Abstract
The type three secretion system (TTSS) encoded by pNGR234a, the symbiotic plasmid of Rhizobium sp. strain NGR234, is responsible for the flavonoid- and NodD1-dependent secretion of nodulation outer proteins (Nops). Abolition of secretion of all or specific Nops significantly alters the nodulation ability of NGR234 on many of its hosts. In the closely related strain Rhizobium fredii USDA257, inactivation of the TTSS modifies the host range of the mutant so that it includes the improved Glycine max variety McCall. To assess the impact of individual TTSS-secreted proteins on symbioses with legumes, various attempts were made to identify nop genes. Amino-terminal sequencing of peptides purified from gels was used to characterize NopA, NopL, and NopX, but it failed to identify SR3, a TTSS-dependent product of USDA257. By using phage display and antibodies that recognize SR3, the corresponding protein of NGR234 was identified as NopP. NopP, like NopL, is an effector secreted by the TTSS of NGR234, and depending on the legume host, it may have a deleterious or beneficial effect on nodulation or it may have little effect.
A variety of prokaryotic organisms are capable of enzymatically reducing atmospheric nitrogen to ammonia. This process, known as biological nitrogen fixation, can be performed either by free-living bacteria (e.g., Klebsiella pneumoniae) or by symbiotic bacteria. Nitrogen-fixing symbioses between plants belonging to the family Leguminosae and soil bacteria collectively called rhizobia contribute substantially to plant productivity. Ultimately, these associations lead to the formation of specialized structures called nodules on the stems or roots of host plants, where infecting rhizobia differentiate into bacteroids that reduce atmospheric nitrogen to ammonia. The ammonia is incorporated into amino acids that are supplied to the host, which return the favor by supplying the microsymbionts with photosynthates. Nodulation, the process that leads to bacterial colonization of root or stem nodules, is highly selective; a continuous exchange of molecular signals between the two symbionts enables the host to distinguish compatible rhizobia from potential pathogens. Initially, lipochitooligosaccharidic nodulation factors (Nod factors) that are secreted by rhizobia in response to plant flavonoids are essential for infection. Later, additional bacterial signals, such as surface polysaccharides or secreted proteins, are also required for efficient nodulation of specific hosts (7, 35).
Among the known microsymbionts, Rhizobium sp. strain NGR234 has the rare ability to nodulate more than 112 genera of legumes (37). The closely related strain Rhizobium fredii USDA257 forms nodules on a smaller subset of plants (>79 genera), but it fixes nitrogen with Glycine max and Glycine soja, two hosts that fail to establish effective symbioses with NGR234. Early work showed that mutations in the cultivar specificity locus nolXWBTUV modified the host range of USDA257 so that it included the improved soybean variety McCall (14, 28). Flavonoid-dependent secretion of extracellular proteins by USDA257 (24) was later shown to be linked to the same region (21). Sequencing the symbiotic plasmid pNGR234a (12), along with molecular analyses of the corresponding nolXWBTUV locus, confirmed the presence of a complete and symbiotically active type III secretion system (TTSS) in NGR234 (42). In plant and animal pathogens, the TTSS deliver into the host cytoplasm various factors often required for virulence. Genes encoding the TTSS of pNGR234a are organized into eight transcription units (34) that are clustered in 30 kb between the transcriptional regulator ttsI (formerly y4xI) and the predicted gene y4yS (12), which are also preserved in USDA257 (23, 26). A flavonoid-dependent regulatory cascade, which includes NodD1 and TtsI as primary regulators, controls expression of TTSS loci in NGR234. In the presence of compatible flavonoids, NodD1, which binds to specific DNA motifs called nod boxes, triggers the expression of ttsI, albeit at much lower levels than the expression of nodulation genes involved in the synthesis of Nod factors (19). In turn, TtsI is thought to bind to tts boxes and to activate the expression of operons that encode secreted proteins, as well as elements of the type III secretion machinery (20; C. Marie, W. J. Deakin, T. Ojanen-Reuhs, E. Diallo, B. Reuhs, W. J. Broughton, and X. Perret, submitted for publication). Primed by flavonoids, this regulation system allows sequential activation of nodulation and other loci. Genes involved in Nod factor synthesis and secretion are activated within a few minutes of the initial flavonoid induction, whereas transcription of TTSS-related functions occurs several hours later (19, 33, 42).
Of the nine nodulation outer proteins (Nops) reported to be secreted in a flavonoid- and TTSS-dependent way (26, 27), only three have been assigned to specific open reading frames (ORFs). NopX (NolX in the old nomenclature) and NopL (encoded by y4xL) were identified by N-terminal amino acid sequencing of proteins isolated from sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) gels (42). NopA (previously NolJ), which was first identified in USDA257, was later shown to be fully conserved in NGR234 (27). All three of these secreted proteins are encoded by distinct, tts box-controlled transcription units in the TTSS clusters (26a, 34). Mutations in the essential rhcN gene (NGRΩrhcN), as well as in ttsI (NGRΩttsI), completely abolished the TTSS-dependent secretion of proteins (42) and considerably modified the host range of the mutant strain (27, 42). Despite a DNA sequence that is 98% identical to that of NGR234 (23, 26), the TTSS cluster of USDA257 (GenBank accession no. AF229441) secretes fewer signal response proteins (SR1 to SR5). SR3 and SR5 were used to raise polyclonal antibodies (21). Although it is now known that SR5 corresponds to NopA (1, 27), SR3 has not been characterized.
To understand the role of TTSS in symbiosis, the function of each Nop has to be defined. NopA is thought to be a key component of the extracellular part of the secretion machinery, whereas NopX is proposed to be part of the translocon that directs effector proteins across the plant plasma membrane (27). By using in vitro assays, as well as transgenic Nicotiana tabacum and Lotus japonicus plants, NopL was shown to be a high-affinity substrate for phosphorylation by plant protein kinases and to impair the signal transduction pathway that leads to plant defense responses (4, 5). Here we describe the identification of a second putative effector secreted by the TTSS of NGR234. By using a phage display technique that was previously established for the isolation of RapA, a cell surface-associated agglutinin of Rhizobium leguminosarum bv. trifolii (2), SR3 was identified as NopP. SDS-PAGE analyses confirmed that secretion of NopP is flavonoid and TTSS dependent. A nopP deletion mutant was also constructed, and its symbiotic properties were compared to those of strains NGR234 and NGRΩrhcN.
MATERIALS AND METHODS
Molecular and microbiological techniques.
Escherichia coli recombinants were grown in or on Luria-Bertani medium or Terrific broth (40). Strains of Rhizobium were grown in or on tryptone-yeast extract medium (6) or Rhizobium minimal medium (8) containing rifampin and kanamycin (50 μg/ml) for selection of Rhizobium sp. strain NGR234 and R. fredii USDA257, respectively. Genomic DNA of NGR234 was prepared as described by Perret and Broughton (30). Plasmid DNA preparation, sequencing of inserts in selected phagemids, and DNA sequence analyses were performed as described previously (2).
Construction of a phage display library of NGR234.
A library was constructed in the pG8SAET phagemid vector essentially as described by Jacobsson and Frykberg (15). Briefly, genomic DNA of NGR234 was sonicated into 0.1- to 5-kb fragments, and the blunt-ended DNA fragments were ligated into the pG8SAET vector that had been cleaved with SnaBI and treated with calf intestine phosphatase by using a Ready-To-Go ligation kit (Amersham Biosciences, Uppsala, Sweden). The ligation mixture was introduced by electroporation into TG1 cells, yielding 108 individual ampicillin-resistant clones. All 24 randomly picked clones contained inserts that were between 0.1 and 2 kb long. Amplification and preparation of the phage library were performed as described previously and resulted in a titer of 2 × 1010 CFU of phagemid particles ml−1.
Selection of SR3-binding phage from the library.
To remove immunoglobulin G antibodies from the polyclonal SR3 antiserum (21) (and thereby reduce the complexity of the binding substrate), each well of a 96-well plate was first coated with 100 μg of recombinant streptococcal protein G (a kind gift from B. Guss) per ml. After washing with Tris-buffered saline (TBS) containing 0.05% (vol/vol) Tween 20 (TBS-T) and blocking with 1% (wt/vol) bovine serum albumin in TBS-T, SR3 antiserum was diluted 1:10 with TBS, added to the wells, and incubated overnight at 4°C. After three subsequent washes, the NGR234 phagemid library was added to the SR3-coated wells, incubated for 4 to 6 h at room temperature, and washed with TBS-T (10 times for the first pannings and 30 times for the repannings). Bound phage particles were eluted in 50 mM citrate buffer (pH 2) and used to infect E. coli TG1 cells. A TG1 culture was also added to the wells after removal of the elution buffer to recover very strongly bound phage particles. In some experiments, a depletion step was used prior to the first panning to reduce the amount of phage expressing epitopes recognized by non-SR3-specific antibodies present in the serum. Then 100 μl of the library was passed along a row of eight wells coated with rabbit preimmune serum. The library was left for 10 min in each well and then was added to the SR3-coated well and panned as described above.
Construction of the NGRΔnopP mutant.
The 3,051-bp SalI restriction fragment of cosmid pXB740 (31) that carries y4yP/nopP (positions 518077 to 518892 of pNGR234a) (12) was cloned into the XhoI site of pJQ200SK (38), yielding pJQyP. Construct pYPSpr was obtained by replacing the adjacent 500- and 628-bp XhoI fragments internal to y4yP/nopP with the spectinomycin-resistant (Spr) interposon (36). The resulting construct was then mobilized into NGR234 by triparental mating using helper plasmid pRK2013 (11). Marker exchange was forced by selection on Rhizobium minimal medium plates containing 5% (wt/vol) sucrose, and putative NGRΔnopP mutants were confirmed by probing Southern blots of restricted genomic DNA by standard methods (40).
Purification and analysis of extracellular proteins.
Extracellular proteins of NGR234, NGRΩrhcN, NGRΔnopP, and USDA257 were prepared as follows. After 40 h of induction with 10−6 M apigenin or genistein, secreted proteins were isolated as described by Viprey et al. and Marie et al. (27, 42). Aliquots of proteins were separated on SDS-12 or 15% (wt/vol) polyacrylamide gels and stained with silver (3). Separated proteins were transferred to Immobilon-P polyvinylidene difluoride membranes (Millipore Corp., Bedford, Mass.) and probed with dilutions of the SR3 (21), SRT (polyclonal antibodies raised against all the extracellular proteins secreted by flavonoid-induced cells of USDA257 [27]), and NopP antibodies. Anti-NopP antiserum was generated by immunizing rabbits with two peptides (Eurogentec, Herstal, Belgium), N-MVLDPKQHPDKWTQT and N-QCSTSCSYETYEDDFM. Peptides were coupled to carrier proteins prior to immunization according to the manufacturer's protocols. The antiserum was diluted 1:2,000 with phosphate-buffered saline containing Tween 20 for immunoblotting. Horseradish peroxidase-labeled goat anti-rabbit immunoglobulin antibodies from an ECL kit (Amersham Biosciences) were used as secondary antibodies. Reaction results were visualized by enhanced chemiluminescence (Amersham Biosciences).
Plant tests.
Nodulation tests were performed in Magenta jars (25). All plants were grown at a day temperature of 26°C and a night temperature of 20°C and with a 16-h light phase. Each plant (Flemingia congesta, Pachyrhizus tuberosus, and Vigna unguiculata cv. Red Caloona [37]) was inoculated with 108 bacteria and harvested 6 to 8 weeks after inoculation.
RESULTS
Both NGR234 and USDA257 secrete SR3.
To verify that antibodies raised against extracellular proteins of USDA257 cross-react with proteins secreted by flavonoid-induced cultures of NGR234, the two strains were grown in identical media and induced for 40 h with either apigenin or genistein (final concentration, 10−6 M). After isolation, proteins found in supernatants of induced cultures were separated on SDS-PAGE gels (Fig. 1A). Although the protein secretion profiles of NGR234 and USDA257 were clearly distinct, induction with either genistein or apigenin had no apparent effect on the type and/or amount of proteins secreted by each strain. Immunostaining of the same protein preparations with antibodies raised against SR3 of USDA257 (24) confirmed that several proteins of NGR234 cross-reacted (Fig. 1B). Immunoprobing of proteins secreted by mutant strains NGRΔnopX, NGRΩnopL, and NGRΔnopXΩnopL (27, 41) induced with apigenin confirmed that two of the three proteins of NGR234 that cross-reacted with SR3 antibodies were NopX and NopL (Fig. 1B). The remaining protein, which was found in supernatants of both USDA257 and NGR234, corresponded to SR3. Previous attempts to characterize the SR3 protein by N-terminal peptide sequencing failed to produce conclusive results, however (24). As an alternative method, a genomic phage display library of NGR234 was constructed and screened by using SR3 antibodies that showed sufficiently strong and specific antigen-antibody binding (21).
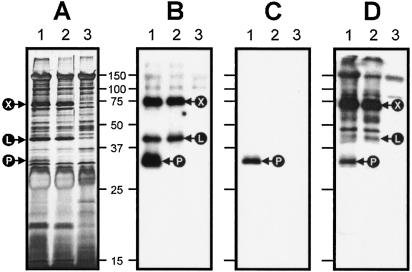
FIG. 1.
Comparison of flavonoid-induced extracellular proteins secreted by Rhizobium sp. strain NGR234, R. fredii USDA257, and various mutant strains. (A) Silver-stained SDS—15% PAGE gel of secreted proteins of NGR234 (lanes 1 and 2) and USDA257 (lanes 3 and 4) collected after 40 h of induction with apigenin (lanes 1 and 3) or genistein (lanes 2 and 4). (B) Western blot of flavonoid-induced secreted proteins of NGR234 (lanes 1 and 2), USDA257 (lanes 3 and 4), NGRΔnopX (lane 5), NGRΩnopL (lane 6), and the double mutant NGRΔnopXΩnopL (lane 7) separated on an SDS—12% PAGE gel and immunostained with SR3 antibodies. Cells were induced with either apigenin (lanes 1, 3, 5, 6, and 7) or genistein (lanes 2 and 4). The positions of NopX, NopL, and SR3 are indicated by arrows. Estimated molecular masses (in kilodaltons) are indicated on the left.
Enrichment of the phage display library with SR3 antibodies.
In the phage display library, peptides encoded by random DNA fragments of the NGR234 genome were displayed on the surface of filamentous phage particles as fusions to the major phage coat protein VIII. Successive rounds of panning enriched the library of phagemid particles expressing epitopes recognized by SR3 antibodies. The enrichment factor for clones encoding the specific binding peptide was as high as 90% in previous experiments when the display vector pG8SAET was used (16). Initially, only a modest increase in the number of bound phage particles (∼10-fold) and a low ligand specificity ratio (typically 2:1 for SR3 antiserum and preimmune serum) were obtained after two consecutive rounds of panning. To estimate the overall efficiency of SR3 panning, candidate clones were screened by colony hybridization using radioactively labeled nopX as a control probe. Although not intended as the primary target for the phage display, NopX cross-hybridized with SR3 antibodies (Fig. 1B), indicating that it should be copurified together with the unknown SR3-encoding sequences. Colony hybridizations confirmed that there was a modest 1 to 2% enrichment in nopX, a value which was too low to allow identification of the unknown SR3 component by random sequencing of the repanned clones.
To decrease the non-SR3-specific enrichment of phage due to the complexity of the antiserum, the phage library was first passed over a row of preimmune serum-coated wells prior to panning in the SR3-coated wells. This step depleted the library of phagemid particles that bound to non-SR3-specific antibodies present in the SR3 antiserum and increased the ligand specificity ratio to 20:1 in the repannings. Inserts of 40 randomly picked clones were sequenced from one panning experiment in which good ligand specificity was obtained (8 × 105 CFU ml−1 in the SR3-coated wells versus 3.4 × 104 CFU ml−1 in the preimmune serum-coated wells). Generally, multiple copies of DNA sequences of genes targeted by phage display are found by panning, and the best candidates are overlapping, nonidentical clones. Four groups of overlapping DNA sequences were identified (Table 1). Two groups corresponded to inserts coding for known genes cloned in the wrong orientation (SR3c21and SR3c22; SR3c1 and SR3cb5), indicating that these results were most probably false positives (Table 1). The ORFs found on the opposite strands of the known genes were properly fused to the phage coat protein and most probably encoded mimotopes, peptides that mimicked the structure of epitopes recognized by a fraction of antibodies in the complex mixture of the antiserum. The remaining two sets of sequences were correctly fused to phage coat protein VIII and coded for peptides that are homologous to FlaB of Agrobacterium tumefaciens (SR3c14 and SR3c11), as well as y4yP of pNGR234a (SR3c13 and SR3ce6). Like nopL and nopX, which are known to encode secreted proteins, y4yP is part of the TTSS clusters of NGR234 and USDA257 (12, 23). It also codes for a predicted product whose size is similar to that of SR3 (ca. 32 kDa), again suggesting that y4yP may be the gene which encodes the protein that cross-reacts with the SR3 antibody.
TABLE 1.
Homologies of overlapping clones determined by phage display with SR3 antibodiesa
| Overlapping clonesb | Best BlastX match
|
Best BlastN match
|
Orientationc | ||
|---|---|---|---|---|---|
| Sequence | % Similarity | Sequence | % Identity | ||
| SR3c13 (81) and SR3ce6 (73) | y4yP (NGR234) | 100 | pNGR234a | 100 | Correct |
| SR3c14 (202) and SR3c11 (52) | FlaB (A. tumefaciens) | 57 | None | Correct | |
| SR3c21 (132) and SR3c22 (118) | Smb21498 (Rhizobium meliloti) | 91 | Rhizobium meliloti pSymB | 90 | Inverted |
| SR3c1 (88) and SR3cb5 (56) | Smc04127 (Rhizobium meliloti) | 100 | Rhizobium meliloti chromosome | 96 | Inverted |
Blast searches were performed by using GenBank nonredundant databases.
The numbers in parentheses are sizes of the inserts (in base pairs).
Compares the orientation of the peptide encoded by the phage insert with that of the best match identified in BlastX searches.
y4yP cross-reacts with SR3 antibodies, and its secretion is TTSS dependent.
To confirm that y4yP encodes an extracellular protein, the y4yP deletion mutant designated NGRΔnopP was constructed by using an omega interposon (36). After 40 h of induction with 10−6 M apigenin, proteins found in the supernatants of cultures of NGR234, NGRΩrhcN, and NGRΔnopP were isolated and separated by SDS-PAGE (Fig. 2A). Comparison of the distinct profiles of the extracellular proteins showed clearly that a single ca. 32-kDa product found in induced cultures of NGR234 was missing in induced cultures of NGRΔnopP. This confirmed that (i) a functional y4yP is required for the flavonoid-dependent secretion of a 32-kDa product and (ii) a polar mutation in y4yP does not prevent secretion via the TTSS of other proteins, such as NopL and NopX (Fig. 2A and B). In contrast, the polar mutation in rhcN that abolishes secretion of protein via the TTSS (42) also prevents accumulation of the 32-kDa protein in the supernatants of induced cells. Western blots showed that the ∼32-kDa protein that neither NGRΔnopP nor NGRΩrhcN secreted strongly cross-reacted with both SR3 antibodies and anti-NopP antiserum (which was raised against two synthetic peptides of the predicted NopP protein [Fig. 2B and C]). These data suggest that NopP, like NopL, is not a part of the secretion machinery and confirm that y4yP encodes NopP, a 32-kDa protein that corresponds to SR3. Furthermore, probing the extracts with the SRT antiserum (Fig. 2D) showed that a cross-reacting band at ∼32 kDa was absent from the supernatants of NGRΔnopP (but not wild-type) cultures. This ∼32-kDa band was identified previously by using the SRT antiserum (27) and was designated Nop34. Nop34 is secreted in a flavonoid- and TTSS-dependent manner and is most probably NopP.
FIG. 2.
NopP cross-reacts with SR3 antibodies, and its secretion requires a functional TTSS. (A) Silver staining of an SDS—12% PAGE gel containing extracellular proteins of apigenin-induced strains NGR234 (lane 1), NGRΔnopP (lane 2), and NGRΩrhcN (lane 3). (B, C, and D) Western blots of identical gels immunostained with SR3, NopP, and SRT antibodies, respectively. The positions of NopP (P), NopL (L), and NopX (X) are indicated by arrows. Estimated molecular masses (in kilodaltons) are indicated.
Symbiotic phenotype of NGRΔnopP.
NGR234 mutants in which either the TTSS or certain secreted proteins are deficient often have altered symbiotic properties that differ depending on the host plant tested (27, 42). To evaluate the effect of NopP on symbiosis, the nodulation abilities of strains NGR234, NGRΩrhcN, and NGRΔnopP with various hosts were compared (Table 2). In contrast to wild-type NGR234, which formed few but often large nodules (up to four nodules per plant) on P. tuberosus, the NGRΩrhcN mutant efficiently nodulated this plant. These data suggest that the TTSS and its secreted proteins have a globally detrimental effect on the nodulation of P. tuberosus (27, 42). The completely Nod− phenotype obtained with NGRΔnopP indicates that NopP is required for formation of the small number of nodules by the wild-type strain, however. On F. congesta, the situation appeared to be reversed; the NGRΩrhcN strain formed fourfold-fewer nodules than NGR234, but inactivation of nopP increased nodule number by more than 15% compared to the number obtained with the parent strain. Thus, in contrast to the positively acting TTSS-dependent effector NopL (27), NopP appeared to negatively affect nodulation of F. congesta. On V. unguiculata, a plant that hardly responded to the TTSS of NGR234, the NGRΔnopP mutant induced 36% more nodules than the parent strain induced (Table 2). In contrast, the shoot dry weight of plants inoculated with the NopP− mutant was reduced (data not shown), suggesting that NopP contributes to the optimal symbiosis of NGR234 with V. unguiculata.
TABLE 2.
Numbers of nodules induced by mutants with mutations in rhcN and nopP on three hosts of NGR234a
| Host | No. of nodulesb
|
||
|---|---|---|---|
| NGR234 | NGRΩrhcN | NGRΔnopP | |
| Flemingia congesta | 30.3 ± 3.6 (18) | 7.8 ± 1.1 (19) | 35 ± 3.1 (20) |
| Pachyrhizus tuberosus | 0.9 ± 0.2 (20) | 31 ± 2.2 (22) | 0 (22) |
| Vigna unguiculata | 36.6 ± 2.5 (16) | 39.8 ± 3 (16) | 49.9 ± 4.1 (15) |
Plant tests were performed in Magenta jars. Nodules of all plants fixed nitrogen (Fix+). P. tuberosus was harvested 56 days postinoculation, whereas F. congesta and V. unguiculata were grown for only 42 days.
Mean ± standard error of the mean. The numbers in parentheses are numbers of plants.
Homologues of NopP in other rhizobia.
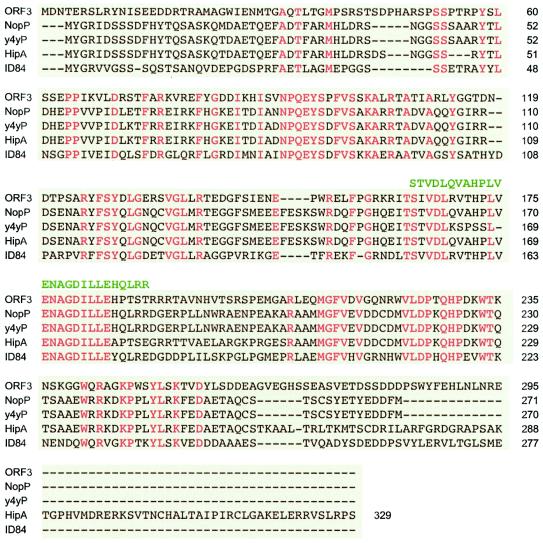
BLAST searches against protein databases showed that NopP has few homologues; all are found in rhizobia, and with the exception of Rhizobium etli strain CNPAF512 (previously called R. leguminosarum bv. phaseoli), the strains that carry nopP also possess a TTSS (Table 3). USDA110 (13) contains four putative proteins (ID84, ID185, ID186, and ID322) that exhibit significant homology with either parts of NopP or the entire NopP protein (Table 3). Only the sequence of ID84 (277 amino acids) matches the complete sequence of y4yP, however. ID186 is homologous to the N-terminal half of NopP, and ID185 and ID322 are homologous to the C-terminal half of NopP (18). When conserved and less conserved amino acids were included, the levels of sequence similarity with NopP ranged from 63% for ORF3 of CNPAF512 to 99% for y4yP of USDA257 (Table 3). ClustalV alignment of the complete sequences of ORF3 of CNPAF512 (29), y4yP of R. fredii USDA257 (GenBank accession no. AF229441), host-inducible protein A (HipA) of R. fredii USDA201 (39), USDA110 predicted product ID84 (13), and NopP of NGR234 showed that these proteins share more than 80 amino acids, mostly grouped in three conserved domains (Fig. 3). Interestingly, the peptide sequence encoded by clone SR3c13 isolated from the phage display library matches the most central domain. As NopP of NGR234 and y4yP of USDA257 differ significantly in the central part of this region (Fig. 3), the epitope recognized by SR3 antibodies probably corresponds to the Glu-Asn-Ala-Gly-Asp-Ile-Leu-Leu-Glu part of the consensus sequence found in most NopP homologues. No other significant matches were identified when any of the three motifs conserved in NopP homologues was used as a pattern to scan the Swiss-Prot and TrEMBL databases.
TABLE 3.
Homologues of NopP in protein databasesa
| Strain | Protein | No. of predicted residues | % Similarity to NopP | % Identity to NopP | Positionb |
|---|---|---|---|---|---|
| R. fredii USDA257 | y4yP | 270 | 99 | 97 | 1-271 |
| R. fredii USDA201 | HipAc | 329 | 93 | 86 | 1-271 |
| R. etli CNPAF512 | ORF3 | 295 | 63 | 31 | 1-271 |
| B. japonicum 110 | ID84 | 277 | 77 | 47 | 1-271 |
| ID322 | 127 | 84 | 54 | 138-271 | |
| ID186 | 140 | 65 | 31 | 1-129 | |
| ID185 | 140 | 79 | 50 | 137-271 |
FIG. 3.
ClustalV alignment of NopP homologues. NopP of NGR234 (NopP), ORF3 of R. etli strain CNPAF512 (ORF3), y4yP of R. fredii USDA257 (y4yP), HipA of R. fredii USDA201 (HipA), and ID84 of Bradyrhizobium japonicum USDA110 (ID84) were aligned by using ClustalV. Note that the predicted N terminus of HipA was extended to meet the same start codon as that in NopP. Amino acids common to all NopP homologues are indicated by red type, and the peptide encoded by the SR3c13 clone is indicated by green type. Putative proteins, such as ID185, ID186, and ID322, that correspond to either the N-terminal half of NopP (ID186) or the C-terminal half of NopP (ID185 and ID322) were not included in the alignment.
DISCUSSION
By using phage display, SR3 (also known as Nop34) of NGR234 was identified as NopP. This 31.2-kDa protein is encoded by y4yP, an ORF that is present in the TTSS clusters of both NGR234 and USDA257 (12, 23). SR3 antibodies cross-react strongly with NopP, but they also bind NopX and NopL that are found in the supernatants of flavonoid-induced strains of NGR234 (Fig. 1B and 2B). This is in agreement with our observation that low enrichment of phage with nopX-containing inserts was achieved in pannings against SR3 antisera. Clones containing nopL-specific inserts were not recovered, however. Perhaps this was due to the low levels of NopL-specific antibodies in the SR3 antiserum, as revealed by the considerably lower intensity of the NopL-specific signal than of the NopX and NopP signals in Western blots (Fig. 2B). Apparently, these three TTSS-secreted proteins do not share obvious features, suggesting that SR3 antibodies recognize a variety of different epitopes. This is probably the case, since SR3-positive clones from the phage display library of NGR234 often carry inserts that do not overlap, although many inserts code for peptides with common motifs (data not shown). Five of the selected clones (SR3c6, SR3c11, SR3c14, SR3c17, and SR3cb11) encode peptides with significant similarity to parts of the N termini of A. tumefaciens flagellins FlaA, FlaB, and FlaC (data not shown). With molecular masses that range from 31.7 kDa for FlaA to 33 kDa for FlaB of A. tumefaciens (9), it is likely that flagellins of USDA257 were copurified with NopP during the initial protein extraction from SDS-PAGE gels (21). The finding that USDA257 is motile on YEM or RMS swarm plates supports this contention (W. J. Deakin, unpublished data).
Thus, SR3 antibodies were probably raised against a mixture of proteins, which explains the variety of epitopes selected. In contrast, the antibodies raised against the two peptides selected from the carboxy terminus of NopP (amino acids 217 to 231 and 256 to 271) are very specific (Fig. 2C). The efficiency with which phage display helps identify a candidate gene(s) depends on the quality and specificity of the antibodies used during the procedure. In this respect, antibodies that recognize many different epitopes yield large numbers of apparently unrelated clones, and complex analyses are required to identify the best target(s) among many candidates. This was observed when the NGR234 phage display library was screened with polyclonal SRT antibodies raised against all the extracellular proteins secreted by flavonoid-induced cells of USDA257. In this case, no noticeable enrichment of target clones was obtained (data not shown), even though SRT cross-reacted with NopP, NopX, and other secreted proteins (Fig. 2D). For these reasons, it seems unlikely that further screening of NGR234 phage display libraries with SRT will lead to identification of other Nops.
NGR234 and USDA257 secrete a number of proteins in a TTSS-dependent manner. In both systems, the secretion occurs after flavonoid induction, and apigenin (a strong inducer of NGR234 nod genes [19]) or genistein (used predominantly with USDA257) appears to activate TTSS-related genes with the same efficiency (Fig. 1A). This is not surprising, since genistein and apigenin have been reported to be potent activators of expression of nodulation genes in NGR234 and R. fredii (10, 22). Although SDS-PAGE techniques are unable to pinpoint subtle differences in protein composition, the profiles of proteins secreted when a strain is induced with apigenin and when a strain is induced with genistein are similar. Nevertheless, the two strains secrete markedly different mixtures of proteins (Fig. 1A), suggesting that despite the many genes of pNGR234a that are also found in R. fredii (32, 33), several loci that are active in only one of the two strains contribute to the extracellular protein signature. Nevertheless, NopA (27), NopL (42), NopX (42), and NopP seem to be secreted in a flavonoid- and TTSS-dependent manner in USDA257 (Fig. 1).
Interestingly, the TTSS cluster of Mesorhizobium loti MAFF303099, which exhibits 75% sequence identity with the TTSS cluster of pNGR234a (17, 26), includes copies of nopX and nopA but lacks homologues of nopL and nopP. Inactivation of NopL and NopP, which are encoded by monocistronic units controlled by tts boxes (26a, 34), does not affect the TTSS-dependent secretion of other Nops. The symbiotic effects of mutants disrupted in NopL or NopP are less pronounced than the effects observed in the nopX mutant and are restricted to fewer plants. Depending on the host plant, secreted proteins have one of the following effects: (i) beneficial (e.g., NopP with P. tuberosus), (ii) detrimental (NopP in F. congesta), or (iii) no effect on nodulation (NopL on P. tuberosus). Interestingly, on certain hosts (e.g., F. congesta) NopP and NopL seem to have antagonistic effects; NGRΔnopP forms more nitrogen-fixing nodules than the wild type forms, whereas the nodulation efficiency of NGRΩnopL is significantly impaired (27). Subtle phenotypes like these are commonly observed when the genes encoding effectors of pathogenic bacteria are mutated and imply that only cocktails of various effectors have drastic effects on specific hosts (26). We are currently examining the roles of the individual effectors in these mixtures and are creating multiple mutations to investigate possible synergistic interactions.
Acknowledgments
We thank Y.-Y. Aung and D. Gerber for their help with many aspects of this work, Lars Frykberg for valuable suggestions during the phage display experiments, and B. Guss for the kind gift of streptococcal protein G.
Financial support for this project was provided by the Fonds National Suisse de la Recherche Scientifique (grants 31-45921.95 and 31-67977.02), the Swedish University of Agricultural Sciences, and the University of Geneva.
REFERENCES
- 1.Annapurna, K., and H. B. Krishnan. 2003. Molecular aspects of soybean cultivar-specific nodulation by Sinorhizobium fredii USDA257. Indian J. Exp. Biol. 41:1114-1123. [PubMed] [Google Scholar]
- 2.Ausmees, N., K. Jacobsson, and M. Lindberg. 2001. A unipolarly located, cell-surface-associated agglutinin, RapA, belongs to a family of Rhizobium-adhering proteins (Rap) in Rhizobium leguminosarum bv. trifolii. Microbiology 147:549-559. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, and J. A. Smith. 1991. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 4.Bartsev, A. V., N. M. Boukli, W. J. Deakin, C. Staehelin, and W. J. Broughton. 2003. Purification and phosphorylation of the effector protein NopL from Rhizobium sp. NGR234. FEBS Lett. 554:271-274. [DOI] [PubMed] [Google Scholar]
- 5.Bartsev, A. V., W. J. Deakin, N. M. Boukli, C. B. McAlvin, G. Stacey, P. Malnoë, W. J. Broughton, and C. Staehelin. 2004. NopL, an effector protein of Rhizobium sp. NGR234, thwarts activation of plant defense reactions. Plant Physiol. 134:871-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beringer, J. E. 1974. R factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 84:188-198. [DOI] [PubMed] [Google Scholar]
- 7.Broughton, W. J., S. Jabbouri, and X. Perret. 2000. Keys to symbiotic harmony. J. Bacteriol. 182:5641-5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Broughton, W. J., C.-H. Wong, A. Lewin, U. Samrey, H. Myint, H. Meyer Z. A., D. N. Dowling, and R. Simon. 1986. Identification of Rhizobium plasmid sequences involved in recognition of Psophocarpus, Vigna, and other legumes. J. Cell Biol. 102:1173-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deakin, W. J., V. E. Parker, E. L. Wright, K. J. Ashcroft, G. J. Loake, and C. H. Shaw. 1999. Agrobacterium tumefaciens possesses a fourth flagellin gene located in a large gene cluster concerned with flagellar structure, assembly and motility. Microbiology 145:1397-1407. [DOI] [PubMed] [Google Scholar]
- 10.Fellay, R., P. Rochepeau, B. Relić, and W. J. Broughton. 1995. Signals to and emanating from Rhizobium largely control symbiotic specificity, p. 199-220. In U. S. Singh, R. P. Singh, and K. Kohmoto (ed.), Pathogenesis & host specificity in plant diseases: histopathological, biochemical, genetic and molecular bases, vol. 1. Prokaryotes. Pergamon/Elsevier Science Ltd, Oxford, United Kingdom.
- 11.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freiberg, C., R. Fellay, A. Bairoch, W. J. Broughton, A. Rosenthal, and X. Perret. 1997. Molecular basis of symbiosis between Rhizobium and legumes. Nature 387:394-401. [DOI] [PubMed] [Google Scholar]
- 13.Göttfert, M., S. Röthlisberger, C. Kündig, C. Beck, R. Marty, and H. Hennecke. 2001. Potential symbiosis-specific genes uncovered by sequencing a 410-kilobase DNA region of the Bradyrhizobium japonicum chromosome. J. Bacteriol. 183:1405-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heron, D. S., T. Ersek, H. B. Krishnan, and S. Pueppke. 1989. Nodulation mutants of Rhizobium fredii USDA257. Mol. Plant-Microbe Interact. 2:4-10. [Google Scholar]
- 15.Jacobsson, K., and L. Frykberg. 1995. Cloning of ligand-binding domains of bacterial receptors by phage display. Bio/Technolgy 18:878-885. [PubMed] [Google Scholar]
- 16.Jacobsson, K., and L. Frykberg. 1999. Gene VIII-based, phage display vectors for selection against complex mixture of ligands, p. 225-238. In M. McClelland and A. B. Pardee (ed.), Expression genetics: high throughput methods. Eaton Publishing, Natick, Mass. [DOI] [PubMed]
- 17.Kaneko, T., Y. Nakamura, S. Sato, E. Asamizu, T. Kato, S. Sasamoto, A. Watanabe, K. Idesawa, A. Ishikawa, K. Kawashima, T. Kimura, Y. Kishida, C. Kiyokawa, M. Kohara, M. Matsumoto, A. Matsuno, Y. Mochizuki, S. Nakayama, N. Nakazaki, S. Shimpo, M. Sugimoto, C. Takeuchi, M. Yamada, and S. Tabata. 2000. Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res. 7:331-338. [DOI] [PubMed] [Google Scholar]
- 18.Kaneko, T., Y. Nakamura, S. Sato, K. Minamisawa, T. Uchiumi, S. Sasamoto, A. Watanabe, K. Idesawa, M. Iriguchi, K. Kawashima, M. Kohara, M. Matsumoto, S. Shimpo, H. Tsuruoka, T. Wada, M. Yamada, and S. Tabata. 2002. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res. 9:189-197. [DOI] [PubMed] [Google Scholar]
- 19.Kobayashi, H., Y. Naciri-Graven, W. J. Broughton, and X. Perret. 2004. Flavonoids induce temporal shifts in gene-expression of nod-box controlled loci in Rhizobium sp. NGR234. Mol. Microbiol. 51:335-347. [DOI] [PubMed] [Google Scholar]
- 20.Krause, A., A. Doerfel, and M. Göttfert. 2002. Mutational and transcriptional analysis of the type III secretion system of Bradyrhizobium japonicum. Mol. Plant-Microbe Interact. 15:1228-1235. [DOI] [PubMed] [Google Scholar]
- 21.Krishnan, H. B., C.-I. Kuo, and S. G. Pueppke. 1995. Elaboration of flavonoid-induced proteins by the nitrogen-fixing symbiont Rhizobium fredii is regulated by both nodD1 and nodD2, and is dependent on the cultivar specificity locus, nolXWBTUV. Microbiology 141:2245-2251. [Google Scholar]
- 22.Krishnan, H. B., A. Lewin, R. Fellay, W. J. Broughton, and S. G. Pueppke. 1992. Differential expression of nodS accounts for the varied abilities of Rhizobium fredii USDA257 and Rhizobium sp. strain NGR234 to nodulate Leucaena spp. Mol. Microbiol. 6:3321-3330. [DOI] [PubMed] [Google Scholar]
- 23.Krishnan, H. B., J. Lorio, W. S. Kim, G. Jiang, K. Y. Kim, M. DeBoer, and S. G. Pueppke. 2003. Extracellular proteins involved in soybean cultivar-specific nodulation are associated with pilus-like surface appendages and exported by a type III protein secretion system in Sinorhizobium fredii USDA257. Mol. Plant-Microbe Interact. 16:617-625. [DOI] [PubMed] [Google Scholar]
- 24.Krishnan, H. B., and S. G. Pueppke. 1993. Flavonoid inducers of nodulation genes stimulate Rhizobium fredii USDA257 to export proteins into the environment. Mol. Plant-Microbe Interact. 6:107-113. [DOI] [PubMed] [Google Scholar]
- 25.Lewin, A., E. Cervantes, C.-H. Wong, and W. J. Broughton. 1990. nodSU, two new nod genes of the broad host range Rhizobium strain NGR234, encode host-specific nodulation of the tropical tree Leucaena leucocephala. Mol. Plant-Microbe Interact. 3:317-326. [DOI] [PubMed] [Google Scholar]
- 26.Marie, C., W. J. Broughton, and W. J. Deakin. 2001. Rhizobium type III secretion systems: legume charmers or alarmers? Curr. Opin. Plant Biol. 4:336-342. [DOI] [PubMed] [Google Scholar]
- 26a.Marie, C., W. J. Deakin, T. Ojanen-Reuhs, E. Diallo, B. Reuhs, W. J. Broughton, and X. Perret. Ttsl, a key regulator of Rhizobium species NGR234, is required for type-III dependent protein secretion and synthesis of rhamnose-rich polysaccharides. Mol. Plant-Microbe Interact., in press. [DOI] [PubMed]
- 27.Marie, C., W. J. Deakin, V. Viprey, J. Kopcinska, W. Golinowski, H. B. Krishnan, X. Perret, and W. J. Broughton. 2003. Characterisation of Nops, nodulation outer proteins, secreted via the type III secretion system of NGR234. Mol. Plant-Microbe Interact. 16:743-751. [DOI] [PubMed] [Google Scholar]
- 28.Meinhardt, L. W., H. B. Krishnan, P. A. Balatti, and S. G. Pueppke. 1993. Molecular cloning and characterization of a sym plasmid locus that regulates cultivar-specific nodulation of soybean by Rhizobium fredii USDA257. Mol. Microbiol. 9:17-29. [DOI] [PubMed] [Google Scholar]
- 29.Michiels, J., H. Pelemans, K. Vlassak, C. Verreth, and J. Vanderleyden. 1995. Identification and characterization of a Rhizobium leguminosarum bv. phaseoli gene that is important for nodulation competitiveness and shows structural homology to a Rhizobium fredii host-inducible gene. Mol. Plant-Microbe Interact. 8:468-472. [DOI] [PubMed] [Google Scholar]
- 30.Perret, X., and W. J. Broughton. 1998. Rapid identification of Rhizobium strains by targeted PCR fingerprinting. Plant Soil 204:21-34. [Google Scholar]
- 31.Perret, X., W. J. Broughton, and S. Brenner. 1991. Canonical ordered cosmid library of the symbiotic plasmid of Rhizobium species NGR234. Proc. Natl. Acad. Sci. USA 88:1923-1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perret, X., R. Fellay, A. J. Bjourson, J. E. Cooper, S. Brenner, and W. J. Broughton. 1994. Subtraction hybridisation and shot-gun sequencing: a new approach to identify symbiotic loci. Nucleic Acids Res. 22:1335-1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perret, X., C. Freiberg, A. Rosenthal, W. J. Broughton, and R. Fellay. 1999. High-resolution transcriptional analysis of the symbiotic plasmid of Rhizobium sp. NGR234. Mol. Microbiol. 32:415-425. [DOI] [PubMed] [Google Scholar]
- 34.Perret, X., H. Kobayashi, and J. Collado-Vides. 2003. Regulation of symbiotic gene expression in Rhizobium sp. NGR234. Indian J. Exp. Biol. 41:1101-1113. [PubMed] [Google Scholar]
- 35.Perret, X., C. Staehelin, and W. J. Broughton. 2000. Molecular basis of symbiotic promiscuity. Microbiol. Mol. Biol. Rev. 64:180-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prentki, P., and H. M. Krisch. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303-313. [DOI] [PubMed] [Google Scholar]
- 37.Pueppke, S. G., and W. J. Broughton. 1999. Rhizobium sp. strain NGR234 and R. fredii USDA257 share exceptionally broad, nested host ranges. Mol. Plant-Microbe Interact. 12:293-318. [DOI] [PubMed] [Google Scholar]
- 38.Quandt, J., and M. F. Hynes. 1993. Versatile suicide vectors which allow direct selection for gene replacement in Gram-negative bacteria. Gene 127:15-21. [DOI] [PubMed] [Google Scholar]
- 39.Sadowsky, M. J., E. R. Olson, V. E. Foster, R. M. Kosslak, and D. P. Verma. 1988. Two host-inducible genes of Rhizobium fredii and characterization of the inducing compound. J. Bacteriol. 170:171-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 41.Viprey, V. 1999. Etude du génome de Rhizobium sp. NGR234 et rôle du système de sécrétion de type III dans la symbiose avec les légumineuses. Ph.D. thesis. University of Geneva, Geneva, Switzerland.
- 42.Viprey, V., A. Del Greco, W. Golinowski, W. J. Broughton, and X. Perret. 1998. Symbiotic implications of type III protein secretion machinery in Rhizobium. Mol. Microbiol. 28:1381-1389. [DOI] [PubMed] [Google Scholar]