 175:3730-3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deutscher, J., and R. Engelmann. 1984. Purification and characterization of an ATP-dependent protein kinase from Streptococcus faecalis. FEMS Microbiol. Lett. 23:157-162. [Google Scholar]
- 14.Deutscher, J., A. Galinier, and I. Martin-Verstraete. 2002. Carbohydrate uptake and metabolism, p. 129-150. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 15.Dossonnet, V., V. Monedero, M. Zagorec, A. Galinier, G. Pérez-Martínez, and J. Deutscher. 2000. Phosphorylation of HPr by the bifunctional HPr kinase/P-Ser-HPr phosphatase from Lactobacillus casei controls catabolite repression and inducer exclusion but not inducer expulsion. J. Bacteriol. 182:2582-2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dufour, A., A. Rincé, P. Uguen, and J. P. Le Pennec. 2000. IS1675, a novel lactococcal insertion element, forms a transposon-like structure including the lacticin 481 lantibiotic operon. J. Bacteriol. 182:5600-5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 18.Fieulaine, S., S. Morera, S. Poncet, I. Mijakovic, A. Galinier, J. Janin, J. Deutscher, and S. Nessler. 2002. X-ray structure of a bifunctional protein kinase in complex with its protein substrate HPr. Proc. Natl. Acad. Sci. USA 99:13437-13441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujita, Y., Y. Miwa, A. Galinier, and J. Deutscher. 1995. Specific recognition of the Bacillus subtilis gnt cis-acting catabolite-responsive element by a protein complex formed between CcpA and seryl-phosphorylated HPr. Mol. Microbiol. 17:953-960. [DOI] [PubMed] [Google Scholar]
- 20.Galinier, A., J. Haiech, M.-C. Kilhoffer, M. Jaquinod, J. Stülke, J. Deutscher, and I. Martin-Verstraete. 1997. The Bacillus subtilis crh gene encodes a HPr-like protein involved in carbon catabolite repression. Proc. Natl. Acad. Sci. USA 94:8439-8444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galinier, A., M. Kravanja, R. Engelmann, W. Hengstenberg, M.-C. Kilhoffer, J. Deutscher, and J. Haiech. 1998. New protein kinase and protein phosphatase families mediate signal transduction in bacterial catabolite repression. Proc. Natl. Acad. Sci. USA 95:1823-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garrett, D. S., Y. J. Seok, A. Peterkofsky, A. M. Gronenborn, and G. M. Clore. 1999. Solution structure of the 40,000 Mr phosphoryl transfer complex between the N-terminal domain of enzyme I and HPr. Nat. Struct. Biol. 6:166-173. [DOI] [PubMed] [Google Scholar]
- 23.Gauthier, M., D. Brochu, L. D. Eltis, S. Thomas, and C. Vadeboncoeur. 1997. Replacement of isoleucine-47 by threonine in the HPr protein of Streptococcus salivarius abrogates the preferential metabolism of glucose and fructose over lactose and melibiose but does not prevent the phosphorylation of HPr on serine-46. Mol. Microbiol. 25:695-705. [DOI] [PubMed] [Google Scholar]
- 24.Giard, J.-C., A. Rince, H. Capiaux, Y. Auffray, and A. Hartke. 2000. Inactivation of the stress- and starvation-inducible gls24 operon has a pleiotrophic effect on cell morphology, stress sensitivity, and gene expression in Enterococcus faecalis. J. Bacteriol. 182:4512-4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gough, J. A., and N. E. Murray. 1983. Sequence diversity among related genes for recognition of specific targets in DNA molecules. J. Mol. Biol. 166:1-19. [DOI] [PubMed] [Google Scholar]
- 26.Greenberg, D. B., J. Stülke, and M. H. Saier, Jr. 2002. Domain analysis of transcriptional regulators bearing PTS regulatory domains. Res. Microbiol. 153:519-526. [DOI] [PubMed] [Google Scholar]
- 27.Henkin, T. M., F. J. Grundy, W. L. Nicholson, and G. H. Chambliss. 1991. Catabolite repression of α-amylase gene expression in Bacillus subtilis involves a trans-acting gene product homologous to the Escherichia coli lacI and galR repressors. Mol. Microbiol. 5:575-584. [DOI] [PubMed] [Google Scholar]
- 28.Jones, B. E., V. Dossonnet, E. Küster, W. Hillen, J. Deutscher, and R. E. Klevit. 1997. Binding of the catabolite repressor protein CcpA to its DNA target is regulated by phosphorylation of its corepressor HPr. J. Biol. Chem. 272:26530-26535. [DOI] [PubMed] [Google Scholar]
- 29.Klaenhammer, T., E. Altermann, F. Arigoni, A. Bolotin, F. Breidt, J. Broadbent, R. Cano, S. Chaillou, J. Deutscher, M. Gasson, M. van de Guchte, J. Guzzo, A. Hartke, T. Hawkins, P. Hols, R. Hutkins, M. Kleerebezem, J. Kok, O. Kuipers, M. Lubbers, E. Maguin, L. McKay, D. Mills, A. Nauta, R. Overbeek, H. Pel, D. Pridmore, M. Saier, D. van Sinderen, A. Sorokin, J. Steele, D. O'Sullivan, W. de Vos, B. Weimer, M. Zagorec, and R. Siezen. 2002. Discovering lactic acid bacteria by genomics. Antonie Leeuwenhoek 82:29-58. [DOI] [PubMed] [Google Scholar]
- 30.Kravanja, M., R. Engelmann, V. Dossonnet, M. Blüggel, H. E. Meyer, R. Frank, A. Galinier, J. Deutscher, N. Schnell, and W. Hengstenberg. 1999. The hprK gene of Enterococcus faecalis encodes a novel bifunctional enzyme: the HPr kinase/phosphatase. Mol. Microbiol. 31:59-66. [DOI] [PubMed] [Google Scholar]
- 31.Leloup, L., S. D. Ehrlich, M. Zagorec, and F. Morel-Deville. 1997. Single-crossover integration in the Lactobacillus sake chromosome and insertional inactivation of the ptsI and lacL genes. Appl. Environ. Microbiol. 63:2117-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lindner, C., A. Galinier, M. Hecker, and J. Deutscher. 1999. Regulation of the activity of the Bacillus subtilis antiterminator LicT by multiple PEP-dependent, enzyme I- and HPr-catalysed phosphorylation. Mol. Microbiol. 31:995-1006. [DOI] [PubMed] [Google Scholar]
- 33.Mahr, K., C. D. Esteban, W. Hillen, F. Titgemeyer, and G. Perez-Martinez. 2002. Cross communication between components of carbon catabolite repression of Lactobacillus casei and Bacillus megaterium. J. Mol. Microbiol. Biotechnol. 4:489-494. [PubMed] [Google Scholar]
- 34.Martin-Verstraete, I., V. Charrier, J. Stülke, A. Galinier, B. Erni, G. Rapoport, and J. Deutscher. 1998. Antagonistic effects of dual PTS-catalysed phosphorylation on the Bacillus subtilis transcriptional activator LevR. Mol. Microbiol. 28:293-303. [DOI] [PubMed] [Google Scholar]
- 35.Martin-Verstraete, I., M. Débarbouillé, A. Klier, and G. Rapoport. 1990. Levanase operon of Bacillus subtilis includes a fructose-specific phosphotransferase system regulating the expression of the operon. J. Mol. Biol. 214:657-671. [DOI] [PubMed] [Google Scholar]
- 36.Martin-Verstraete, I., M. Débarbouillé, A. Klier, and G. Rapoport. 1992. Mutagenesis of the Bacillus subtilis“−12, −24” promoter of the levanase operon and evidence for the existence of an upstream activating sequence. J. Mol. Biol. 226:85-99. [DOI] [PubMed] [Google Scholar]
- 37.Martin-Verstraete, I., J. Deutscher, and A. Galinier. 1999. Phosphorylation of HPr and Crh by HprK, early steps in the catabolite repression signalling pathway for the Bacillus subtilis levanase operon. J. Bacteriol. 181:2966-2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mijakovic, I., S. Poncet, A. Galinier, V. Monedero, S. Fieulaine, J. Janin, S. Nessler, J. A. Marquez, K. Scheffzek, S. Hasenbein, W. Hengstenberg, and J. Deutscher. 2002. Pyrophosphate-producing protein dephosphorylation by HPr kinase/phosphorylase: a relic of early life? Proc. Natl. Acad. Sci. USA 99:13442-13447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Monedero, V., M. J. Gosalbes, and G. Perez-Martinez. 1997. Catabolite repression in Lactobacillus casei ATCC 393 is mediated by CcpA. J. Bacteriol. 179:6657-6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Monedero, V., S. Poncet, I. Mijakovic, S. Fieulaine, V. Dossonnet, I. Martin-Verstraete, S. Nessler, and J. Deutscher. 2001. Mutations lowering the phosphatase activity of HPr kinase/phosphatase switch off carbon metabolism. EMBO J. 20:3928-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nicholson, W. L., and G. H. Chambliss. 1985. Isolation and characterization of a cis-acting mutation conferring catabolite repression resistance to α-amylase synthesis in Bacillus subtilis. J. Bacteriol. 161:875-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nicholson, W. L., Y.-K. Park, T. M. Henkin, M. Won, M. J. Weickert, J. A. Gaskell, and G. H. Chambliss. 1987. Catabolite repression-resistant mutations of the Bacillus subtilis alpha-amylase promoter affect transcription levels and are in an operator-like sequence. J. Mol. Biol. 198:609-618. [DOI] [PubMed] [Google Scholar]
- 43.Nölling, J., G. Breton, M. V. Omelchenko, K. S. Makarova, Q. Zeng, R. Gibson, H. M. Lee, J. Dubois, D. Qiu, J. Hitti, GTC Sequencing Center Production, Finishing, and Bioinformatics Teams, Y. I. Wolf, R. L. Tatusov, F. Sabathe, L. Doucette-Stamm, P. Soucaille, M. J. Daly, G. N. Bennett, E. V. Koonin, and D. R. Smith. 2001. Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J. Bacteriol. 183:4823-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pittard, A. J., and B. E. Davidson. 1991. TyrR protein of Escherichia coli and its role as repressor and activator. Mol. Microbiol. 5:1585-1592. [DOI] [PubMed] [Google Scholar]
- 45.Postma, P. W., J. W. Lengeler, and G. R. Jacobson. 1993. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol. Rev. 57:543-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roossien, F. F., J. Brink, and G. T. Robillard. 1983. A simple procedure for the synthesis of [32P]phosphoenolpyruvate via the pyruvate kinase exchange reaction at equilibrium. Biochim. Biophys. Acta 760:185-187. [DOI] [PubMed] [Google Scholar]
- 47.Schmalisch, M. H., S. Bachem, and J. Stülke. 2003. Control of the Bacillus subtilis antiterminator protein GlcT by phosphorylation. Elucidation of the phosphorylation chain leading to inactivation of GlcT. J. Biol. Chem. 278:51108-51115. [DOI] [PubMed] [Google Scholar]
- 48.Studholme, D. J., and R. Dixon. 2003. Domain architectures of σ54-dependent transcriptional activators. J. Bacteriol. 185:1757-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stülke, J., M. Arnaud, G. Rapoport, and I. Martin-Verstraete. 1998. PRD—a protein domain involved in PTS-dependent induction and carbon catabolite repression of catabolic operons in bacteria. Mol. Microbiol. 28:865-874. [DOI] [PubMed] [Google Scholar]
- 50.Thompson, J., and D. A. Torchia. 1984. Use of 31P nuclear magnetic resonance spectroscopy and 14C fluorography in studies of glycolysis and regulation of pyruvate kinase in Streptococcus lactis. J. Bacteriol. 158:791-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Viana, R., V. Monedero, V. Dossonnet, C. Vadeboncoeur, G. Pérez-Martínez, and J. Deutscher. 2000. Enzyme I and HPr from Lactobacillus casei: their role in sugar transport, carbon catabolite repression and inducer exclusion. Mol. Microbiol. 36:570-584. [DOI] [PubMed] [Google Scholar]
- 52.Yeh, J. I., V. Charrier, J. Paulo, L. Hou, D. Parsonage, E. Darbon, A. Claiborne, W. G. J. Hol, and J. Deutscher. 2004. Structures of enterococcal glycerol kinase in the absence and presence of glycerol: correlation of conformation to substrate binding and mechanism of activation by phosphorylation. Biochemistry 43:362-373. [DOI] [PubMed] [Google Scholar]