Abstract
Escherichia coli strain 397c carries a temperature-sensitive mutation, rpoC397, that removes the last 50 amino acids of the RNA polymerase β′ subunit and is nonpermissive for plating of bacteriophage P2. P2 gor mutants productively infect 397c and define a new gene, lysC, encoded by a reading frame that extensively overlaps the P2 lysis accessory gene, lysB. The unusual location of lysC with respect to lysB is reminiscent of the Rz/Rz1 lysis gene pair of phage λ. Indeed, coexpression of lysB and lysC complemented the growth defect of λ Rz/Rz1 null mutants, indicating that the LysB/C pair is similar to Rz/Rz1 in both gene arrangement and function. Cells carrying the rpoC397 mutation exhibited an early onset of P2-induced lysis, which was suppressed by the gor mutation in lysC. We propose that changes in host gene expression resulting from the rpoC397 mutation result in changes in the composition of the bacterial cell wall, making the cell more susceptible to P2-mediated lysis and preventing accumulation of progeny phage sufficient for plaque formation.
During bacteriophage infection, the machinery for macromolecular synthesis in the cell is recruited to serve the needs of the virus, and systematic changes in viral gene expression take place in a defined sequence. Bacterial DNA-dependent RNA polymerase (RNAP; subunit composition, a2ββ′ωσ), the enzyme responsible for most host transcription, is a major target of this regulation (14). Thus, mutations in RNAP rpo genes often specifically prevent bacteriophage development. To date, mutations that interfere with bacteriophage development have been identified in genes coding for all RNAP subunits except the smallest subunit, ω (reviewed in reference 34). Some of these mutations define RNAP sites that interact directly with viral regulators (25, 26, 40), while others affect phage gene expression indirectly by altering the properties of RNAP, such as the efficiency of transcription termination (39). The power of phage genetics, allowing isolation of suppressor mutations that overcome the blocks conferred by the changes in RNAP, has been invaluable in the study of these phenomena, and the results have greatly enriched our understanding of the basic processes of transcription (27).
From this perspective, the block against plaque formation for phages P2 (10) and N4 (25) conferred by the rpoC397 mutation has long been provocative. This mutation removes 16 bp close to the end of rpoC, resulting in the replacement of the last 50 amino acids of RNAP subunit β′ with 23 incorrect residues (10). In addition to being nonpermissive for the two phages, Escherichia coli strain 397c, carrying rpoC397, has a gross temperature-sensitive (Ts) growth phenotype. Based on biochemical experiments, the N4 block appears to be due to the loss of a contact between N4 SSB (single-strand DNA binding protein) and β′ that is required for activation of viral late transcription (25). However, the mechanism by which the rpoC397 mutation prevents P2 growth has not been determined. Here, we report genetic and physiological experiments addressing this issue and discuss the surprising results in terms of the timing of host lysis.
MATERIALS AND METHODS
Bacterial strains, plasmids, phages, and molecular cloning.
The strains, plasmids, phages, and oligonucleotides used throughout this work are listed in Table 1. Luria-Bertani (LB) medium (4) was the standard culture medium, supplemented as appropriate where indicated.
TABLE 1.
Bacterial strains, plasmids, phages, and primers used in this study
| Strain, plasmid, bacteriophage, or primer | Genotype/relevant features | Source or reference |
|---|---|---|
| Strains | ||
| E. coli K-12 | ||
| P90A5c | Prototrophic; parent of 397c | Christie et al. (10) |
| 397c | Ts; carries rpoC397 deletion | Christie et al. (10) |
| SA1030 | his strA gal-3 | Adhya and Shapiro (1) |
| AD1600 | his strA gal-3 rho(Ts)15 | Das et al. (13) |
| DH5α | Auxotrophic rK− mK+ | BRL Life Technologies |
| MC4100 | araD139Δ(argF-lac)U169 thiA1 rpsL150 relA1 deoC1 PtsF25 rbsR flbB5301 | Zhang and Young (43) |
| RW4206 | Met+rpoC+ transductant of MG1655 metA | Robert Weisberg |
| RW4204 | Met+rpoC397 transductant of MG1655 metA | Robert Weisberg |
| E. coli C | ||
| C-1a | Prototrophic F− | Sasaki and Bertani (32) |
| C-520 | supD F+; P2 indicator | Sunshine et al. (36) |
| C-4508 | Auxotrophic F− | King et al. (20) |
| C-4517 | C4508 supD | King et al. (20) |
| C-4518 | C4508 supF | King et al. (20) |
| Plasmids | ||
| pCYB2 | Expression vector | New England Biolabs |
| pCYB2lysB+ | This work | |
| pCYB2lysC+ | This work | |
| pCYB2lysC(trl) | This work | |
| pCYB2lysC(gor) | This work | |
| pCYB2lysB+lysC+ | This work | |
| pCYB2lysB+lysC(gor) | This work | |
| pCYB2β′ | rpoC+ expression vector | Nedea et al. (28) |
| pCYB2β′397c | rpoC397 expression vector | This work |
| pBBRV7 | P2 EcoRV fragment (6290-8378) in pUC18/HincII | Ziermann et al. (44) |
| pDEB50 | Delta expression vector with modified T7A1 (PA1/04/03) promoter and lacIq | Christie et al. (9) |
| pFCAT100 | P2 pF-cat expression plasmid | Grambow et al. (15) |
| pGC160 | pUCF4 with wild-type P2 lysis cassette | This work |
| pGC163 | pUCF4 with P2 gor lysis cassette | This work |
| pGCRV7gor-1 | P2 gor-1 EcoRV fragment (6290-8378) in pUC18/HincII | This work |
| pNL130 | P2 BstEII fragment (7455-8862) in pUC9/HincII | Linderoth et al. (24) |
| pSL130 | Ptac cat expression plasmid | Li et al. (22) |
| pTG257 | Ogr expression vector with modified T7A1 (PA1/04/03) promoter and lacIq | This work |
| pTG605 | pNL130 with lysC(Am) mutation | This work |
| pUC18 | Cloning vector | Norrander et al. (29) |
| pUCF4 | pUC8 carrying P2 pF promoter | Christie and Calendar (12) |
| pRG1 | pACYC177/PstI with 1.2-kb PstI fragment carrying lacIq | Robert Garcea |
| Bacteriophages | ||
| P2 vir-22 | Immunity insensitive | Chattoraj and Inman (8) |
| P2 vir-22 gor-1 | Spontaneous mutant; plates on AD1600, 397c | This work |
| P2 vir-22 trl-21 | Spontaneous mutant, plates on 397c | This work |
| P2 vir-22 trl-22 | Spontaneous mutant, plates on 397c | This work |
| P2 vir-1 | Clear plaque mutant | Bertani (5) |
| P2 vir-1 R(Am)3 | Tail assembly defect | Lindahl (23) |
| P2 vir-1 lysC(Am) | This work | |
| 186 c(Ts) | Baldwin et al. (2) | |
| Hy5 | P2/186 hybrid | Hocking and Egan (17) |
| λ | stf::cat::tfa cI857 | Zhang and Young (43) |
| λRz(Am) Rz1+ | Zhang and Young (43) | |
| λRz+Rz1(Am) | Zhang and Young (43) | |
| λRz(Am) Rz1(Am) | Zhang and Young (43) | |
| Primersa | ||
| MR2 | GCACGCCAGTGATTGTC | Linderoth et al. (24) |
| 28am | ACGGCGATTTATAGGCCGATATCCGG | This work |
| lysB_Up | CATATGTCAAGGCTGATGATTG | This work |
| lysC_Up | CATATGAGAACGAAGATTTTCG | This work |
| lysC_Down | CTCGAGTCAGTCAGCGCCCTGCGCA | This work |
| Lys3 | GAACTGCAGTCAATCCACGAATATCCGCAG | This work |
| Lys5 | CGAGGATCCGTCAATCTGTGGGAGTAAC | This work |
Primer sequences are given 5′→3′.
P2 lysC, or lysB/lysC, lysB/lysC(gor) or lysB/lysC(Am) pairs were amplified by PCR from P2 lysates and cloned between the NdeI and XhoI sites of pCYB2. Since pCYB2 allows substantial expression of structural genes even in the absence of induction (28), all experiments were conducted without IPTG (isopropyl-β-d-thiogalactopyranoside).
pUCF4 is a pUC8 derivative carrying the P2 late promoter pF (12). Plasmid pGC160 carries the P2 lysis region (nucleotides [nt] 6695 to 8544) under pF control. This region was amplified from P2 vir-1 with primers Lys5 and Lys3, using Pfu Turbo DNA polymerase, digested with BamHI and PstI, and ligated with pUCF4 cleaved with the same two enzymes. Plasmid pGC163 was made the same way, using the equivalent fragment amplified from P2 gor-1.
To construct pTG257, a plasmid that carries an IPTG-inducible copy of the P2 ogr gene, a 400-bp XhoI-HindIII fragment containing the ogr gene under the control of a variant T7A1 promoter with two lac operators (PA1/03/04) (21) was isolated from plasmid pBJ49 (18). This fragment was inserted at the unique BamHI site of pRG1, a derivative of pACYC177 (7) containing a 1.2-kb PstI fragment encoding lacIq (Robert Garcea, unpublished observation). Both the fragment and linearized pRG1 were filled-in with Klenow polymerase prior to ligation.
Activation of a cloned P2 late promoter.
Chloramphenicol acetyltransferase (CAT) expression was assayed in cultures of P90A5c and 397c carrying either the P2 late promoter expression plasmid pFCAT100 (15) and the IPTG-inducible P4 Delta plasmid pDEB50 (9) or carrying pSL130 (22), a control plasmid expressing cat from the tac promoter, and the compatible lacIq plasmid pRG1. Cultures were grown in LB medium supplemented with ampicillin (100 μg/ml) and kanamycin (60 μg/ml) to an A600 of 0.5 and induced by the addition of IPTG to 1 mM. Forty-five minutes after induction, duplicate cultures were lysed by sonication and CAT activity was determined spectrophotometrically and normalized to the protein concentration, as described previously (15). The values given represent the average of at least three determinations on two separate cultures.
P2 complementation by plasmid-encoded proteins.
Overnight E. coli cultures were inoculated into 100 volumes of LB medium containing 2 mM CaCl2 and 2 mM MgCl2 and grown at 30°C to an optical density at 600 nm (OD600) of 0.5 (ca. 3 to 5 h). One hundred microliters of cell culture was mixed with 100 μl of 10 mM Tris-HCl (pH 7.9), 1% (wt/vol) NaCl, 2 mM MgCl2, and 2 mM CaCl2 containing ca. 500 to 1,000 PFU of P2 and incubated for 7 min at 30°C. Cell-phage suspensions were mixed with 2 ml of molten 0.6% LB top agar and poured over the surface of 60-mm-diameter plates containing 10 ml of hardened 1.5% LB bottom agar. Both top agar and bottom agar contained 2 mM CaCl2. For plasmid-carrying strains, ampicillin was added to the medium at a final concentration of 200 μg/ml. Plates were incubated face up overnight at various temperatures. Phage plaques and cleared zones were recorded with a Nikon SMZ-U binocular microscope attached to a Hitachi KP-D50 digital camera (zoom, 1:10).
Construction of a P2 lysC(Am) mutant.
An amber mutation in the putative lysC open reading frame was introduced into the sequence between lysB and gene R, at P2 nt 8370 to 8372, by incorporation of a phosphorylated mutant oligonucleotide during PCR amplification. DNA was amplified from pNL130 by using 100 ng each of the P2 primer MR2 and the lacZ primer 1212 (New England Biolabs), and 1,000 ng of phosphorylated mutagenic primer 28am. PCR was carried out with Vent DNA polymerase in the presence of Taq DNA ligase (both from New England Biolabs). The full-length PCR product was gel purified, digested with EcoRI and SphI, and used to replace the corresponding EcoRI-SphI fragment in pNL130. The mutation in the resulting plasmid, pTG605, was verified by sequence analysis and introduced into the genome of P2 vir-1 by homologous recombination, using rescue of P2 vir-1 R(Am)3 (which has a Ts phenotype on supD strains), as described by Ziermann et al. (44).
Marker rescue.
For rescue of the cloned gor-1 or lysC(Am) mutations, P2 phages were grown in permissive strains carrying the plasmid with the desired mutation. Phage lysates were treated with UV light to 50% survival prior to infection to enhance recombination. Following infection, the progeny phage was then plated on both permissive and nonpermissive strains to assess the frequency of rescue and to select the desired recombinant phages.
Burst size determination.
E. coli strain C-1a was grown with shaking at 37°C in LB medium supplemented with 0.2% glucose (LBglc) to a value of 50 Klett units. Cells were concentrated twofold in LBglc supplemented with 5 mM CaCl2 and infected with P2 or P2 lysC(Am) at a multiplicity of infection of 5. After 8 min of incubation, the infected cells were diluted 10-fold into LBglc containing anti-P2 serum (K value of approximately 6 [first-order inactivation constant]), incubated for an additional 8 min, and then diluted 10−4 into LBglc. An aliquot was removed immediately, and the titer was determined on strain C-520 to determine the number of infected cells. The culture was incubated with shaking at 37°C for 60 min, and the titer was determined again on C-520.
λ prophage construction and induction.
The λ prophages containing Rz and/or Rz1(Am) mutations carry chloramphenicol resistance and are thermally inducible (43). Phage lysates were prepared from logarithmically growing lysogens of E. coli MC4100 by induction for 15 min at 43°C, followed by aeration at 37°C for 40 to 60 min or until visible lysis occurred. After the addition of 1% (vol/vol) chloroform and 10 mM MgCl2, cultures were vigorously vortexed for 20 s, incubated at 37°C for 30 min, and clarified by low-speed centrifugation. The cleared phage lysates were stored at 4°C.
Lysis induction by the cloned P2 lysis cassette.
E. coli strains transformed with the Ogr expression plasmid pTG257 and a compatible lysis region plasmid or vector control were grown at 30°C in LB medium containing ampicillin (100 μg/ml) and kanamycin (60 μg/ml) to a value of ∼50 Klett units. Expression of ogr was induced by the addition of IPTG to 1 mM, and lysis was monitored by measuring the OD of the culture. Each lysis plasmid was assayed at least six times from a minimum of three independent transformants.
RESULTS
The P2 growth defect depends on the Ts phenotype of the rpoC deletion.
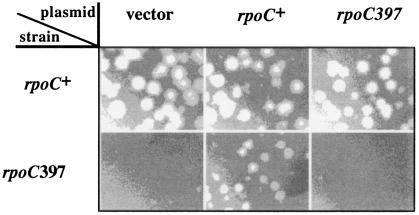
To assess whether the rpoC397 mutation was sufficient for the growth defect of P2 on strain 397c, isogenic plasmids constitutively expressing the wild-type rpoC allele or the rpoC397 allele were introduced into both the wild-type and the 397c hosts and the transformants were tested for plaque-formation by P2. The experiment presented in Fig. 1 shows that wild-type rpoC expressed from a plasmid complemented the rpoC397 block to P2, whereas a plasmid carrying the mutant rpoC gene did not. P2 growth in the wild-type rpoC host was unaffected by the presence of the rpoC397 plasmid, indicating that the rpoC defect is recessive. Thus, the lesion in RNAP is solely responsible for the P2 growth defect exhibited by the 397c cells and is recessive to the wild type.
FIG. 1.
The rpoC397 mutation interferes with bacteriophage P2 growth. A plasmid-encoded RNAP β′ subunit complements the P2 plating defect on a 397c host. Bacteriophage P2 was used to infect lawns of 397c or its rpoC+ parent, P90A5c, harboring plasmids expressing wild-type rpoC, rpoC carrying the 397c mutation or control vector plasmid. The results of overnight plating at 30°C are presented.
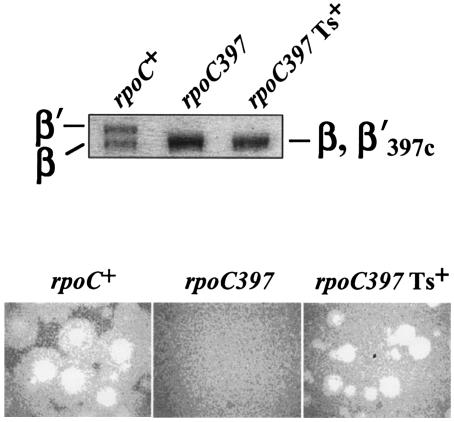
If the nonpermissive phenotype for P2 was due to a lack of an interaction between a phage transcription factor and the mutant RNAP present in 397c, then Ts+ revertants of the rpoC deletion mutant should retain the plating defect. However, a spontaneous Ts+ derivative of 397c efficiently plated P2 vir-1, but still contained the deletion, as indicated by the presence of the shortened β′ subunit (Fig. 2). The result therefore suggests that in contrast to N4 (25), P2 is unlikely to encode a transcription factor that directly interacts with the portion of β′ removed by the rpoC397 deletion.
FIG. 2.
P2 plates on a Ts+ pseudorevertant of 397c. A spontaneous revertant of E. coli 397c able to form colonies at 42°C was isolated. (Top panel) Proteins from whole-cell lysates of rpoC+ strain P90A5c, the 397c mutant, and the 397c Ts+ revertant were resolved by electrophoresis on a 5% sodium dodecyl sulfate gel and visualized by Coomassie staining. The portion of the gel containing the β and β′ subunits is shown. (Bottom panel) Bacteriophage P2 was used to infect lawns of the indicated cells. The results of overnight growth at 30°C are presented.
Initial efforts to elucidate the P2 growth defect conferred by the rpoC397 mutation included assessing the effects of this mutation on P2 DNA replication and transcription. Formation of pulse-labeled, covalently closed circular P2 DNA following infection at 33°C was assayed by CsCl-ethidium bromide density gradient centrifugation, and the levels were found to be similar in both P90A5c and 397c; the identity of this newly synthesized DNA was further confirmed by EcoRI restriction (33; data not shown). Since P2 DNA replication is dependent upon P2 early gene products, this implies that P2 early transcription is also not impaired in 397c. P2 late gene transcription is normally activated by the P2 Ogr protein, used during P2 lytic infection, or by the related Delta protein of satellite phage P4, which stimulates high levels of P2 late gene expression when P2 is serving as a helper for P4 lytic growth (reviewed in reference 11). To examine the effect of the rpoC397 mutation on initiation of P2 late transcription, Delta-dependent expression of cat from the P2 late promoter PF was assayed in P90A5c and 397c and compared to that of a control plasmid in which cat was expressed from the tac promoter. For the tac promoter, cat expression was reduced slightly in 397c, to 74% of the level seen in P90A5c. A slightly larger effect on expression from PF was seen; in 397c, the CAT activity was 44% of that obtained in P90A5c. It is likely that this larger reduction is due to a combined nonspecific effect on expression of P4 Delta from pDEB50 and on subsequent Delta-dependent expression from PF. We conclude that in contrast to the late transcription defect imposed by the rpoA109 mutation, which prevents an essential interaction between the RNAP α subunit and Ogr or Delta and reduces expression from PF by about 2 orders of magnitude (15), the rpoC397 mutation does not appear to confer a specific block to P2 late transcription.
P2 mutations that allow growth on 397c define a new gene.
To determine the basis of the P2 plating defect, spontaneous suppressor mutants that formed plaques on 397c lawns were obtained. One class of suppressors, called trl, was isolated directly by plating on 397c lawns at 30°C. A second class of mutants, called gor, were originally isolated based on their ability to overcome a block to P2 growth imposed by the rho(Ts)15 mutation in E. coli strain AD1600 (13) and were found to plate on 397c as well. The suppression was not reciprocal: while the P2 gor mutants formed plaques on 397c lawns, P2 trl mutants did not form plaques on AD1600 lawns (Table 2).
TABLE 2.
Plating of P2 and related phages on strains carrying the rpoC397 and rho(Ts)15 mutations
| Phage | Efficiency of platinga
|
|||
|---|---|---|---|---|
| P90A5c (rpoC+) | 397c (rpoC397) | SA1030 (rho+) | AD1600 [rho(Ts)15] | |
| P2 vir-1 | 1.0 | 1 × 10−7 | 1.0 | 4 × 10−9 |
| 186 cI(Ts) | 1.0 | 1.0 | 1.0 | 1.0 |
| Hy5 (P2/186 hybrid) | 1.0 | 1 × 10−7 | 1.0 | <1 × 10−9 |
| P2 vir-1 trl-1 | 1.0 | 1.0 | 1.0 | <1 × 10−7 |
| P2 vir-22 L(Ts)37 gor-3b | 1.0 | 1.0 | 1.0 | 0.21 |
All platings were performed at 33°C.
The vir-22 and L(Ts)37 mutations do not affect P2 growth under the conditions used and were employed to facilitate mapping of the gor mutation. Hy5 also carries the 186 cI(Ts) mutation.
The rpoC397 mutation did not affect growth of the P2-related phage 186, but growth of the hybrid phage Hy5, which carries the early region of 186 and the late region of P2 (17), was blocked (Table 2), suggesting that the trl mutations map to the P2 late genes. Indeed, three factor crosses (33; data not shown) placed the trl and gor mutations between P2 genes K and R, in a region encoding host lysis and tail functions. A cloned 2.1-kb EcoRV fragment spanning the lysis region (P2 coordinates 6290 to 8378; GenBank accession no. AF063097; see also reference 43) from P2 gor1 rescued the plating of wild-type P2 on 397c (data not shown). The DNA sequence of this region from gor1 and from two independent trl isolates was determined and compared to the wild-type P2 sequence. The two trl mutants had a single, identical nucleotide change, from C to A at nt 8274, and the gor1 mutant also had a single-nucleotide change, which was immediately adjacent to the trl substitution, from C to T at nt 8275. These changes fall within lysB, encoding a nonessential lysis gene (44), and also within an overlapping +1 reading frame, spanning P2 coordinates 8202 to 8492. This reading frame is preceded by a good Shine-Dalgarno sequence and potentially encodes a proline-rich protein (10 Pro residues in 96 amino acids). The first half of this open reading frame is embedded within the last 48 codons of lysB; the end of this reading frame extends into the tail gene R (Fig. 3A). Originally designated orf28 by Portelli et al. (31), this reading frame was predicted to be another nonessential lysis gene, lysC, and to encode a homolog of the λ Rz1 lysis protein (43). Since the gor mutation is silent within lysB, the suppression of the Gro− phenotype of 397c by the gor and trl mutations must be due to the changes in lysC (Pro 25 Leu and Pro 25 Thr for gor and trl, respectively; Fig. 3A). This result indicates that lysC does encode a P2 protein. Although sequence similarity between LysC and Rz1-like proteins is not significant, inspection of the LysC primary structure reveals a consensus signal peptidase II cleavage site (Fig. 3A) and a high proportion of Pro residues, both characteristic of all Rz1 protein sequences (43). These considerations, the unique intragenic embedding of lysC and Rz1, and the results of functional analysis presented below strongly indicate that LysC and Rz1 are evolutionarily homologous, as first suggested by Zhang et al. (43).
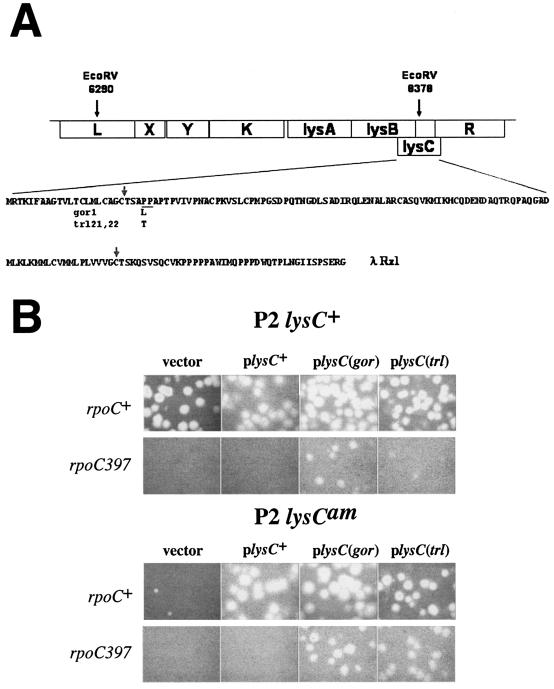
FIG. 3.
P2 mutants that grow on 397c define a new gene, lysC. (A) Map of the bacteriophage P2 lysis region. The P2 genome coordinates of the EcoRV fragment used to rescue the trl and gor mutations are indicated, as are the relative locations of the genes in this region. The amino acid sequence of the lysC reading frame is shown below the map, and the amino acid sequence changes resulting from the trl and gor mutations are indicated below the sequence. The PP dipeptide characteristic of all Rz1 proteins is underlined. Also shown for comparison is the amino acid sequence of λ Rz1. Gray arrows indicate the putative signal peptidase II cleavage sites in the two proteins. (B) Overexpression of lysC harboring gor or trl mutations, but not wild-type lysC, is sufficient to overcome the plating defect of P2. 397c or P90A5c cells harboring plasmids expressing wild-type lysC, lysC carrying the gor or trl mutations, or control vector plasmid pCYB2 were infected with wild-type P2 (top) or a P2 mutant carrying an amber mutation in lysC (bottom). The results of overnight growth at 30°C are presented.
In order to obtain further evidence as to whether lysC represented a protein-coding gene, we introduced an amber mutation in place of codon 57 (P2 coordinates 8370 to 8372). This nonsense mutation does not alter the predicted amino acid sequence of LysB, yet it showed a distinct suppressible plating phenotype, plating with normal plaque size on supD or supF strains but with a tiny plaque size on nonsuppressing hosts, indicating that orf28 encodes a protein. A comparison of the burst size of this mutant with that of P2 vir-1 in a nonsuppressing host showed less than a twofold reduction (124 for the amber mutant versus 200 for P2 vir-1), suggesting that the plating phenotype might be related to host lysis, rather than the intracellular production of virions. Taken together, these data further strengthen the conclusion that lysC, previously orf28, is a P2 gene encoding an Rz1-like protein product.
To confirm that the lysC mutations are sufficient to overcome the P2 plating defect on 397c cells, we created plasmids that expressed the wild-type, trl, or gor alleles of lysC from the tac promoter. These plasmids were introduced into 397c cells, and their ability to complement the plating defect of wild-type P2 was determined. Expression of the wild-type lysC had no effect on P2 growth. In contrast, expression of lysC(gor), and to a lesser degree lysC(trl), allowed plaque formation (Fig. 3B). The suppressing effect of the plasmid-borne lysC(trl) allele was more pronounced in P2lysC(Am) infections (Fig. 3B). This suggests that the wild-type LysC may be inhibiting the function of the LysC(trl) mutant.
P2 LysB/C complements λ Rz/Rz1 lysis defects.
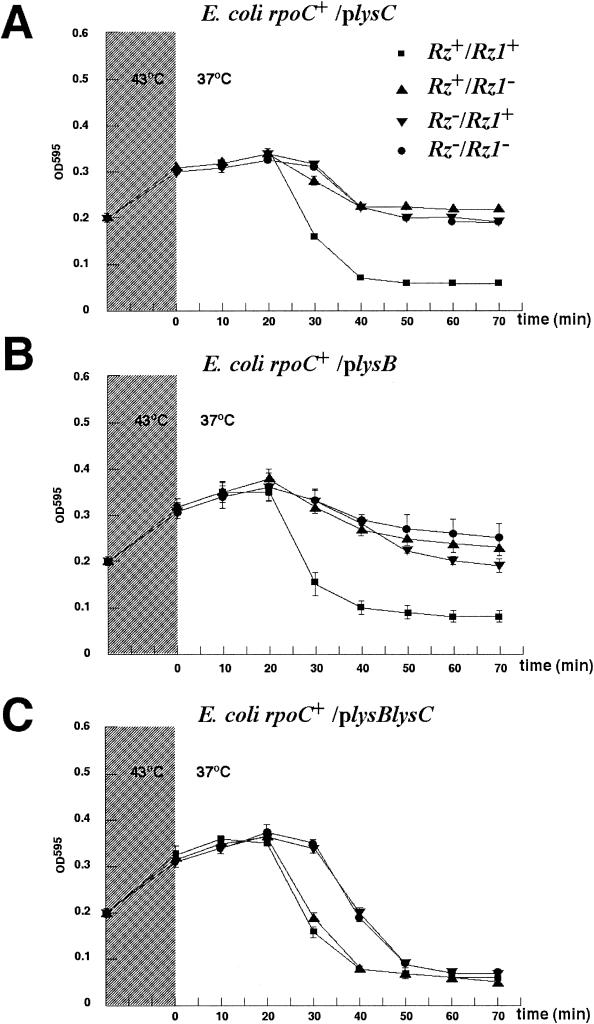
The lysC gene and its overlapping arrangement with lysB are conserved in closely related P2-like phages, including 186, Wφ, L-413C, Fels-2, SopEφ, and PSP3, as well as in the more distantly P2-related Pseudomonas aeruginosa phage φCTX. No significant sequence similarity is found with other genes in the database. Rz1 genes are always found embedded within Rz genes (30, 41, 43), so lysC and lysB may be similar to Rz1 and Rz genes, respectively. In λ and P22, Rz and Rz1 are both required for lysis if the host envelope is stabilized by millimolar concentrations of divalent cations. Rz1 is a lipoprotein, processed by signal peptidase II, and LysC has an appropriately placed Cys residue, which could serve as part of a signal peptidase II motif. Moreover, although there is no significant amino acid sequence similarity between LysC and Rz1, both are rich in Pro residues. To establish whether LysB/C and Rz/Rz1 have analogous functions, we introduced plasmids carrying lysB, lysC, or both genes into E. coli cells lysogenic for wild-type λ or various λ Rz/Rz1 mutants (43) and subjected the transformants to thermal induction in the presence of 10 mM MgCl2. As expected, lysis was observed in the induction of the Rz+ Rz1+ lysogen, but not in Rz(Am), Rz1(Am), and Rz(Am) Rz1(Am) (data not shown; see also reference 43). This lysis defect was complemented by a plasmid coexpressing lysB and lysC, but not by plasmids expressing lysB or lysC alone (Fig. 4). In the lysB- and lysC-complemented inductions, lysis kinetics were somewhat delayed for both Rz(Am) lysogens, whereas the Rz+ Rz1(Am) lysogen underwent lysis at the same time as the parental lysogen (Fig. 4). We conclude that P2 LysB/LysC can functionally substitute for λ Rz/Rz1, but neither LysB nor LysC alone can complement either an Rz or Rz1 defect.
FIG. 4.
Co-overexpression of bacteriophage P2 lysB and lysC complements the lysis defect caused by bacteriophage λ Rz and/or Rz1 mutations. Logarithmically growing rpoC+ P90A5c cells lysogenic for λ Rz+/Rz1+ or the indicated Rz and/or Rz1 λ mutants were thermally induced for 15 min at 43°C (hatched area) and then transferred to 37°C. Cell lysis was monitored spectrophotometrically. The values presented are mean values from three independent experiments. Different panels represent lysis curves obtained with induced lysogens harboring plasmids expressing lysC (pCYB2lysC) (A), lysB (pCYB2lysB) (B), or coexpressing wild-type lysB and lysC (pCYB2lysB, lysC) (C).
lysC mutations delay host lysis.
Efforts to monitor P2-induced lysis directly in strains carrying rpoC397 are complicated by the relatively poor adsorption of P2 to E. coli K-12 strains, which precludes analysis of one-step growth curves (our unpublished data), and by the apparent incompatibility of rpoC397 with E. coli C, in which P2 is normally studied (33). In order to circumvent these technical limitations, we assayed P2-induced host cell lysis by using expression plasmids carrying the entire lysis gene cassette (Y K lysABC). Tight control of these otherwise toxic genes was achieved by regulation of this cassette by using the P2 late promoter PF. Expression from PF was regulated by the P2 late transcription factor Ogr, which was supplied in trans from compatible plasmid pTG257, carrying an IPTG-inducible copy of the P2 ogr gene. The ogr gene in this construct was expressed from the same promoter used for expression of P4 Delta in the cat assays described above, and, accordingly, should be expressed in rpoC397 cells, albeit at a slightly reduced level. Ogr was chosen instead of Delta for these experiments because it is a less efficient activator of PF, thus minimizing the potentially lethal effects of any leaky activator expression in the absence of IPTG induction.
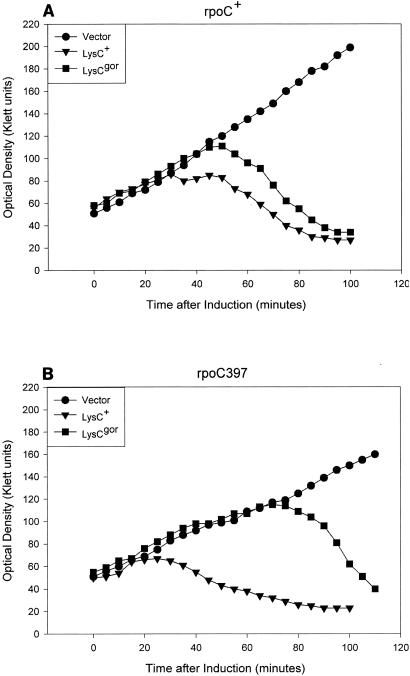
The otherwise isogenic E. coli strains RW4206 (rpoC+) and RW4204 (rpoC397) carrying pTG257 and plasmids with either the wild-type (pGC160) or gor-1 (pGC163) alleles of lysC were grown to early logarithmic phase at 30°C and induced with IPTG (Fig. 5). In the rpoC+ host, the wild-type lysis cassette caused a cessation of growth at about 30 min after induction, and overt lysis was observed beginning at about 45 min. The same plasmid caused cessation of growth significantly earlier in the rpoC397 host, and gradual lysis was detectable by 25 min after induction. This was surprising, especially in light of the fact that we expected expression of the plasmid-encoded lysis cassette to be slightly reduced in this strain compared to that in the rpoC strain. The early cessation of growth was reproducible and may indicate that some cells started to lyse at this point, while others continued to grow. In contrast, the expression of the lysis cassette carrying lysC(gor) resulted in an extended growth period and delayed lysis in both the parental and rpoC397 backgrounds—especially in the latter, where growth continued until lysis was apparent at about 70 min after induction.
FIG. 5.
lysC gor delays host lysis. Logarithmically growing RW4206 (rpoC+) (A) or RW4204 (rpoC397) (B) E. coli cells carrying the ogr expression plasmid pTG257 and either pUCF4 (vector control), pGC160 (lysC+), or pGC163 (lysCgor) plasmids with P2 lysis genes under the control of the P2 pF promoter were induced with IPTG, and cell lysis was monitored spectrophotometrically. Curves representative of the results from six independent experiments are shown.
We conclude that the primary effect of the lysC(gor) allele is a delay in the onset of lysis. Accordingly, we propose that early onset of lysis observed with the wild-type lysis cassette in the rpoC397 background underlies the plating defect of P2 on this host, and that the suppressor effect of the gor mutation derives from a delay in lysis that allows accumulation of phage progeny sufficient for plaque formation.
DISCUSSION
Identification of the P2 lysB and lysC genes as functional homologs of Rz and Rz1.
We report here the genetic identification and initial characterization of a new P2 gene, lysC. P2 lysC mutants suppress the defect to P2 growth conferred by the E. coli rpoC397 mutation in RNAP. A defect in interaction between a viral activator of late transcription and the mutant RNAP has been proposed to explain the reported inability of bacteriophage N4 to grow on 397c (25). We therefore expected that P2 phages that grow on 397c would also carry mutations in genes whose products interact with RNAP and affect viral gene expression. However, the experiments presented here strongly suggest that LysC is not involved in transcription regulation but instead plays a role, together with its partner protein LysB, in host lysis.
It was proposed previously by one of us that the LysB/LysC pair is homologous to Rz/Rz1 based on unusual amino acid composition and the overlapping organization of these genes (43). Both Rz1 and LysC are relatively short polypeptides containing an unusually high proportion of Pro residues (11 of 79 and 10 of 96, respectively). The N-terminal domains of both proteins contain putative signal peptidase II motifs, indicating that both are processed to lipoprotein forms, resulting in short (60 and 76 amino acids, respectively) polypeptides predicted to be attached to the inner leaflet of the outer membrane by lipid residues on the N-terminal Cys of the mature polypeptide (43). However, neither LysB nor LysC is significantly related by standard or reiterative BLAST searches to Rz or Rz1 or in fact to any proteins except those encoded by closely related P2-like phages. Moreover, the λ Rz1 reading frame is completely embedded within the Rz gene, while the longer lysC reading frame spans the end of lysB, the beginning of gene R, and the intervening 107 nt. Nevertheless, we show here that production of P2 LysB and LysC efficiently suppresses the λ cation-sensitive lysis defect caused by either Rz or Rz1 mutations, indicating that P2 LysC and LysB and λ Rz1 and Rz have similar functions. Production of either LysB or LysC alone does not correct the lysis defect caused by Rz and/or Rz1 mutations. This strongly implies that LysB and LysC form heteromeric complexes. A similar conclusion was reached about Rz and Rz1 interaction while comparing host lysis defects caused by Rz, Rz1, and Rz/Rz1 double mutants (43). Moreover, two-hybrid analysis has shown that the Rz and Rz1 homologs from phage T7 interact (3). Our data suggest that interspecies complexes (i.e., LysB/Rz1) either do not form or are not functional. The presence of the λ Rz1 protein (in the absence of Rz) results in a retardation of lysis mediated by LysB/LysC complexes in the presence of high magnesium ion concentrations (Fig. 4C), perhaps because nonfunctional LysB/Rz1 complexes reduce the amount of functional LysB/LysC heteromers. Presumably, the unique overlapping organization of these genes imposes severe restrictions on their evolutionary variation and helps to ensure that they coevolve.
The functional role of Rz/Rz1 and their homologues, LysB/LysC, is not clear. Sequence analysis reveals that genes whose products are similar to Rz/Rz1 and/or LysB/LysC are widespread in double-stranded DNA phage genomes. The unusual nested or overlapping arrangement of the two genes, as well as their localization immediately after the main host lysis genes, is also conserved, suggesting that the two proteins play an important, evolutionarily conserved function. In vitro analysis by the Taylor group demonstrated that λ Rz1 is found in the outer membrane and that, as predicted from the primary structure analysis, Rz1 is processed and lipoylated (16, 19, 37). More recently, the same group reported that Rz1 promotes membrane fusion in vitro (6). However, the lysis phenotype in lambda remains unexplained. In vivo, bacteriophage λ mutants with lesions in either Rz or Rz1 are defective in host lysis, but only in the presence of millimolar levels of divalent cations (43). In contrast, the lysC(Am) mutation severely affects plating efficiency and plaque morphology of P2 (Fig. 3), and P2 lysB(Am) mutants show a slightly retarded lysis phenotype (44), and in neither case is the presence of millimolar concentrations of divalent cations required. A model for Rz function has been proposed in which Rz, and by extension, Rz/Rz1, is involved in attacking links between the outer membrane and the murein (42). Under standard laboratory conditions, the phage endolysin degrades the peptidoglycan sufficiently to cause bursting of the infected cell, but in the presence of high levels of divalent cations, the outer membrane would be significantly stabilized, thus making further degradation of the envelope necessary. The lack of a cation effect in P2 may simply reflect that the K endolysin is not as efficient at murein degradation and thus has a constitutive partial requirement for the LysB/C function.
lysC alleles, the suppression of RNAP defects, and implications for lysis control.
How does the rpoC397 mutation result in defective P2 development, and how do lysC missense mutations overcome this defect? Our initial working model was that the RNAP mutation directly affected transcription of viral genes, leading somehow to a lysis defect. However, the locations of the compensatory trl and gor mutations in a gene located near the 3′ end of a long late transcription unit, coupled with the lack of a block to initiation of phage late transcription in an rpoC397 strain, argue that the effect of the RNAP mutation would have to be on elongation or some kind of attenuation mechanism affecting lysis gene expression. Since cells carrying the rpoC397 mutation appear susceptible to accelerated lysis rather than impaired lysis, one would have to further propose that the change in transcription affected the relative levels of the various lysis proteins to somehow bring about the observed change in timing and that one of the lysis proteins modulates this effect. We cannot formally rule out such a model, but it requires invoking a complicated hypothetical regulatory mechanism for which there is no direct evidence or precedent. An attractive alternative model envisions that rpoC397 causes changes in host gene expression that alter bacterial physiology, including properties of the cell membrane and/or cell wall, and thus affects the timing of host lysis during bacteriophage P2 infection. Indeed, whole-genome transcription profiling of 397c cells shows that genes responsible for type I extracellular polysaccharide synthesis are dramatically overproduced (38), a typical stress response when the bacterial envelope is compromised (35). Thus, in this scenario, 397c cells, with defective envelopes, lyse prematurely when infected with wild-type P2, reducing the burst size below the threshold required for plaque formation. The gor mutation dramatically delays the lysis of rpoC397 cells, presumably allowing a sufficient number of progeny to accumulate in each round of infection to generate visible plaques.
Both lysC mutations that we describe here affect the same residue of LysC: a Pro at position 25, which is replaced by either Leu (gor) or Thr (trl). Unlike P2, the closely related coliphage 186 does form plaques on strain 397c and the AD1600 [rho(Ts)15] strain (33) (Table 2). The predicted phage 186 LysC has a Gln at residue 25 in place of Pro. Thus, 186 may be naturally gor (or trl). At present, it is premature to speculate how gor and trl mutants retard host lysis, lacking understanding of the function of the wild-type Rz/Rz1 and thus LysB/LysC, protein complexes. However, the results reported here are very significant in terms of the general picture of the phage-mediated lysis of gram-negative bacteria. Heretofore, the timing of lysis has been thought to be the exclusive province of the holin proteins, which are produced by all double-stranded DNA phages (41). In this sense, timing refers to the termination of the infection cycle, because when the holin triggers, the host membrane is disrupted and all macromolecular synthesis and assembly stops. Here, timing is strongly affected by the the lysC(gor) allele, which allows an extended growth period and thus compensates for the abortive, truncated infective cycle in P2 infections of the rpoC397 host. Thus for the first time, a gene other than a holin gene is implicated in lysis timing. Although it is premature to speculate on the mechanistic details of this effect, nevertheless these results may be evidence for the formation of a “lysis-some,” a multiprotein complex spanning the envelope and including all the lysis proteins, the holin Y, endolysin K, and the LysB/LysC complexes (and perhaps LysB, a putative antiholin; T. Park and R. Young, unpublished observation). Future experiments defining biological and biochemical functions, as well as interacting molecular partners, of LysB/LysC and Rz/Rz1 should help resolve these issues.
Acknowledgments
We thank Tina Goodwin for technical assistance and Robert Weisberg for the MG1655 rpoC derivatives.
This work was supported by NIH grant GM59295 and a Burroughs Wellcome Career Award to K.S., ACS grant RPG-92-008NP to G.E.C., and NIH grant GM27099 to R.Y. D.M. was postdoctoral fellow of the Charles and Johanna Busch Memorial Fund.
REFERENCES
- 1.Adhya, S. L., and J. A. Shapiro. 1969. The galactose operon of E. coli K-12. I. Structural and pleiotropic mutations of the operon. Genetics 62:231-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldwin, R. L., P. Barrand, A. Fritsch, D. A. Goldthwait, and F. Jacob. 1966. Cohesive sites on the deoxyribonucleic acids from several temperate coliphages. J. Mol. Biol. 17:343-357. [DOI] [PubMed] [Google Scholar]
- 3.Bartel, P. L., J. A. Roecklein, D. SenGupta, and S. Fields. 1996. A protein linkage map of Escherichia coli bacteriophage T7. Nat. Genet. 12:72-77. [DOI] [PubMed] [Google Scholar]
- 4.Bertani, G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62:293-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertani, L. E. 1957. The effect of the inhibition of protein synthesis on the establishment of lysogeny. Virology 4:53-71. [DOI] [PubMed] [Google Scholar]
- 6.Bryl, K., S. Kedzierska, M. Laskowska, and A. Taylor. 2000. Membrane fusion by proline-rich Rz1 lipoprotein, the bacteriophage lambda Rz1 gene product. Eur. J. Biochem. 267:794-799. [DOI] [PubMed] [Google Scholar]
- 7.Chang, A. C. Y., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chattoraj, D. K., and R. B. Inman. 1972. Position of two deletion mutations on the physical map of bacteriophage P2. J. Mol. Biol. 66:423-434. [DOI] [PubMed] [Google Scholar]
- 9.Christie, G. E., D. L. Anders, V. McAlister, T. S. Goodwin, B. Julien, and R. Calendar. 2003. Identification of upstream sequences essential for activation of a bacteriophage P2 late promoter. J. Bacteriol. 185:4609-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christie, G. E., S. B. Cale, L. A. Isaksson, D. J. Jin, M. Xu, B. Sauer, and R. Calendar. 1996. Escherichia coli rpoC397 encodes a temperature-sensitive C-terminal frameshift in the β′ subunit of RNA polymerase that blocks growth of bacteriophage P2. J. Bacteriol. 178:6991-6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christie, G. E., and R. Calendar. 1990. Interactions between satellite bacteriophage P4 and its helpers. Annu. Rev. Genet. 24:465-490. [DOI] [PubMed] [Google Scholar]
- 12.Christie, G. E., and R. Calendar. 1985. Bacteriophage P2 late promoters. II. Comparison of the four late promoter sequences. J. Mol. Biol. 181:373-382. [DOI] [PubMed] [Google Scholar]
- 13.Das, A., D. Court, and S. Adhya. 1976. Isolation and characterization of conditional lethal mutants of E. coli deficient in transcription termination factor rho. Proc. Natl. Acad. Sci. USA 73:1959-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geiduschek, E. P., and G. A. Kassavetis. 1988. Changes in RNA polymerase, p. 93-109. In R. Calendar (ed.), The bacteriophages, vol. 1. Plenum Press, New York, N.Y.
- 15.Grambow, N. J., N. K. Birkeland, D. L. Anders, and G. E. Christie. 1990. Deletion analysis of a bacteriophage P2 late promoter. Gene 95:9-15. [DOI] [PubMed] [Google Scholar]
- 16.Hanych, B., S. Kedzierska, B. Walderich, B. Uznanski, and A. Taylor. 1993. Expression of the Rz gene and the overlapping Rz1 reading frame present at the right end of the bacteriophage lambda genome. Gene 129:1-8. [DOI] [PubMed] [Google Scholar]
- 17.Hocking, S. M., and J. B. Egan. 1982. Genetic characterization of twelve P2-186 hybrid bacteriophages. Mol. Gen. Genet. 187:174-176. [DOI] [PubMed] [Google Scholar]
- 18.Julien, B., and R. Calendar. 1996. Bacteriophage PSP3 and φR73 activator proteins: analysis of promoter specificities. J. Bacteriol. 178:5668-5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kedzierska, S., A. Wawrzynow, and A. Taylor. 1996. The Rz1 gene product of bacteriophage lambda is a lipoprotein localized in the outer membrane of Escherichia coli. Gene 168:1-8. [DOI] [PubMed] [Google Scholar]
- 20.King, R. A., D. L. Anders, and G. E. Christie. 1992. Site-directed mutagenesis of an amino acid residue in the bacteriophage P2 Ogr protein implicated in interaction with Escherichia coli RNA polymerase. Mol. Microbiol. 6:3313-3320. [DOI] [PubMed] [Google Scholar]
- 21.Lanzer, M., and H. Bujard. 1988. Promoters largely determine the efficiency of repressor action. Proc. Natl. Acad. Sci. USA 85:8973-8977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, S. C., C. L. Squires, and C. Squires. 1984. Antitermination of E. coli rRNA transcription is caused by a control region segment containing lambda nut-like sequences. Cell 38:851-860. [DOI] [PubMed] [Google Scholar]
- 23.Lindahl, G. 1971. On the control of transcription in bacteriophage P2. Virology 46:620-633. [DOI] [PubMed] [Google Scholar]
- 24.Linderoth, N. A., B. Julien, K. Flick, R. Calendar, and G. E. Christie. 1994. Molecular cloning and characterization of bacteriophage P2 genes R and S involved in tail completion. Virology 200:347-359. [DOI] [PubMed] [Google Scholar]
- 25.Miller, A., D. Wood, R. H. Ebright, and L. B. Rothman-Denes. 1997. RNA polymerase beta′ subunit: a target of DNA binding-independent activation. Science 275:1655-1657. [DOI] [PubMed] [Google Scholar]
- 26.Nechaev, S., and K. Severinov. 1999. Inhibition of E. coli RNA polymerase by bacteriophage T7 gene 2 protein. J. Mol. Biol. 289:815-826. [DOI] [PubMed] [Google Scholar]
- 27.Nechaev, S., and K. Severinov. 2003. Bacteriophage-induced modifications of host RNA polymerase. Annu. Rev. Microbiol. 57:301-322. [DOI] [PubMed] [Google Scholar]
- 28.Nedea, E. C., D. Markov, T. Naryshkina, and K. Severinov. 1999. Localization of Escherichia coli rpoC mutations that affect RNA polymerase assembly and activity at high temperature. J. Bacteriol. 181:2663-2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Norrander, J., T. Kempe, and J. Messing. 1983. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene 26:101-106. [DOI] [PubMed] [Google Scholar]
- 30.Pedulla, M. L., M. E. Ford, T. Karthikeyan, J. M. Houtz, R. W. Hendrix, G. F. Hatfull, A. R. Poteete, E. B. Gilcrease, D. A. Winn-Stapley, and S. R. Casjens. 2003. Corrected sequence of the bacteriophage P22 genome. J. Bacteriol. 185:1475-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Portelli, R., B. Dodd, Q. Xue, and J. B. Egan. 1998. The late-expressed region of the temperate coliphage 186 genome. Virology 248:117-130. [DOI] [PubMed] [Google Scholar]
- 32.Sasaki, I., and G. Bertani. 1965. Growth abnormalities in Hfr derivatives of Escherichia coli strain C. J. Gen. Microbiol. 40:365-376. [DOI] [PubMed] [Google Scholar]
- 33.Sauer, B. L. 1979. Regulation of late gene expression in the temperate coliphage P2. Ph.D. thesis. University of California, Berkeley, Calif.
- 34.Severinov, K. 2000. RNA polymerase structure/function: insights into points of transcriptional regulation. Curr. Opin. Microbiol. 3:118-125. [DOI] [PubMed] [Google Scholar]
- 35.Sledjeski, D. D., and S. Gottesman. 1996. Osmotic shock induction of capsule synthesis in Escherichia coli K-12. J. Bacteriol. 178:1204-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sunshine, M. G., M. Thorn, W. Gibbs, R. Calendar, and B. Kelly. 1971. P2 phage amber mutants: characterization by use of a polarity suppressor. Virology 46:691-702. [DOI] [PubMed] [Google Scholar]
- 37.Taylor, A., S. Kedzierska, and A. Wawrzynow. 1996. Bacteriophage lambda lysis gene product modified and inserted into Escherichia coli outer membrane: Rz1 lipoprotein. Microb. Drug Resist. 2:147-153. [DOI] [PubMed] [Google Scholar]
- 38.Van Dyk, T. K., Y. Wei, M. K. Hanafey, M. Dolan, M. J. Reeve, J. A. Rafalski, L. B. Rothman-Denes, and R. A. LaRossa. 2001. A genomic approach to gene fusion technology. Proc. Natl. Acad. Sci. USA 98:2555-2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weisberg, R. A., and M. E. Gottesman. 1999. Processive antitermination. J. Bacteriol. 181:359-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wood, L. F., N. Y. Tszine, and G. E. Christie. 1997. Activation of P2 late transcription by P2 Ogr protein requires a discrete contact site on the C terminus of the α subunit of Escherichia coli RNA polymerase. J. Mol. Biol. 274:1-7. [DOI] [PubMed] [Google Scholar]
- 41.Young, R. 1992. Bacteriophage lysis: mechanism and regulation. Microbiol. Rev. 56:430-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Young, R., S. Way, J. Yin, and M. Syvanen. 1979. Transposition mutagenesis of bacteriophage lambda: a new gene affecting cell lysis. J. Mol. Biol. 132:307-322. [DOI] [PubMed] [Google Scholar]
- 43.Zhang, N., and R. Young. 1999. Complementation and characterization of the nested Rz and Rz1 reading frames in the genome of bacteriophage lambda. Mol. Gen. Genet. 262:659-667. [DOI] [PubMed] [Google Scholar]
- 44.Ziermann, R., B. Bartlett, R. Calendar, and G. E. Christie. 1994. Functions involved in bacteriophage P2-induced host cell lysis and identification of a new tail gene. J. Bacteriol. 176:4974-4984. [DOI] [PMC free article] [PubMed] [Google Scholar]