Abstract
Hepatocellular carcinoma (HCC) ranks third in overall global cancer-related mortality. Symptomatic presentation often means advanced disease where potentially curative treatment options become very limited. Numerous international guidelines propose the routine monitoring of those with the highest risk factors for the condition in order to diagnose potential tumourigenesis early. To aid this, the fields of metabonomic- and proteomic-based biomarker discovery have applied advanced tools to identify early changes in protein and metabolite expression in HCC patients vs controls. With robust validation, it is anticipated that from these candidates will rise a high-performance non-invasive test able to diagnose early HCC and related conditions. This review gathers the numerous markers proposed by studies using mass spectrometry and proton nuclear magnetic resonance spectroscopy and evaluates areas of consistency as well as discordance.
Hepatocellular carcinoma (HCC) ranks third in overall global cancer-related mortality (Ferlay et al, 2010) accounting for 85–90% of all tumours emerging from the liver in high-incidence areas and between 70 and 75% of cases in lower incidence regions. In a 2008 survey, overseen by the World Health Organisation (WHO), it was reported that there were ∼748 000 new cases of liver cancer diagnosed worldwide, in that year alone, with an estimated 695 000 reported deaths in the same period (Ferlay et al, 2010). These figures reflect the high mortality rate of this disease owing to multiple contributing factors; most important of which include the absence of monitoring in high-risk populations, insufficient diagnostic resources, and very limited treatment options, many of which require early tumour identification for any potential of curative intervention. As a consequence, the HCC biomarker discovery field is rapidly expanding with new and ongoing research continuing to propose a fast growing list of biomarker candidates. Many of these candidate biomarkers await further validation before being used in conjunction with, or in place of ultrasound scanning as the primary non-invasive test for HCC determination.
The main risk and causative factors for HCC are well described in the literature with a distinct geographical separation observed between established infectious agents such as the hepatitis B and C viruses (HBV and HCV) vs an increasing number of lifestyle-related risk factors such as chronic alcohol abuse (Morgan et al, 2004) non-alcoholic steatohepatitis, diabetes and obesity (Regimbeau et al, 2004; El-Serag and Rudolph, 2007). The focus of this review is to summarise and explore the new potential candidate biomarkers arising from the surge of proteomic and metabolic profiling studies infiltrating the literature. The advance of these markers through the implementation of systems biology approaches will bring to the forefront robust candidates for enhanced diagnostics and therapies for chronic liver diseases (CLDs; Bertino et al, 2014; Malaguarnera et al, 2014). The key selection criteria for an ideal biomarker for any disease are for the:
Target molecule to be measurable in a non-invasive sample source such as blood or urine.
Marker to have excellent diagnostic and/or prognostic abilities for condition of interest (i.e., high sensitivity and specificity).
Marker should be amenable to measurement techniques that are reliable, robust, and reproducible and, for use in field analysis should ideally take a simple kit-based format not requiring additional equipment for interpretation.
Assay should be cheap and thus accessible to all the populations requiring it.
The biomarker should be validated across a broad range of populations.
Currently the most widely used biomarker for HCC, particularly in the developing world where disease burden is greatest is alpha-fetoprotein (AFP). Alpha-fetoprotein ticks several of the boxes in relation to the points mentioned above but fails dramatically in its diagnostic performance and ability, thus significantly reducing its reliability in clinical settings.
The role of AFP in diagnosing HCC and monitoring those at high risk of its development is heavily constrained, with revised guidelines reflecting its significant limitations in this regard. A publication by the American Association for the Study of Liver Diseases (AASLD) summarises that AFP ‘lacks adequate sensitivity and specificity for effective surveillance and for diagnosis' of HCC (Bruix and Sherman, 2011). Similarly, the European Association for the Study of the Liver (EASL) does not recommend AFP within the panel of tests for HCC diagnosis (European Association for the Study of the Liver and European Organisation for Research and Treatment of Cancer, 2012). Not much has been reported on the independent diagnostic ability of AFP, but it is approximated at 0.70, with at least one publication showing area under the curve (AUC) values as low as <0.60 when used at a cut-off of 100 ng ml−1 (Giannini et al, 2012). To circumvent this problem, advanced radiological methods are increasingly being relied on as the best non-invasive tools for accurate diagnosis and monitoring of CLD patients. However, as CLDs and their sequelae of conditions inflict their biggest burden in the developing world, this shift has meant that many of the patients mostly in need of monitoring and diagnosis are left to rely on the poor performance of AFP. The AASLD and EASL both recommend routine surveillance of patients with chronic hepatitis infections and fibrosis, to detect HCC at the early disease stage, when curative treatment options such as tumour resection (Bruix and Sherman, 2011), ablation (Livraghi et al, 2008), transarterial chemoembolisation (Llovet and Bruix, 2003), and liver transplant, recommended for patients with localised tumours on a background of advanced cirrhosis are still prospective. To make this a reality, there is an urgent need for the identification and conversion into clinical use of affordable, non-invasive, and high-performance diagnostic tools deployable in both the developing and developed worlds.
With the availability of novel state-of-the-art technologies and approaches, medical researchers have in recent years used the tools of mass spectrometry (MS) and proton nuclear magnetic resonance (1H NMR) spectroscopy to delve deeper into the human proteome and metabolome from accessible body fluids in order to identify high-performance screening and diagnostic markers capable of detecting and or predicting HCC development. However, there are currently no clinically approved alternatives to AFP that could form robust, non-invasive routine tests available to confidently detect HCC or its main precursor condition of liver cirrhosis (LC) at their early stages. This review aims to summarise and assess literature reports of the application of 1H NMR spectroscopy and MS methods to identify and validate the differential expression of proteins, and metabolites potentially exploitable as biomarkers of HCC. Some degree of focus will be given to markers of earlier stages of CLD such as liver fibrosis and LC where they have been reported. However, due to the high degree of variability in research approaches used to validate the expression trends of putative biomarkers and the small sample sizes generally utilised in profiling, a true meta-analysis of all published work is as yet not possible. Part of the selection criteria for inclusion of papers in this review was that all initial discovery of proposed markers was to be conducted using spectroscopic methodologies, it may thus be striking that some of the more widely proposed markers associated with HCC and CLDs are absent. These have mainly been proposed in experiments utilising more classical protein and metabolite identification and measurement methods, and thus do not form the focus of this work. Many of these classical markers have been widely discussed and reviewed in the context of liver diseases (Bertino et al, 2012; Masuzaki et al, 2012). Several worthy of mention include des-carboxy prothrombin, squamous cell carcinoma antigen–immunoglobulin M complexes, and chromogranin A, which have been proposed and/or compared with AFP in key publications (Bertino et al, 2010, 2011; Biondi et al, 2012). As the list of candidate markers being identified and proposed byspectroscopic methods increases, it is important that reports are comprehensively amalgamated in order to allow researchers to focus more on validating their robustness and expression patterns using independent methods and platforms. This review is an attempt to accelerate this agenda and to ascertain which panel of metabolites and proteins would offer a parsimonious and robust solution for an effective diagnosis.
Materials and Methods
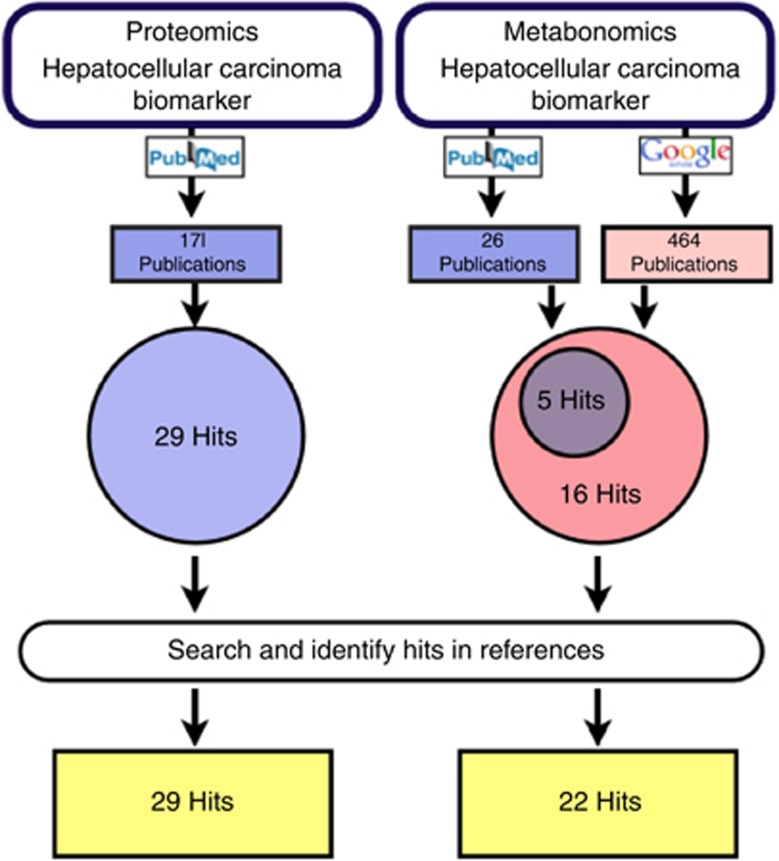
This review focuses on biomarkers proposed from sources searched in the databases PubMed and Google Scholar. The exact search procedure is outlined in Figure 1. For the identification of proteomic papers, the PubMed database was searched with the term ‘(((Hepatocellular Carcinoma) AND Biomarker) AND Proteomics)'. For the metabonomics part, a similar PubMed search was conducted (but with the word ‘metabonomics' instead of ‘proteomics'). All returned publications shortlisted for adherence to the inclusion/exclusion criteria stated below were considered for analyses. Since only 10 metabonomic publications were identified on PubMed, an additional Google Scholar search was conducted, which identified a further eight publications. Additional publications were identified by screening all references associated with these primary papers.
Figure 1.
The search terms ‘Proteomics' or ‘Metabonomics' and ‘Hepatocellular Carcinoma' and ‘Biomarker' were entered into the search engine PubMed (http://www.ncbi.nlm.nih.gov/pubmed). For the identification of metabonomic publications, an additional Google Scholar search was conducted for the time range 2003–2013 (http://scholar.google.co.uk/).
The criteria for inclusion in this review were:
HCC biomarker studies using plasma, serum, or (for metabonomic papers) urine.
HCCs linked to established viral and dietary risk factors with no reports of metastases.
Initial discovery conducted on a MS or 1H NMR spectroscopy-based platform.
Exclusion criteria were non-English literature reports, studies on tissues, cell lines, or animals. Studies including subjects who underwent surgery or transplant were also excluded in order to avoid cases of recurrent HCC following treatment or intervention – these recurrences may have unique developmental profiles that could influence scope of targets identified. Once this initial filtering was applied, a secondary criterion including biomarker candidates with multiple reports, that is, the biomarker should have been reported by at least two independent methods or research groups, was applied.
HCC markers proposed by metabonomic studies
The liver is the metabolic hub of humans as most compounds absorbed by the intestine pass through it at least once. Owing to this, it is able to regulate the expression levels of numerous metabolites, which makes metabolomic analyses approaches particularly relevant for the investigation of liver diseases, such as LC and HCC. In recent years, a lot of effort has been put into metabonomic research on CLDs mainly using urine, blood, or hepatic tissue sections. A number of differential metabolites have been reported by the few studies on HCC tumour tissues, suggesting that there are major metabolic changes taking place during disease development and progression. A common finding of these studies is a glycolytic HCC phenotype (Yang et al, 2007b; Beyoğlu et al, 2013), indicating that the Warburg effect may take place in liver cancer. Although this and other findings give important insight into HCC tumour biology from a clinical perspective, a more important question is whether there exist specific metabolites detectable in biofluids, such as blood or urine, which can serve as biomarkers for the diagnosis of early HCC.
Up to the submission of this review, 22 metabonomic studies matched the inclusion criteria described above and investigated the blood and/or urine metabolome in the context of HCC (Table 1). Most of these studies are MS based and investigate HCC in the context of HBV or HCV (Table 2), which reflects the global trend of primary HCC risk factors (Boyle and Levin, 2008). In contrast, the patient cohorts' ethnicities in these studies did not reflect the geographical distribution of HCC incidence. Although the majority of studies were performed in China, where HCC is a major health issue, only a single study investigated the metabolome of HCC in a sub-Saharan African cohort (Table 2), where HCC is an equally important health burden (Ferlay et al, 2010).
Table 1. Number of metabonomic publications broken down to platform and specimen type.
Abbreviations: 1H NMR=proton nuclear magnetic resonance; MS=mass spectrometry.
Number of studies that included a validation cohort.
Table 2. Summary of metabonomic studies that were reviewed.
|
HCC group |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Reference | Biofluid | Platform | Cohort sizes | Validation cohort | Aetiology | Sex (m : f) | Background CLD | Ethnicity | Staging |
| Baniasadi et al, 2013 | Serum | HPLC–MS/MS (MRM) | HCC: 30 CIR: 22 | No | HCV | 3.3 : 1 | Yes | Mainly Caucasian | No |
| Chen et al, 2011a | Serum | UPLC–MS/MS | HCC: 41 HCON: 38 | No | HBV | 2 : 1 | No information | Chinese | No |
| Chen et al, 2013b | Serum | UFLC–IT–TOF/MS | HCC: 30 CIR: 30 CHB: 30 HCON: 30 | No | HBV | No information | No information | Chinese | No |
| Chen et al, 2013a | Serum | UHPLC–TQ–MS (MRM) | HCC: 29 HCON: 30 | No | No information | No information | No information | No information | No |
| Chen et al, 2011b | Serum, urine | GC–TOF–MS, UPLC–qTOF–MS | HCC: 82 Benign LD: 24 HCON: 71 | Yes | Mainly HBV | 2 : 1 | Majority yes | Chinese | TMN |
| Chen et al, 2009 | Urine | RPLC–qTOF–MS, HILIC–qTOF–MS | HCC: 21 HCON: 24 | Yes | No information | Partially | No information | Chinese | No |
| Gao et al, 2009 | Serum | 1H NMR | HCC: 39 CIR: 36 HCON: 63 | No | HCC: HCV CIR: No information | No information | No information | Chinese | No |
| Huang et al, 2013 | Serum | UHPLC–MS CE–MS | HCC: 139 CIR: 78 CHB: 81 | Yes | HCC: Mainly HBV CIR: HBV | 5.3 : 1 | No information | No information | No |
| Nahon et al, 2012a | Serum | 1H NMR | HCC: 61 CIR: 93 | Yes | Alcohol | 1 : 0 | Yes | Caucasian or residence in France | Small HCC Large HCC |
| Patterson et al, 2011 | Plasma | UPLC–qTOF–MS UPLC–TQ–MS (MRM) GC–MS | HCC: 20 CIR: 7 AML: 22 HCON: 6 | No | HCC: Mixed CIR: No information | 5.6 : 1 | Majority yes | No information | Barcelona criterion |
| Ressom et al, 2012 | Serum | UPLC–qTOF–MS, UPLC–MS (SRM) | HCC: 78 CIR: 184 | No | HCC: Mainly HCV CIR: Mixed | 5.5 : 1 | Yes | Mainly Caucasian | Yes (system not specified) |
| Shariff et al, 2011 | Urine | 1H NMR | HCC: 16 CIR: 14 HCON: 17 | No | Mainly HCV | 15 : 1 | Majority yes | Egyptian | Okuda |
| Shariff et al, 2010 | Urine | 1H NMR | HCC: 18 CIR: 10 HCON: 14 | No | HCC: Mainly HBV CIR: HBV | 2 : 1 | No information | Nigerian | Okuda |
| Wang et al, 2012 | Serum | UPLC–MS | HCC: 23 CIR: 28 HCON: 70 | Yes | HBV | 12 : 1 | Partially | Chinese | AJCC |
| Wei et al, 2012 | Serum | 1H NMR | HCC: 40 CHC: 22 | No | HCV | 3.3 : 1 | No information | Mainly Caucasian | No |
| Wu et al, 2009 | Urine | GC–MS | HCC: 20 HCON: 20 | No | No information | 1 : 0 | No information | Chinese | No |
| Xiao et al, 2012 | Serum | UPLC–qTOF–MS UPLC–MS/MS (SRM) | HCC: 40 CIR: 49 | No | HCC: HCV CIR: Mainly HCV | 3.4 : 1 | No information | Egyptian | TMN |
| Xue et al, 2008 | Serum | GC–MS | HCC: 20 HCON: 20 | No | No information | 1 : 0 | No information | Chinese | No |
| Yin et al, 2009 | Serum | RPLP–qTOF–MS, HILIC–qTOF–MS | HCC: 24 CIR: 25 HCON: 25 | No | HCC: No information CIR: HBV | No information | Partially | Chinese | No |
| Zhang et al, 2013 | Urine | UPLC–qTOF–HDMS | HCC: 25 HCON: 12 | No | No information | 1 : 1 | No information | Chinese | No |
| Zhou et al, 2012a | Serum | UPLC–qTOF–MS (MRM) | HCC: 69 CIR: 28 HCON: 31 | No | HBV and HCV | 4.3 : 1 | No information | Chinese | No |
| Zhou et al, 2012b | Serum | RPLC–qTOF–MS | HCC: 30 CIR: 30 CHB: 30 HCON: 30 | No | No information | 2.3 : 1 | Yes | Chinese | No |
Abbreviations: AJCC=American Joint Committee on Cancer; AML=acute myeloid leukaemia; CE=capillary electrophoresis; CHB=chronic hepatitis B; CHC=Carcinome HepatoCellulaire; CIR=cirrhosis cohort; CLD=chronic liver disease; GC=gas chromatography; HBV=hepatitis B virus; HCC=hepatocellular carcinoma; HCON=healthy control cohort; HCV=hepatitis C virus; HDMS=high-definition mass spectrometry; HILIC=hydrophobic interaction chromatography; IT=ion trap; MRM=multiple reaction monitoring; MS=mass spectrometry; NMR=nuclear magnetic resonance; qTOF=quadrupole time of flight; RPLC=reversed phase liquid chromatography; SRM=single reaction monitoring; TOF=time of flight; TQ=triple quadrupole; UFLC=ultra-fast liquid chromatography; UHPLC=ultra-high-performance liquid chromatography; UPLC=ultra-performance liquid chromatography.
Most studies inferred differential expression of metabolites by comparing either serum or urine profiles of HCC patients with control groups, mainly made up of healthy volunteers or cirrhotics. This was accomplished with univariate and/or multivariate statistical methods. Multivariate methods used in these publications not only include partial least squares discriminant analysis (PLS-DA), but also support vector machine and random forest approaches. On average 18 biomarker candidates were identified per study, with only 32% of these using established diagnostic models for comprehensive assessment of diagnostic performance. This low assessment rate makes a meta-analysis impossible at this stage.
The following section is structured according to compound classes to provide an overview of the metabolites that were reported in more than one study. In addition, the diagnostic models that have been reported to perform extremely well in diagnosing HCC will be mentioned at the end of this section.
Bile acids
Bile acids constitute one of the most frequently reported compound classes suggested as discriminating between HCC patients and a control group, be it healthy or CLD. An increase in conjugated bile acids has long been recognised in patients with hepatobiliary diseases in general, such as viral hepatitis, cirrhosis, HCC, and cholangiocarcinoma (Neale et al, 1971). Many metabonomic publications reviewed here confirm an increase in particular conjugated serum bile acids in HCC patients when compared to healthy individuals (Table 3).
Table 3. Bile acid compounds discriminatory between HCC and comparison group.
|
HCC: Healthy |
HCC: Cirrhosis |
|||||
|---|---|---|---|---|---|---|
| Compound | HMDB ID | Association | Serum | Urine | Serum | Urine |
| GCA | 00138 | Bile acid | ↑ (Yin et al, 2009; Chen et al, 2011b; Zhou et al, 2012a, 2012b) | ↑ (Chen et al, 2011b; Zhang et al, 2013) | ↓ (Ressom et al, 2012; Xiao et al, 2012) | |
| GCDCA | 00637 | Bile acid | ↑ (Yin et al, 2009; Chen et al, 2011b; Wang et al, 2012; Zhou et al, 2012b)† | ↓ (Wang et al, 2012; Xiao et al, 2012) | ||
| GDCA | 00631 | Bile acid | ↑ (Patterson et al, 2011; Zhou et al, 2012a) | ↓ (Ressom et al, 2012; Xiao et al, 2012) | ||
| TCA | 00036 | Bile acid | ↑ (Yin et al, 2009; Chen et al, 2011b)† | |||
| TCDCA | 00951 | Bile acid | ↓ (Ressom et al, 2012; Xiao et al, 2012) | |||
| Glycine | 00123 | Bile acid conjugate | ↓ (Shariff et al, 2011) ↑ (Wu et al, 2009) | |||
| Taurine | 00251 | Bile acid conjugate | ↓ (Yin et al, 2009) ↑ (Chen et al, 2011b) | |||
Abbreviations: GCA=glycocholic acid; GCDCA=glycochenodeoxycholic acid; GDCA=glycodeoxycholic acid; HCC=hepatocellular carcinoma; HCV=hepatitis C virus; HMDB=The Human Metabolome Database; TCA=taurocholic acid; TCDCA=taurochenodeoxycholic acid.
↓ indicates decrease in HCC; ↑ indicates increase in HCC; † indicates control group of the study (Yin et al, 2009) comprised chronic HCV carriers.
Interestingly, higher bile acid serum levels (glycochenodeoxycholic acid, glycocholic acid, CA, and deoxycholic acid (DCA)) have been found in HCC patients with cirrhosis, than without (Chen et al, 2011b), which may indicate that the bile acid concentration is primarily associated with cirrhosis or overall hepatic performance. Evidence for this is given by Chen et al, 2012, who found serum bile acids increased in patients with hepatic decompensation, when compared to compensated patients. A closer look at the HCC cohorts' characteristics of the matched studies reveals that only a few assessed and included the extent of liver disease in the background of HCC or the hepatic compensation/decompensation status in their analyses (Table 2). This may have significantly biased the results. Other confounding factors that have not been controlled for many studies include (1) the prandial state of the patients (elevated serum levels of certain bile acids have been reported previously in patients after food intake (LaRusso et al, 1978; Ponz De Leon et al, 1978)) and (2) the size of the tumour mass. Large tumour masses may compromise the ability of non-tumourous liver tissue to produce bile acids, thereby lowering the serum bile acid levels. Although some studies report the fraction of HCC patients in different tumour stages, the majority of studies did not include that information in their data analyses.
Altogether, the level of serum bile acids reported here seems to be associated primarily with cirrhosis and not HCC and we do not recommend this compound group as biomarkers for early HCC without further directed analysis of the bile acids across different stages of cirrhosis. From a mechanistically viewpoint, bile acids may play a role in the development of HCC, for example, by the production reactive oxygen species, thereby producing oxidative stress and DNA damage (Baptissart et al, 2013). Some of the bile acids have also been described as versatile signalling molecules (Thomas et al, 2008, 2009; Gadaleta et al, 2011), for instance, lithocholic acid and DCA are both involved in the promotion of energy expenditure and participation in glucidic metabolism by acting on the G-protein-coupled receptor TRG5 (Baptissart et al, 2013).
Conflicting urinary concentrations of free glycine and taurine, two bile acid conjugates, have also been reported. These equivocal reports may also be explained by the recruitment of heterogeneous study cohorts (which differ in the degree of background liver disease or tumour masses). Since, both of the amino acids, glycine and taurine exhibit a wide range of metabolic activity, it is likely that their serum and urine concentrations in HCC patients reflect multiple metabolic processes, which further complicates the interpretation of these results. Less frequently reported bile compounds include various other bile acid species.
Lysophosphatidylcholines
When compared to a healthy control group, nine lysophosphatidylcholines (LPCs) have been reported to be significantly decreased in the sera of HCC patients in multiple publications (Table 4). The most frequently reported species include LPC C16 : 0, LPC C18 : 0, and LPC C18 : 2. Two of these (LPC 16 : 0 and LPC 18 : 0) have also been reported to be differentially expressed in the sera of HCC patients when compared with cirrhotic patients, with increased serum levels of LPC C18 : 0 and inconclusive findings for LPC C16 : 0. The depletion of LPC C16 : 0 in HCC tumours has recently been described by two independent tissue metabonomic studies (Beyoğlu et al, 2013; Huang et al, 2013). Transcriptomic analyses confirmed the overexpression of lysophosphatidylcholine acyltransferase 1 (LPCAT1), which converts LPC C16 : 0 to phosphatidylcholine 18 : 1 (Morita et al, 2013). This upregulation of LPCAT1 could account for the depletion of LPC C16 : 0. However, some LPC species, including 16 : 0, LPC 18 : 0, LPC 20 : 4, and LPC 20 : 5 have also been described as differentially expressed between hepatic decompensated and compensated patients (Chen et al, 2012). As mentioned before, many studies did not control for the background liver disease, that is, hepatic performance, which could have biased the described findings.
Table 4. LPCs reported to be discriminatory between HCC and comparison groups.
|
HCC: Healthy |
HCC: Cirrhosis |
|||||
|---|---|---|---|---|---|---|
| Compound | HMDB ID | Association | Serum | Urine | Serum | Urine |
| LPC C14 : 0 | 10379 | Phospholipid catabolism | ↓ (Patterson et al, 2011; Zhou et al, 2012b; Chen et al, 2013a) | |||
| LPC C16 : 0 | 10382 | Phospholipid catabolism | ↓ (Patterson et al, 2011; Wang et al, 2012; Zhou et al, 2012a; Chen et al, 2013a, 2013b) | ↓ (Patterson et al, 2011; Zhou et al, 2012a) ↑ (Ressom et al, 2012; Wang et al, 2012) | ||
| LPC C18 : 0 | 10384 | Phospholipid catabolism | ↓ (Yin et al, 2009; Patterson et al, 2011; Wang et al, 2012; Zhou et al, 2012a; ) | ↑ (Ressom et al, 2012; Wang et al, 2012) | ||
| LPC C18 : 1 | 02815/10385/10408 | Phospholipid catabolism | ↓ (Patterson et al, 2011; Zhou et al, 2012a; Chen et al, 2013a) | |||
| LPC C18 : 2 | 10386 | Phospholipid catabolism | ↓ (Yin et al, 2009; Zhou et al, 2012a; Baniasadi et al, 2013; Chen et al, 2013a, 2013b) | |||
| LPC C18 : 3 | 10386 | Phospholipid catabolism | ↓ (Yin et al, 2009; Patterson et al, 2011) | |||
| LPC C20 : 3 | 10393/10394 | Phospholipid catabolism | ↓ (Patterson et al, 2011; Zhou et al, 2012b) | |||
| LPC C20 : 4 | 10395/10396 | Phospholipid catabolism | ↓ (Patterson et al, 2011; Zhou et al, 2012a) | |||
| LPC C20 : 5 | 10397 | Phospholipid catabolism | ↓ (Patterson et al, 2011; Zhou et al, 2012b) | |||
Abbreviations: HCC=hepatocellular carcinoma; HMDB=The Human Metabolome Database; LPCs=lysophosphatidylcholines.
↓ indicates decrease in HCC; ↑ indicates increase in HCC.
LPCs, together with arachidonic acid, may play important roles in the progression of HCC, as they have been described as important players in endothelial cell migration, that is, vascularisation processes (Linkous et al, 2010). This finding matches with the observation that hypervascularisation is a frequent feature of HCC. A positive relationship has been described between LPC concentrations and both body weight and inflammatory processes in malignant diseases (Taylor et al, 2007). The detection of urinary LPCs in the context of HCC has not been reported.
Free fatty acids
Although numerous free fatty acid (FFA) species have been described as being discriminatory in serum between HCC patients and a control group, there was no common trend in serum FFA levels observable with some studies reporting increased levels and others reporting a decrease in FFA concentrations (Table 5). FFA species reported frequently include FFA C16 : 0, FFA C18 : 0, FFA C20 : 4, and FFA C24 : 1. The conflicting reports on the serum concentrations may be a result of heterogenic patient groups, in terms of different diets, gender, comorbidities (such as cardiovascular diseases), and of course the extent of liver damage, that is the hepatic compensation/decompensation status. As was the case for bile acids and LPCs, FFA levels seem to be also influenced by background liver disease (Chen et al, 2012). The association of FFA 16 : 0 and FFA 18 : 0 with hepatitis B and cirrhosis has been described by Chen et al, 2011b, who investigated HCC patients with and without LC and hepatitis separately. In contrast, increased concentrations of FFA C16 : 0 and FFA C18 : 0 have been confirmed by a tissue metabonomic study on HCC (Huang et al, 2013), indicating that there may be quantitative differences between serum FFA concentrations of HCC and cirrhosis patients.
Table 5. Discriminative FFAs between HCC and comparison groups.
|
HCC: Healthy |
HCC: Cirrhosis |
|||||
|---|---|---|---|---|---|---|
| Compound | HMDB ID | Association | Serum | Urine | Serum | Urine |
| FFA C24 : 1 (nervonic acid) | 02368 | Lipid metabolism/energy metabolism | ↓ (Chen et al, 2011b; Patterson et al, 2011) | |||
| Oleamide | 02117 | Lipid metabolism/energy metabolism | ↓ (Chen et al, 2011b) ↑ (Wang et al, 2012) | ↓ (Wang et al, 2012; Xiao et al, 2012) | ||
| FFA C16 : 0 (palmitic acid) | 00220 | Lipid metabolism/energy metabolism | ↓ (Chen et al, 2011b) ↑ (Zhou et al, 2012b) • (Xue et al, 2008) | |||
| FFA C18 : 0 (stearic acid) | 00827 | Lipid metabolism/energy metabolism | ↓ (Chen et al, 2011b) ↑ (Zhou et al, 2012b) • (Xue et al, 2008) | |||
| FFA C20 : 4 (arachidonic acid) | 01043 | Lipid metabolism/energy metabolism | ↓ (Chen et al, 2011b) ↑ (Zhou et al, 2012a, Zhou et al, 2012b) | |||
Abbreviations: FFAs=free fatty acids; HCC=hepatocellular carcinoma; HMDB=The Human Metabolome Database.
↓ indicates decrease in HCC; ↑ indicates increase in HCC; • indicates not reported whether decreased or increased.
Other serum lipid compounds found to be discriminative between HCC and healthy controls are FFA C24 : 1 and oleamide. Although FFA 24 : 1 has not been found to discriminate between cirrhosis and HCC patients, oleamide has. Oleamide, the amide of FFA C18 : 1 (oleic acid), exhibits a variety of neuropharmacological effects, including increased food intake (Martínez-González et al, 2004) and relaxation of blood vessels (Hoi and Hiley, 2006). Since Oleamide has not been associated with CLDs, nor previously been reported in cancer literature, it may represent a specific marker for HCC. However, further validation studies are required.
Carnitine and related acylesters
A compound class closely associated to fatty acids and their catabolism, is carnitine and its acylesters. Carnitine is mainly derived from dietary sources and its main function is to transport activated long-chain fatty acids from the cytosol into the mitochondria for energy production via beta-oxidation. Physiologically, almost all free carnitine is intracellular (Cave et al, 2008) and the renal absorption rate is high (Bellinghieri et al, 2003). Interestingly, two urinary NMR studies report a trend towards increased urinary levels of free carnitine HCC patients, when compared with a healthy control or cirrhosis group (Table 6). Since renal function was not assessed in any of the studies considered in this review, and renal diseases, such as the hepatorenal syndrome, are common secondary to cirrhosis and HCC, increased urinary-free carnitine may indicate kidney dysfunction, instead of being specific for HCC. Increased urinary acylcarnitines (esterified acyl groups with carnitine) have been reported in specific FFA oxidation disturbances and after extensive workout (Flanagan et al, 2010). Urinary acetylcarnitine, however, has not been associated with HCC before and may therefore be a specific HCC biomarker candidate.
Table 6. Discriminative carnitines between HCC and comparison groups.
|
HCC: Healthy |
HCC: Cirrhosis |
|||||
|---|---|---|---|---|---|---|
| Compound | HMDB ID | Association | Serum | Urine | Serum | Urine |
| Carnitine | 00062 | Energy metabolism | ↑ (Yin et al, 2009; Chen et al, 2011b) ↓ (Zhou et al, 2012b; Huang et al, 2013) | ↑ (Chen et al, 2009; Shariff et al, 2010, 2011) | ↑ (Shariff et al, 2010, 2011) | |
| Acetylcarnitine | 00201 | Energy metabolism | ↑ (Yin et al, 2009) ↓ (Zhou et al, 2012b) | |||
| Acylcarnitine C3 : 0 | 00824 | Energy metabolism | ↓ (Zhou et al, 2012b; Huang et al, 2013) | |||
| Acylcarnitine C8 : 0 | 00791 | Energy metabolism | ↓ (Zhou et al, 2012b; Chen et al, 2013a) | |||
| Acylcarnitine C8 : 1 | 00791 | Energy metabolism | ↓ (Zhou et al, 2012b; Chen et al, 2013a) | |||
| Acylcarnitine C10 : 0 | 00651 | Energy metabolism | ↓ (Zhou et al, 2012b; Chen et al, 2013a) | |||
| Acylcarnitine C10 : 1 | 13205 | Energy metabolism | ↓ (Zhou et al, 2012b; Chen et al, 2013a) | |||
| Acylcarnitine C18 : 1 | 13338 | Energy metabolism | ↑ (Zhou et al, 2012a, 2012b; Chen et al, 2013a) | ↓ (Xiao et al, 2012) ↑ (Zhou et al, 2012a) | ||
| Acylcarnitine C18 : 2 | 13212 | Energy metabolism | ↑ (Zhou et al, 2012a, 2012b; Chen et al, 2013a) | ↓ (Xiao et al, 2012; Zhou et al, 2012a) | ||
Abbreviation: HBV=hepatitis B virus; HCC=hepatocellular carcinoma; HMDB=The Human Metabolome Database.
↓ indicates decrease in HCC; ↑ indicates increase in HCC; † indicates control group of the study (Huang et al, 2013) comprised chronic HBV carriers.
The differential serum levels of carnitines, when HCC is compared to a healthy control cohort, seem to exhibit a specific pattern, with mostly increased free carnitine levels, decreased short to medium-chain acylcarnitines and increased levels of long-chain acylcarnitine C18 : 1 and C18 : 2. A major role of short- and medium-chain acylcarnitines is to remove organic acids from organelles such as mitochondria, and eventual excretion in urine and bile. The decrease in serum, however, indicates either an increased excretion rate or a decreased accumulation rate. In contrast, long-chain acylcarnitines are formed intracellularly for the purpose of energy production. Therefore, elevated long-chain acylcarnitine levels could possibly permit an increased FFA transport into mitochondria and hence increase energy production.
The interpretation of these findings should be taken with caution, since carnitine levels can be influenced by many other factors, such as by diet, renal dysfunction or altered biosynthesis rates. Blood acylcarnitines levels may also be influenced by the hepatic fatty acid metabolism, that is, increased (or decreased) acylcarnitine levels due to the increased (or decreased) production of acyl groups in the liver or other tissues.
There is a body of evidence that patients with different cirrhosis aetiologies show specific carnitine profiles. For instance, when compared to healthy volunteers, patients with viral hepatitis-induced cirrhosis (HBV and HCV) showed an increase in plasma long-chain acylcarnitines, whereas patients with alcohol-induced cirrhosis had elevated levels of both, long- and short-chain acylcarnitines (Krähenbühl, 1996). Clinical studies on L-carnitine and acetyl-L-carnitine as a dietary supplement found positive effects on cognitive function (Malaguarnera, 2012), indicating that treatment with carnitine may provide useful information on energy metabolism in cirrhosis and HCC.
Increased urinary and plasma carnitine and acylcarnitine levels have also been described in kidney diseases (Calabrese et al, 2006; Ganti et al, 2012) and in patients treated with certain chemotherapeutic drugs, such as cisplatin (Dodson et al, 1989; Heuberger et al, 1998).
Energy, nucleotide, and amino acid metabolism
Other energy metabolism-related compounds found to be discriminatory between HCC and healthy cohorts include 2-oxoglutarate, succinate, and glycerol (Table 7). The elevation of 2-oxoglutarate, a key component of the TCA cycle, may be a consequence from a decreased mitochondrial respiration, that is, the Warburg effect. This hypothesis is strengthened by tissue metabolomics studies that report glycolytic phenotypes of HCC (Huang et al, 2013). Contradictory urinary concentrations were reported for succinate, another TCA intermediate. A factor that may convolve the biological signal in urinary metabolite profiles is the extent of which a urinary metabolite concentration reflects the metabolite's blood concentration. Although the urinary metabolome is a subset of the total body metabolome, urinary metabolite concentrations depend on the renal excretion rate, which is metabolite specific and tightly regulated, depending on many factors such as blood pH. Evidence that supports this hypothesis is given in Chen et al, 2011b and Bouatra et al, 2013. Furthermore, a number of amino acids were found to discriminate between HCC and healthy controls in serum and/or urine. An amino acid imbalance has been recognised previously in the serum of patients with CLD; specifically the decrease in branched chain amino acids (BCAAs: leucine, isoleucine, and valine) and the increase in aromatic amino acids (AAAs: phenylalanine, tryptophan, tyrosine, and histidine); in particular tyrosine (Michitaka et al, 2010). This mainly has been supported by a metabonomic study, with an exception of phenylalanine, which was found increased in the sera of HCC patients, when compared to healthy controls. Decreased serum BCAAs may also be linked to elevated serum carnitine levels, since they are able to oxidise BCAAs (Hoppel, 2003).
Table 7. Additional metabolites associated with TCA cycle, and protein and nucleotide metabolism.
|
HCC: Healthy |
HCC: Cirrhosis |
|||||
|---|---|---|---|---|---|---|
| Compound | HMDB ID | Association/compound class | Serum | Urine | Serum | Urine |
| Isoleucine | 00687 | AA/protein synthesis | ↓ (Chen et al, 2013a) • (Xue et al, 2008) | |||
| Leucine | 00687 | AA/protein synthesis | ↓ (Chen et al, 2011b, 2013a) | |||
| Methionine | 00696 | AA/protein synthesis | ↓ (Chen et al, 2011b; Huang et al, 2013) †) ↑ (Chen et al, 2013a) | |||
| Phenylalanine | 00159 | AA/protein synthesis | ↓ (Chen et al, 2011b) ↑ (Gao et al, 2009; Wang et al, 2012; Zhou et al, 2012a; Chen et al, 2013a) | ↓ (Wang et al, 2012; Zhou et al, 2012a; Baniasadi et al, 2013) | ||
| Threonine | 00167 | AA/protein synthesis | ↑ (Wu et al, 2009; Chen et al, 2011b) | |||
| Tryptophan | 00929 | AA/protein synthesis | ↓ (Chen et al, 2011b, 2013a; Zhou et al, 2012a, 2012b) | |||
| Tyrosine | 00158 | AA/protein synthesis | ↑ (Gao et al, 2009; Chen et al, 2013a) ↓ (Chen et al, 2011b) | ↑ (Wu et al, 2009; Chen et al, 2011b) | ||
| Valine | 00883 | AA/protein synthesis | ↓ (Gao et al, 2009; Chen et al, 2013a) ↑ (Wei et al, 2012) † | |||
| Creatinine | 00562 | Alanine and proline metabolism | ↓ (Chen et al, 2011b; Wei et al, 2012) † | ↓ (Chen et al, 2009; Shariff et al, 2010, 2011) | ↑ (Shariff et al, 2010, 2011) | |
| Choline | 00097 | Lipid metabolism | ↑ (Wei et al, 2012) †) ↓ (Gao et al, 2009) | |||
| Glycerol | 00131 | Lipid metabolism/energy metabolism | ↓ (Chen et al, 2011b) ↑ (Gao et al, 2009) • (Xue et al, 2008) | |||
| Trimethylamine N-oxide | 00925 | Microbial co-metabolite | ↓ (Shariff et al, 2011; Chen et al, 2011b) | |||
| Creatine | 00064 | Other | ↓ (Chen et al, 2011b) ↑ (Shariff et al, 2010, 2011) | |||
| Hypoxanthine | 00157 | Purine nucleoside | ↓ (Wu et al, 2009) ↑ (Chen et al, 2009, 2011b) | |||
| 2-Oxoglutarate | 00208 | TCA cycle | ↑ (Gao et al, 2009, Chen et al, 2011b) | |||
| Succinate | 00254 | TCA cycle | ↓ (Wu et al, 2009; Chen et al, 2011b) | |||
Abbreviation: AA=amino acid; HBV=hepatitis B virus; HCC=hepatocellular carcinoma; HCV=hepatitis C virus; HMDB=The Human Metabolome Database; TCA=tricarboxylic acid.
↓ indicates decrease in HCC; ↑ indicates increase in HCC; † indicates control group of the study (Huang et al, 2013; Wei et al, 2012) comprised chronic HCV or HBV carriers, respectively.
In addition, the urinary concentration of the purine nucleoside hypoxanthine has been found to be significantly higher in HCC when compared to healthy individuals. However, another study reports a decreased urine concentration, potentially due to the different HCC aetiologies of the cohorts. A tissue metabonomic study reported increased hypoxanthine concentrations in HCC tissue, compared to matched non-tumourous tissue of HCC patients (Huang et al, 2013).
Other relevant metabolites, shown to be reduced in the urine of HCC patients, include creatinine and trimethylamine-N-oxide (TMAO). Urinary creatinine excretion is related to muscle mass (Oterdoom et al, 2009) and therefore may be a manifestation of cancer cachexia, instead of being specifically HCC related. Trimethylamine-N-oxide is generated in the liver from oxidation of the gut microbiota co-metabolite trimethylamine (TMA) by flavin monooxygenases. The production of TMA by the human gut microbiota may represent a link between HCC and interactions of host – gut microbiota, which have previously been suggested to have the capacity to promote hepatocarcinogenesis (Mederacke et al, 2012). In addition, increased TMAO levels may be partially responsible for decrease of serum bile acids, since TMAO is involved in cholesterol metabolism (Koeth et al, 2013).
Diagnostic performance of metabolite biomarker candidates
From 22 studies reviewed, only 6 assessed the diagnostic performance of the biomarker candidates proposed (Xue et al, 2008; Chen et al, 2011a; Wang et al, 2012; Zhou et al, 2012a; Baniasadi et al, 2013; Huang et al, 2013). However, the performance of these metabonomic models to detect and differentiate HCC from healthy volunteers or cirrhosis patients is consistently better than the performance of AFP. The model with the best performance in discriminating HCC from cirrhosis cohort was constructed with full-serum LC-MS data (without variable selection), yielding 100% sensitivity and specificity (ROC=100% Wang et al, 2012). A heavily reduced model, constructed with only canavaninosuccinate and AFP, achieved a sensitivity and specificity of 96.4 and 100%, respectively (Wang et al, 2012). Another high-performance model for the discrimination between HCC and cirrhosis patients was obtained with acetylcarnitine C3 (propionylcarnitine) and betaine, yielding in an AUC of 0.98 (Huang et al, 2013). A comparably high ROC value was obtained for a four-compound PLS-DA model for the discrimination between HCV infected cirrhosis patients with and without HCC, including methionine, 5-hydroxymethyl-2′-deoxyuridine, N2,N2-dimethylguanosine and uric acid (Baniasadi et al, 2013). The best model performance for the discrimination between HCC and healthy controls was based on serum levels of 1-methyladenosine combined with AFP, yielding in an AUC of 0.95 (Chen et al, 2011a). Very similar, in terms of performance, but without AFP was a model that included endocannabinoids anandamide and palmitylethanolamide yielding in an AUC of 0.94 (Zhou et al, 2012a). The same model was used to discriminate between HCC and cirrhotics with an AUC of 0.88. A different model that included 13 serum components had an overall accuracy of 75% in discriminating HCC from healthy controls (Xue et al, 2008). Only one urinary study evaluated the performance of an established PCA model for discriminating between HCC patients and healthy controls. Their model included 18 urinary compounds plus serum AFP and performed much better than AFP, with an AUC of 0.928 (Wu et al, 2009). Taken together, these models seem to suggest that a robust diagnostic of HCC could be produced with a reduced panel of biomarkers rather than using the whole spectroscopic dataset. However, given that the panels of biomarker thus far reported in the literature have not been assessed in independent validation cohorts, there is now a pressing requirement for driving a full validation study for a selected biomarker panel.
HCC markers proposed by MS-based proteomics
State of the science
The PubMed search conducted using the search term stated in the Methods section resulted in the selection of 171 references. These were initially screened by methodology and all publications not utilising MS as a primary discovery approach were filtered out. The remaining references were read for adherence to the specified inclusion criteria leading to the exclusion of a further 31, leaving 29 to be used in this paper. Early studies applying MS techniques to clinical proteomics biomarker research exerted a greater focus on the utilisation of SELDI/MALDI ionisation platforms, occasionally coupled with weak/strong ion-exchange chromatography techniques for the identification of protein marker candidates. The MS analysis was typically applied in concert with two-dimensional electrophoresis methods where proteins separated according to their isoelectric points and molecular weights were observed on a global scale. Notable points of differentiation in spot patterns correlating with disease groupings would be excised for identification following in-gel digestion. Alternative approaches to marker discovery, which have since become more popular include untargeted analyses using tandem MS fragmentation methods to break up chromatographically separated proteins expressed in clinical samples followed by database-driven identification (Nesvizhskii, 2007). Both these approaches are applicable to pre-fractionated or un-fractionated samples as well as those which have undergone pre-enrichment steps for particular post-translational modifications (PTMs) of interest; a major form of this being glycosylation changes. This general shift from the coupling of gel-based classical proteomic approaches with MS to standalone chromatography-MS platforms has enabled researchers a greater detail in the analysis of the human proteome as well as increased flexibility and unmatched depth in finding biomarker targets.
Protein families
Proteomic exploration using MS is widely focussed on serum and plasma, which form the key body fluids with the greatest dynamic range of protein expression. This review of protein markers for HCC determined by MS in various populations identifies candidates from a wide range of protein families. Acute-phase and transport proteins, enzymes as well as proteins involved in various complementary and lipid metabolic pathways form some of the most commonly reported classes of biomarker candidates. Alongside these, a smaller number of proteins involved in numerous functions ranging from apoptosis, ion transport, host immunity, iron homoeostasis, protection from toxins and oxidative stress, cell adhesion, differentiation, regeneration, and death are reflected in the list of reported markers (Table 8). As the most frequently proposed protein subtype for biomarker candidates, the suggestion of acute-phase proteins as biomarker candidates is often eyed with scepticism as they are seen to lack specificity for particular disease processes. These proteins, however, constitute a well validated subset of proposed markers suggested from experiments run on numerous MS platforms in the context of HCC and CLDs. A1AT and alpha-1-antichymotrypsin (AACT) are major acute-phase proteins, which serve as protease inhibitors protecting tissues from the enzymatic action of inflammatory cells. Five independent publications identified these proteins as elevated in HCC patients; with a highest reported fold change of 5.3. Another large subset of proteins often reported following MS interrogation of HCC samples are apolipoproteins. These are proteins that bind lipids and cholesterols in the blood and lymphatic systems. Several isoforms, namely Apo A1, Apo J, Apo L1, and Apo B-100 have been consistently reported as downregulated in HCC. In particular, Apo J in its association with HCC has been implicated as a possible indicator of the metastatic potential of primary tumours (Lau et al, 2006). In overall performance reports, some of the most consistently highlighted proteins include those with functional properties such as ion transport and scavenging, in particular – iron homoeostasis. Haptoglobin and haemopexin are key players in the maintenance of haemoglobin and free haem in the blood. Haptoglobin binds free haemoglobin with high affinity and thus protects cells from its oxidative effects. Haemopexin on the other hand binds free haem released in the turnover of erythrocytes and preserves the body's iron stores. Like most of the proteins with unique expression trends concordant with liver disease, the specific mechanisms linking these proteins to CLD are not understood. However, amalgamated reports demonstrate that haptoglobins are largely increased in blood from HCC patients. Haemopexin in its whole and fucosylated forms follow the same trend with a suggestion in one report of a decrease in LC (Fye et al, 2013).
Table 8. Summary of proteomic studies that were reviewed.
| Protein (key role) | UniProt ID | Biofluid | Relation to HCC (↑, ↓) | HCC LB rate | Cohort sizes | Aetiology | AUROC | Fold change | Platform | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| AGP1 (AP: Transport) | P02763 | Plasma | ↑ HCC:CON | No information | *10 HCC; 30 controls (healthy/LC/HBV) | HBV | 0.73 | 1.6* | Lectin MRM–UPLC–ESI–MS | Ahn et al, 2012a, 2012b* |
| Plasma | ↓ HCC:LC | No information | Disc: 10 HCC/2 HCON Val: 18 HCC, 10 HCON | No information | 0.65 | — | nUPLC–ESI–QTOF–MS and QQQ | Lee et al, 2011 | ||
| Serum | ↑ HCC:LC | 100% ALL | Disc: 9 HCC/9 LC Val: 52 HCC/40 LC | HBV | — | 1.8† | ICAT–LC–ESI–MS/MS | Kang et al, 2010 | ||
| A1AT (AP: Protease inhibitor) | P01009 | Plasma | ↑ HCC:CON | Disc 24.2% Val 4.8% | Disc: 120 HCC, 99 LC, 120 HCON Val: 21 HCC, 6 LC, 18 ASC, 10 HCON | HBV/aflatoxin | 0.84 | — | QTOF–LC–MS, ELISA | Fye et al, 2013 |
| Plasma | ↑ HCC:CON | No information | *10 HCC; 30 controls (healthy/LC/HBV) | HBV | 0.92 | 2.4** | Lectin MRM–UPLC–ESI–MS | Ahn et al, 2012a, 2012b* | ||
| Serum | ↑ HCC:CON | No information | Disc: 20 HCC, 20 HBV, 20 HCON Val: 20 HCC, 20 HBV, 20 HCON | HBV | — | — | MALDI–TOF–MS/MS, 2DE | Feng et al, 2005 | ||
| A2M (Protease inhibitor) | P01023 | Plasma | ↑ HCC:CON | Disc 24.2% Val 4.8% | Disc: 120 HCC, 99 LC, 120 HCON Val: 21 HCC, 6 LC, 18 ASC, 10 HCON | HBV/aflatoxin | — | 1.13* | QTOF–nUPLC–ESI–MS | Fye et al, 2013 |
| Serum | ↓ HCC:LC | 100% of HCCs | Disc: 9 HCC/9 LC Val: 52 HCC/40 LC | HBV | — | 0.26† | ICAT–LC–ESI–MS/MS | Kang et al, 2010 | ||
| AACT (AP: Protease inhibitor) | P01011 | Plasma | ↑ HCC:CON | No information | *10 HCC; 30 controls (healthy/LC/HBV) | HBV | 0.93 | 3.7** | Lectin MRM–UPLC–ESI–MS | Ahn et al, 2012a, 2012b* |
| Plasma | ↑ HCC:CON | No information | 6 HCC, 3 HCON | No information | — | 5.3* | 2D LC–MALDI–TOF/TOF | Ishihara et al, 2011 | ||
| AFM (Vitamin transport) | P43652 | Plasma | ↑ HCC:CON | No information | 6 HCC, 3 HCON | No information | — | 33.1† | 2D LC–MALDI–TOF/TOF | Ishihara et al, 2011 |
| Plasma | ↓ HCC:CON | No information | 28 HCC, 10 HCON | No information | 0.72 | — | nUPLC–ESI–QTOF–MS and QQQ | Lee et al, 2011 | ||
|
Apolipoproteins | ||||||||||
| Apo A1 (Cholesterol metabolism) | P02647 | Plasma | ↓ HCC:CON | Disc 24.2% Val 4.8% | Disc: 120 HCC, 99 LC, 120 HCON Val: 21 HCC, 6 LC, 18 ASC, 10 HCON | HBV/aflatoxin | 0.83 | — | QTOF–nUPLC–ESI–MS | Fye et al, 2013 |
| Serum | ↓ HCC:CON | 100% of HCCs | 20 HCC, 20 HCON | No information | — | −3.59* | 2DE-coupled MALDI–TOF–MS | Sun et al, 2010b | ||
| Serum | ↓ HCC:CON | 100% of HCCs | 8 HCC, 21 HBV, 7 HCON | HBV | — | −3.10† | 2DE-coupled MALDI–TOF–MS | Steel et al, 2003 | ||
| Apo B-100 (Cholesterol metabolism) | P04114 | Plasma | ↓ HCC:CON | No information | 6 HCC, 3 HCON | No information | — | −3.7* | 2D LC–MALDI–TOF/TOF | Ishihara et al, 2011 |
| Apo L1 (Cholesterol metabolism) | O14791 | Plasma | ↓ HCC:CON | No information | *10 HCC; 30 controls (healthy/LC/HBV) | HBV | 0.68 | 0.5ns | Lectin MRM–UPLC–ESI–MS | Ahn et al, 2012a, 2012b* |
| B2M (Immunity) | P61769 | Plasma | ↑ HCC:CON | No information | 6 HCC, 16 LC/HBV/HCV, 8 HCON | HCV/HBV | — | — | SELDI–TOF–MS | Nakatsura, 2010 |
| Serum | ↑HCC:pre-HCC | Some Histo. % not stated | 38 Pre-HCC; 35 samples upon clinical diagnosis of HCC; 18 samples post treatment (longitudinal collection) | HCV | — | 1.82** | SELDI–TOF–MS | Ward et al, 2006 | ||
| C4B-α (Complement pathway) | P04003 | Plasma | ↑ HCC:CON | Disc 24.2% Val 4.8% | Disc: 120 HCC, 99 LC, 120 HCON Val: 21 HCC, 6 LC, 18 ASC, 10 HCON | HBV/aflatoxin | — | 1.25ns | QTOF–nUPLC–ESI–MS | Fye et al, 2013 |
| ↑ HCC:CON | No information | *10 HCC; 30 HCON/LC/HBV | HBV | 0.66 | 1.3ns | Lectin MRM–UPLC–ESI–MS | Ahn et al, 2012a, 2012b* | |||
| CC3 (Complement pathway) | P01024 | Plasma | ↑ HCC:CON | No information | 10 HCC; 30 HCON/LC/HBV | HBV | 0.67 | 2.0ns | Lectin MRM–ESI–UPLC–MS | Ahn et al, 2012b |
| Plasma | ↑ HCC:LC | Disc 24.2% Val 4.8% | Disc: 120 HCC, 99 LC, 120 HCON Val: 21 HCC, 6 LC, 18 ASC, 10 HCON | HBV/aflatoxin | 0.70 | — | QTOF–nUPLC–ESI–MS, ELISA | Fye et al, 2013 | ||
| Serum | ↓ HCC:CON | 100% of HCCs | 8 HCC, 21 HBV, 7 HCON | HBV | — | −1.87** | 2DE-coupled MALDI–TOF–MS | Steel et al, 2003 | ||
| CC3-α | Serum | ↑HCC:CLD/CON | No LBs done | 45 HCC, 42 CLD, 21 HCON | HCV | 0.70 | — | SELDI–TOF–MS | Kanmura et al, 2010 | |
| ↑HCC:CLD/CON | No information | 28 HCC, 10 HCON | HCV | — | n/r but P of Δ 0.00001 | SELDI–TOF–MS, WB | Lee et al, 2006 | |||
| CERU (Cu/ion transport) | P00450 | Plasma | ↑ HCC:CON | No information | 10 HCC; 30 HCON/LC/HBV) | HBV | 0.77 | 1.5** | Lectin MRM–UPLC–ESI–MS | Ahn et al, 2012b |
| Plasma | ↑ HCC:CON | No information | 6 HCC, 3 HCON | No information | — | 6.8† | 2D LC–MALDI–TOF/TOF | Ishihara et al, 2011 | ||
| Serum | HCC only | 100% of HCCs | 5 HCC, 5 HCON | HCV/HBV | — | — | nanoHPLC–ESI–MS/MS | Yang et al, 2007a | ||
| Serum | ↑ HCC:CON | No information | Disc: 20 HCC, 20 HBV, 20 HCON Val: 20 HCC, 20 HBV, 20 HCON | HBV | — | — | 2DE and MALDI–TOF–MS/MS | Feng et al, 2005 | ||
| CLU (apoptosis/complement pathway) | P10909 | Plasma/serum | ↓ HCC:CON | No information | No information | No information | — | — | SID–MRM–MS | Zhao et al, 2010 |
| Plasma | ↓ HCC:CON | Disc 24.2% Val 4.8% | Disc: 120 HCC, 99 LC, 120 HCON Val: 21 HCC, 6 LC, 18 ASC, 10 HCON | HBV/aflatoxin | — | 0.59* | QTOF–nUPLC–ESI–MS | Fye et al, 2013 | ||
| Plasma | ↓ HCC:CON | No information | 10 HCC; 30 HCON/LC/HBV | HBV | 0.70 | 0.8ns | Lectin MRM–UPLC–ESI–MS | Ahn et al, 2012a, 2012b | ||
| Serum | ↑ HCC:CON | No information | Disc: 20 HCC, 20 HBV, 20 HCON Val: 20 HCC, 20 HBV, 20 HCON | HBV | — | — | 2DE and MALDI–TOF–MS/MS | Feng et al, 2005 | ||
| ERBB3 (Signalling pathway) | P21860 | Serum | ↑HCC:LC/CON | Disc and Val 1–100% Val 2; Unclear | Disc: 10 total, HCC and non-HCC Val 1: 113 HCC, 47 LC, 64 HBV/HCV Val 2: 57 HCC, 35 HBV/HCV | HBV/HCV | 0.93/0.71 | — | MALDI–TOF–MS, WB, ELISA | Hsieh et al, 2011 |
| GFAP (cell differentiation/regeneration) | P14136 | Serum | ↑ HCC:CON | 100% HCCs | Disc: 20 HCC, 20 HCON Val: 5 HCC, 5 HCON | No information | — | — | 2DE-coupled MALDI–TOF, WB | Wu et al, 2012 |
| hCE1 (response to toxins) | P23141 | Plasma | ↑ HCC:CON | No information | 24 HCC, 14 LC, 7 CH, 8 CCa, 15 stomach cancer, 16 pancreatic cancer | No information | 0.80 | — | 2DE MALDI–TOF/TOF, nano-LC MS/MS, WB | Na et al, 2009 |
| HP (AP: Immunity and Fe homoeostasis) | P00738 | Serum | ↑ HCC:CON | 100% of HCCs | 5 HCC, 5 HCON | HCV/HBV | — | — | 2DE LC–ESI–MS/MS | Yang et al, 2007a |
| Serum | ↑ HCC:CLD | 100% of HCCs | 56 HCC, 40 CLD | No information | 0.73 | 2.57** | ELISA | Ang et al, 2006 | ||
| Serum | ↑ HCC:CON | No information | Disc: 20 HCC, 20 HBV, 20 HCON Val: 20 HCC, 20 HBV, 20 HCON | HBV | — | — | 2DE and MALDI–TOF–MS/MS | Feng et al, 2005 | ||
| Serum | ↑ HCC:CON | 100% of HCCs | 20 HCC, 20 HCON | No information | — | 5.10** | 2DE-coupled MALDI–TOF–MS | Sun et al, 2010b | ||
| Plasma | ↑ CON:LC | Disc 24.2% Val 4.8% | Disc: 120 HCC, 99 LC, 120 HCON Val: 21 HCC, 6 LC, 18 ASC, 10 HCON | HBV/aflatoxin | — | 1.35* | QTOF–nUPLC–ESI–MS | Fye et al, 2013 | ||
| HPR (metabolic process) | P00739 | Serum | ↑ HCC:CON | 100% of HCCs | 5 HCC, 5 HCON | HCV/HBV | — | — | 2DE LC–ESI–MS/MS | Yang et al, 2007a |
| Plasma | ↓ HCC:CON | Disc 24.2% Val 4.8% | Disc: 120 HCC, 99 LC, 120 HCON Val: 21 HCC, 6 LC, 18 ASC, 10 HCON | HBV/aflatoxin | — | 0.65* | QTOF–nUPLC–ESI–MS | Fye et al, 2013 | ||
| HPX (Fe homoeostasis) | P02790 | Plasma | ↑ HCC:CON | No information | 10 HCC; 30 HCON/LC/HBV | HBV | 0.61 | 1.2ns | Lectin MRM–UPLC–ESI–MS | Ahn et al, 2012b |
| Plasma | ↑ CON:LC | Disc 24.2% Val 4.8% | Disc: 120 HCC, 99 LC, 120 HCON Val: 21 HCC, 6 LC, 18 ASC, 10 HCON | HBV/aflatoxin | 0.81 | — | QTOF–nUPLC–ESI–MS | Fye et al, 2013 | ||
| Fu-HPX | Plasma | ↑ HCC:CON | Some Histo. % not stated | 72 HCC, 32 LC, 33 HBV, 133 HCV, 62 other liver diseases, 20 CON | HCV/HBV | 0.95 | 1.4** | Lectin LC–MS/MS | Comunale, 2009 | |
| HSP90 (stress response) | P08238 | Serum | ↑ HCC:CON | 100% of HCCs | 20 HCC, 20 HCON | No information | — | 7.04** | MALDI–TOF–MS, ELISA | Sun et al, 2010b |
| OPN (biomineralisation/cell adhesion) | P10451 | Plasma | ↑HCC:CLD/CON | Val: None | Disc: 17 HCC, 18 LC Val: 131 HCC, 76 LC, 52 CHCV/HBV, 53 HCON | HCV/HBV | 0.76 | — | LC–ESI–MS/MS with 2D nHPLC-coupled LTQ OrbiTrap | Shang et al, 2012 |
| Plasma | ↑HCC:CON/LC | Some Histo. % not stated | 30 HCC, 30 LC, 20 HCON | HCV | 0.92 | 4.33† | ELISA | El-Din Bessa et al, 2010 | ||
| PON-1 (antioxidant/hydrolase) | P27169 | Serum | ↑ HCC:CON | 100% of HCCs | 5 HCC, 5 HCON | HCV/HBV | — | — | 2DE LC–ESI–MS/MS | Yang et al, 2007a |
| Plasma | ↓ HCC:CON | Disc 24.2% Val 4.8% | Disc: 120 HCC, 99 LC, 120 HCON Val: 21 HCC, 6 LC, 18 ASC, 10 HCON | HBV/aflatoxin | — | 0.63** | QTOF–nUPLC–ESI–MS | Fye et al, 2013 | ||
| Prx-II (antioxidant/oxidoreductase/peroxidase) | P32119 | Plasma | ↑ LC:CON | 100% of cases | Disc: 27 HBV, 7 HCON Val: 68 HBV, 42 HCON | HBV | 1.00 | — | MALDI–TOF–MS | Lu et al, 2010 |
| Tissue | ↑ HCC:CON | 100% of HCCs | 6 HCC, 6 non-malignant adjacent tissue | HBV | — | 2.1† | 2DE-coupled MALDI–TOF–MS | Matos et al, 2009 | ||
| SAA (AP and innate response/cell adhesion) | P0DJI8 | Serum | ↑ HCC:CON | No information | 67 HCC, 53 CHBV, 44 HCON | HBV | — | 4.5† | 2DE MALDI–TOF–MS, SELDI–TOF–MS | He et al, 2008 |
| Serum | HCC only | 100% of HCCs | 5 HCC, 5 HCON | HCV/HBV | — | — | 2DE LC ESI–MS/MS | Yang et al, 2007a | ||
| SGP-2 (cell death) | Q6LDQ3 | Serum | ↑ HCC:CON | 100% HCCs | Disc: 20 HCC, 20 HCON Val: 5 HCC, 5 HCON | No information | — | — | 2DE MALDI–TOF–MS, WB | Wu et al, 2012 |
| TGM2 (apoptosis/transferase) | P21980 | Tissue/serum | ↑ HCC:CON | ALL: 100% of HCCs | Tissue disc: 61 HCC, 61 adjacent non-tumour tissue Val: 109 HCC, 42 HCON | HBV/HCV | — | — | LTQ–FT–MS/MS, ELISA | Sun et al, 2008 |
| TTR (Transport) | P02766 | Serum | ↓ HCC:CON | No information | Disc: 20 HCC, 20 HBV, 20 HCON Val: 20 HCC, 20 HBV, 20 HCON | HBV | — | — | 2DE and MALDI–TOF–MS/MS | Feng et al, 2005 |
| Serum | ↓ HCC:CON | 100% of HCCs | 5 HCC, 5 HCON | HCV/HBV | — | — | 2DE LC–ESI–MS/MS | Yang et al, 2007a | ||
| VIM (host-virus interaction) | P08670 | Tissue/serum | ↑HCC:LC/CON | Disc: 100% of HCCs Val: No information | Disc (tissue): 40 HCC, 36 CON Val: 88 HCC, 64 CON | HBV | 0.69 | 3.27† | 2DE MALDI–TOF/TOF, ELISA | Sun et al, 2010a |
| VIT (cell adhesion) | P04004 | Serum | ↑ HCC:LC | 91% of HCCs | 44 HCC, 38 LC | No information | 0.85 | — | SELDI–TOF–MS | Paradis et al, 2005 |
| Serum | ↓ HCC:CON | No information | 10 HCC, 10 HCON | No information | — | — | SID–MRM–MS | Zhao et al, 2010 | ||
| Plasma | ↑ HCC:CON | No information | 10 HCC; 30 HCON/LC/HBV | HBV | 0.54 | 1.1ns | Lectin MRM–UPLC–ESI–MS | Ahn et al, 2012b | ||
| Plasma | ↓ HCC:CON | No information | 28 HCC, 10 HCON | No information | 0.98 | — | nUPLC–ESI–QTOF–MS and QQQ | Lee et al, 2011 | ||
Abbreviations: 2D=two dimensional; 2DE=two-dimensional electrophoresis; ASC=asymptomatic carrier; AUROC=area under receiver operating characteristics; CCa=cholangiocarcinoma; CHBV/CHCV=chronic HBV or HCV; CLD=chronic liver disease; ELISA=enzyme-linked immunosorbent assay; ESI=electrospray ionisation; HBV=hepatitis B virus; HCC=hepatocellular carcinoma; HCON=healthy control; HCV=hepatitis C virus; Histo=histology; ICAT=isotope-coded affinity tag; LB=liver biopsy; LC=liquid chromatography; MALDI=matrix-assisted laser desorption/ionization; MRM=multiple reaction monitoring; MS=mass spectrometry; n/r=not recorded; nanoHPLC=nano high-performance liquid chromatography; nUPLC=nano ultra-performance liquid chromatography; QQQ=triple quadropole mass spectrometer; qTOF=quadrupole time of flight; WB=western blot; SELDI=surface-enhanced laser desorption/ionization; TOF=time-of-flight. *P<0.05; **P<0.01; ns, non-significant; †, no P-value reported.
Challenging discrepancies
This review identifies 31 individual proteins as showing differential signatures associated with a stage of liver disease. Each of these have undergone at least one level of validation whether in the context of the same publication or in an independent report. Overall, the direction of the alterations in protein expression are uniform, there however are some specific proteins, namely alpha-1-acid glycoprotein, afamin, complement component 3, and haptoglobin-related protein that demonstrate discrepancies in behaviour in selected case–control groupings. Alpha-1-acid glycoprotein, for example, is reported to be downregulated in HCC's in comparison to cirrhotics by one group (Lee et al, 2011) and upregulated in HCC's vs LC's by another (Kang et al, 2010). Complement component 3 shows a similar discrepancy in two studies (Steel et al, 2003; Ahn et al, 2012b). In each of these cases, key differences related to HCC assessment, sample pretreatment and MS platform used are likely contributors to the lack of consistency observed. Numerous publications included in this review and directly implicated in these discrepant examples have little to no clinical information on how LC or HCC cases were diagnosed (Ishihara et al, 2011; Ahn et al, 2012b), not to mention an almost uniform lack of staging data. For proteomic analyses in particular, different approaches exist to sample pretreatment, with options to deplete key abundant proteins, enrich for, or exclude (Lee et al, 2011) particular PTMs. With albumin constituting ∼60% of the total protein content of blood (Shen et al, 2004) and forming the backbone of many protein–protein interactions and complexes, its depletion could contribute to the loss of important endogenous signatures. Varying depletion protocol efficiencies may also impact directly on the accuracy of downstream quantitation. Additional separation by isoelectric or electrophoretic analysis according to molecular weight or isoelectric point whether ‘in' or ‘off-gel' can be applied to neat or pre-treated samples meaning that the final matrix injected into the MS instrument may already be severely biased towards identification of selected protein subtypes or sizes. This phenomenon along with varying MS platforms and ionisation methods will all contribute to the differences observed in shortlisted marker profiles at final analyses. Thus, the quantitative data assembled, though important cannot always be taken as conclusively reflective of endogenous expression. This can only be adequately assessed by targeted measurement of whole proteins using highly specific antibody-based methods or absolute quantitation by S/MRM MS analysis of unique peptide sequences in well-characterised sample cohorts.
Glycosylation and HCC
An area of particular focus in terms of identifying unique exploitable changes for differential CLD diagnosis are those which occur in the glycosylation of many proteins during liver disease progression (Blomme et al, 2009). Glycosylation is essential to the tertiary and quaternary structures of many proteins bearing direct impact on their solubility and transport. The carbohydrate moieties bound to these modified proteins can be directly impacted by changes in host immunity and carcinogenesis. The surface of hepatocytes contain various receptors that bind proteins via their carbohydrate groups; when changes to the liver surface occur as a result of fibrosis, cirrhosis, or HCC, alterations in receptor distribution can lead to the accumulation of certain glycoproteins in the blood, which can be picked up by methods such as MS. As asparagine-linked (N-linked) glycosylation is the predominant form found in human sera, much research has been focussed on this particular subclass of glycoproteins. Investigations looking at the glycosylation patterns of key serum proteins associated with HCC and LC reports evidence of hyperfucosylation for 19 glycoproteins (Comunale et al, 2006) in the context of liver disease. Mass spectrometric assays conducted on lectin-enriched samples are seen to represent a sizable fraction of the protein markers identified in this review. Fucosylated haemopexin stands out as one of the highest performing candidate markers for HCC diagnosis with a reported AUC of 0.95 (Comunale, 2009). Numerous additional publications using lectin enrichment steps prior to MS analysis have identified various glycosylated proteins such as alpha-1-acid glycoprotein 1 (A1AG1), AACT, Apo L1, Apo J (Ahn et al, 2012a, 2012b), and several complement proteins as differentially expressed in HCC vs other disease stages. The multiply reported decline in the blood levels of various isoforms of the apolipoproteins involved in lipid and cholesterol metabolism have also been demonstrated to hold significant association with HCC.
Critical assessment and performance comparisons
In the presentation of candidates as potential biomarkers for HCC or any other condition; there has to be a widely applicable tool of measurement offering universal assessment of the potential usefulness and validity of a marker. Area under the curve reports with its achievable sensitivity and specificity at a chosen cut-off and fold change statistics form the most widely employed tools, and have become the benchmarks used for comparative assessment of candidates. Few studies report both AUC and fold change statistics as is seen in the comprehensive tables presented (Tables 2 and 8). As AFP is the only established non-invasive tool used for HCC diagnosis – all biomarker studies are in effect in competition with its performance. One major gap, which exists in making comparative assessments with AFP within and across studies is that oftentimes the test forms an intrinsic part of the initial diagnostic profile used to classify subjects into the various clinical groups under evaluation. Without direct comparison of AFP and the gold standard in HCC diagnosis of liver biopsy within the same subject populations, no true evaluation of AFP performance can be accepted as an accurate reference point. In the absence of this, most publications are either mute on this point or address it by basing their performance evaluations on reported estimates for AFP performance often ranging between 0.6 and 0.7 with some dipping lower (Giannini et al, 2012).
Discussion and conclusion
This review serves as a focus for summarizing and highlighting metabonomic and proteomic literature on biomarkers for HCC in the urine and blood. A multitude of biomarker candidates have been proposed by comparative 1H NMR or MS analysis of urine and/or blood from HCC patients and various CLD groups. A review of these studies has shown that noteworthy changes occur at both the metabolite and protein levels, which correlate with disease aetiology and/or progression. Of significant note however are the multitudes of candidates, which do not appear to be reproducible but nevertheless weigh heavily on literature reports. These are vast in number and were filtered from this review, as they detracted from its stated aims. In order to move forward and accelerate the conversion of biomarker candidates into routine and robust tests to be used in clinics and the field, particularly in the developing world, this area of research must be held to a minimum standard of uniformity (Table 9) and from this facilitate a momentum shift from discovery to validation-based experiments. A noteworthy example is a recently conducted imaging MS study, which validated an altered phospholipid composition in HCC tumour tissues by transcriptomic analyses, thereby identifying LPCAT1 as a potential target molecule to inhibit HCC progression. A key limitation identified from the amalgamation of these reports, which is of particular significance in investigations targeting metabolites is the limited availability of descriptive statistics such as ROC analyses and fold change ratios offering quantitative measures of assessing the performance of a putative marker. Also missing in many studies are any efforts at validating the proposed biomarkers in distinct population sets. In both the proteomic and metabonomic papers reviewed, <15% of publications included any attempts at independent validation of results. Without a consistent ability to reproduce primary discovery results in diverse populations, biomarkers will not accelerate on the pipeline towards serious consideration.
Table 9. Recommendations for future metabonomic and proteomic studies on HCC.
| Key point | Recommendation |
|---|---|
| Use of published guidelines to inform case definitions | CLD diagnosis should be made using agreed international guidelines (e.g., EASL and AASLD). New guidelines must be developed/adapted for areas of HCC endemicity in the developing world |
| Measure of total protein expression | Use commercial assays (e.g., QuantiPro BCA) to quantify and normalise total protein expression in sampled blood |
| Prandial state | 8–12 h Pre-prandial |
| Physical exercise | Should be avoided immediately prior to sample collection |
| Overall liver function | Assessment of Child-Pugh score: A: Hepatic compensated B: Slightly decompensated liver state C: Hepatic decompensated |
| Tumour size and nodularity | Tumour staging, for example, TMN classification |
| Comorbidities | Clinical assessment of cirrhosis in the background of HCC Clinical assessment of renal impairment, for example, kidney function tests such as glomerular filtration rate (important for urinary analyses) |
| Tumour size and nodularity | Tumour staging, for example, TMN classification |
| Validation of the diagnostic model | Inclusion of external validation cohorts, for example, early HCCs, different HCC aetiologies, and tumour controls |
| Performance assessment of the diagnostic model | AUROC statistic, enables direct comparison to other models and AFP |
Abbreviations: AASLD=American Association for the Study of Liver Diseases; AFP=alpha-fetoprotein; AUROC=area under receiver operating characteristics; BCA=bicinchoninic acid; CLD=chronic liver disease; EASL=European Association for the Study of the Liver; HCC=hepatocellular carcinoma; TNM=tumour, nodes, and metastasis.
It is recognised that routine liver biopsy assessment of suspected HCCs is not readily available in many populations where the condition is endemic. The relevance of biopsy sample retrieval and definitive tumour staging however, cannot be overstated. For any new HCC diagnostic test to have a direct impact on reducing mortality from end-stage liver disease, it must be able to discern those at highest risk of progression to HCC from a background of LC or fibrosis as well as reflect early changes in the liver related to tumourigenesis. Most of the markers included in this review have been proposed from heterogeneous case groups where considerations such as background health of liver, tumour stage, and severity of cirrhosis or fibrosis have not been assessed or are not reported. Thus, when these candidates are proposed for validation studies, accurate case profiles must be presented that clearly stratify HCC stages in order to home in on how candidates perform in the diagnostic groups most difficult to distinguish. Best practice would warrant the clear and comprehensive distinction of these at the discovery stage but this is yet to be reflected in current practice. Some metabolites are correlated with the degree of cirrhosis or the hepatic compensation status of patients. For example, the metabolic profile of patients with small HCC tumours on a background of severe cirrhosis differs significantly from late HCC with minor background cirrhosis (Nahon et al, 2012b). Group heterogeneity can thus bias results and impact directly on the range of proteins, metabolites, or pathways detected as significantly different. Such comprehensive descriptions and assessments are largely absent from current HCC biomarker literature and must be incorporated and prioritised in order to identify biomarker signatures of the highest impact.
Although the majority of studies presented provide detailed demographic data for participants, there was an observed lack of consistency in which clinical indices were reported in published works. Reports on liver biopsy rates and AFP levels are arguably of primary relevance to any HCC biomarker studies yet figures for these were present only in a fraction of papers. For AFP in particular, the cut-off taken as indicative of HCC, where reported, was still highly variable, ranging from 20 to 500 ng ml−1 depending on the specific study population or chosen guidelines. Alongside this, details on overall liver health, liver compensation status, clinical chemistry tests (e.g., liver enzymes and bilirubin), and the treatment status of participants were at best inconsistently reported. The absence or lack of consideration of these key clinical parameters will undoubtedly have significant implications on participant selection and classification as well as the interpretation of disease-specific signatures. Thus, a key recommendation to be made following this review is the need for some degree of uniformity in the rationale and level of detail forming the basis of case and control selection. Providing detailed case definitions would reduce or at least allow for the contextualisation of marked variations, which exist in the selection of cohorts. This could perhaps contribute to a wider overlap in identified proteins and metabolites associated with CLDs across different investigative platforms and thus offer greater consensus on which targets to validate.
In order to move some of the many suggested HCC biomarker targets forward and accelerate their conversion into routine and robust tests usable in clinics and the field, particularly in the developing world, the momentum of research must now shift from discovery to validation-based experiments – focused both on expression level and mechanistic changes between clinically distinct groups. The populations involved in these studies must be well characterised to dissect biomarkers that are specific for (early) HCC and if these are still valid, in the presence of cirrhosis or other comorbidities. Below, we summarise key points that can significantly bias the results of a biomarker study on HCC. At the same time, we give recommendations, which with careful consideration could raise the likelihood of successful conversion of proposed biomarkers in the field of HCC research.
Although the consideration of these points and recommendations will not lead to uniform impact for all researchers in this field due to the diversity of challenges faced, it is however important that they are recognised as highly relevant and begin to be discussed on a larger scale.
References
- Ahn YH, Ji ES, Shin PM, Kim KH, Kim YS, Ko JH, Yoo JS. A multiplex lectin-channel monitoring method for human serum glycoproteins by quantitative mass spectrometry. Analyst. 2012;137:691–703. doi: 10.1039/c1an15775b. [DOI] [PubMed] [Google Scholar]
- Ahn YH, Shin PM, Oh NR, Park GW, Kim H, Yoo JS. A lectin-coupled, targeted proteomic mass spectrometry (MRM MS) platform for identification of multiple liver cancer biomarkers in human plasma. J Proteomics. 2012;75:5507–5515. doi: 10.1016/j.jprot.2012.06.027. [DOI] [PubMed] [Google Scholar]
- Ang IL, Poon TC, Lai PB, Chan AT, Ngai SM, Hui AY, Johnson PJ, Sung JJ. Study of serum haptoglobin and its glycoforms in the diagnosis of hepatocellular carcinoma: a glycoproteomic approach. J Proteome Res. 2006;5:2691–2700. doi: 10.1021/pr060109r. [DOI] [PubMed] [Google Scholar]
- Baniasadi H, Gowda GA, Gu H, Zeng A, Zhuang S, Skill N, Maluccio M, Raftery D. Targeted metabolic profiling of hepatocellular carcinoma and hepatitis C using LC-MS/MS. Electrophoresis. 2013;34:2910–2917. doi: 10.1002/elps.201300029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baptissart M, Vega A, Maqdasy S, Caira F, Baron S, Lobaccaro JM, Volle DH. Bile acids: from digestion to cancers. Biochimie. 2013;95:504–517. doi: 10.1016/j.biochi.2012.06.022. [DOI] [PubMed] [Google Scholar]
- Bellinghieri G, Santoro D, Calvani M, Mallamace A, Savica V. Carnitine and hemodialysis. Am J Kidney Dis. 2003;41:S116–S122. doi: 10.1053/ajkd.2003.50099. [DOI] [PubMed] [Google Scholar]
- Bertino G, Ardiri A, Malaguarnera M, Malaguarnera G, Bertino N, Calvagno GS. Hepatocellualar carcinoma serum markers. Semin Oncol. 2012;39:410–433. doi: 10.1053/j.seminoncol.2012.05.001. [DOI] [PubMed] [Google Scholar]
- Bertino G, Ardiri AM, Calvagno GS, Bertino N, Boemi PM. Prognostic and diagnostic value of des-gamma-carboxy prothrombin in liver cancer. Drugs News Perspect. 2010;23:498–508. doi: 10.1358/dnp.2010.23.8.1444236. [DOI] [PubMed] [Google Scholar]
- Bertino G, Demma S, Ardiri A, Proiti M, Gruttadauria S, Toro A, Malaguarnera G, Bertino N, Malaguarnera M, Malaguarnera M, Di Carlo I. Hepatocellular carcinoma: novel molecular targets in carcinogenesis for future therapies. Biomed Res Int. 2014;2014:203693. doi: 10.1155/2014/203693. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Bertino G, Neri S, Bruno CM, Ardiri AM, Calvagno GS, Malaguarnera M, Toro A, Malaguarnera M, Clementi S, Bertino N, Di Carlo I. Diagnostic and prognostic value of alpha-fetoprotein, des-gamma-carboxy prothrombin and squamous cell carcinoma antigen immunoglobulin M complexes in hepatocellular carcinoma. Minerva Med. 2011;102:363–371. [PubMed] [Google Scholar]
- Beyoğlu D, Imbeaud S, Maurhofer O, Bioulac-Sage P, Zucman-Rossi J, Dufour JF, Idle JR. Tissue metabolomics of hepatocellular carcinoma: tumor energy metabolism and the role of transcriptomic classification. Hepatology. 2013;58:229–238. doi: 10.1002/hep.26350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biondi A, Malaguarnera G, Vacante M, Berretta M, D'Agata V, Malaguarnera M, Basile F, Drago F, Bertino G. Elevated serum levels of chromogranin A in hepatocellular carcinoma. BMC Surg. 2012;12 Suppl 1:S7. doi: 10.1186/1471-2482-12-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomme B, Van Steenkiste C, Callewaert N, Van Vlierberghe H. Alteration of protein glycosylation in liver diseases. J Hepatol. 2009;50:592–603. doi: 10.1016/j.jhep.2008.12.010. [DOI] [PubMed] [Google Scholar]
- Bouatra S, Aziat F, Mandal R, Guo AC, Wilson MR, Knox C, Bjorndahl TC, Krishnamurthy R, Saleem F, Liu P, Dame ZT, Poelzer J, Huynh J, Yallou FS, Psychogios N, Dong E, Bogumil R, Roehring C, Wishart DS. The human urine metabolome. PLoS One. 2013;8:e73076. doi: 10.1371/journal.pone.0073076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle P, Levin B.2008. World cancer report 2008.
- Bruix J, Sherman M, American Association for the Study of Liver Diseases Management of hepatocellular carcinoma: an update. Hepatology. 2011;53:1020–1022. doi: 10.1002/hep.24199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese V, Giuffrida Stella AM, Calvani M, Butterfield DA. Acetylcarnitine and cellular stress response: roles in nutritional redox homeostasis and regulation of longevity genes. J Nutr Biochem. 2006;17:73–88. doi: 10.1016/j.jnutbio.2005.03.027. [DOI] [PubMed] [Google Scholar]
- Cave MC, Hurt RT, Frazier TH, Matheson PJ, Garrison RN, McClain CJ, McClave SA. Obesity, inflammation, and the potential application of pharmaconutrition. Nutr Clin Pract. 2008;23:16–34. doi: 10.1177/011542650802300116. [DOI] [PubMed] [Google Scholar]
- Chen F, Xue J, Zhou L, Wu S, Chen Z. Identification of serum biomarkers of hepatocarcinoma through liquid chromatography/mass spectrometry-based metabonomic method. Anal Bioanal Chem. 2011;401:1899–1904. doi: 10.1007/s00216-011-5245-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Wang W, Lv S, Yin P, Zhao X, Lu X, Zhang F, Xu G. Metabonomics study of liver cancer based on ultra performance liquid chromatography coupled to mass spectrometry with HILIC and RPLC separations. Anal Chim Acta. 2009;650:3–9. doi: 10.1016/j.aca.2009.03.039. [DOI] [PubMed] [Google Scholar]
- Chen S, Kong H, Lu X, Li Y, Yin P, Zeng Z, Xu G. Pseudotargeted metabolomics method and its application in serum biomarker discovery for hepatocellular carcinoma based on ultra high-performance liquid chromatography/triple quadrupole mass spectrometry. Anal Chem. 2013;85:8326–8333. doi: 10.1021/ac4016787. [DOI] [PubMed] [Google Scholar]
- Chen S, Yin P, Zhao X, Xing W, Hu C, Zhou L, Xu G. Serum lipid profiling of patients with chronic hepatitis B, cirrhosis, and hepatocellular carcinoma by ultra fast LC/IT-TOF MS. Electrophoresis. 2013;34:2848–2856. [PubMed] [Google Scholar]
- Chen T, Xie G, Wang X, Fan J, Qiu Y, Zheng X, Qi X, Cao Y, Su M, Wang X, Xu LX, Yen Y, Liu P, Jia W. Serum and urine metabolite profiling reveals potential biomarkers of human hepatocellular carcinoma. Mol Cell Proteomics. 2011;10:M110 004945. doi: 10.1074/mcp.M110.004945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Xu Z, Kong H, Chen N, Chen J, Zhou L, Wang F, Dong Y, Zheng S, Chen Z, Xu G, Li L. Differences between the metabolic profiles of decompensated and compensated cirrhosis patients with hepatitis B virus infections under high-performance liquid chromatography-mass spectrometry. Metabolomics. 2012;8 (5:845–853. [Google Scholar]
- Comunale MA, Wang M, Hafner J, Krakover J, Rodemich L, Kopenhaver B, Long RE, Junaidi O, Bisceglie AM, Block TM, Mehta AS. Identification and development of fucosylated glycoproteins as biomarkers of primary hepatocellular carcinoma. J Proteome Res. 2009;8 (2:595–602. doi: 10.1021/pr800752c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comunale MA, Lowman M, Long RE, Krakover J, Philip R, Seeholzer S, Evans AA, Hann HW, Block TM, Mehta AS. Proteomic analysis of serum associated fucosylated glycoproteins in the development of primary hepatocellular carcinoma. J Proteome Res. 2006;5:308–315. doi: 10.1021/pr050328x. [DOI] [PubMed] [Google Scholar]
- Dodson WL, Sachan DS, Krauss S, Hanna W. Alterations of serum and urinary carnitine profiles in cancer patients: hypothesis of possible significance. J Am Coll Nutr. 1989;8:133–142. doi: 10.1080/07315724.1989.10720288. [DOI] [PubMed] [Google Scholar]
- El-Din Bessa SS, Elwan NM, Suliman GA, El-Shourbagy SH. Clinical significance of plasma osteopontin level in Egyptian patients with hepatitis C virus-related hepatocellular carcinoma. Arch Med Res. 2010;41:541–547. doi: 10.1016/j.arcmed.2010.10.007. [DOI] [PubMed] [Google Scholar]
- El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- European Association for the Study of the Liver. European Organisation for Research and Treatment of Cancer EASL-EORTC clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol. 2012;56:908–943. doi: 10.1016/j.jhep.2011.12.001. [DOI] [PubMed] [Google Scholar]
- Feng JT, Liu YK, Song HY, Dai Z, Qin LX, Almofti MR, Fang CY, Lu HJ, Yang PY, Tang ZY. Heat-shock protein 27: a potential biomarker for hepatocellular carcinoma identified by serum proteome analysis. Proteomics. 2005;5:4581–4588. doi: 10.1002/pmic.200401309. [DOI] [PubMed] [Google Scholar]
- Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- Flanagan JL, Simmons PA, Vehige J, Willcox MD, Garrett Q. Role of carnitine in disease. Nutr Metab. 2010;7:30. doi: 10.1186/1743-7075-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fye HK, Wright-Drakesmith C, Kramer HB, Camey S, Nogueira da Costa A, Jeng A, Bah A, Kirk GD, Sharif MI, Ladep NG, Okeke E, Hainaut P, Taylor-Robinson SD, Kessler BM, Mendy ME. Protein profiling in hepatocellular carcinoma by label-free quantitative proteomics in two west african populations. PLoS One. 2013;8:e68381. doi: 10.1371/journal.pone.0068381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadaleta RM, van Erpecum KJ, Oldenburg B, Willemsen EC, Renooij W, Murzilli S, Klomp LW, Siersema PD, Schipper ME, Danese S, Penna G, Laverny G, Adorini L, Moschetta A, van Mil SW. Farnesoid X receptor activation inhibits inflammation and preserves the intestinal barrier in inflammatory bowel disease. Gut. 2011;60:463–472. doi: 10.1136/gut.2010.212159. [DOI] [PubMed] [Google Scholar]
- Ganti S, Taylor SL, Kim K, Hoppel CL, Guo L, Yang J, Evans C, Weiss RH. Urinary acylcarnitines are altered in human kidney cancer. Int J Cancer. 2012;130:2791–2800. doi: 10.1002/ijc.26274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Lu Q, Liu X, Cong H, Zhao L, Wang H, Lin D. Application of 1H NMR-based metabonomics in the study of metabolic profiling of human hepatocellular carcinoma and liver cirrhosis. Cancer Sci. 2009;100:782–785. doi: 10.1111/j.1349-7006.2009.01086.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannini EG, Marenco S, Borgonovo G, Savarino V, Farinati F, Del Poggio P, Rapaccini GL, Anna Di Nolfo M, Benvegnù L, Zoli M, Borzio F, Caturelli E, Chiaramonte M, Trevisani F, Italian Liver Cancer (ITA.LI.CA) group Alpha-fetoprotein has no prognostic role in small hepatocellular carcinoma identified during surveillance in compensated cirrhosis. Hepatology. 2012;56:1371–1379. doi: 10.1002/hep.25814. [DOI] [PubMed] [Google Scholar]
- He QY, Zhu R, Lei T, Ng MY, Luk JM, Sham P, Lau GK, Chiu JF. Toward the proteomic identification of biomarkers for the prediction of HBV related hepatocellular carcinoma. J Cell Biochem. 2008;103:740–752. doi: 10.1002/jcb.21443. [DOI] [PubMed] [Google Scholar]
- Heuberger W, Berardi S, Jacky E, Pey P, Krähenbühl S. Increased urinary excretion of carnitine in patients treated with cisplatin. Eur J Clin Pharmacol. 1998;54:503–508. doi: 10.1007/s002280050504. [DOI] [PubMed] [Google Scholar]
- Hoi PM, Hiley CR. Vasorelaxant effects of oleamide in rat small mesenteric artery indicate action at a novel cannabinoid receptor. Br J Pharmacol. 2006;147:560–568. doi: 10.1038/sj.bjp.0706643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppel C. The role of carnitine in normal and altered fatty acid metabolism. Am J Kidney Dis. 2003;41:S4–S12. doi: 10.1016/s0272-6386(03)00112-4. [DOI] [PubMed] [Google Scholar]
- Hsieh SY, He JR, Yu MC, Lee WC, Chen TC, Lo SJ, Bera R, Sung CM, Chiu CT. Secreted ERBB3 isoforms are serum markers for early hepatoma in patients with chronic hepatitis and cirrhosis. J Proteome Res. 2011;10:4715–4724. doi: 10.1021/pr200519q. [DOI] [PubMed] [Google Scholar]
- Huang Q, Tan Y, Yin P, Ye G, Gao P, Lu X, Wang H, Xu G. Metabolic characterization of hepatocellular carcinoma using non-targeted tissue metabolomics. Cancer Res. 2013;73:4992–5002. doi: 10.1158/0008-5472.CAN-13-0308. [DOI] [PubMed] [Google Scholar]
- Ishihara T, Fukuda I, Morita A, Takinami Y, Okamoto H, Nishimura S, Numata Y. Development of quantitative plasma N-glycoproteomics using label-free 2-D LC-MALDI MS and its applicability for biomarker discovery in hepatocellular carcinoma. J Proteomics. 2011;74:2159–2168. doi: 10.1016/j.jprot.2011.06.010. [DOI] [PubMed] [Google Scholar]
- Kang X, Sun L, Guo K, Shu H, Yao J, Qin X, Liu Y. Serum protein biomarkers screening in HCC patients with liver cirrhosis by ICAT-LC-MS/MS. J Cancer Res Clin Oncol. 2010;136:1151–1159. doi: 10.1007/s00432-010-0762-6. [DOI] [PubMed] [Google Scholar]
- Kanmura S, Uto H, Sato Y, Kumagai K, Sasaki F, Moriuchi A, Oketani M, Ido A, Nagata K, Hayashi K, Stuver SO, Tsubouchi H. The complement component C3a fragment is a potential biomarker for hepatitis C virus-related hepatocellular carcinoma. J Gastroenterol. 2010;45:459–467. doi: 10.1007/s00535-009-0160-5. [DOI] [PubMed] [Google Scholar]
- Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, Britt EB, Fu X, Wu Y, Li L, Smith JD, DiDonato JA, Chen J, Li H, Wu GD, Lewis JD, Warrier M, Brown JM, Krauss RM, Tang WH, Bushman FD, Lusis AJ, Hazen SL. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med. 2013;19:576–585. doi: 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krähenbühl S. Carnitine metabolism in chronic liver disease. Life Sci. 1996;59:1579–1599. doi: 10.1016/0024-3205(96)00343-8. [DOI] [PubMed] [Google Scholar]
- LaRusso NF, Hoffman NE, Korman MG, Hofmann AF, Cowen AE. Determinants of fasting and postprandial serum bile acid levels in healthy man. Am J Dig Dis. 1978;23:385–391. doi: 10.1007/BF01072919. [DOI] [PubMed] [Google Scholar]
- Lau SH, Sham JS, Xie D, Tzang CH, Tang D, Ma N, Hu L, Wang Y, Wen JM, Xiao G, Zhang WM, Lau GK, Yang M, Guan XY. Clusterin plays an important role in hepatocellular carcinoma metastasis. Oncogene. 2006;25:1242–1250. doi: 10.1038/sj.onc.1209141. [DOI] [PubMed] [Google Scholar]
- Lee IN, Chen CH, Sheu JC, Lee HS, Huang GT, Chen DS, Yu CY, Wen CL, Lu FJ, Chow LP. Identification of complement C3a as a candidate biomarker in human chronic hepatitis C and HCV-related hepatocellular carcinoma using a proteomics approach. Proteomics. 2006;6:2865–2873. doi: 10.1002/pmic.200500488. [DOI] [PubMed] [Google Scholar]
- Lee JY, Kim JY, Park GW, Cheon MH, Kwon KH, Ahn YH, Moon MH, Lee HJ, Paik YK, Yoo JS. Targeted mass spectrometric approach for biomarker discovery and validation with nonglycosylated tryptic peptides from N-linked glycoproteins in human plasma. Mol Cell Proteomics. 2011;10:M111 009290. doi: 10.1074/mcp.M111.009290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkous AG, Yazlovitskaya EM, Hallahan DE. Cytosolic phospholipase A2 and lysophospholipids in tumor angiogenesis. J Natl Cancer Inst. 2010;102:1398–1412. doi: 10.1093/jnci/djq290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livraghi T, Meloni F, Di Stasi M, Rolle E, Solbiati L, Tinelli C, Rossi S. Sustained complete response and complications rates after radiofrequency ablation of very early hepatocellular carcinoma in cirrhosis: is resection still the treatment of choice. Hepatology. 2008;47:82–89. doi: 10.1002/hep.21933. [DOI] [PubMed] [Google Scholar]
- Llovet JM, Bruix J. Systematic review of randomized trials for unresectable hepatocellular carcinoma: chemoembolization improves survival. Hepatology. 2003;37:429–442. doi: 10.1053/jhep.2003.50047. [DOI] [PubMed] [Google Scholar]
- Lu Y, Liu J, Lin C, Wang H, Jiang Y, Wang J, Yang P, He F. Peroxiredoxin 2: a potential biomarker for early diagnosis of hepatitis B virus related liver fibrosis identified by proteomic analysis of the plasma. BMC Gastroenterol. 2010;10:115. doi: 10.1186/1471-230X-10-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malaguarnera G, Bertino G, Vacante M, Malaguarnera M. Hepatocellular carcinoma markers in the omics era: the glycomic analysis. Hepatobiliary Surg Nutr. 2014;3:407–409. doi: 10.3978/j.issn.2304-3881.2014.07.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malaguarnera M. Carnitine derivatives: clinical usefulness. Curr Opin Gastroenterol. 2012;28:166–176. doi: 10.1097/MOG.0b013e3283505a3b. [DOI] [PubMed] [Google Scholar]
- Martínez-González D, Bonilla-Jaime H, Morales-Otal A, Henriksen SJ, Velázquez-Moctezuma J, Prospéro-García O. Oleamide and anandamide effects on food intake and sexual behavior of rats. Neurosci Lett. 2004;364:1–6. doi: 10.1016/j.neulet.2004.03.080. [DOI] [PubMed] [Google Scholar]
- Masuzaki R, Karp SJ, Omata M. New serum markers of hepatocellular carcinoma. Semin Oncol. 2012;39:434–439. doi: 10.1053/j.seminoncol.2012.05.009. [DOI] [PubMed] [Google Scholar]
- Matos JM, Witzmann FA, Cummings OW, Schmidt CM. A pilot study of proteomic profiles of human hepatocellular carcinoma in the United States. J Surg Res. 2009;155:237–243. doi: 10.1016/j.jss.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mederacke I, Mencin A, Lefkowitch Jay H, Sartor RB, Dapito Dianne H, Rabadan R, Jang M-K, Pradere J-P, Caviglia Jorge M, Khiabanian H, Schwabe Robert F, Friedman R, Bower M, Gwak G-Y, Bataller R, Adeyemi A.2012. Promotion of Hepatocellular Carcinoma by the Intestinal Microbiota and TLR4. [DOI] [PMC free article] [PubMed]
- Michitaka K, Hiraoka A, Kume M, Uehara T, Hidaka S, Ninomiya T, Hasebe A, Miyamoto Y, Ichiryu M, Tanihira T, Nakahara H, Ochi H, Tanabe A, Uesugi K, Tokumoto Y, Mashiba T, Abe M, Hiasa Y, Matsuura B, Onji M. Amino acid imbalance in patients with chronic liver diseases. Hepatol Res. 2010;40:393–398. doi: 10.1111/j.1872-034X.2009.00614.x. [DOI] [PubMed] [Google Scholar]
- Morgan TR, Mandayam S, Jamal MM. Alcohol and hepatocellular carcinoma. Gastroenterology. 2004;127:S87–S96. doi: 10.1053/j.gastro.2004.09.020. [DOI] [PubMed] [Google Scholar]
- Morita Y, Sakaguchi T, Ikegami K, Goto-Inoue N, Hayasaka T, Hang VT, Tanaka H, Harada T, Shibasaki Y, Suzuki A, Fukumoto K, Inaba K, Murakami M, Setou M, Konno H. Lysophosphatidylcholine acyltransferase 1 altered phospholipid composition and regulated hepatoma progression. J Hepatol. 2013;59:292–299. doi: 10.1016/j.jhep.2013.02.030. [DOI] [PubMed] [Google Scholar]
- Na K, Lee EY, Lee HJ, Kim KY, Lee H, Jeong SK, Jeong AS, Cho SY, Kim SA, Song SY, Kim KS, Cho SW, Kim H, Paik YK. Human plasma carboxylesterase 1, a novel serologic biomarker candidate for hepatocellular carcinoma. Proteomics. 2009;9:3989–3999. doi: 10.1002/pmic.200900105. [DOI] [PubMed] [Google Scholar]
- Nahon P, Amathieu R, Triba MN, Bouchemal N, Nault JC, Ziol M, Seror O, Dhonneur G, Trinchet JC, Beaugrand M, Le Moyec L. Identification of serum proton NMR metabolomic fingerprints associated with hepatocellular carcinoma in patients with alcoholic cirrhosis. Clin Cancer Res. 2012;18:6714–6722. doi: 10.1158/1078-0432.CCR-12-1099. [DOI] [PubMed] [Google Scholar]
- Nahon P, Amathieu R, Triba MN, Bouchemal N, Nault JC, Ziol M, Seror O, Dhonneur G, Trinchet JC, Beaugrand M, Le Moyec L. Identification of serum proton NMR metabolomic fingerprints associated with hepatocellular carcinoma in patients with alcoholic cirrhosis. Clin Cancer Res. 2012;18:6714–6722. doi: 10.1158/1078-0432.CCR-12-1099. [DOI] [PubMed] [Google Scholar]
- Saito Y, Oba N, Nishinakagawa S, Mizuguchi Y, Kojima T, Nomura K, Nakatsura T. Identification of β2-microgloblin as a candidate for early diagnosis of imaging-invisible hepatocellular carcinoma in patient with liver cirrhosis. Oncol Rep. 2010;23:1325–1330. doi: 10.3892/or_00000767. [DOI] [PubMed] [Google Scholar]
- Neale G, Lewis B, Weaver V, Panveliwalla D. Serum bile acids in liver disease. Gut. 1971;12:145–152. doi: 10.1136/gut.12.2.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii AI. Protein identification by tandem mass spectrometry and sequence database searching. Methods Mol Biol. 2007;367:87–119. doi: 10.1385/1-59745-275-0:87. [DOI] [PubMed] [Google Scholar]
- Oterdoom LH, Gansevoort RT, Schouten JP, de Jong PE, Gans RO, Bakker SJ. Urinary creatinine excretion, an indirect measure of muscle mass, is an independent predictor of cardiovascular disease and mortality in the general population. Atherosclerosis. 2009;207:534–540. doi: 10.1016/j.atherosclerosis.2009.05.010. [DOI] [PubMed] [Google Scholar]
- Paradis V, Degos F, Dargère D, Pham N, Belghiti J, Degott C, Janeau JL, Bezeaud A, Delforge D, Cubizolles M, Laurendeau I, Bedossa P. Identification of a new marker of hepatocellular carcinoma by serum protein profiling of patients with chronic liver diseases. Hepatology. 2005;41:40–47. doi: 10.1002/hep.20505. [DOI] [PubMed] [Google Scholar]
- Patterson AD, Maurhofer O, Beyoglu D, Lanz C, Krausz KW, Pabst T, Gonzalez FJ, Dufour JF, Idle JR. Aberrant lipid metabolism in hepatocellular carcinoma revealed by plasma metabolomics and lipid profiling. Cancer Res. 2011;71:6590–6600. doi: 10.1158/0008-5472.CAN-11-0885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponz De Leon M, Murphy GM, Dowling RH. Physiological factors influencing serum bile acid levels. Gut. 1978;19:32–39. doi: 10.1136/gut.19.1.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regimbeau JM, Colombat M, Mognol P, Durand F, Abdalla E, Degott C, Degos F, Farges O, Belghiti J. Obesity and diabetes as a risk factor for hepatocellular carcinoma. Liver Transpl. 2004;10:S69–S73. doi: 10.1002/lt.20033. [DOI] [PubMed] [Google Scholar]
- Ressom HW, Xiao JF, Tuli L, Varghese RS, Zhou B, Tsai TH, Ranjbar MR, Zhao Y, Wang J, Di Poto C, Cheema AK, Tadesse MG, Goldman R, Shetty K. Utilization of metabolomics to identify serum biomarkers for hepatocellular carcinoma in patients with liver cirrhosis. Anal Chim Acta. 2012;743:90–100. doi: 10.1016/j.aca.2012.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang S, Plymoth A, Ge S, Feng Z, Rosen HR, Sangrajrang S, Hainaut P, Marrero JA, Beretta L. Identification of osteopontin as a novel marker for early hepatocellular carcinoma. Hepatology. 2012;55:483–490. doi: 10.1002/hep.24703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shariff MI, Gomaa AI, Cox IJ, Patel M, Williams HR, Crossey MM, Thillainayagam AV, Thomas HC, Waked I, Khan SA, Taylor-Robinson SD. Urinary metabolic biomarkers of hepatocellular carcinoma in an Egyptian population: a validation study. J Proteome Res. 2011;10:1828–1836. doi: 10.1021/pr101096f. [DOI] [PubMed] [Google Scholar]
- Shariff MI, Ladep NG, Cox IJ, Williams HR, Okeke E, Malu A, Thillainayagam AV, Crossey MM, Khan SA, Thomas HC, Taylor-Robinson SD. Characterization of urinary biomarkers of hepatocellular carcinoma using magnetic resonance spectroscopy in a Nigerian population. J Proteome Res. 2010;9:1096–1103. doi: 10.1021/pr901058t. [DOI] [PubMed] [Google Scholar]
- Shen Y, Jacobs JM, Camp DG, 2nd, Fang R, Moore RJ, Smith RD, Xiao W, Davis RW, Tompkins RG. Ultra-high-efficiency strong cation exchange LC/RPLC/MS/MS for high dynamic range characterization of the human plasma proteome. Anal Chem. 2004;76:1134–1144. doi: 10.1021/ac034869m. [DOI] [PubMed] [Google Scholar]
- Steel LF, Shumpert D, Trotter M, Seeholzer SH, Evans AA, London WT, Dwek R, Block TM. A strategy for the comparative analysis of serum proteomes for the discovery of biomarkers for hepatocellular carcinoma. Proteomics. 2003;3:601–609. doi: 10.1002/pmic.200300399. [DOI] [PubMed] [Google Scholar]
- Sun S, Poon RT, Lee NP, Yeung C, Chan KL, Ng IO, Day PJ, Luk JM. Proteomics of hepatocellular carcinoma: serum vimentin as a surrogate marker for small tumors (<or=2 cm) J Proteome Res. 2010;9:1923–1930. doi: 10.1021/pr901085z. [DOI] [PubMed] [Google Scholar]
- Sun Y, Mi W, Cai J, Ying W, Liu F, Lu H, Qiao Y, Jia W, Bi X, Lu N, Liu S, Qian X, Zhao X. Quantitative proteomic signature of liver cancer cells: tissue transglutaminase 2 could be a novel protein candidate of human hepatocellular carcinoma. J Proteome Res. 2008;7:3847–3859. doi: 10.1021/pr800153s. [DOI] [PubMed] [Google Scholar]
- Sun Y, Zang Z, Xu X, Zhang Z, Zhong L, Zan W, Zhao Y, Sun L. Differential proteomics identification of HSP90 as potential serum biomarker in hepatocellular carcinoma by two-dimensional electrophoresis and mass spectrometry. Int J Mol Sci. 2010;11:1423–1433. doi: 10.3390/ijms11041423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor LA, Arends J, Hodina AK, Unger C, Massing U. Plasma lyso-phosphatidylcholine concentration is decreased in cancer patients with weight loss and activated inflammatory status. Lipids Health Dis. 2007;6:17. doi: 10.1186/1476-511X-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C, Gioiello A, Noriega L, Strehle A, Oury J, Rizzo G, Macchiarulo A, Yamamoto H, Mataki C, Pruzanski M, Pellicciari R, Auwerx J, Schoonjans K. TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metab. 2009;10:167–177. doi: 10.1016/j.cmet.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C, Pellicciari R, Pruzanski M, Auwerx J, Schoonjans K. Targeting bile-acid signalling for metabolic diseases. Nat Rev Drug Discov. 2008;7:678–693. doi: 10.1038/nrd2619. [DOI] [PubMed] [Google Scholar]
- Wang B, Chen D, Chen Y, Hu Z, Cao M, Xie Q, Chen Y, Xu J, Zheng S, Li L. Metabonomic profiles discriminate hepatocellular carcinoma from liver cirrhosis by ultraperformance liquid chromatography-mass spectrometry. J Proteome Res. 2012;11:1217–1227. doi: 10.1021/pr2009252. [DOI] [PubMed] [Google Scholar]
- Ward DG, Cheng Y, N'Kontchou G, Thar TT, Barget N, Wei W, Martin A, Beaugrand M, Johnson PJ. Preclinical and post-treatment changes in the HCC-associated serum proteome. Br J Cancer. 2006;95:1379–1383. doi: 10.1038/sj.bjc.6603429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei S, Suryani Y, Gowda GA, Skill N, Maluccio M, Raftery D. Differentiating hepatocellular carcinoma from hepatitis C using metabolite profiling. Metabolites. 2012;2:701–716. doi: 10.3390/metabo2040701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Xue R, Dong L, Liu T, Deng C, Zeng H, Shen X. Metabolomic profiling of human urine in hepatocellular carcinoma patients using gas chromatography/mass spectrometry. Anal Chim Acta. 2009;648:98–104. doi: 10.1016/j.aca.2009.06.033. [DOI] [PubMed] [Google Scholar]
- Wu W, Li J, Liu Y, Zhang C, Meng X, Zhou Z. Comparative proteomic studies of serum from patients with hepatocellular carcinoma. J Invest Surg. 2012;25:37–42. doi: 10.3109/08941939.2011.603816. [DOI] [PubMed] [Google Scholar]
- Xiao JF, Varghese RS, Zhou B, Nezami Ranjbar MR, Zhao Y, Tsai TH, Di Poto C, Wang J, Goerlitz D, Luo Y, Cheema AK, Sarhan N, Soliman H, Tadesse MG, Ziada DH, Ressom HW. LC-MS based serum metabolomics for identification of hepatocellular carcinoma biomarkers in Egyptian cohort. J Proteome Res. 2012;11:5914–5923. doi: 10.1021/pr300673x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue R, Lin Z, Deng C, Dong L, Liu T, Wang J, Shen X. A serum metabolomic investigation on hepatocellular carcinoma patients by chemical derivatization followed by gas chromatography/mass spectrometry. Rapid communications in mass spectrometry: RCM. 2008;22:3061–3068. doi: 10.1002/rcm.3708. [DOI] [PubMed] [Google Scholar]
- Yang MH, Tyan YC, Jong SB, Huang YF, Liao PC, Wang MC. Identification of human hepatocellular carcinoma-related proteins by proteomic approaches. Anal Bioanal Chem. 2007;388:637–643. doi: 10.1007/s00216-007-1263-6. [DOI] [PubMed] [Google Scholar]
- Yang Y, Li C, Nie X, Feng X, Chen W, Yue Y, Tang H, Deng F. Metabonomic studies of human hepatocellular carcinoma using high-resolution magic-angle spinning 1H NMR spectroscopy in conjunction with multivariate data analysis. J Proteome Res. 2007;6:2605–2614. doi: 10.1021/pr070063h. [DOI] [PubMed] [Google Scholar]
- Yin P, Wan D, Zhao C, Chen J, Zhao X, Wang W, Lu X, Yang S, Gu J, Xu G. A metabonomic study of hepatitis B-induced liver cirrhosis and hepatocellular carcinoma by using RP-LC and HILIC coupled with mass spectrometry. Mol Biosyst. 2009;5:868–876. doi: 10.1039/b820224a. [DOI] [PubMed] [Google Scholar]
- Zhang A, Sun H, Yan G, Han Y, Ye Y, Wang X. Urinary metabolic profiling identifies a key role for glycocholic acid in human liver cancer by ultra-performance liquid-chromatography coupled with high-definition mass spectrometry. Clin Chim Acta. 2013;418:86–90. doi: 10.1016/j.cca.2012.12.024. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Jia W, Sun W, Jin W, Guo L, Wei J, Ying W, Zhang Y, Xie Y, Jiang Y, He F, Qian X. Combination of improved (18)O incorporation and multiple reaction monitoring: a universal strategy for absolute quantitative verification of serum candidate biomarkers of liver cancer. J Proteome Res. 2010;9:3319–3327. doi: 10.1021/pr9011969. [DOI] [PubMed] [Google Scholar]
- Zhou L, Ding L, Yin P, Lu X, Wang X, Niu J, Gao P, Xu G. Serum metabolic profiling study of hepatocellular carcinoma infected with hepatitis B or hepatitis C virus by using liquid chromatography-mass spectrometry. J Proteome Res. 2012;11:5433–5442. doi: 10.1021/pr300683a. [DOI] [PubMed] [Google Scholar]
- Zhou L, Wang Q, Yin P, Xing W, Wu Z, Chen S, Lu X, Zhang Y, Lin X, Xu G. Serum metabolomics reveals the deregulation of fatty acids metabolism in hepatocellular carcinoma and chronic liver diseases. Anal Bioanal Chem. 2012;403:203–213. doi: 10.1007/s00216-012-5782-4. [DOI] [PubMed] [Google Scholar]